
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci. , 25 July 2024
Sec. Veterinary Epidemiology and Economics
Volume 11 - 2024 | https://doi.org/10.3389/fvets.2024.1426384
 Yanyan Jiang1†
Yanyan Jiang1† Shanshan Zhou1,2†
Shanshan Zhou1,2† Zhongying Yuan1
Zhongying Yuan1 Xinyu Hu1
Xinyu Hu1 Zhen Li1
Zhen Li1 Yaxue Wang1
Yaxue Wang1 Yujuan Shen1*
Yujuan Shen1* Jianping Cao1,2*
Jianping Cao1,2*Introduction: Enterocytozoon bieneusi, an obligatory intracellular fungus, is prevalent among animals and humans. Due to their close interaction with humans and their extensive regional distribution, brown rats (Rattus norvegicus) are important pathogen reservoirs. To assess the zoonotic transmission potential of E. bieneusi, a molecular investigation was conducted on 817 R. norvegicus from four cities in Heilongjiang Province, China.
Methods: A total of 817 R. norvegicus were collected from four cities in Heilongjiang Province, China. The genotyping of E. bieneusi was conducted through PCR amplification of the small subunit ribosomal RNA (SSU rRNA)’s internal transcribed spacer (ITS) segments. Phylogenetic and similarity analyses were used to examine zoonotic potential and genetic characteristics of the E. bieneusi-positive specimens.
Results: Among the 817 R. norvegicus, the total infection rate was 33.3% (272/817). Seventy-five genotypes were identified, including 14 known genotypes D (n = 167), A (n = 15), HLJ-CP1 (n = 12), WR8 (n = 6), EbpC (n = 2), BEB6 (n = 1), CS-4 (n = 1), CHPM1 (n = 1), Henan-II (n = 1), HNH-22 (n = 1), HNH-25 (n = 1), I (n = 1), JLD-XI (n = 1), SDD5 (n = 1), and 61 novel genotypes designated as SHWR1 (n = 10), SYSWR1 (n = 2), and SHWR2 to SHWR17, SYSWR2 to SYSWR36 and QTHWR1 to QTHWR8 (n = 1, each). Moreover, 10 samples exhibited mixed genotype infections, including D + A (n = 3), D + EbpC (n = 1), D + HLJ-CP1 (n = 1), D + SHWR1 (n = 1), D + SHWR16 (n = 1), D + SHWR17 (n = 1), SDD5 + WR8 (n = 1), and CS-4 + SYSWR36 (n = 1). Phylogenetic analysis grouped the genotypes into three main groups: group 1 (n = 67), group 2 (n = 5), and group 9 (n = 3).
Discussion: The high prevalence and genetic diversity of E. bieneusi in Heilongjiang Province’s R. norvegicus imply that these animals spread the pathogen. The R. norvegicus that E. bieneusi carries can spread zoonotic disease, making it a serious hazard to the local human population. Therefore, it is imperative to raise awareness about the dangers posed by R. norvegicus and implement measures to reduce their population to prevent environmental contamination.
Microsporidia, a vast and diverse group of intracellular fungi, encompass more than 1,500 species. While most of these species are innocuous to human beings, 17 species have been identified as human pathogens (1). Enterocytozoon bieneusi is the most common cause of human microsporidial infections, making up more than 90% of cases (2). The spore, which is the infectious form of E. bieneusi, is expelled from the feces of the host and dispersed into the environment, thereby ensuring its widespread presence (3). Transmission of spores to humans can occur via direct contact, including person-to-person or animal-to-person transmission, as well as indirectly through contaminated food or water sources (4). Once contaminated, pathogenic spores may spread easily among individuals and other animals, exacerbating environmental contamination. There is an example of a food-borne outbreak linked to E. bieneusi infection, as well as a reported large outbreak of waterborne diseases caused by lake contamination (5, 6). Regular surveillance of E. bieneusi infections in both human and animal populations is crucial for implementing preventive interventions to reduce the spread of these diseases and minimize the probability of outbreaks. Accurate identification of the causal agent is essential for implementing effective monitoring and control strategies.
The advent of molecular typing tools provides the possibility for accurate identification of E. bieneusi. Currently, the main technique used to identify E. bieneusi is by sequencing analysis of the internal transcriptional spacer (ITS) within the small subunit ribosomal RNA (SSU rRNA) gene of this pathogen (7). The combination of homology and phylogenetic analysis provides an important basis for assessing genotype transmission across species and host adaptability characteristics among different hosts (8). Recently, researchers have identified around 900 different genotypes of E. bieneusi. Out of them, more than 150 genotypes are found only in humans, around 700 genotypes are found only in animals, and 67 genotypes are shared by both species (9–11). Notably, the total number of genotypes continues to grow as more genotypes are discovered, along with a concurrent increase in the number of zoonotic genotypes. For instance, in 2011, there were 93 genotypes: 34 exclusive to humans, and 11 shared between humans and various animals (12). The overall number of genotypes had expanded nine times by 2021, while the number of genotypes associated with zoonotic diseases had increased six times (9). If this pattern continues, thousands of novel genotypes might be documented in the next decade. Evolutionary analysis has also been considered as another effective means of assessing the possibility of zoonotic transmission. Currently, the genotypes that have been found are categorized into 15 distinct classifications (8, 13). The majority of these genotypes, which infect humans or animals, belong to the first and second groups. This indicates their significance in the formation of possible clusters of zoonotic diseases. The remaining 13 clusters are mainly present in specific hosts and wastewater (13).
In humans, genotypes D, EbpC, Type IV, and EbpA are the most common, and all have a broad host range (3, 9). Thus, there is great interest in determining the sources of infection for these zoonotic genotypes. Investigation suggests that these genotypes are not only widespread in humans and animals but also frequently identified in aqueous environments (3). This implies that it is necessary to develop comprehensive health strategies to tackle the high occurrence of E. bieneusi. Given that they are freely roaming and extensively dispersed, wild rodents have become a crucial connection between humans, animals, and the environment. E. bieneusi has been documented in at least 44 rodent species across 25 studies which conducted in China, the Czech Republic, Iran, Japan, Peru, Poland, Slovakia, and the United States (11, 14, 15). At present, there are 117 genotypes of E. bieneusi known to exist in rodents, including at least 18 zoonotic genotypes. The most common genotype is D, Type IV, which is similar to the prevalence in humans (9, 11, 14, 15). This overlap underscores the crucial role rodents play in transmitting E. bieneusi to humans, emphasizing the need to incorporate rodent control into the eradication efforts against this pathogen.
China has become an influential player in molecular epidemiology data regarding E. bieneusi in wild rodents. The research carried out in at least 13 provinces, has yielded vital information on ways in which E. bieneusi is transmitted and the sources of its infection (Table 1) (11, 13–20, 22–24). However, significant data gaps persist in certain regions of China, particularly in regions such as Heilongjiang, where the presence of E. bieneusi in humans and other animals has been widely reported (20, 25–28). It would be advantageous to monitor rodents in these locations to get insight into the origins and means of infection transmission among humans and other animals. Therefore, the objective of this study was to ascertain the infection rates and genotype composition of E. bieneusi carried by wild rodents in Heilongjiang Province, China, ultimately evaluating the zoonotic transmission risk through homology and phylogenetic analysis.
