- 1Optipharm, Inc., Cheongju, Republic of Korea
- 2Optipharm Animal Disease Diagnostic Center, Cheongju, Republic of Korea
Introduction: Research quality can be improved with reliable and reproducible experimental results when animal experiments are conducted using laboratory animals with guaranteed microbiological and genetic quality through health monitoring. Therefore, health monitoring requires the rapid and accurate diagnosis of infectious diseases in laboratory animals.
Methods: This study presents a performance evaluation of a commercially available multiplex real-time PCR (mRT-PCR) assay for the rapid detection of 12 infectious pathogens (Set 1: Sendai virus [SeV, formally murine respirovirus], Mycoplasma spp., Rodentibacter pneumotropicus, and Rodentibacter heylii; Set 2: Helicobacter spp., Murine norovirus [MNV], Murine hepatitis virus [MHV], and Salmonella spp.; Set 3: Staphylococcus aureus, Streptobacillus moniliformis, Corynebacterium kutscheri, and Pseudomonas aeruginosa). To evaluate the efficacy of the mRT-PCR assay, 102 clinical samples encompassing fecal and cecal specimens were analyzed. The resulting data were then compared with the findings from sequence analysis for validation.
Results: The assay’s detection limit ranged from 1 to 100 copies per reaction. Specificity testing involving various viruses and bacteria indicated no cross-reactivity between strains. Additionally, the assay exhibited good reproducibility, with mean coefficients of variation for inter- and intra assay variation below 3%. The overall positive rate was 52.9% (n = 54), with the mRT-PCR assay findings matching sequence analysis results (κ = 1). MHV (n = 29, 28.4%) was the most prevalent pathogen, followed by Helicobacter spp. (n = 28, 27.5%), R. heylii (n = 18, 17.6%), Mycoplasma spp. (n = 14, 13.7%), MNV (n = 12, 11.8%), S. aureus (n = 9, 8.8%), P. aeruginosa (n = 4, 3.9%), and R. pneumotropicus (n = 1, 0.9%).
Discussion: This assay offers a rapid turnaround time of 100 min, including 30 min for DNA preparation and 70 min for target DNA/RNA amplification. It ensures accuracy, minimizing false positives or negatives, making it a convenient tool for the simultaneous detection of infectious diseases in many samples. Overall, the propose‑d assay holds promise for the effective detection of the most important pathogens in laboratory animal health monitoring.
Introduction
Laboratory mice and rats are now widely used in animal experiments in various fields of research. To obtain reliable and reproducible results from animal experiments, laboratory animals must be maintained in a protected environment to avoid microbiological interference, and quality must be guaranteed through health monitoring (1). Therefore, microbiological quality control should be performed regularly to determine whether laboratory animals are free from various pathogens, including viruses, bacteria, fungi, and parasites (2).
Guidelines for the health monitoring of experimental animals were published by the Federation of European Laboratory Animal Science Associations (FELASA) (3). Until now, the health monitoring of laboratory animals has relied primarily on microscopic and culture-based methods (4). Serological test methods, such as enzyme-linked immunosorbent assay for primary screening or immunofluorescence assay (IFA) for confirmation, are mainly used to detect viruses or bacteria that are difficult to culture (5, 6). Due to the propensity for nonspecific reactions leading to false positives with these analysis methods, microbial quality control has been concurrently conducted alongside molecular diagnostic assays like polymerase chain reaction (PCR) or reverse transcription-polymerase chain reaction (RT-PCR) for cross-validation purposes (7–10). These assays are expensive, exhibit low sensitivity, require electrophoresis of the amplification products, and pose a contamination risk, potentially compromising result accuracy. Real-time (RT) PCR assay, the so-called quantitative PCR, has been a powerful analytical tool for detecting pathogenic bacteria or viruses since their development (11). Given its remarkable speed, heightened sensitivity, and precision, RT-PCR finds extensive use in pathogen diagnosis spanning animals, humans, and plants, and is additionally employed in laboratory animal studies (12–20).
Pathogens are commonly found in various systems, such as the respiratory, digestive, nervous, and other systems of laboratory animals. Sendai virus (SeV, formally murine respirovirus), Mycoplasma spp., murine hepatitis virus (MHV), Murine norovirus (MNV), and Helicobacter spp. are the primary pathogens found in the respiratory and digestive systems that pose marked interference with experimental results (2, 21, 22). In this study, we developed three sets of multiplex real-time PCR (mRT-PCR) assays that can simultaneously detect four different viral or bacterial pathogens causing respiratory (Opti SeV/MP/Rpn/Rhey Multi-qPCR; Sendai virus, Mycoplasma spp., Rodentibacter pneumotropicus, R. heylii), digestive (Opti Hel/MNV/MHV/Sal Multi-qPCR; Helicobacter spp., MNV, MHV, Salmonella spp.), and abscess or sepsis (Opti Sau/Smo/Cku/Pae Multi-qPCR; S. aureus, S. moniliformis, C. kutscheri, P. aeruginosa) diseases. We also confirmed their performance evaluations. The clinical performance of the mRT-PCR assay was evaluated using clinical samples requested for health monitoring of mice and rats, and the results were confirmed by sequence analysis.
Methods
Sample preparation
Seven viruses (SeV, MNV, MHV, Murine adenovirus, Ectromelia virus, Mouse minute virus, and Murine rotavirus) and thirty-five bacterial strains for specificity testing were provided by Xpressbio, the International Council for Laboratory Animal Science (ICLAS), and the American Type Culture Collection (ATCC). Provided by the Optipharm Animal Disease Diagnostic Center commissioned from 2020 to 2023, 102 fecal/cecal samples and health statuses were used to evaluate the diagnostic performance of the mRT-PCR assay. According to the manufacturer’s recommendation, DNA/RNA was extracted from 100 μL of phosphate-buffered saline (PBS)-pretreated samples using a commercial automated system (Miracle-AutoXT Automated Nucleic Acid Extraction System, Intronbio, Seongnam, Republic of Korea). To avoid cross-contamination, all samples were individually processed and stored at −20°C. The content and purity of the extracted DNA/RNA were analyzed by measuring the absorbance at 260 and 280 nm using a spectrophotometer (Infinite 200 NanoQuant; Tecan, Switzerland).
