
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Vet. Sci. , 12 April 2024
Sec. Veterinary Infectious Diseases
Volume 11 - 2024 | https://doi.org/10.3389/fvets.2024.1374430
N6-methyladenosine (m6A) methylation is an internal post-transcriptional modification that has been linked to viral multiplication and pathogenicity. To elucidate the conservation patterns of potential 5′-DRACH-3′ motifs in avian leukosis virus subgroup J (ALV-J), 149 ALV-J strains (139 isolates from China; ALV-J prototype HPRS-103 from the UK; and 9 strains from the USA, Russia, India, and Pakistan) available in GenBank before December 2023 were retrieved. According to the prediction results of the SRAMP web-server, these ALV-J genomes contained potential DRACH motifs, with the total number ranging from 43 to 64, which were not determined based on the isolation region and time. Conservative analysis suggested that 37 motifs exhibited a conservation of >80%, including 17 motifs with a grading above “high confidence.” Although these motifs were distributed in the U5 region of LTRs and major coding regions, they were enriched in the coding regions of p27, p68, p32, and gp85. The most common m6A-motif sequence of the DRACH motif in the ALV-J genome was GGACU. The RNA secondary structure of each conserved motif predicted by SRAMP and RNAstructure web-server was mainly of two types—A–U pair (21/37) and hairpin loop (16/37)—based on the core adenosine. Considering the systematic comparative analysis performed in this study, future thorough biochemical research is warranted to determine the role of m6A modification during the replication and infection of ALV-J. These conservation and distribution analysis of the DRACH motif for potential m6A sites in ALV-J would provide a foundation for the future intervention of ALV-J infection and m6A modification.
As a representative member of the 11 subgroups of avian leukosis virus (ALV-A–K) that have been recognized to date, ALV subgroup J (ALV-J) is an oncogenic virus that is associated with significant morbidity and causes avian leukosis, thus affecting the poultry industry worldwide (1). The genome of ALV comprises a single positive-strand linear RNA dimer, and its provirus presents a gene structure characteristic of C-type retroviruses, spanning a total length of 7–8 kilobases (2). The genes encoding the group-specific antigen (gag), polymerase (pol), and envelope glycoprotein (env) are located within the central coding region of the viral genome molecules (3). The noncoding region, mainly contained the long terminal repeat (LTR), is a pair of identical sequences of DNA situated at both ends of the genome, which comprises the 3′ unique region (U3), a short repeated region (R), and the 5′ unique region (U5) (4). These regions play crucial roles in viral replication, RNA processing, and integration of the virus into the host genomes (5, 6).
Currently, several research projects are dedicated to the elucidation of the pathogenesis of ALV-J, which is mainly related to its tumorigenic effect, immunosuppression, and decreased performance in chickens and layers (7). After the integration of the virus genome sequence into the host genome, the viral LTR plays a key role in the activation of the cellular proto-oncogenes, a process known as promoter insertion, which ultimately leads to multiorgan hemangioma and myeloid leukosis (8). Recent studies have shown that the aberrant expression of endogenous microRNAs (miRNAs), circular RNAs (circRNAs), and long noncoding RNAs (lncRNAs) caused by ALV-J infection is closely related to avian leukosis virus-induced tumorigenesis (9–12). These regulating effectors are mainly related to various activate/inactivate signaling pathways and differential expression change of genes at mRNA and protein levels.
Historically, DNA has been acknowledged as the carrier of genetic information, whereas mRNA has been identified as the central molecule that connects DNA to proteins and is crucial for the transmission of genetic information during various biological processes (13). To date, >100 chemically distinct modifications have been reported in cellular RNAs. Notably, in eukaryotic organisms, the presence of a modified 5′-cap at the 5′-end and a polyadenylate (poly(A)) tail at the 3′-end of mRNAs is crucial for the regulation of mRNA transcription (14). The prevalent alterations observed in mRNA mainly include N1-methyladenosine (m1A), 7-methylguanosine (m7G), N5-methylcytosine (m5C), and the prominent modification N6-methyladenosine (m6A) (15).