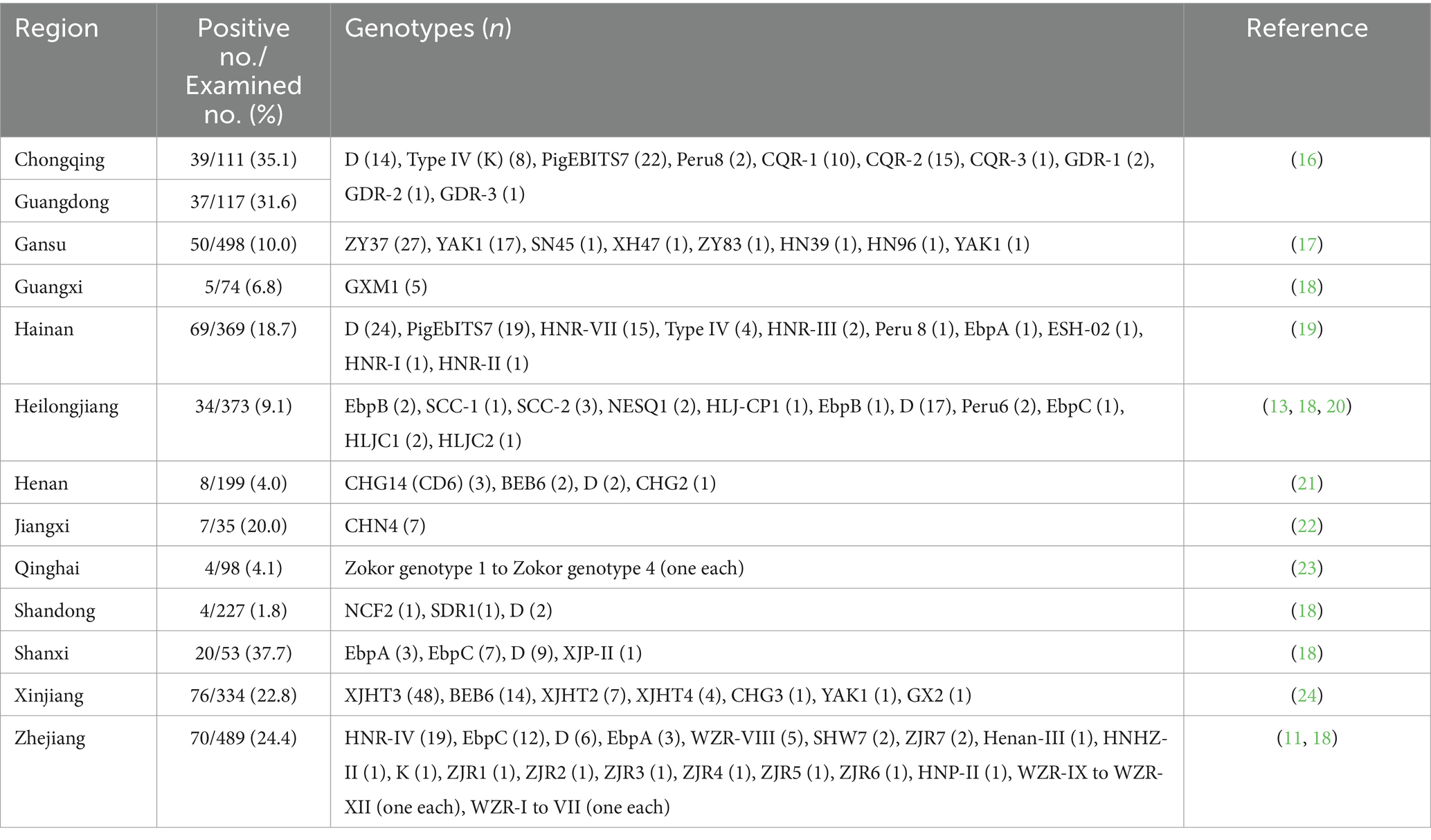
Table 1. Occurrence and distribution of Enterocytozoon bieneusi genotypes in wild rodents in China by Region.
All rodents included in the current study underwent capture and humane euthanasia procedures that strictly adhered to the Chinese Laboratory Animal Administration Act (2017) and the National Institutes of Health’s (NIH, 2020) guidelines for euthanasia of rodents using carbon dioxide (CO2). No rodent species that were endangered or protected were found in the regions that were researched. The objectives and procedures of the present study were reviewed and approved by the National Institute of Parasitic Diseases, Chinese Center for Disease Control and Prevention, China (Reference no. IPD-2021-21).
Between November 2022 and December 2023, a total of 2,000 traps were set up across residential communities in rural areas of Suihua, Qitaihe, Shuangyashan, and Harbin in Heilongjiang Province of China (Figure 1). Each trap was baited with sunflower seeds and peanut/sesame butter to attract the rodents. Throughout the study, a total of 910 rodents were captured, of which 817 were brown rats (Rattus norvegicus). This prompted us to focus solely on this species for the subsequent investigations. The captured brown rats were euthanized by CO2 inhalation. After euthanasia, the animals were transferred to the laboratory while being kept at low temperatures. Fecal samples were carefully taken from the intestinal and rectal areas of each rodent in the biosafety cabinets, following Level 2 containment protocols. Subsequently, these samples were stored in frozen conditions at a temperature of −80°C to maintain their integrity for subsequent analysis.
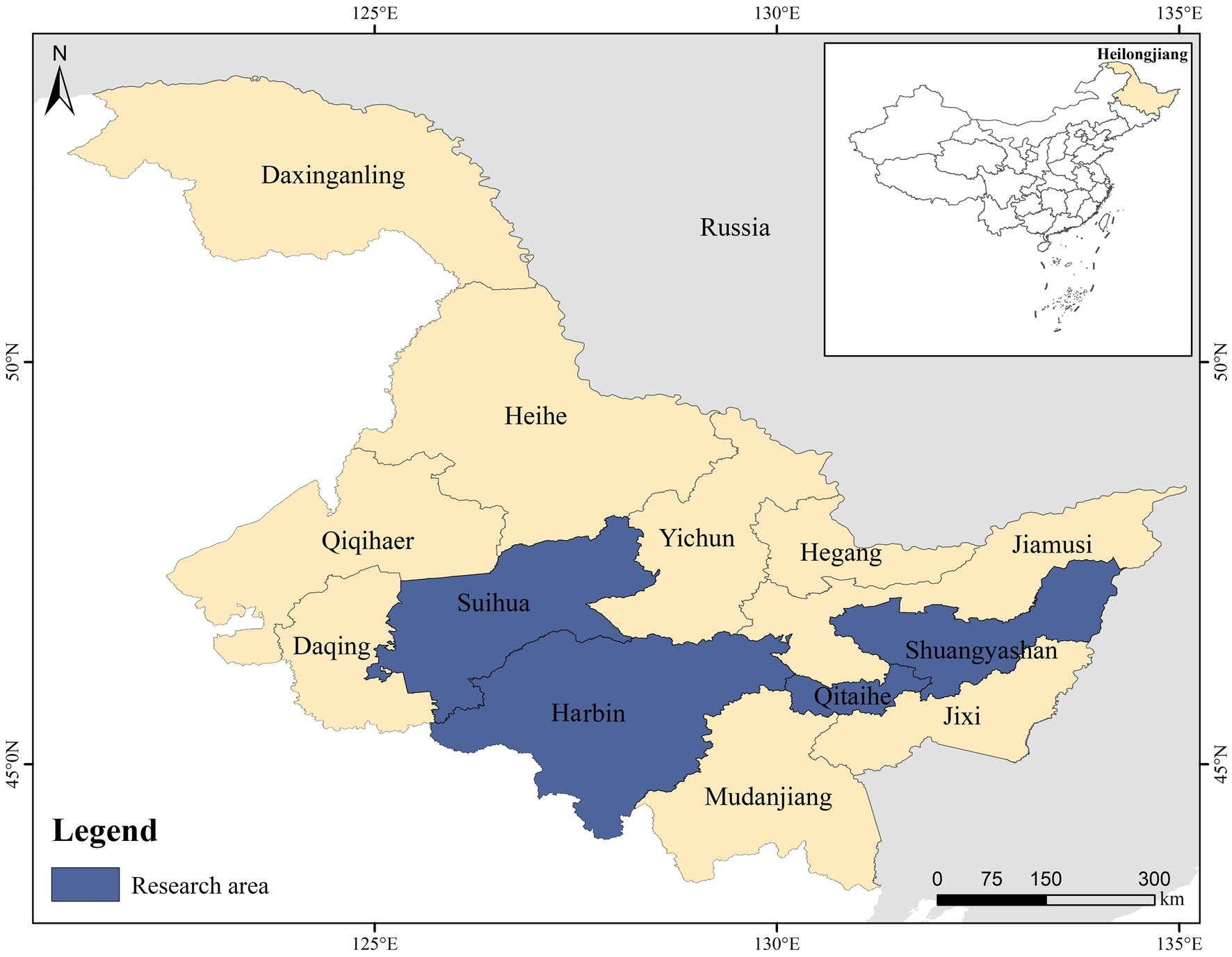
Figure 1. A map that illustrates the locations where Rattus norvegicus was sampled in Heilongjiang Province, China.