Single RT-PCR assay
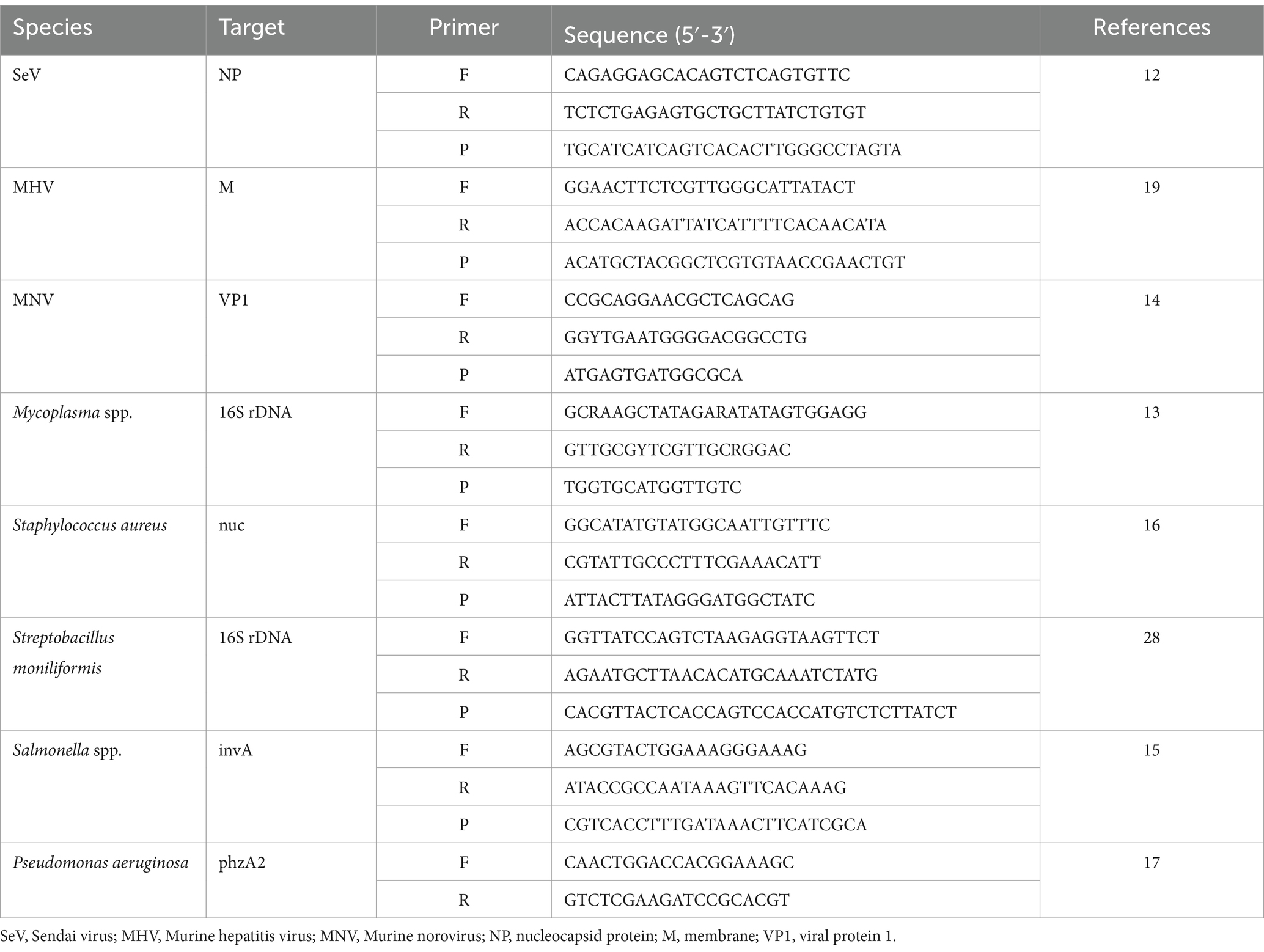
The effectiveness of the mRT-PCR assay was evaluated by comparing it with a single RT-PCR assay from a previously published reference. Table 1 contains the primers and probes used for the single RT-PCR. Each RT-PCR was performed according to the methods described in the paper.
mRT-PCR assay
Twelve oligonucleotide primers and probes corresponding to two strands were designed using Primer3Plus1 at the positions of each gene (see Figure 1). Primers were prepared as probes corresponding to the complementary strands and used exclusively thereafter. To verify the efficiency of the selected primers and probes, a positive control DNA sample was synthesized using Bioneer (Daejeon, Republic of Korea). The resulting product was mutagenized after subcloning with a pBHA vector. Clinical samples were screened for the presence of 12 diseases using Opti Multi-qPCR kits (Set 1: Opti SeV/MP/Rpn/Rhey; Set 2: Opti Hel/MNV/MHV/Sal; and Set 3: Opti Sau/Smo/Cku/Pae, Optipharm, Cheongju, Republic of Korea). These kits utilize a quantitative mRT-PCR-based assay and were processed using a CFX-96 RT-PCR system (Bio-Rad, Hercules, CA, USA) for both thermal cycling and fluorescence detection. RT-PCR amplification was performed in a total reaction volume of 20 μL containing 10 μL of 2× Thunderbird probe qPCR mix (Toyobo, Osaka, Japan), 2.5 μL of a mixture of 5 pmol each primer and 5 pmol TaqMan probe that were labeled with fluorophores (FAM/HEX-BHQ1, CalRed610/Cy5-BHQ2), and 3 μL template DNA/RNA. Positive (plasmid DNA) and negative controls comprising molecular grade (DNAse/RNAse-free) water (Ultra-pure water; Welgene, Gyeongsan, Republic of Korea) without template DNA/RNA were included in each assay. The assay was performed under the following conditions: For DNA, 95°C for 3 min, then 10 cycles of 3 s at 95°C and 30 s at 60°C, and then by 40 cycles of 3 s at 95°C and 30 s at 55°C; for RNA, 50°C for 2 min, 95°C for 2 min, then 10 cycles of 3 s at 95°C and 30 s at 60°C, and then by 40 cycles of 3 s at 95°C and 30 s at 55°C. Each sample was tested in duplicate by running the PCR cycle twice, and a positive result was obtained when the CT value was below 35.
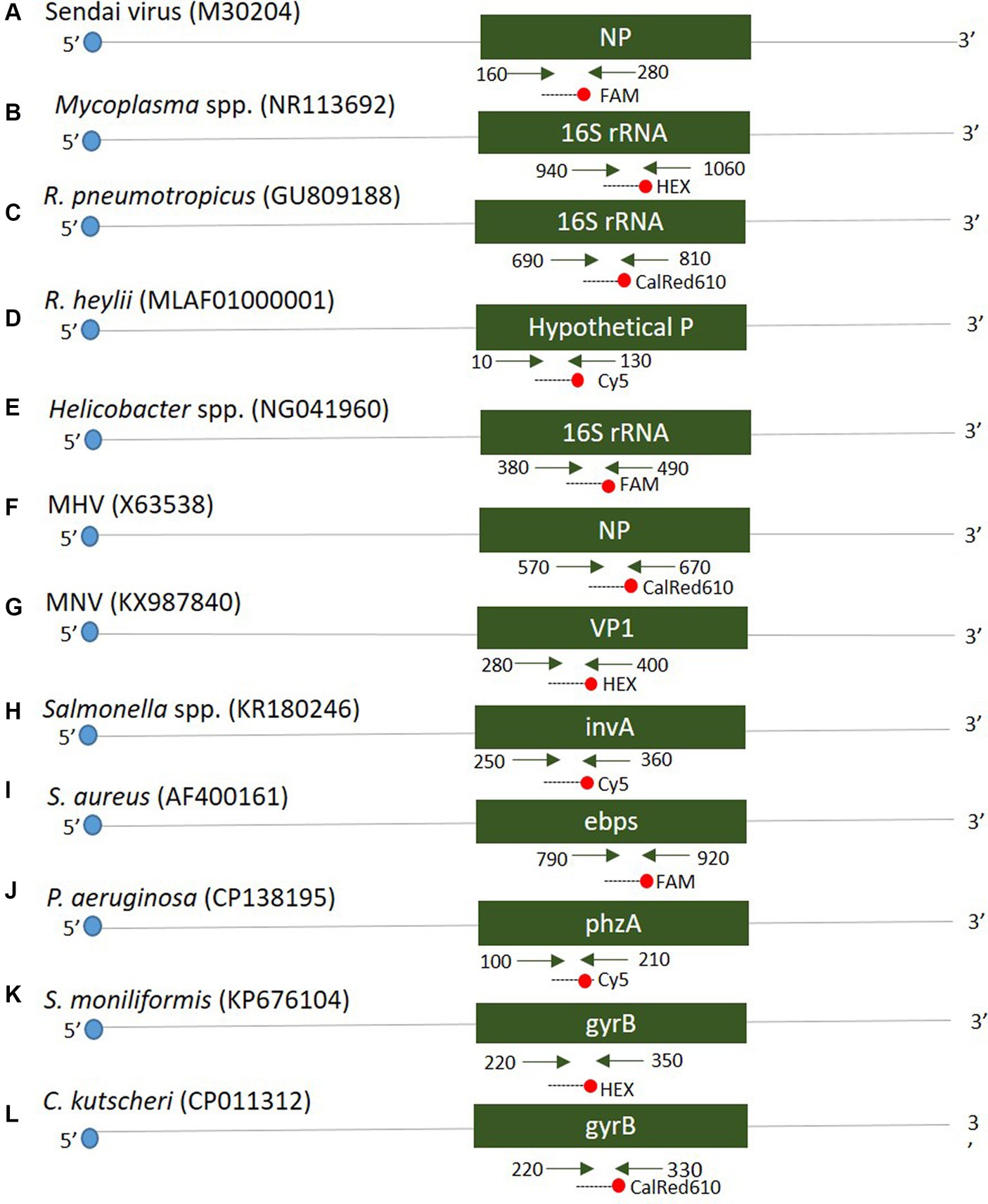
Figure 1. Schematic diagram distinguishing the positions of primers and probes of 12 target genes. A positive control was synthesized to verify the efficiency of the developed primers and probes. The mark in parentheses is the NCBI accession number.