Generally, the m6A modification of RNAs occurs via a hetero-multimeric complex of nuclear methyltransferases (writers); in turn, this dynamic modification can be reversed by a group of demethylases (erasers), whereas a third group of proteins, known as readers, preferentially recognize the binding of m6A (readers) to methylated RNA and confer downstream functions (16). Therefore, aberrant m6A modifications and their related enzymes/proteins may be the best strategy for understanding the epigenetic mechanisms underlying the expression of important viral genes during the course of viral infection (17). Recently, there has been an increasing amount of research on the relevance of the m6A changes in viral replication and virus–host interactions in relation to virus induced illnesses in humans and animals (17). However, the functional characteristics of the m6A-mediated regulation of the RNAs/transcripts of DNA/RNA viruses remain unclear. According to the modification process of m6A, the core step consists in the recognition of the target genes, which calls for the accurate identification of RNA m6A sites (18). Numerous independent reports have revealed that the 5′-DRA*CH-3′ (where D stands for non-cytosine base; R for purine; H for non-guanine base; and A* for the methylatable adenosine) consensus motif is restricted for m6A binding; however, the mapping of these sites in viral RNAs/transcripts remains challenging and cumbersome (19, 20). Consequently, we aimed to use publicly accessible ALV-J genome sequences to predict m6A locations and study their evolutionary conservation, to stimulate research on m6A modification in ALV-J.
The complete genome sequences of 149 ALV-J strains submitted before December 2023 were retrieved from GenBank (Supplementary Table 1). These ALV-J strains included 139 isolates from China, the ALV-J prototype HPRS-103 from the UK, and nine strains from the USA, Russia, India, and Pakistan.
The genome of ALV-J comprises a positive-strand linear RNA, and the coding regions of the three primary genes (gag, pol, and env) are continuous or overlapping. Moreover, previous studies have shown that m6A modification is primarily distributed within the coding regions, close to the start or stop codons and near the beginning of 3′-untranslated region (3′-UTR) or ending of 5′-UTR in mRNA transcripts derived from chickens that were negative/positive for ALV-J (21, 22). Therefore, the m6A-modification sites in the genome sequences and coding sequences of the 149 ALV-J strains were predicted via the SRAMP1 online server using the “Full transcript” and “Mature mRNA” modes, respectively (23). For additional validation, the RNA secondary structure for each motif predicted in this manner was also summarized.
Multiple sequence alignments of the genome and coding gene sequences of ALV-J strains were performed using MEGA version 11 (24). Based on a comparison of the prototype strains HPRS-103 and NX0101 (GenBank accession no.: DQ115805, representative Chinese ALV-J strain caused myeloma), the conservativeness and diversity of all predicted DRACH motifs were visualized using TBtools-II software (v2.028) (25). The distribution of conserved m6A-modification sites in ALV-J genomes was visualized via lollipop-plotting using ChiPlot2, and the genome structure of ALV-J was illustrated using IBS software (26).
According to the conservation analysis and rating scores of the predicted motifs, to further evaluate the accuracy and potential modification mode of each m6A motif, the RNA secondary structure of the flanking sequences of each m6A motif was re-predicted using the RNAstructure web-server3 (27).
Based on the prediction results obtained for the 149 ALV-J genome sequences, the genomes of these strains contained potential DRACH motifs, with the total number ranging from 43 (GD1411-1; accession NO., KU500038) to 64 (GD16FS01; accession NO., MT538237) (Supplementary Table 1), which were not determined based on the isolation region and time. According to the rating grades obtained via SRAMP prediction, the proportion of motifs with “low confidence,” “medium confidence,” “high confidence,” and “very high confidence” grading was 23.76, 36.85, 34.53, and 4.87%, respectively.
Genome construction revealed that the potential m6A modification sites were distributed throughout the entire genome, as displayed in Supplementary Table 1 and Figure 1. In terms of conservation analysis and rating grade of the predicted DRACH motifs, 37 motifs exhibiting a conservation of >80% are summarized and listed in Table 1; these motifs comprise 17 motifs with a grading above “high confidence,” as indicated by SRAMP. Although these motifs were distributed in the U5 region of LTRs and major coding regions, they were enriched in the coding regions of p27, p68, p32, and gp85 (Table 1).