For DNA extraction, each fecal sample was washed with distilled water using centrifugation at room temperature for 10 min at a speed of 1,500×g. Genomic DNA was extracted from approximately 200 mg of each processed fecal specimen using the QIAamp DNA Stool Mini Kit (QIAgen, Hilden, Germany) according to the manufacturer’s instructions. DNA was eluted in 200 μl of ATE buffer and was stored at −80°C until required for polymerase chain reaction (PCR)-based analyses.
The R. norvegicus was identified at the species level by amplification of a 762 bp region of the partial mitochondrial cytochrome b (Cytb) gene from fecal DNA using PCR (29). Moreover, each of the DNA specimens was analyzed for E. bieneusi by PCR amplification of the partial SSU rRNA gene (fragment: approximately 390 bp; containing 243 bp of the ITS region), with the primers and the cycle parameters being described previously by Buckholt et al. (30). The PCR reactions were performed using 2 × TransTaq®-T PCR SuperMix (+dye) from TransGen Biotech Co., Beijing, China. A negative control, devoid of any DNA, was included in all PCR experiments. Each DNA sample underwent two replicated PCR reactions to identify and genotype E. bieneusi. The final PCR results were subjected to electrophoresis using a 1.5% agarose gel stained with GelStrain from TransGen Biotech Co., Beijing, China. The gel was then examined, photographed, and recorded using a Gel Doc ™ XR + Imaging System from Bio-Rad, USA.
All the final PCR products of the expected size were sequenced with their respective secondary PCR primers in Shanghai Saiheng Biotechnology Company Limited (Shanghai, China) by an ABI 3730XL Genetic Analyzer (Applied Biosystems, Foster City, CA). To verify sequence accuracy, bidirectional sequencing was employed. The Chromas Pro 1.5 software (Technelysium, Pty, Ltd.) was used to edit the raw sequences used in this study. The sequences were then aligned with each other and reference sequences were downloaded from the GenBank database using the Basic Local Alignment Search Tool (BLASTl; http://blast.ncbi.nlm.nih.gov/Blast.cgi) and MEGA71 to ascertain the genotypes of E. bieneusi. The two PCR products of the expected size were both sequenced from the same sample to make sure accuracy. When the sequences obtained were different, these samples were designed as mixed infection by re-sequencing to confirm the accuracy of the sequences. If the sequences obtained were identical to those published in GenBank, they were considered known genotypes and given the first published name. If not, they were deemed as novel genotypes. To ensure accuracy, two distinct PCR amplicons from the identical DNA preparation were sequenced in the event that the acquired nucleotide sequences deviated from the published ones, including any variations, insertions, or deletions in the nucleotide sequence. Following the established nomenclature scheme, the 243 base pairs of the ITS gene section of the rRNA gene were used to identify each genotype of E. bieneusi (7).
Using the program MEGA 7,2 a neighbor-joining (NJ) tree of the ITS region of all the nucleotide sequences was created to identify the phylogenetic groups and evaluate the genetic relationship and zoonotic potential of novel genotypes of E. bieneusi obtained in this study to the known ones. Evolutionary distances among the nucleotide sequences of the ITS region of the SSU rRNA gene were calculated using the Kimura-2-parameter algorithm. The reliability of branches in trees was assessed by the bootstrap analysis with 1,000 replicates.
Statistical analyses were performed using the Statistical Package for the Social Sciences (SPSS) version 22.0, a software developed by SPSS Inc. in Chicago, Illinois, USA. The chi-square test was utilized to assess the prevalence of E. bieneusi in R. norvegicus across various regional groups, and statistical significance was determined when the p-values were below 0.05.
The nucleotide sequences obtained in this study have been deposited in the GenBank database and can be accessed using accession numbers ranging from PP719220 to PP719286.
During this study, 817 R. norvegicus were screened based on morphological characteristics. Following that, a sequential examination of the partial Cytb gene was carried out to determine the molecular identity of all the animals. Of these, 385 were sampled from Suihua, 210 from Qitaihe, 200 from Shuangyashan, and 22 from Harbin (Table 2). Moreover, analysis of the ITS region of the SSU rRNA gene revealed the presence of E. bieneusi in 33.3% of the brown rats (272/817) sampled across multiple regions. Specifically, the infection rates were 27.5% (106/385) in Suihua, 29.5% (62/210) in Qitaihe, 45.5% (91/200) in Shuangyashan, and 59.1% (13/22) in Harbin (Table 2). These results imply that the infection rates of brown rats from various geographic locations vary significantly (χ2 = 27.1, df = 3, p < 0.001).
From a total of 272 E. bieneusi-positive specimens, 75 representative sequences were obtained, encompassing 75 genotypes. Among these genotypes, 14 were known (A, BEB6, CHPM1, CS-4, D, EbpC, Henan-II, HJL-CP1, HNH-22, HNH-25, I, JLD-X1, SDD5, and WR8), while 61 were novel, designated as SHWR1 to SHWR17, QTHWR1 to QTHWR8, and SYSWR1 to SYSWR36. These genotypes showed significant genetic diversity, with 88 polymorphic sites identified (Figure 2). Of the genotypes identified, genotype D was the most prevalent, encompassing 61.4% (167/272) of the total specimens. Genotype A contributed 5.5% (15/272), genotype HLJ-CP1 accounted for 4.4% (12/272), genotype SHWR1 represented 3.7% (10/272), genotype WR8 contributed 2.2% (6/272), genotypes SYSWR1 and EbpC each contributed 0.7% (2/272), while the remaining genotypes were each represented by a single specimen. Notably, genotype D exhibited the widest geographical distribution, being detected in all four surveyed cities. In contrast, other genotypes demonstrated varying geographical patterns. Genotypes HLJ-CP1 and SHWR1 were found in three cities, except for Harbin. Genotype A was observed in the cities of Suihua and Qitaihe, while genotype EbpC was found in Shuangyashan and Harbin. Exclusive to Suihua were the genotypes WR8, SDD5, and SHWR2 to SHWR17. Similarly, genotypes CHPM1, Henan-II, HNH-22, I, JLD-XI, CS-4, and SYSWR1 to SYSWR36 were distinctively detected in Shuangyashan. Lastly, genotypes BEB6, HNH-25, and QTHWR1 to QTHWR8 were isolated in Qitaihe (Table 2). Meanwhile, 10 samples demonstrated infections with mixed genotypes, specifically, two samples from Qitaihe displayed the D + A genotype, one from Harbin exhibited the D + EbpC genotype, one from Shuangyashan presented with the CS-4 + SYSWR36 genotype, and six samples from Suihua were infected with the D + A, D + HLJ-CP1, D + SHWR1, D + SHWR16, D + SHWR17, and SDD5 + WR8 genotypes (Table 2).