Interfering reactions and reproducibility analysis
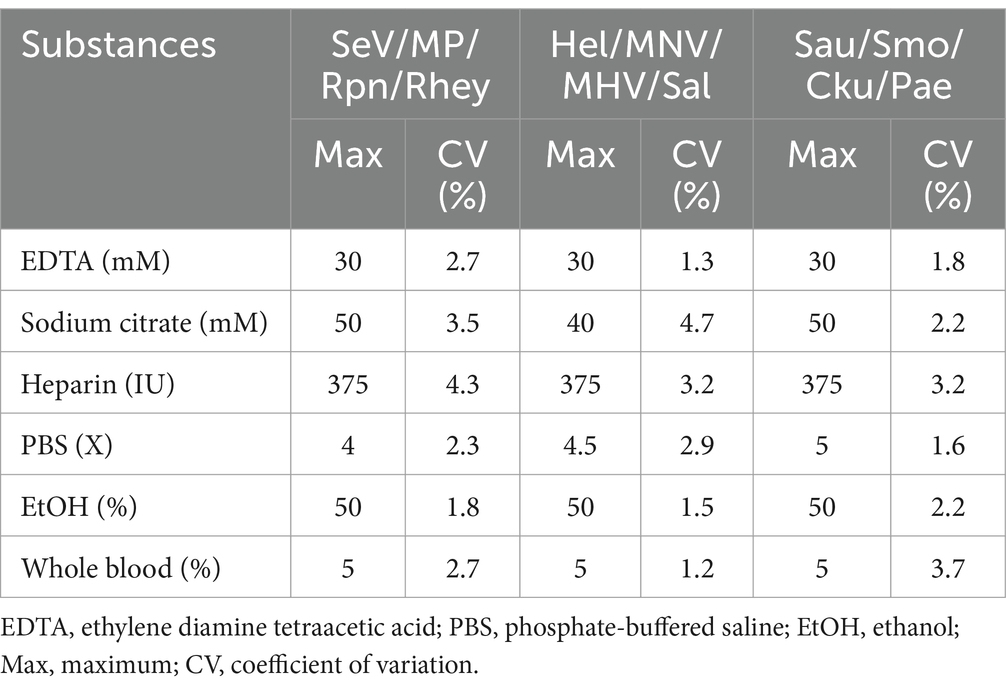
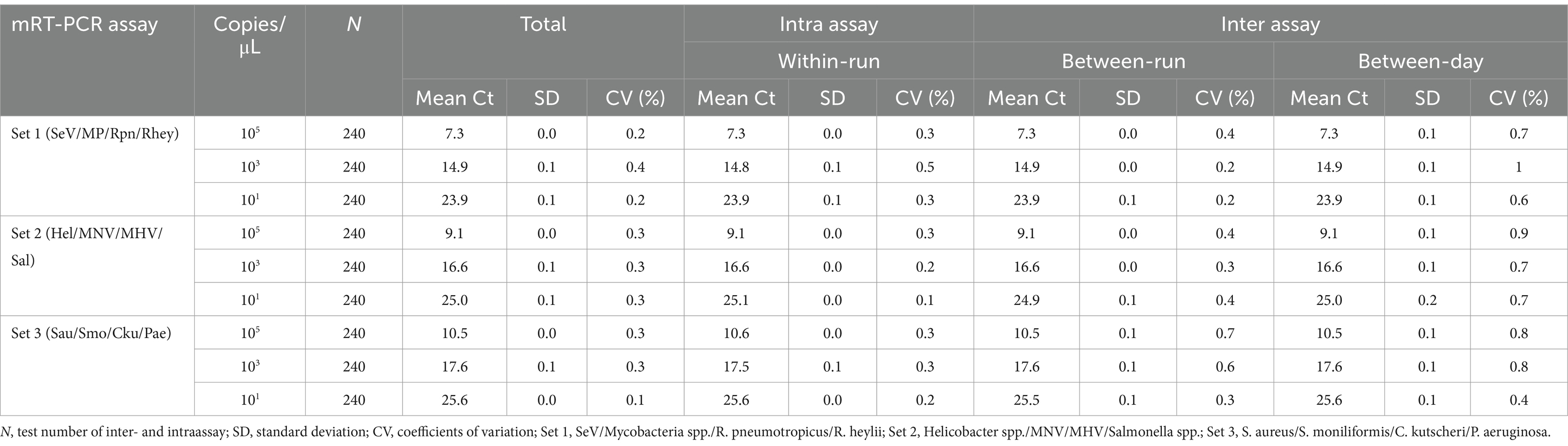
For interfering reactions, we used the following six substances by concentration: ethylene diamine tetraacetic acid (EDTA), sodium citrate (1, 10, 20, and 50 mM), heparin (250, 300, 375, and 500 IU) for anticoagulants, PBS (1X, 5X, 10X, and 20X) for tissue emulsion, EtOH and xylene (1, 5, 10, 20, and 50%), and blood (1, 5, and 10%). Assay repeatability and reproducibility were evaluated through 240 tests (10 days × 2 runs/day × 4 replicates × 3 lots). The coefficient of variation (CV) was calculated according to the mean CT values/standard deviation (SD).
Sequence analysis
To confirm the mRT-PCR assay results, PCR amplicons of positive clinical isolates were sequenced using an ABI 3,730 automated DNA sequencer (Applied Biosystems, Foster City, CA, USA) and the ABI Prism BigDye Terminator (Applied Biosystems) system (CosmoGenetech, Deajeon, Republic of Korea). The obtained sequence was compared with that of the National Center for Biotechnology Information GenBank database.