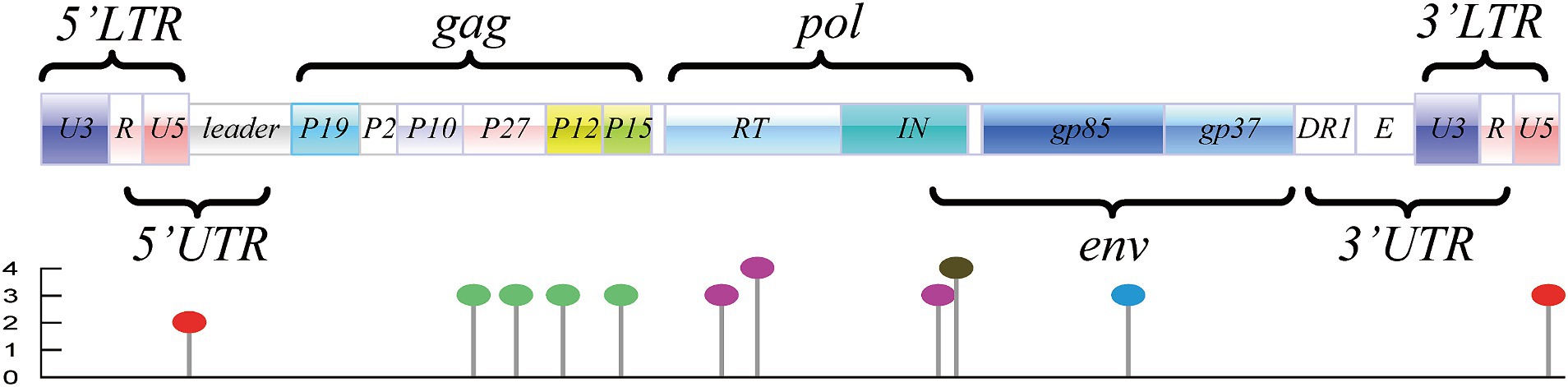
Figure 1. Genome structure and distribution of potential m6A motifs of ALV-J (represented by the ALV-J prototype strain of HPRS-103; GenBank accession number: Z46390.1). Motifs located in LTRs are marked with “red lollipop,” motifs located in the coding region of gag are marked with “green lollipop,” motifs located in the coding region of pol are marked with “purple lollipop,” motifs located in the coding region of env are marked with “blue lollipop,” and the motif in the overlapped coding region is marked with “black lollipop.”
Meanwhile, the diversity of all motifs, motifs with “high/very high confidence,” motifs with a conservation of >80%, and motifs with a conservation of >80% and grading of “high/very high confidence” are displayed in Figure 2, which revealed that GGACU was the most common m6A-motif sequence.