The investigation of 61 novel E. bieneusi genotypes revealed that genotype SYSWR21 exhibited the highest significant genetic diversity. This genotype varied by 10 bases from its closest relative, genotype SDD2. Following this, genotype SYSWR13 exhibited a 9-base difference from its most similar genotype WR7. Genotype SYSWR7 and SYSWR4 had an 8-base difference from the genotypes WR7 and A, respectively. Genotypes SYSWR2 and SYSWR3 differed from genotype WR7 by 7 bases, while SYSWR5 differed from genotype SDD2 by the same margin. Genotypes SYSWR10, SYSWR14, and SYSWR15 each differed from WR7 by 5 bases. Genotypes SYSWR6, SYSWR8, SYSWR9, SYSWR11, and SYSWR12 all differed from genotype WR7 by 4 bases. Moreover, the genotypes SHWR1, SYSWR17, and SYSWR33 differed from genotypes WR7, HLJ-CP1 and D by 3 bases, respectively (Table 3). The genotypes that remained showed minor alterations, differing from the known genotypes by only 1 or 2 bases (Table 4). Notably, this study also identified length variations in the ITS region of E. bieneusi, with genotypes QTHWR7, SYSWR28, SYSWR29, and SYSWR15 exhibiting 242 bp, and genotype SYSWR21 exhibiting 244 bp.
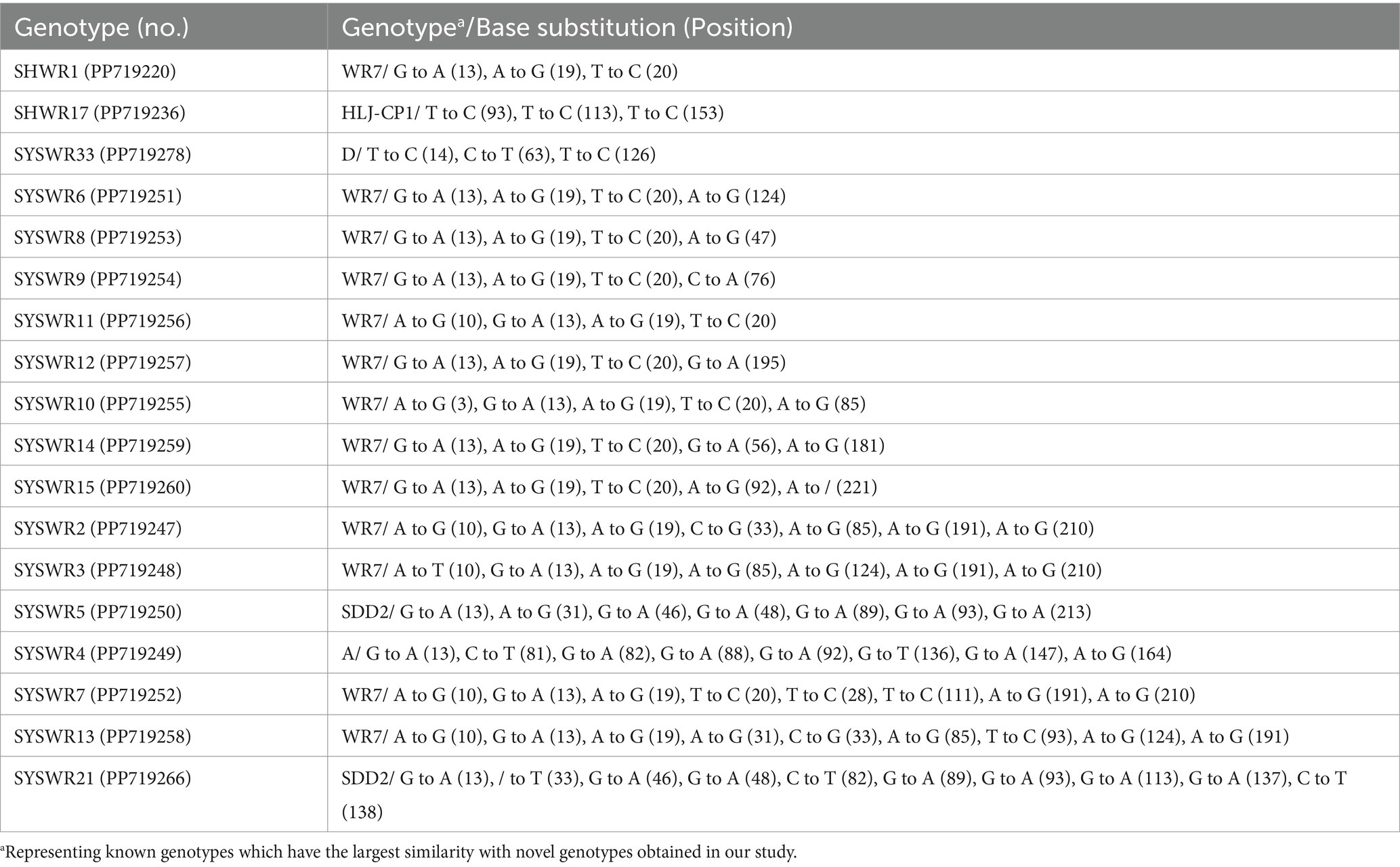
Table 3. The results of similarity analysis comparing novel E. bieneusi genotypes which existence of three or more base differences in the ITS region of the rRNA gene.
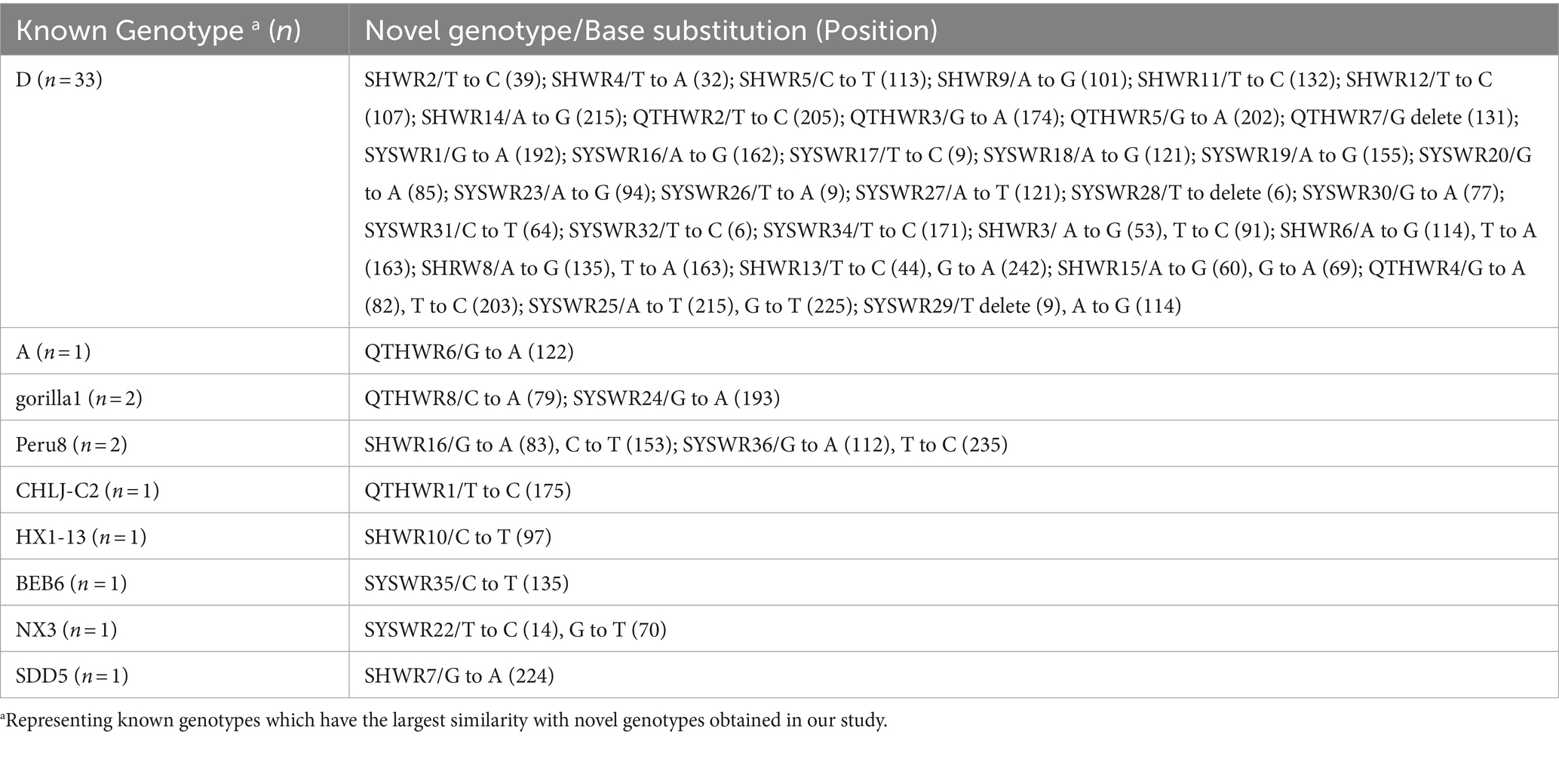
Table 4. The results of similarity analysis comparing novel E. bieneusi genotypes which existence of one or two base differences in the ITS region of the rRNA gene.