Results
Analytical sensitivity and specificity of the mRT-PCR assay
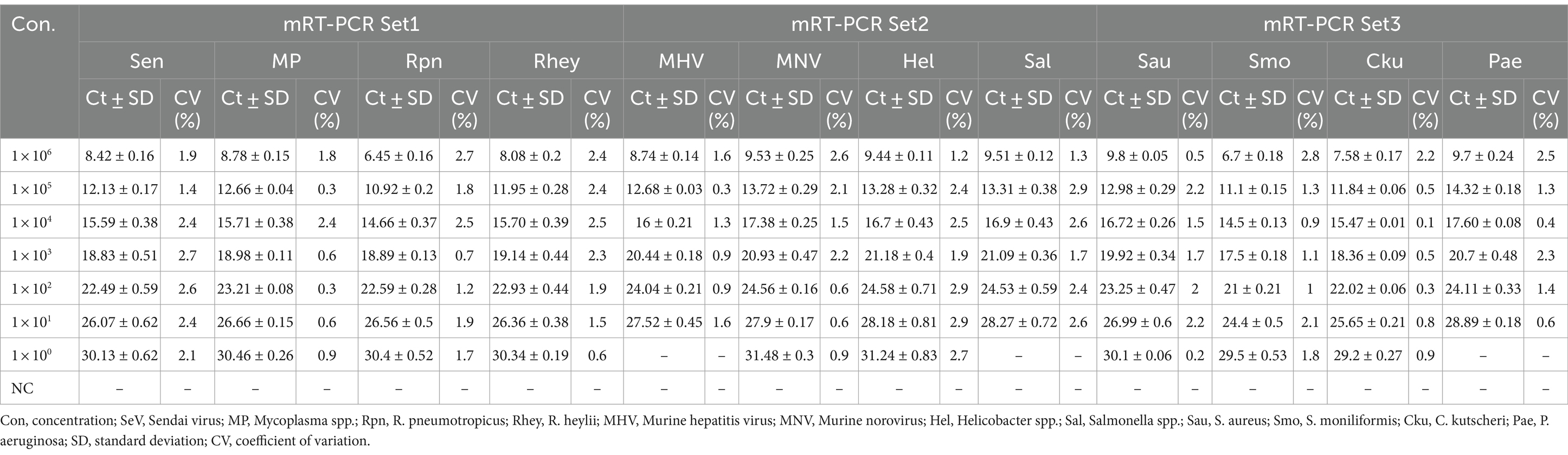
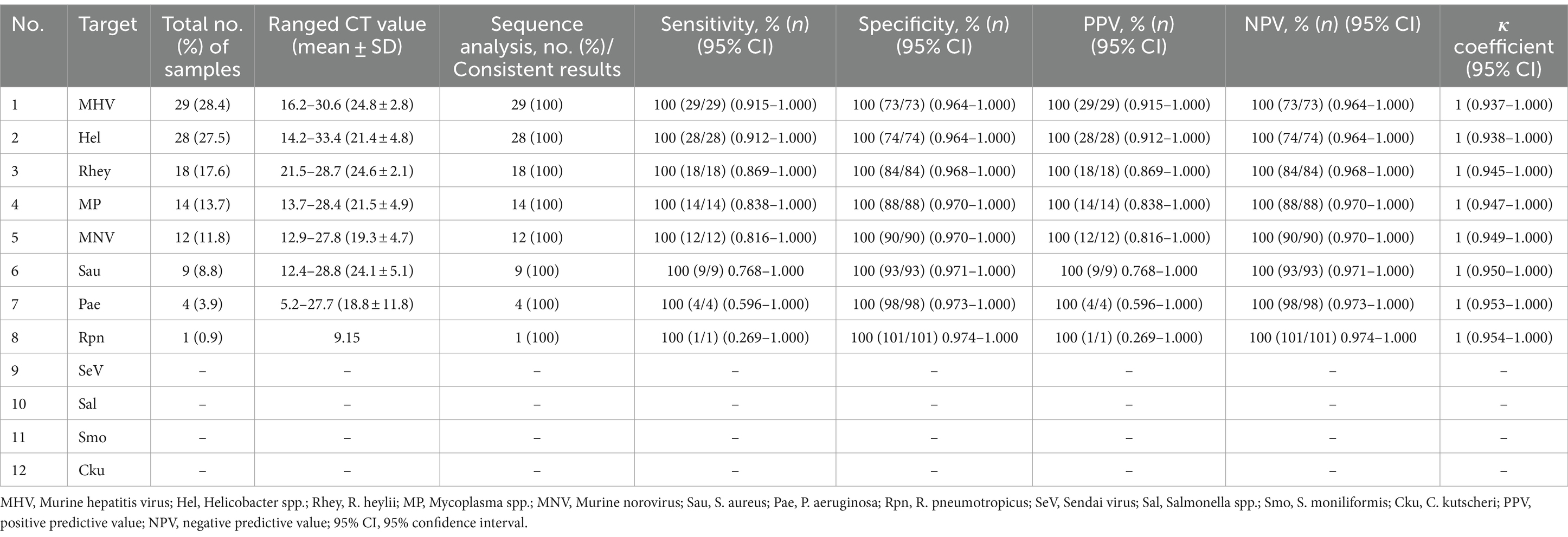
The lower detection limit of the mRT-PCR assay for detecting three viruses (SeV, MNV, and MHV) and nine bacteria (Mycoplasma spp., R. pneumotropicus, R. heylii, Helicobacter spp., Salmonella spp., S. aureus, S. moniliformis, C. kutscheri, and P. aeruginosa) was measured with serial 10-fold dilutions (106 copies to 1 copy) of the plasmid DNA stocks that contained each target gene sequence. Analytical sensitivity was estimated as the lowest number of gene copies yielding a positive result in all 10 replicates, and the corresponding CT value was selected as the analysis cutoff. A standard curve was generated by plotting the log quantity of each plasmid DNA versus the corresponding CT value, and the coefficient of determination (R2) for linear regression was 0.993–1.0, with a slope ranging from −3.193 to −3.934 (Figure 2). The detection limit of the mRT-PCR assay was detected at a concentration of approximately 1–100 copies per reaction. The mean CT values of each DNA concentration ranged from 6.3 to 31.8, and the mean CT (Standard deviation, SD) values were 6.45 ± 0.1 [95% confidence interval (CI), 6.4–6.5] to 31.4 ± 0.3 (95% CI, 31.1–31.8), and the CV was below 3% (Table 2).
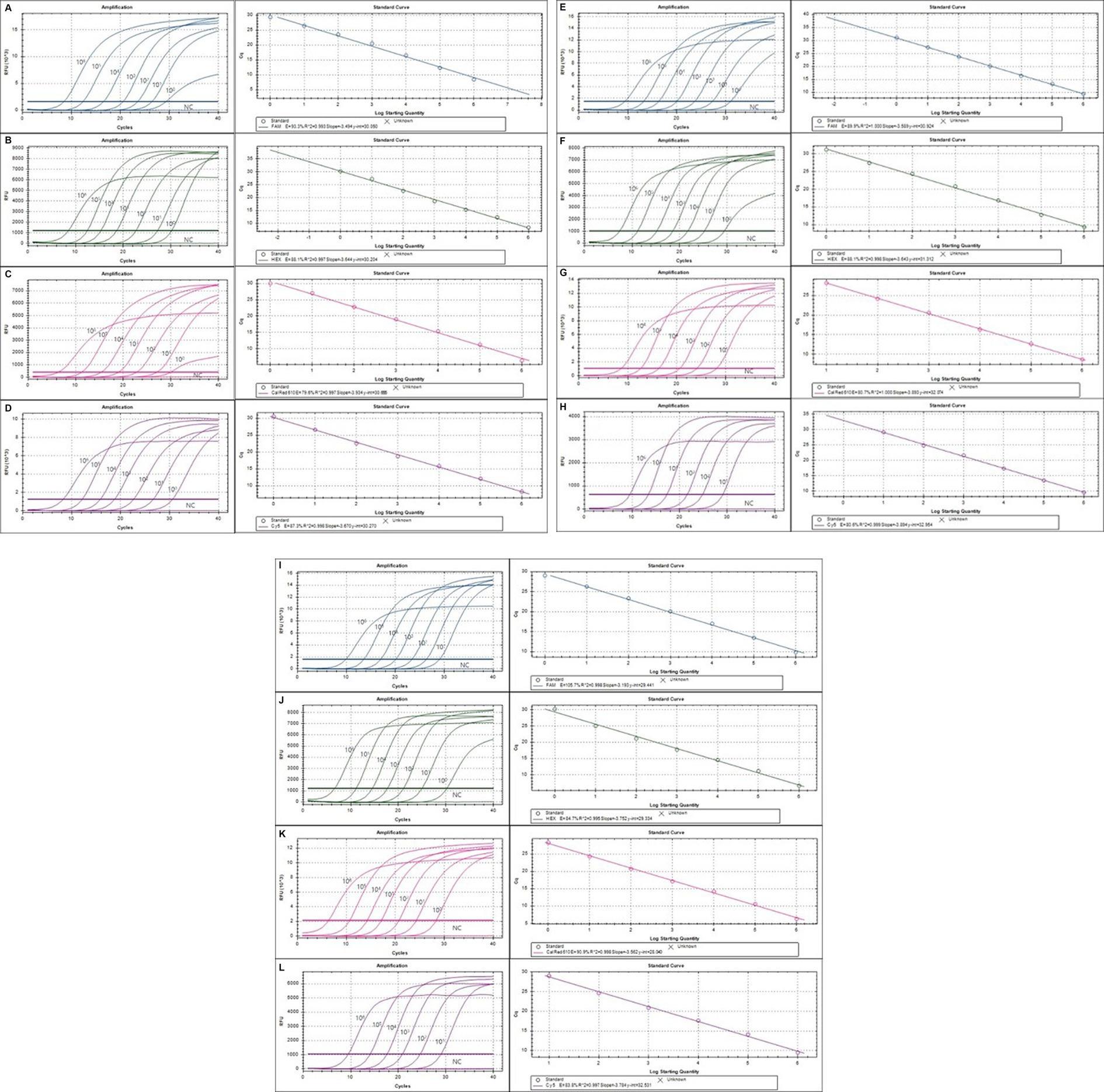
Figure 2. The detection limit of the multiplex real-time PCR (mRT-PCR) assay. The detection limit of the assay was evaluated using 10-fold serially diluted plasmid DNA. Each serially diluted control DNA, ranging from 106 copies to 1 copy per reaction, was used to determine the detection limit of the RT-PCR assay. In the RT-PCR assay, the amplification curve of the specific probe for detecting SeV (A), Mycoplama spp. (B), R. pneumotropicus (C), R. heylii (D), Helicobacter spp. (E), MNV (F), MHV (G), Salmonella spp. (H), S. aureus (I), S. moniliformis (J), C. kutscheri (K), and P. aeruginosa (L) is shown. The overall detection limit of this assay for each control DNA ranged from approximately 1 to 100 copy DNA per reaction. CT was plotted against the input of the quantity of each DNA (repeated 10 times). The linearity was generated by plotting the log quantity of each DNA versus the corresponding CT value, and the coefficient of determination of the linear regression was 0.993–1.0, with a slope ranging from −3.193 to −3.934. The fluorescence intensity is displayed on the Y-axis (R2 = reporter signal/passive reference signal). RFU, relative fluorescence unit; R2, fluorescence units.