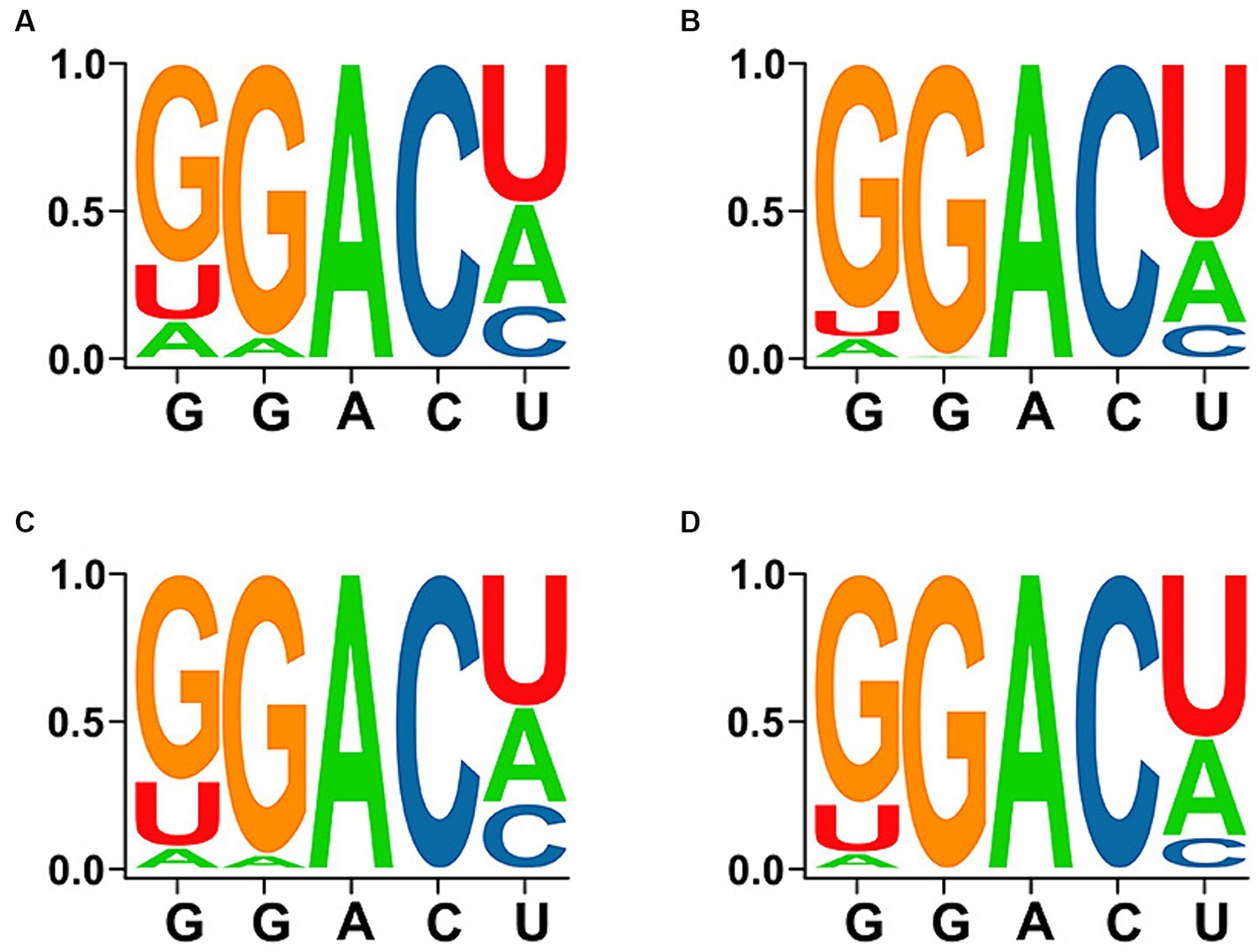
Figure 2. Diversity and conservation of DRACH sequences in the motifs. (A) Total predicted motifs. (B) Motifs with “high/very high confidence.” (C) Motifs with a conservation of >80%. (D) Motifs with a conservation of >80% and “high/very high confidence.”
The prediction results afforded by the SRAMP web-server indicated that the RNA secondary structure was mainly of two types, i.e., an “A–U pair” and a “hairpin loop,” based on the core adenosine in the m6A motif. Among the 37 conserved motifs, 21 were of the A–U pair type, with the remaining motifs of the hairpin loop type, which included the loop adjacent to the G-C pair. The RNA secondary structures for each motif predicted by the SRAMP and RNAstructure web-servers were mostly consistent, such as the prediction of an A–U pair type for the motifs located at positions 273 (5′-LTR [U5]), 2,444 (coding region of gag [p 15]), and 6,220 (coding region of env [gp85]), although a hairpin loop type was predicted for the motif located at position 1,383 (coding region of gag [p27]). Moreover, the RNA secondary structures of the motif located at position 5,342 (overlapping coding regions of pol and env) were predicted to be of the A–U pair and hairpin loop types, respectively. The complex RNA secondary structures also indicated whether the structural change was related to the m6A modification (Figure 3).