In a phylogenetic analysis conducted on a NJ tree of E. bieneusi genotypes, 61 novel genotypes were categorized into three distinct phylogenetic groups. Specifically, 58 genotypes were grouped into Group 1, alongside known genotypes A, CHPM1, CS-4, D, EbpC, Henan-II, HJL-CP1, HNH-22, and HNH-25. Two genotypes were allocated to Group 2, along with genotypes BEB6, JLD-XI, and I. Finally, genotype SHWR7 was assigned to group 9, alongside genotypes SDD5 and WR8. Figure 3 provides a visual representation of the specific results of this phylogenetic analysis, with an emphasis on the novel genotypes found in this investigation.
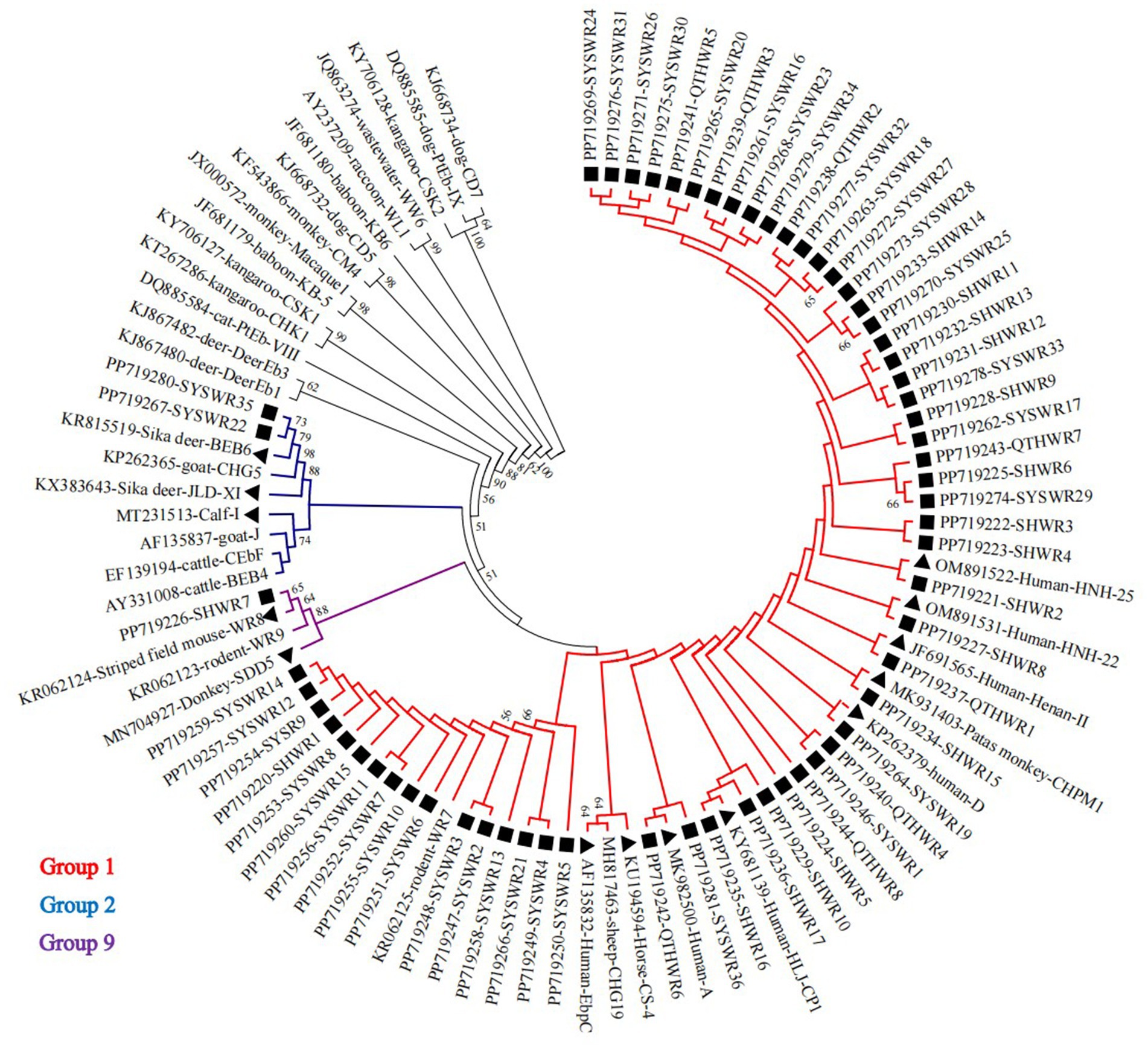
Figure 3. A phylogenetic tree was constructed, utilizing the Kimura-2-parameter model and the neighboring-joining method, to depict the genetic relationships among diverse E. bieneusi genotypes based on their ITS sequences. To ensure the reliability of the tree, bootstrap values were computed from 1,000 replicates. In this tree, genotypes are distinguished by triangle and squares filled in black, signifying known sequences and novel sequences identified in this study, respectively.
Rattus norvegicus is one of the most common species of rodents that have adapted well to anthropized ecosystems, present on all continents except Antarctica. These rats exploit various urban, rural, and sylvatic environments across diverse regions and climates. Their extensive distribution and the ease with which they adapt to various environments and resources might account for the abundance of zoonotic pathogens, including E. bieneusi (15). However, the occurrence of E. bieneusi in R. norvegicus has only been investigated in eight published investigations, all of which were conducted in China (11, 18–20, 22, 37). Except for Shanxi, which reported a slightly higher rate of 37.7% (18), all other studies reported infection rates that were lower than the 33.3% as observed in the present analysis. Even when compared to other rodent species, the present study’s infection rate remains high, given that the global average infection rate of E. bieneusi in rodents is 13.6% (15). Reported infection rates vary among rodent species, with higher rates in chipmunks (71.4%), prairie dogs (48.3%), striped field mice (42.9%), coypus (41.2%), and lesser rice-field rats (36.4%), while lower rates in bamboo rats (5.1%), pet chinchillas (3.6%), and muskrats (8.4%) [(15); (14)]. Poland has a greater prevalence of E. bieneusi infection in rodents compared to other countries (31). The endemicity levels of E. bieneusi infection in other countries are as follows: 8.8% in Iran (32), 21.1% in Spain (33), 18.0% in the United States (34), 10.7% in the border between the Czech Republic and Germany (35), 13.0% in Japan (38), and 1.1% in Slovakia (36). Overall, there is still limited knowledge about E. bieneusi infections in wild rodents worldwide. In general, the prevalence of E. bieneusi in different rodent species across various regions can be attributed to factors such as different rodent species, habitat environment, animal age, health status, population density of the hosts, and other unidentified variables of the animals tested. It is therefore recommended that further studies be conducted to elucidate the global prevalence of E. bieneusi in rodents, as this would aid in implementing effective public health interventions.