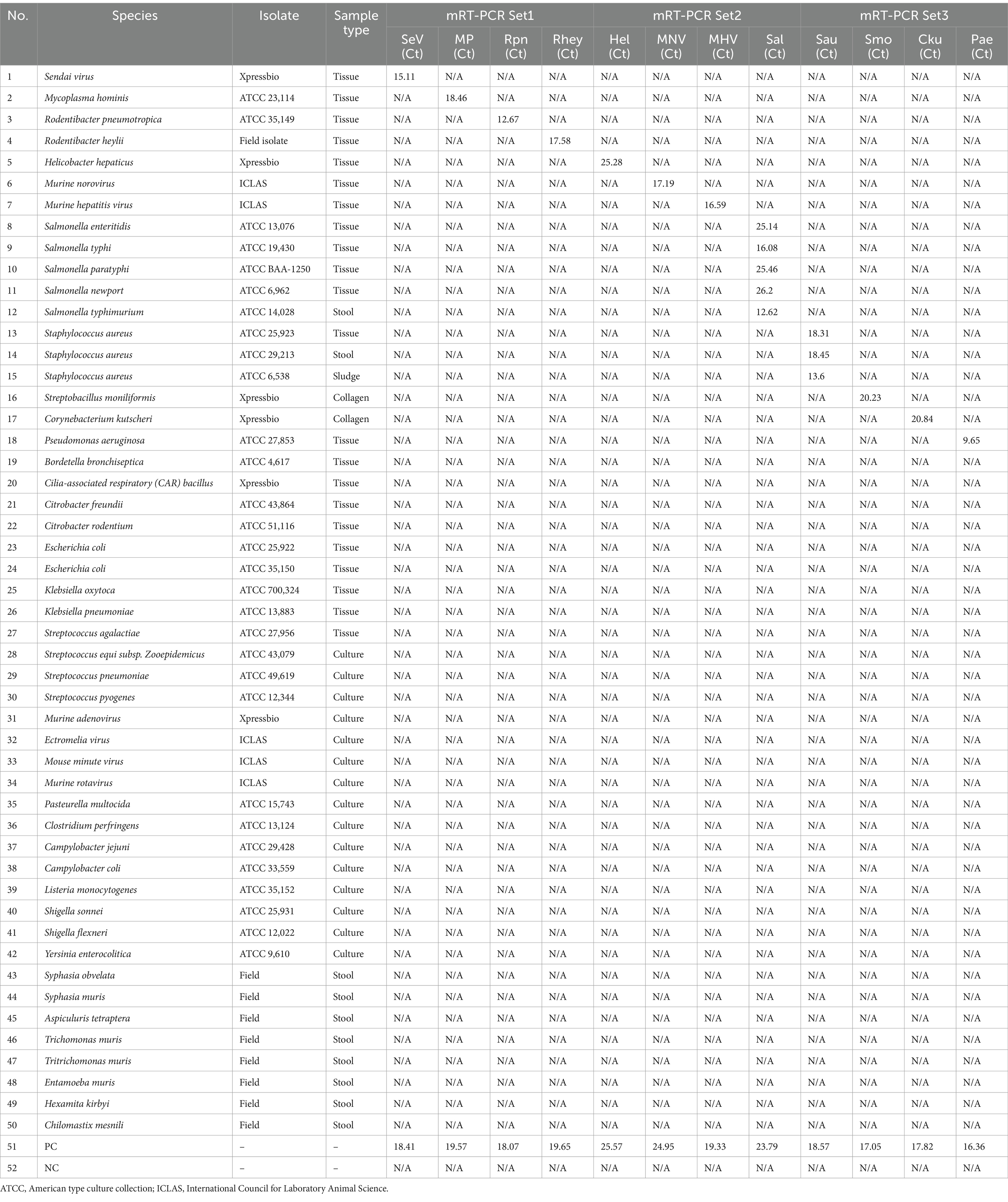
To assess the potential cross-reactivity, analytical specificity analysis was performed using 50 strains, including 7 viruses, 35 bacteria, and 8 parasites. Three sets of mRT-PCR assays showed negative results in all strains, except for control DNA or RNA (Supplementary Figure S1). Hence, these three sets of 12 primers and probes did not react with any viral or bacterial strains (Table 3).
Results of interfering reactions and reproducibility analysis using the mRT-PCR assay
We tested six substances at different concentrations to check for potential interference with the sample preparation, extraction, or qPCR progress. No interference was observed below these values: 30 mM EDTA, 40–50 mM sodium citrate, 375 IU heparin, 4-5X PBS, 50% EtOH, and 5% blood (Table 4). For repeatability and reproducibility, the measured number for the 3 concentrations (105, 103, and 101) of each positive control DNA was 240 (10 days × 2 runs/day × 4 replicates × 3 lots). The CV for intra- and inter assay variability ranged from 0.1 to 0.5% and 0.1 to 1%, respectively (see Table 5), which were all below 3%. These experimental results suggest that the three sets of developed mRT-PCR assays may have stable results.
Result comparison between single RT-PCR and mRT-PCR assays
To confirm the analytical performance of the three sets of mRT-PCR assays, the concentration of each positive sample was compared with that of the single RT-PCR assay across 10 previous studies (SeV, MNV, MHV, Mycoplasma spp., S. moniliformis, Salmonella spp., S. aureus, P. aeruginosa, R. pneumotropicus, and R. heylii) (Table 1). The mRT-PCR assay demonstrated higher sensitivity than the single RT-PCR assay, except for MNV, S. moniliformis, and R. pneumotropicus. The single qPCR did not detect MNV and R. pneumotropicus, and S. moniliformis appeared similarly (Figure 3). Because research on Helicobacter spp. and C. kutscheri by single RT-PCR remains unavailable, comparative experiments could not be performed in this study.