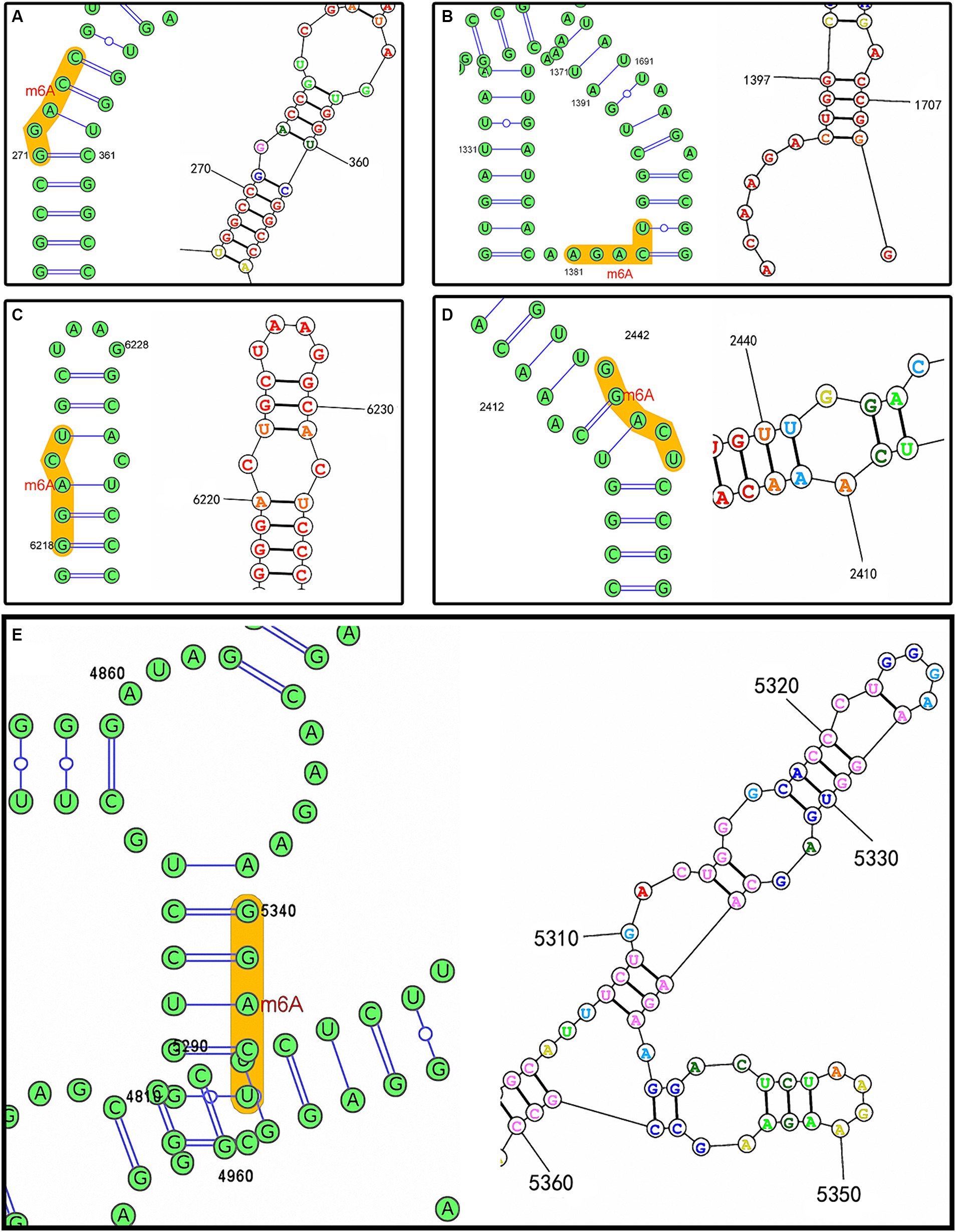
Figure 3. RNA secondary structures predicted by the SRAMP (left) and RNAstructure web-servers (right) based on the ALV-J prototype strain of HPRS-103 (GenBank accession number: Z46390.1). (A) Predicted m6A motif located at position 273. (B) Predicted m6A motif located at position 1,383. (C) Predicted m6A motif located at position 6,220. (D) Predicted m6A motif located at position 2,444. (E) Predicted m6A motif located at position 5,342.