The present study has identified a remarkable degree of genetic diversity of E. bieneusi among R. norvegicus, revealing the presence of 75 genotypes. Of these, 14 have been previously characterized, while 61 are newly identified. It is worth mentioning that 10 of the identified genotypes, namely genotypes D, A, EbpC, HLJ-CP1, BEB6, Henan-II, HNH-22, HNH-25, CS-4 and I, have been previously documented in humans (3, 9). The 10 genotypes mentioned in this study make up 71.6% (202/282) of all the genotypes detected. Among these, genotype D is present in all four studied cities, suggesting its extensive dispersion and accounting for 59.2% (167/282) of the total genotypes. Furthermore, genotype D has garnered significant attention in numerous research, not only due to its presence among humans from 40 distinct nations but also because it has been detected in 63 animal hosts across 25 countries (3, 9). Additionally, its presence in certain water and vegetable samples indicates a broader ecological footprint (3). Considering the widespread occurrence of this genotype among rodents, it is not unexpected that it was found in all rodent species examined in this study. Overall, the present results provide further evidence that brown rats play a crucial role in the dissemination of E. bieneusi.
In the current study, genotype A emerged as the second most frequent genotype, trailing genotype D, with a frequency of 5.3% (15/282). This genotype has been identified to have a wide prevalence among individuals from several nations, including China, Cameroon, Gabon, Germany, Netherlands, Niger, Peru, Switzerland, and Thailand (12, 39). Genotype A has been sporadically found in animals such as baboons and dogs (40, 41). Importantly, this study presents the initial finding of this genotype in R. norvegicus, revealing its potentially vast host range. This discovery raises concerns about its potential transmission from R. norvegicus to humans, emphasizing the importance of conducting comprehensive surveys to gain a deeper understanding of its distribution and impact.
The current work found 12 R. norvegicus rats infected with the HLJ-CP1 genotype. This genotype was first found in a tumor patient and then detected in a wild rat from the same study region (13, 42). This observation strongly hints at the potential risk of interspecies and zoonotic transmission of this genotype within the same region. Consequently, it suggested that the genotype in tumor patients may have originated from rodents, and vice versa. Although EbpC was detected in only two R. norvegicus, this genotype was also identified to be widespread in native children, adult HIV patients, and multiple animal hosts, especially pigs (25, 26, 28, 43, 44). This further suggests the important role of rodents in the transmission of EbpC between humans and other animals.
Among the zoonotic genotypes identified, genotypes BEB6, Henan-II, HNH-22, HNH-25, CS-4 and I were only identified in a limited number of samples. These genotypes have been reported in only a few human cases from China. For instance, genotype BEB6 infected a child, Henan-II infected an HIV-positive patient, genotype I infected a child with diarrhea and an HIV-positive individual, while HNH-22 (OM891522) and HNH-25 (OM891531) were detected in humans residing in Hainan Province, China (39, 45–47). It is worth mentioning that, in addition to the genotype BEB6, additional genotypes were found in R. norvegicus and even rodents for the first time. This finding suggests that these genetic types may have a similar wide range of hosts, which requires more research to determine the exact source of human infection. As demonstrated in the present study, the existence of a novel host was verified, highlighting the need for continued research in this area.
No reported cases of human infections have been associated with the other genotypes (CHPM1, WR8, JLD-XI, and SDD5) identified in this study. The majority of these genotypes are restricted to certain animal hosts: for example, genotype JLD-XI exists in sika deer (48), genotype SDD5 (49) is identified in donkeys, and genotype CHPM1 is found in Patas monkeys (50). This study observed the presence of these genotypes in R. norvegicus, indicating a potentially significant role of these small rodents in the dissemination of E. bieneusi among wildlife, the environment, and domesticated animals. However, there remains uncertainty regarding the host range of these genotypes and the probability of zoonotic transmission. While this study offers novel insights, more thorough research is necessary to fully comprehend these problems.
A total of 61 previously novel genotypes were discovered in this investigation, which is quite high. Out of these genotypes, 54.1% (33/61) exhibited a difference of only one or two bases compared to genotype D (Table 4), suggesting that they may have evolved from genotype D. Therefore, they may possess a broad host range and a high potential for zoonotic transmission, similar to genotype D. Moreover, the evolutionary analysis revealed that nearly all of the novel genotypes, excluding SHWR7, belonged to either Group 1 or 2, with a significant preponderance of 95.1% (58/61) belonging to Group 1, indicating a potential for zoonotic transmission. However, it is becoming increasingly apparent that depending only on ITS sequence data is insufficient to provide a strong phylogenetic signal across the entire tree (9). Therefore, future studies necessitate the utilization of additional genetic markers to comprehensively understand the genetic affinities among these genotypes. Additionally, conclusive evidence of cross-species and zoonosis transmission requires further study.
The current study has uncovered a high infection rate of E. bieneusi in R. norvegicus in Heilongjiang Province, China. Notably, genotyping analysis revealed an exceptionally diverse genotype composition, consisting of 14 known genotypes and 61 novel genotypes. Remarkably, zoonotic genotypes constituted a significant share of 71.6 and 98.4% novel genotypes were classified into Group 1 and Group 2. These findings underscore the crucial role of R. norvegicus in the transmission of E. bieneusi and highlight the potential public health risks in this region. Therefore, it is crucial to increase awareness regarding the hazards presented by these creatures and establish strategies to reduce their abundance, thereby minimizing environmental contamination. Additionally, the identification of 61 novel genotypes expands the understanding of the vast genetic diversity present in E. bieneusi. Further studies are necessary to better comprehend the genetic variety of the parasite and uncover its secrets.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
The animal study was approved by the National Institute of Parasitic Diseases, Chinese Center for Disease Control and Prevention, China (Reference no. IPD-2021-21). The study was conducted in accordance with the local legislation and institutional requirements.
YJ: Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Writing – original draft, Writing – review & editing. SZ: Formal analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. ZY: Formal analysis, Methodology, Visualization, Writing – original draft, Writing – review & editing. XH: Investigation, Methodology, Writing – original draft, Writing – review & editing. ZL: Formal analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. YW: Formal analysis, Methodology, Writing – original draft, Writing – review & editing. YS: Conceptualization, Resources, Supervision, Writing – original draft, Writing – review & editing. JC: Conceptualization, Resources, Supervision, Validation, Writing – original draft, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Key Research and Development Program of China (2021YFC2300902) and the National Natural Science Foundation of China (82273693).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Fadhilah, A, Gabbar, A, and Bokhari, AA. Microsporidium. Treasure Island (FL): StatPearls Publishing (2023).