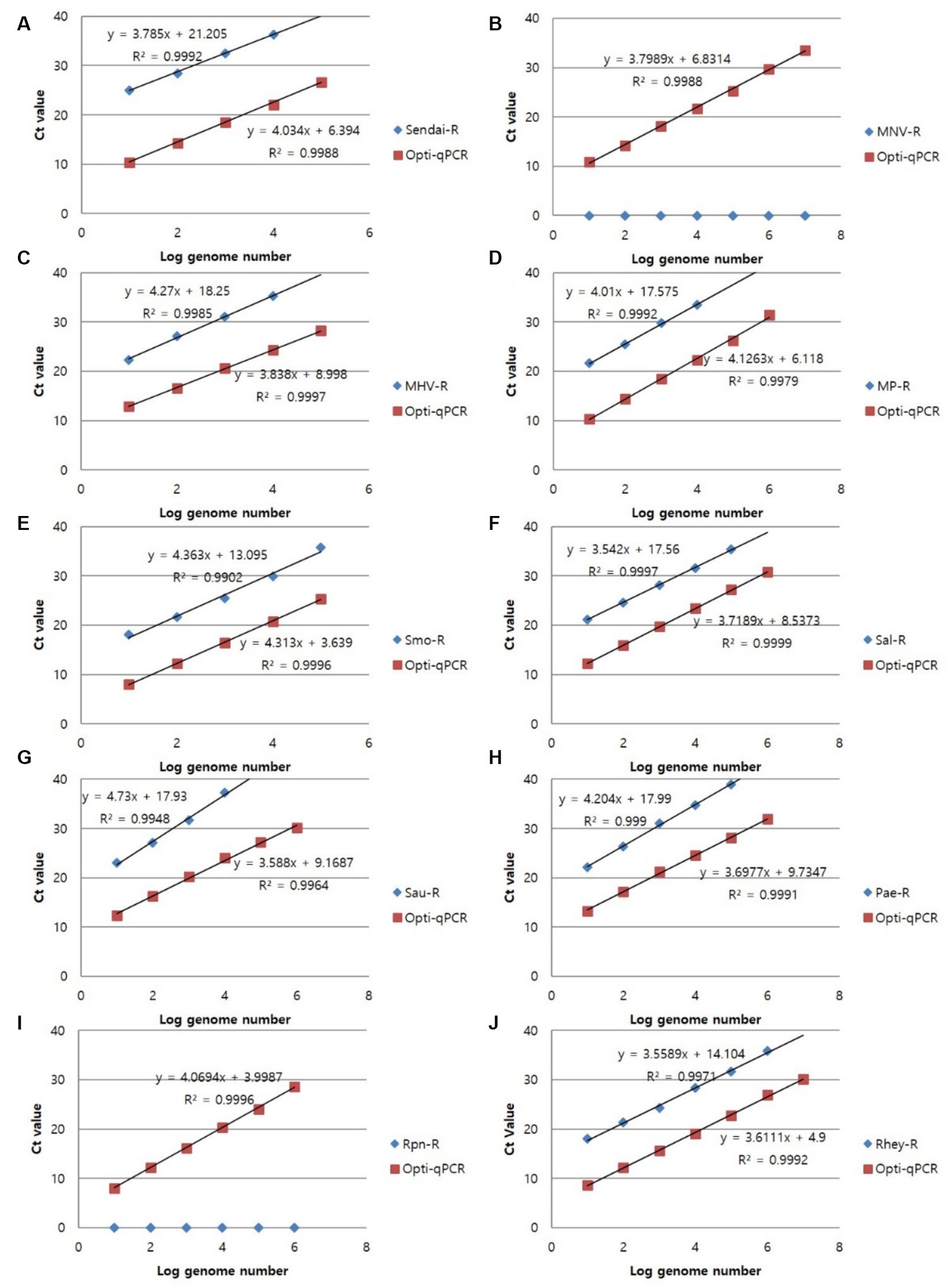
Figure 3. Representative results of the single RT-PCR and mRT-PCR assays according to the concentration of positive samples. Representative results of the two assays with positive samples at concentrations of 1 ng, 100 pg., 10 pg., 1 pg., 100 fg, and 10 fg. (A) SeV; (B) MNV; (C) MHV; (D) Mycoplasma spp.; (E) S. moniliformis; (F) Salmonella spp.; (G) S. aureus; (H) P. aeruginosa; (I) R. pneumotropicus; (J) R. heylii.
Detection of target genes using an mRT-PCR assay in clinical samples
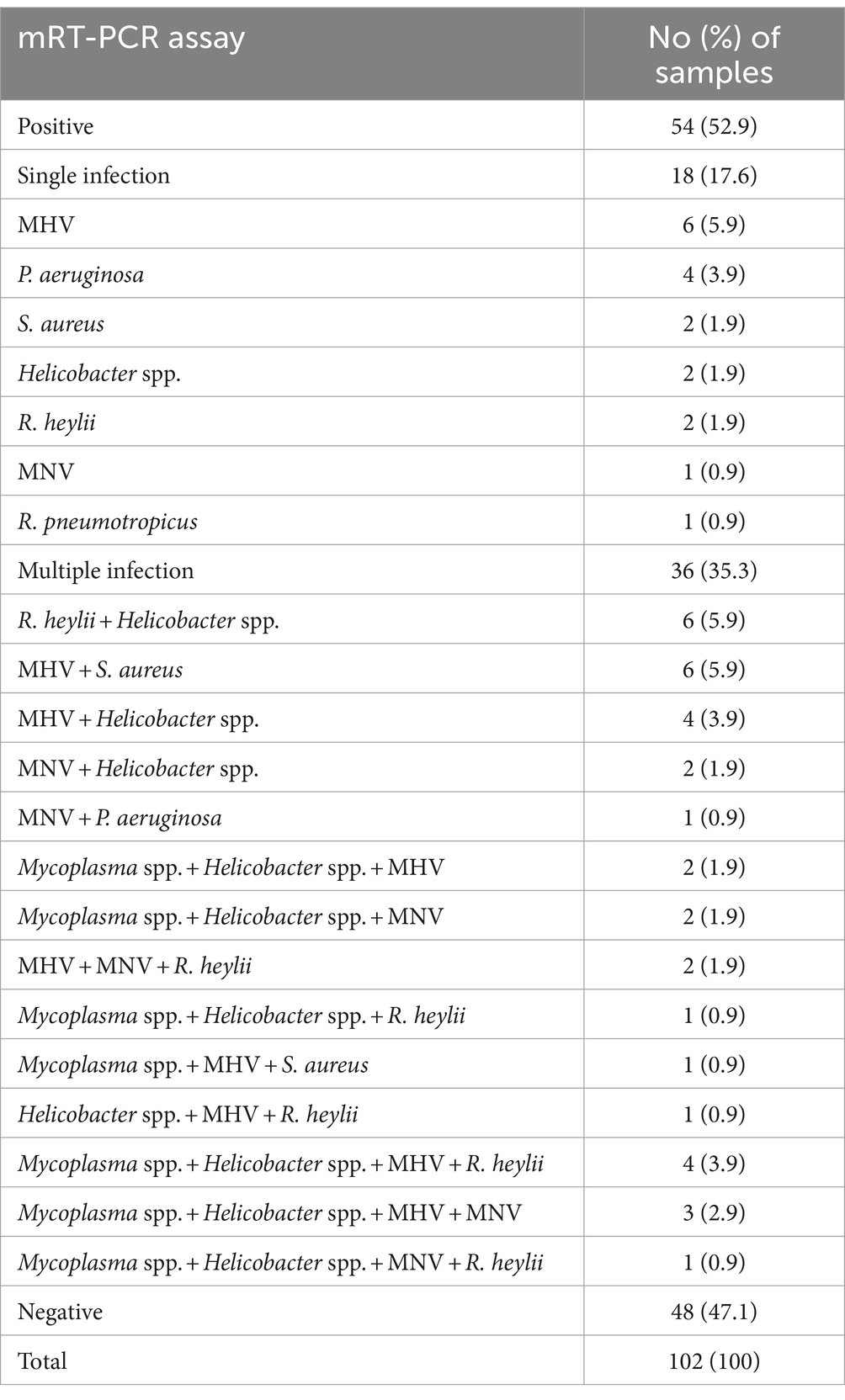
In a pilot study, we tested the detection of three sets of mRT-PCR assays using 102 clinical samples, including feces and ceca. Among the 102 clinical samples, 54 (52.9%) samples were positive for one or more than seven target genes, whereas 48 (47.1%) samples were negative for all 12 target genes as detected by mRT-PCR. Of the 54 positive samples, 29 (28.4%) were MHV, 28 (27.5%) were Helicobacter spp., 18 (17.6%) were R. heylii, 14 (13.7%) were Mycoplasma spp., 12 (11.8%) were MNV, 9 (8.8%) were S. aureus, 4 (3.9%) were P. aeruginosa, and 1 (0.9%) were R. pneumotropicus (Table 6). The CT values of positive samples ranged from 16.2 to 30.6 (mean 24.8, SD ± 2.8) in MHV, 14.2 to 33.4 (mean 21.4, SD ± 4.8) in Helicobacter spp., 21.5 to 28.7 (mean 24.6, SD ± 2.1) in R. heylii, 13.7 to 28.4 (mean 21.5, SD ± 4.9) in Mycoplasma spp., 12.9 to 27.8 (mean 19.3, SD ± 4.7) in MNV, 12.4 to 28.8 (mean 24.1, SD ± 5.1) in S. aureus, 5.2 to 27.7 (mean 18.8, SD ± 11.8) in P. aeruginosa, and 9.15 in R. pneumotropicus, respectively. To validate the mRT-PCR-derived results, a sequence analysis was performed using the same clinical samples. All 54 samples detected as 8-positive target genes by the mRT-PCR assay were consistent with the sequencing results (Table 6). Further investigation of single or multiple infections of the eight target genes identified as positive revealed that 18 (17.6%) had a single infection, and 36 (35.3%) were positive for two or more multiple infections (Table 7).