In recent years, the emergence of sequencing methods such as methylated RNA immunoprecipitation (MeRIP) and cross-linking and immunoprecipitation (CLIP) sequencing has given rise to a new layer of gene-expression regulation termed “epitranscriptomics,” which primarily focuses on the chemical alterations occurring on RNA molecules (28). However, these techniques can detect massive sequence segments containing m6A from the transcriptome, but cannot precisely identify the specific adenosine that undergoes methylation. Therefore, the use of computational tools to predict m6A sites from sequences would facilitate the rapid study of the m6a modification. From this viewpoint, the m6A motifs harbored in the genome of ALV-J strains available from GenBank were predicted using the SRAMP web-server. Consequently, more than 40 potential DRACH motifs were predicted in the genomes of these strains, among which 34.53 and 4.87% of the motifs had a “high confidence” and “very high confidence” grading, respectively. Studies related to the avian m6A modification revealed that “GGACU” was consistently regarded as the best and most highly enriched motif (29, 30). Moreover, “GGACU” was also preferred in the study of the transcriptome-wide dynamics of m6A methylation in chicken liver malignancies caused by ALV-J infection (22). Even though m6A motifs have not been determined for other subtypes of ALV, the consensus m6A sites across them would assist in unveiling the critical aspects of m6A modification in viral infection. According to the diversity analysis, the most common m6A-motif sequence of the DRACH motif of the ALV-J genome was also “GGACU,” which suggests the accuracy of this prediction, to some extent. Subsequently, although the 37 conserved motifs were predicted to be distributed in the U5 region of LTRs and all the major coding regions, they were enriched in the coding regions of the p27, p68, p32, and gp85 genes. This high conservation further confirmed the prediction accuracy for the m6A motif.
In the ALV-J genome, the gag gene is relatively conserved and mainly encodes glycosylated proteins, including p19, p10, p27, p12, and p15 (7). p19, which is alternatively referred to as the viral matrix protein (MA), plays a crucial role in the viral outgrowth process; p12, also known as the nucleocapsid protein (NC), is involved in the processing and packaging of RNA; p15 is a protease (PR) that is involved in the cleavage of the precursors of viral genome-encoded proteins; and p27 is a capsid protein (CA) that serves as a common group-specific antigen of ALV, which distinguishes ALV from other oncogenic viruses because of its high conservation (4, 31, 32). Fourteen potential m6A motifs were predicted in the coding region of p10, p12, and p15, and eight were predicted for p27. Although p27 is relatively conserved between the different groups of ALV, whether the m6A modification occurred in all ALVs via these motifs warrants further analysis.
The pol gene exhibits conservation across the different ALV subgroups and encodes two essential enzymes, i.e., the reverse transcriptase (p68) and the integrase (p32): p68 can use viral RNA as a template for reverse transcription, to produce proviral DNA, which is then inserted into the host chromosome under the integrative action of p32 (33, 34). Moreover, 14 potential m6A motifs were predicted, half each for p68 and p32, including the motif locate at 3740 with a conservation percentage of 95.30% and a rating of “very high confidence.” The motifs enriched in p68 and p32 also indicated that the m6A modification may play a vital role in the reverse transcriptase integrase process during the infection course of ALV-J.