2. Han, B, Pan, G, and Weiss, LM. Microsporidiosis in humans. Clin Microbiol Rev. (2021) 34:e0001020. doi: 10.1128/CMR.00010-20
3. Ruan, Y, Xu, X, He, Q, Li, L, Guo, J, Bao, J, et al. The largest meta-analysis on the global prevalence of microsporidia in mammals, avian and water provides insights into the epidemic features of these ubiquitous pathogens. Parasit Vectors. (2021) 14:186. doi: 10.1186/s13071-021-04700-x
4. Li, W, and Xiao, L. Ecological and public health significance of Enterocytozoon bieneusi. One Health. (2020) 12:100209. doi: 10.1016/j.onehlt.2020.100209
5. Bourli, P, Eslahi, AV, Tzoraki, O, and Karanis, P. Waterborne transmission of protozoan parasites: a review of worldwide outbreaks - an update 2017-2022. J Water Health. (2023) 21:1421–47. doi: 10.2166/wh.2023.094
6. Decraene, V, Lebbad, M, Botero-Kleiven, S, Gustavsson, AM, and Löfdahl, M. First reported foodborne outbreak associated with microsporidia, Sweden, October 2009. Epidemiol Infect. (2012) 140:519–27. doi: 10.1017/S095026881100077X
7. Santín, M, and Fayer, R. Enterocytozoon bieneusi genotype nomenclature based on the internal transcribed spacer sequence: a consensus. J Eukaryot Microbiol. (2009). 56:34–8. doi: 10.1111/j.1550-7408.2008.00380.x
8. Li, W, Feng, Y, and Santin, M. Host specificity of Enterocytozoon bieneusi and public health implications. Trends Parasitol. (2019) 35:436–51. doi: 10.1016/j.pt.2019.04.004
9. Koehler, AV, Zhang, Y, and Gasser, RB. A perspective on the molecular identification, classification, and epidemiology of Enterocytozoon bieneusi of animals. Exp Suppl. (2022) 114:389–415. doi: 10.1007/978-3-030-93306-7_14
10. Wang, Y, Li, XM, Yang, X, Wang, XY, Wei, YJ, Cai, Y, et al. Global prevalence and risk factors of Enterocytozoon bieneusi infection in humans: a systematic review and meta-analysis. Parasite. (2024) 31:9. doi: 10.1051/parasite/2024007
11. Zhang, T, Yu, K, Xu, J, Cao, W, Wang, Y, Wang, J, et al. Enterocytozoon bieneusi in wild rats and shrews from Zhejiang Province, China: occurrence, genetic characterization, and potential for zoonotic transmission. Microorganisms. (2024) 12:811. doi: 10.3390/microorganisms12040811
12. Santín, M, and Fayer, R. Microsporidiosis: Enterocytozoon bieneusi in domesticated and wild animals. Res Vet Sci. (2011) 90:363–71. doi: 10.1016/j.rvsc.2010.07.014
13. Jiang, S, Yu, S, Feng, Y, Zhang, L, Santin, M, Xiao, L, et al. Widespread distribution of human-infective Enterocytozoon bieneusi genotypes in small rodents in Northeast China and phylogeny and zoonotic implications revisited. Acta Trop. (2024) 253:107160. doi: 10.1016/j.actatropica.2024.107160
14. Zhao, W, Wang, T, Ren, G, Li, J, Tan, F, Li, W, et al. Molecular detection of Enterocytozoon bieneusi in farmed Asiatic brush-tailed porcupines (Atherurus macrourus) and bamboo rats (Rhizomys pruinosus) from Hainan Province, China: Common occurrence, wide genetic variation and high zoonotic potential. Acta Trop. (2023) 242:106915. doi: 10.1016/j.actatropica.2023.106915
15. Taghipour, A, Bahadory, S, Abdoli, A, and Javanmard, E. A systematic review and meta-analysis on the global molecular epidemiology of microsporidia infection among rodents: a serious threat to public health. Acta Parasitol. (2022) 67:18–30. doi: 10.1007/s11686-021-00447-8
16. Gui, BZ, Zou, Y, Chen, YW, Li, F, Jin, YC, Liu, MT, et al. Novel genotypes and multilocus genotypes of Enterocytozoon bieneusi in two wild rat species in China: potential for zoonotic transmission. Parasitol Res. (2020) 119:283–90. doi: 10.1007/s00436-019-06491-8
17. Xu, J, Wang, X, Jing, H, Cao, S, Zhang, X, Jiang, Y, et al. Identification and genotyping of Enterocytozoon bieneusi in wild himalayan marmots (Marmota himalayana) and alashan ground squirrels (Spermophilus alashanicus) in the Qinghai-Tibetan plateau area (QTPA) of Gansu Province, China. Parasit Vectors. (2020) 13:367. doi: 10.1186/s13071-020-04233-9
18. Ni, HB, Sun, YZ, Qin, SY, Wang, YC, Zhao, Q, Sun, ZY, et al. Molecular detection of Cryptosporidium spp. and Enterocytozoon bieneusi infection in wild rodents from six provinces in China. Front Cell Infect Microbiol. (2021) 11:783508. doi: 10.3389/fcimb.2021.783508
19. Zhao, W, Zhou, H, Yang, L, Ma, T, Zhou, J, Liu, H, et al. Prevalence, genetic diversity and implications for public health of Enterocytozoon bieneusi in various rodents from Hainan Province, China. Parasit Vectors. (2020) 13:438. doi: 10.1186/s13071-020-04314-9
20. Zhao, W, Wang, J, Ren, G, Yang, Z, Yang, F, Zhang, W, et al. Molecular characterizations of Cryptosporidium spp. and Enterocytozoon bieneusi in brown rats (Rattus norvegicus) from Heilongjiang province, China. Parasit Vectors. (2018) 11:313. doi: 10.1186/s13071-018-2892-7
21. Yu, F, Qi, M, Zhao, Z, Lv, C, Wang, Y, Wang, R, et al. The potential role of synanthropic rodents and flies in the transmission of Enterocytozoon bieneusi on a dairy cattle farm in China. J Eukaryot Microbiol. (2019) 66:435–41. doi: 10.1111/jeu.12687
22. Yu, F, Cao, Y, Wang, H, Liu, Q, Zhao, A, Qi, M, et al. Host-adaptation of the rare Enterocytozoon bieneusi genotype CHN4 in Myocastor coypus (Rodentia: Echimyidae) in China. Parasit Vectors. (2020) 13:578. doi: 10.1186/s13071-020-04436-0
23. Hu, B, Wang, J, Zhang, S, Wang, B, Xing, Y, Han, S, et al. Novel genotypes of Cryptosporidium and Enterocytozoon bieneusi detected in plateau zokors (Myospalax baileyi) from the Tibetan plateau. Int J Parasitol Parasites Wildl. (2022) 19:263–8. doi: 10.1016/j.ijppaw.2022.11.002
24. Tuo, H, Zhang, B, He, Y, Zhao, A, Zhang, Z, Qi, M, et al. Molecular characterization of Enterocytozoon bieneusi genotypes in wild Altai marmot (Marmota baibacina) in Xinjiang, China: host specificity and adaptation. Parasitol Res. (2023) 123:7. doi: 10.1007/s00436-023-08057-1
25. Yang, J, Song, M, Wan, Q, Li, Y, Lu, Y, Jiang, Y, et al. Enterocytozoon bieneusi genotypes in children in Northeast China and assessment of risk of zoonotic transmission. J Clin Microbiol. (2014) 52:4363–7. doi: 10.1128/JCM.02295-14
26. Zhao, W, Yao, L, Zhuang, M, Lin, YL, Chen, XH, Wang, L, et al. A baseline epidemiological study of the co-infection of enteric protozoans with human immunodeficiency virus among men who have sex with men from Northeast China. PLoS Negl Trop Dis. (2022) 16:e0010712. doi: 10.1371/journal.pntd.0010712
27. Zhao, W, Zhang, W, Yang, F, Cao, J, Liu, H, Yang, D, et al. High prevalence of Enterocytozoon bieneusi in asymptomatic pigs and assessment of zoonotic risk at the genotype level. Appl Environ Microbiol. (2014) 80:3699–707. doi: 10.1128/AEM.00807-14
28. Zhou, K, Liu, M, Wu, Y, Zhang, R, Wang, R, Xu, H, et al. Enterocytozoon bieneusi in patients with diarrhea and in animals in the northeastern Chinese city of Yichun: genotyping and assessment of potential zoonotic transmission. Parasite. (2022) 29:40. doi: 10.1051/parasite/2022041