Discussion
Health monitoring of laboratory animals is a fundamental aspect of healthcare, serving as a crucial prerequisite for both animal welfare and scientifically rigorous research (23). Obtaining reliable and reproducible experimental results requires using animals with guaranteed microbiological and genetic quality (24). Therefore, health monitoring necessitates accurate and rapid diagnosis of infectious diseases in laboratory animals.
This study aimed to assess the analytical performance and clinical effectiveness of three newly developed mRT-PCR assays, leveraging both conventional PCR and multiplex PCR techniques to enable rapid and precise simultaneous detection. The development of the mRT-PCR assay for 12 pathogens affecting respiratory (Set 1, SeV/Mycoplasma spp./R. pneumotropicus/R. heylii) and digestive systems (Set 2, Helicobacter spp./MNV/MHV/Salmonella spp.) and abscess/sepsis (Set 3, S. aureus/S. moniliformis/C. kutscheri/P. aeruginosa) was prompted by recommendations outlined in the Korea Laboratory Animal Microbiological Standards and Monitoring (1108-01, 2021) guidelines. These guidelines recommend regular testing for these pathogens every three months, particularly for SPF-grade laboratory rats, mice, guinea pigs, and rabbits. Although conventional PCR or single RT-PCR has been used to isolate these diseases, mRT-PCR assays for the simultaneous differentiation of these viruses or bacteria remain lacking. An mRT-PCR assay enables the detection of various multiple genes in the same reaction tube by using 4–5 fluorescent dyes in RT-PCR equipment. It has the same advantages as conventional RT-PCR, including ease of use, rapid turnaround time of 70 min, accuracy, reproducibility, low risk of cross-contamination due to sequential reactions in the same tube, and high-throughput capacity to enable quick screening of multiple samples (25, 26). This assay is also a rapid method with a total turnaround time of approximately 100 min, including 30 min for DNA preparation and 70 min for target DNA amplification. In particular, RNA samples are also processed in one step, including cDNA synthesis, and the overall amplification time is similar to that of DNA samples. Considering the cost and availability in commercial laboratories, the cost of a single RT-PCR to diagnose one gene is usually $10 per test. If you would like to diagnose four genes with a single RT-PCR, you should need $40, whereas the mRT-PCR assay that diagnoses four genes simultaneously costs $15 per test. Therefore, we believe that the cost can be reduced by about 62.5%, making it highly usable in commercial laboratories.
The analytical performance evaluations of sensitivity, specificity, interference response, and reproducibility conducted in this study are the performance evaluation data required for product approval in Korea. The results of the analytical performance evaluation of the developed mRT-PCR are summarized as follows: (i) the detection limit of this assay was measured from 1 to 100 copies, suggesting that pathogens can be detected even at low amounts; (ii) in the specificity test using various viruses and bacteria, no cross-reactivity was observed between strains. The presence of other samples within the 12 infected strains did not affect the assay performance. (iii) Regarding interference reactions, some substances differed from the results of previous studies (26), and the variations in concentration causing interference were sample-specific. (iv) This assay showed high reproducibility with mean CVs of inter- and intra assay variation of below 3%. Furthermore, this assay demonstrated consistent results over a 12 month period, as verified by an external accelerated aging test conducted by KTC (Gunpo, Republic of Korea) (data not shown). A comparison analysis of the results of single RT-PCR assays used in previous studies validated the high sensitivity of the mRT-PCR assay.
In this study, the mRT-PCR-derived results were consistent with sequence analysis findings as per the clinical samples used, indicating a high level of agreement (κ = 1). Using sequence analysis as the gold standard, the sensitivity, specificity, and positive and negative predictive values of the results by mRT-PCR assay were 100% (n = 54, 95% CI 0.952–1.000, p < 0.001), 100% (n = 48, 95% CI 0.946–1.000, p < 0.001), 100% (95% CI 0.952–1.000, p < 0.001), and 100% (95% CI 0.946–1.000, p < 0.001), respectively. In a previous study, MNV (25.9%) and MHV (3.9%) for viruses and Helicobacter spp. (21%), Pasteurella pneumotropica (18.3%), and S. aureus (9.1%) for bacteria were the most prevalent pathogens identified in mice. Our results showed similar results, with MHV (28.4%) and MNV (11.8%) for viral, Helicobacter spp. (27.5%), Rodentibacter spp. (18.5%, R. heylii 17.6% and R. pneumotropicus 0.9%), Mycoplama spp. (13.7%), and S. aureus (8.8%) (27, 28). P. pneumotropica was recently reclassified to a new genus Rodentibacter, with R. pneumotropicus and R. heylii as the most commonly found species in laboratory mouse colonies (20). Until now, differentiation between R. pneumotropicus and R. heylii by culture or PCR has proven challenging, but we believe that the mRT-PCR assay will enable testing to distinguish between the two species. Multiple infections (n = 36, 35.3%) with two or more pathogens exhibited a higher positivity rate than single infections (n = 18, 17.6%). Confirming the ratio was difficult owing to the lack of simultaneous diagnosis of multiple pathogens, unlike our results. However, because the detection rate of Mycoplama spp. was high (13.7%) compared to the previous study’s 0.47%, the contaminated environment of experimental animals is assumed to have been affected.
This study has certain limitations. (i) Because our clinical results were tested on random samples requested for health monitoring, 4 (SeV, Salmonella spp., S. moniliformis, and C. kutscheri) out of 12 pathogens were not detected. Hence, future studies will require additional tests with larger sample sizes to further validate these findings. (ii) Endo and ecto parasites are a significant threat to the biosecurity of rodent research colonies. In pilot study, we tested the detection of three sets of mRT-PCR assays using 32 parasite samples, including Syphacia spp. and Aspiculuris tetreptera, known as pinworms, confirmed by microscopy and sequencing. As a result, these three sets of mRT-PCR assays did not react in all 32 parasite samples (data not shown). Likewise, not all samples related to endo and ecto parasites were tested, so additional testing is required.
Conclusion
The developed mRT-PCR assay consistently showed high agreement and specificity with the sequence analysis. This assay offers rapidity and accuracy, effectively minimizing the risk of false positives or false negatives. It is a convenient tool for simultaneously detecting the presence of infectious diseases in numerous samples. Therefore, the use of the newly developed mRT-PCR assay will prove beneficial in detecting the most crucial diseases during the health monitoring of laboratory animals. We believe that this assay can serve as a sensitive and specific tool to complement or replace traditional methods because it can reduce the labor and time required for diagnosis in the field of laboratory animals.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
Ethics statement
The animal studies were approved by written informed consent was obtained from the owners for the participation of their animals in this study. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
H-yW: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Validation, Writing – original draft, Writing – review & editing. JA: Writing – review & editing. JL: Writing – original draft, Methodology. SK: Writing – original draft, Investigation. HK: Writing – original draft, Conceptualization, Supervision.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was supported by “Study for establishment of a quality control system for high quality care of laboratory animals” (Project no.: 21184MFDS326) from “Ministry of Food and Drug Safety,” and we appreciate this support.
Acknowledgments
We thank Professor Byeong-Cheol Kang of Seoul National University college of Medicine Bionedical Research Institute (SNUH) for providing research funding.
Conflict of interest
H-yW, JA, JL, SK, and HK were employed by Optipharm, Inc.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2024.1421427/full#supplementary-material
Footnotes
References
1. Hansen, AK, Farlov, H, and Bollen, P. Microbiological monitoring of laboratory pigs. Lab Anim. (1997) 31:193–200. doi: 10.1258/002367797780596248
2. Raj, M, Shinde, A, and Ingle, A. Microbiological monitoring of rodent pathogens using multiplex method of PCR. J Lab Ani Sci. (2022) 7:18–24.