29. Robins, JH, Hingston, M, Matisoo-Smith, L, and Ross, HA. Identifying Rattus species using mitochondrial DNA. Mol Ecol Notes. (2007) 7:717–29. doi: 10.1111/j.1471-8286.2007.01752.x
30. Buckholt, MA, Lee, JH, and Tzipori, S. Prevalence of Enterocytozoon bieneusi in swine: an 18-month survey at a slaughterhouse in Massachusetts. Appl Environ Microbiol. (2002) 68:2595–9. doi: 10.1128/AEM.68.5.2595-2599.2002
31. Perec-Matysiak, A, Buńkowska-Gawlik, K, Kváč, M, Sak, B, Hildebrand, J, and Leśniańska, K. Diversity of Enterocytozoon bieneusi genotypes among small rodents in southwestern Poland. Vet Parasitol. (2015) 214:242–6. doi: 10.1016/j.vetpar.2015.10.018
32. Tavalla, M, Kazemi, F, Mardani - Kateki, F, and Abdizadeh, R Molecular Diagnosis of Enterocytozoon bieneusi and Encephalitozoon spp. in Wild Rats of Southwest of Iran. Jundishapur J Microbiol. (2018) 11:e55961
33. Vioque, F, Dashti, A, Santín, M, Ruiz-Fons, F, Köster, PC, Hernández-Castro, C, et al. Wild micromammal host spectrum of zoonotic eukaryotic parasites in Spain. Occurrence and genetic characterisation. Transbound Emerg Dis. (2022) 69:e2926–e2942. doi: 10.1111/tbed.14643
34. Guo, Y, Alderisio, KA, Yang, W, Cama, V, Feng, Y, and Xiao, L. Host specificity and source of Enterocytozoon bieneusi genotypes in a drinking source watershed. Appl Environ Microbiol. (2014) 80:218–25. doi: 10.1128/AEM.02997-13
35. Sak, B, Kváč, M, Květoňová, D, Albrecht, T, and Piálek, J. The first report on natural Enterocytozoon bieneusi and Encephalitozoon spp. infections in wild East-European House Mice (Mus musculus musculus) and West-European House Mice (M. m. domesticus) in a hybrid zone across the Czech Republic-Germany border. Vet Parasitol. (2011) 178:246–50. doi: 10.1016/j.vetpar.2010.12.044
36. Danišová, O, Valenčáková, A, Stanko, M, Luptáková, L, and Hasajová, A. First report of Enterocytozoon bieneusi and Encephalitozoon intestinalis infection of wild mice in Slovakia. Ann Agric Environ Med. (2015) 22:251–2. doi: 10.5604/12321966.1152075
37. Wang, J, Lv, C, Zhao, D, Zhu, R, Li, C, and Qian, W. First detection and genotyping of Enterocytozoon bieneusi in pet fancy rats (Rattus norvegicus) and guinea pigs (Cavia porcellus) in China. Parasite. (2020) 27:21. doi: 10.1051/parasite/2020019
38. Masuda, A, Wada, M, Saho, H, Tokunaga, K, Kikuchi, Y, Yamasaki, F, et al. Prevalence and molecular characterization of the zoonotic enteric protozoans Cryptosporidium spp., Enterocytozoon bieneusi, and Blastocystis from pallas's squirrels (Callosciurus erythraeus) in Kanagawa prefecture, Japan. Microbiol Spectr. (2021) 9:e0099021. doi: 10.1128/Spectrum.00990-21
39. Jiang, Y, Liu, L, Yuan, Z, Liu, A, Cao, J, and Shen, Y. Molecular identification and genetic characteristics of Cryptosporidium spp., Giardia duodenalis, and Enterocytozoon bieneusi in human immunodeficiency virus/acquired immunodeficiency syndrome patients in Shanghai, China. Parasit Vectors. (2023) 16:53. doi: 10.1186/s13071-023-05666-8
40. Galván-Díaz, AL, Magnet, A, Fenoy, S, Henriques-Gil, N, Haro, M, Gordo, FP, et al. Microsporidia detection and genotyping study of human pathogenic E. bieneusi in animals from Spain. PLoS One. (2014) 9:e92289. doi: 10.1371/journal.pone.0092289
41. Li, W, Kiulia, NM, Mwenda, JM, Nyachieo, A, Taylor, MB, Zhang, X, et al. Cyclospora papionis, Cryptosporidium hominis, and human-pathogenic Enterocytozoon bieneusi in captive baboons in Kenya. J Clin Microbiol. (2011) 49:4326–9. doi: 10.1128/JCM.05051-11
42. Zhang, W, Ren, G, Zhao, W, Yang, Z, Shen, Y, Sun, Y, et al. Genotyping of Enterocytozoon bieneusi and subtyping of Blastocystis in cancer patients: relationship to diarrhea and assessment of zoonotic transmission. Front Microbiol. (2017) 8:1835. doi: 10.3389/fmicb.2017.01835
43. Li, W, Li, Y, Li, W, Yang, J, Song, M, Diao, R, et al. Genotypes of Enterocytozoon bieneusi in livestock in China: high prevalence and zoonotic potential. PLoS One. (2014) 9:e97623. doi: 10.1371/journal.pone.0097623
44. Wan, Q, Xiao, L, Zhang, X, Li, Y, Lu, Y, Song, M, et al. Clonal evolution of Enterocytozoon bieneusi populations in swine and genetic differentiation in subpopulations between isolates from swine and humans. PLoS Negl Trop Dis. (2016) 10:e0004966. doi: 10.1371/journal.pntd.0004966
45. Wang, L, Xiao, L, Duan, L, Ye, J, Guo, Y, Guo, M, et al. Concurrent infections of Giardia duodenalis, Enterocytozoon bieneusi, and Clostridium difficile in children during a cryptosporidiosis outbreak in a pediatric hospital in China. PLoS Negl Trop Dis. (2013) 7:e2437. doi: 10.1371/journal.pntd.0002437
46. Wang, L, Zhang, H, Zhao, X, Zhang, L, Zhang, G, Guo, M, et al. Zoonotic Cryptosporidium species and Enterocytozoon bieneusi genotypes in HIV-positive patients on antiretroviral therapy. J Clin Microbiol. (2013) 51:557–63. doi: 10.1128/JCM.02758-12
47. Zhang, X, Wang, Z, Su, Y, Liang, X, Sun, X, Peng, S, et al. Identification and genotyping of Enterocytozoon bieneusi in China. J Clin Microbiol. (2011) 49:2006–8. doi: 10.1128/JCM.00372-11
48. Huang, J, Zhang, Z, Yang, Y, Wang, R, Zhao, J, Jian, F, et al. New genotypes of Enterocytozoon bieneusi isolated from sika deer and red deer in China. Front Microbiol. (2017) 8:879. doi: 10.3389/fmicb.2017.00879
49. Li, F, Wang, R, Guo, Y, Li, N, Feng, Y, and Xiao, L. Zoonotic potential of Enterocytozoon bieneusi and Giardia duodenalis in horses and donkeys in northern China. Parasitol Res. (2020) 119:1101–8. doi: 10.1007/s00436-020-06612-8
50. Zhang, K, Zheng, S, Wang, Y, Wang, K, Wang, Y, Gazizova, A, et al. Occurrence and molecular characterization of Cryptosporidium spp., Giardia duodenalis, Enterocytozoon bieneusi, and Blastocystis sp. in captive wild animals in zoos in Henan, China. BMC Vet Res. (2021) 17:332. doi: 10.1186/s12917-021-03035-0
Keywords: Enterocytozoon bieneusi, Rattus norvegicus, zoonotic, genotype, China
Citation: Jiang Y, Zhou S, Yuan Z, Hu X, Li Z, Wang Y, Shen Y and Cao J (2024) Brown rats (Rattus norvegicus) as potential reservoirs of Enterocytozoon bieneusi in Heilongjiang Province, China: high prevalence, genetic heterogeneity, and potential risk for zoonotic transmission. Front. Vet. Sci. 11:1426384. doi: 10.3389/fvets.2024.1426384
Received: 23 May 2024; Accepted: 08 July 2024;
Published: 25 July 2024.
Edited by:
Fuchang Yu, Tarim University, ChinaReviewed by:
Rongjun Wang, Henan Agricultural University, ChinaCopyright © 2024 Jiang, Zhou, Yuan, Hu, Li, Wang, Shen and Cao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianping Cao, Y2FvanBAY2hpbmFjZGMuY24=; Yujuan Shen, c2hlbnlqQG5pcGQuY2hpbmFjZGMuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.