4. Compton, SR . PCR and RT-PCR in the diagnosis of laboratory animal infections and in health monitoring. J Am Assoc Lab Anim Sci. (2020) 59:458–68. doi: 10.30802/AALAS-JAALAS-20-000008
5. Kunita, S, Kato, K, Ishida, M, Hagiwara, K, Kameda, S, Ishida, T, et al. Simultaneous detection of antibodies to mouse hepatitis virus recombinant structural proteins by a microsphere-based multiplex fluorescence immunoassay. Clin Vaccine Immunol. (2011) 18:758–66. doi: 10.1128/CVI.00467-10
6. Kitagawa, Y, Tohya, Y, Ike, F, Kajita, A, Park, SJ, Ishii, Y, et al. Indirect ELISA and indirect immunofluorescent antibody assay for detecting the antibody against murine norovirus S7 in mice. Exp Anim. (2010) 59:47–55. doi: 10.1538/expanim.59.47
7. Taylor, K, and Copley, CG. Detection of rodent RNA viruses by polymerase chain reaction. Lab Anim. (1994) 28:31–4. doi: 10.1258/002367794781065861
8. Wang, RF, Campbell, WL, Cao, WW, Colvert, RM, Holland, MA, and Cerniglia, CE. Diagnosis of mouse hepatitis virus contamination in mouse population by using nude mice and RT-PCR. Mol Cell Probes. (1999) 13:29–33. doi: 10.1006/mcpr.1998.0211
9. Andre, JM, Freydiere, AM, Benito, Y, Rousson, A, Lansiaux, S, Kodjo, A, et al. Rat bite fever caused by Streptobacillus moniliformis in a child: human infection and rat carriage diagnosed by PCR. J Clin Pathol. (2005) 58:1215–6. doi: 10.1136/jcp.2005.026401
10. Jeong, ES, Lee, KS, Heo, SH, Seo, JH, and Choi, YK. Rapid identification of Klebsiella pneumoniae, Corynebacterium kutscheri, and Streptococcus pneumoniae using triplex polymerase chain reaction in rodents. Exp Anim. (2013) 62:35–40. doi: 10.1538/expanim.62.35
11. Wang, HY, Song, JK, Shin, S, and Kim, H. Comparison of multiplex real-time PCR and PCR-reverse blot hybridization assays for the direct and rapid detection of porcine circovirus type 2 genotypes. Front Vet Sci. (2020) 7:200. doi: 10.3389/fvets.2020.00200
12. Wagner, AM, Loganbill, JK, and Besselsen, DG. Detection of Sendai virus and pneumonia virus of mice by use of fluorogenic nuclease reverse transcriptase polymerase chain reaction analysis. Comp Med. (2003) 53:173–7.
13. Janetzko, K, Rink, G, Hecker, A, Bieback, K, Klüter, H, and Bugert, P. A single-tube real-time PCR assay for mycoplasma detection as a routine quality control of cell therapeutics. Transfus Med Hemother. (2014) 41:83–9. doi: 10.1159/000357096
14. Hanaki, K, Ike, F, Kajita, A, Yasuno, W, Yanagiba, M, Goto, M, et al. Detection of murine norovirus by reverse transcription loop-mediated isothermal amplification. J Virol Methods. (2014) 204:17–24. doi: 10.1016/j.jviromet.2014.03.025
15. Kasturi, KN, and Drgon, T. Real-time PCR method for detection of Salmonella spp. in environmental samples. Appl Environ Microbiol. (2017) 83:e00644–17. doi: 10.1128/AEM.00644-17
16. Galia, L, Ligozzi, M, Bertoncelli, A, and Mazzariol, A. Real-time PCR assay for detection of Staphylococcus aureus, Panton-valentine leucocidin and methicillin resistance directly from clinical samples. AIMS Microbiol. (2019) 5:138–46. doi: 10.3934/microbiol.2019.2.138
17. Wang, C, Ye, Q, Jiang, A, Zhang, J, Shang, Y, Li, F, et al. Pseudomonas aeruginosa detection using conventional PCR and quantitative real-time PCR based on species-specific novel gene targets identified by pangenome analysis. Front Microbiol. (2022) 13:820431. doi: 10.3389/fmicb.2022.820431
18. Kelly, AJ, Ivey, ML, Gulvik, CA, Humrighouse, BW, and McQuiston, JR. A real-time multiplex PCR assay for detection of the causative agents of rat bite fever, Streptobacillus moniliformis and zoonotic Streptobacillus species. Diagn Microbiol Infect Dis. (2021) 100:115335. doi: 10.1016/j.diagmicrobio.2021.115335
19. Fawzy, A, Giel, AS, Fenske, L, Bach, A, Herden, C, Engel, K, et al. Development and validation of a triplex real-time qPCR for sensitive detection and quantification of major rat bite fever pathogen Streptobacillus moniliformis. J Microbiol Methods. (2022) 199:106525. doi: 10.1016/j.mimet.2022.106525
20. Buchheister, S, Roegener, F, Zschemisch, NH, Talbot, SR, Christensen, H, and Bleich, A. One for two: a novel and highly sensitive virulence factor-based quantitative polymerase chain reaction assay for the simultaneous detection of Rodentibacter pneumotropicus and Rodentibacter heylii in environmental sample material. Lab Anim. (2020) 54:239–50. doi: 10.1177/0023677219853600
21. Compton, SR, and Riley, LK. Detection of infectious agents in laboratory rodents: traditional and molecular techniques. Comp Med. (2001) 51:113–9.
22. Buchheister, S, and Bleich, A. Health monitoring of laboratory rodent colonies-talking about (R)evolution. Animals. (2021) 11:1410. doi: 10.3390/ani11051410
23. National Research Council (US) . Committee for the Update of the guide for the care and use of laboratory animals. Guide for the care and use of laboratory animals. 8th ed. Washington (DC): National Academies Press (US) (2011).
24. Hayashimoto, N, Morita, H, Ishida, T, Yasuda, M, Kameda, S, Uchida, R, et al. Current microbiological status of laboratory mice and rats in experimental facilities in Japan. Exp Anim. (2013) 62:41–8. doi: 10.1538/expanim.62.41
25. Lim, HJ, Kang, ER, Park, MY, Kim, BK, Kim, MJ, Jung, S, et al. Development of a multiplex real-time PCR assay for the simultaneous detection of four bacterial pathogens causing pneumonia. PLoS One. (2021) 16:e0253402. doi: 10.1371/journal.pone.0253402
26. Wang, HY, Song, JK, Shin, S, Choi, KM, and Kim, H. One-tube nested real-time PCR assay for rapid screening of porcine cytomegalovirus in clinical samples. Front Vet Sci. (2020) 7:586045. doi: 10.3389/fvets.2020.586045
27. Hsu, CC, Wobus, CE, Steffen, EK, Riley, LK, and Livingston, RS. Development of a microsphere-based serologic multiplexed fluorescent immunoassay and a reverse transcriptase PCR assay to detect murine norovirus 1 infection in mice. Clin Diagn Lab Immunol. (2005) 12:1145–51. doi: 10.1128/CDLI.12.10.1145-1151.2005
Keywords: mouse, health monitoring, multiplex real-time PCR, diagnosis, sequence analysis
Citation: Wang H-y, Ahn J, Lee J, Kang SC and Kim H (2024) Performance of three multiplex real-time PCR assays for simultaneous detection of 12 infectious pathogens in mice affected with respiratory and digestive diseases. Front. Vet. Sci. 11:1421427. doi: 10.3389/fvets.2024.1421427
Edited by:
Enrico Radaelli, University of Pennsylvania, United StatesReviewed by:
Angela Brice, University of Pennsylvania, United StatesKevin O’Brien, University of Pennsylvania, United States
Copyright © 2024 Wang, Ahn, Lee, Kang and Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hye-young Wang, c2FwcGhpcmUxMTFAZGF1bS5uZXQ=
 Hye-young Wang
Hye-young Wang Jaeil Ahn1
Jaeil Ahn1