The env gene is highly variable across subtypes and encodes glycosylated vesicle membrane proteins, including the outer membrane protein gp85 and transmembrane protein gp37, linked by hydrogen and disulfide bonds; thus, env is a key gene in ALV tumorigenesis (35). In the current study, the number of conserved motifs of the env gene was relatively low possibly because of the mutability of the gene. However, motifs located in the coding region of env were also predicted in different ALV-J strains. It remains unclear whether these non-conservedly predicted motifs are useful and correlated with differences in strain pathogenicity. gp37 protein has two structurally important hydrophobic regions for fusion with cell membranes. Therefore, it has a significant impact on the fusion of viral vesicle membranes with cell membranes; gp85 contains a viral receptor determinant that recognizes a specific receptor on the membrane of target cells for infection. As the major antigen of viruses, gp85 not only determines the subgroup identity and host range of ALV but also induces specific neutralizing antibodies in chickens (36). Furthermore, another motif located at position 3,740 with a rating of “very high confidence” was detected in the overlapped coding region of pol and env. The RNA secondary structures of this motif were predicted to be A–U pair or hairpin loop, which also indicated the complex RNA secondary structures induced by the sequences flanking each m6A motif. As reported previously, m6A stacking at the end of a hairpin loop stabilizes the loop, whereas the N6-methylation of an A–U pair in the middle of a helix would reduce the stability of helix folding (27). Refer to the conserved motifs predicted in ALV-J genomes, they mainly contained A–U pair or hairpin loop, and m6A modification of the unpaired adenosines attached in each helix have been confirmed to increase folding stability (27). Therefore, secondary structure prediction may help determine the structure–function relationships, thus promoting the understanding of the roles of m6A modifications and improving their prediction accuracy.
In this study, m6A modification motifs were predicted in different ALV-J strains, and their conservation was analyzed. Moreover, the RNA secondary structure of some representative motifs was determined. Although these motifs warrant further identification, a systematic comparative analysis would facilitate the rapid investigation of m6a modifications in ALV-J.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
JJ: Investigation, Writing – original draft. XM: Data curation, Writing – original draft. SX: Software, Writing – original draft. XX: Data curation, Methodology, Writing – original draft. ZZ: Methodology, Visualization, Writing – original draft. LY: Supervision, Writing – review & editing. QX: Writing – review & editing. YB: Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was supported by the National Natural Science Foundation of China (Grant no. 31802185), Program for Science & Technology Innovation Talents in Universities of Henan Province (Grant no. 22HASTIT042), and Cultivation Project of Nanyang Normal University for NSFC (Grant no. 2024CX004).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2024.1374430/full#supplementary-material
1. ^ http://www.cuilab.cn/sramp/
2. ^ https://www.chiplot.online/lollipop_plot.html
3. ^ https://rna.urmc.rochester.edu/RNAstructureWeb/index.html
1. Deng, Q, Li, Q, Li, M, Zhang, S, Wang, P, Fu, F, et al. The emergence, diversification, and transmission of subgroup J avian Leukosis virus reveals that the live chicken trade plays a critical role in the adaption and Endemicity of viruses to the yellow-chickens. J Virol. (2022) 96:e0071722. doi: 10.1128/jvi.00717-22
2. Payne, LN, Howes, K, Gillespie, AM, and Smith, LM. Host range of Rous sarcoma virus pseudotype RSV (HPRS-103) in 12 avian species: support for a new avian retrovirus envelope subgroup, designated J. J Gen Virol. (1992) 73:2995–7. doi: 10.1099/0022-1317-73-11-2995
3. Bai, J, Howes, K, Payne, LN, and Skinner, MA. Sequence of host range determinants in the env gene for a full length, infectious provirus clone of exogenous avian leukosis HPRS-103 confirm that it represents a new subgroup (designated j). J Gen Virol. (1995) 76:181–7. doi: 10.1099/0022-1317-76-1-181
4. Benson, SJ, Ruis, BL, Fadly, AM, and Conklin, KF. The unique envelope gene of the subgroup J avian leukosis virus derives from ev/J proviruses, a novel family of avian endogenous viruses. J Virol. (1998) 72:10157–64. doi: 10.1128/JVI.72.12.10157-10164.1998
5. Xu, MQK, Shao, HYY, Nair, VYJ, and Qin, A. 3'UTR of ALV-J can affect viral replication through promoting transcription and mRNA nuclear export. J Virol. (2023) 97:e0115223. doi: 10.1128/jvi.01152-23
6. Cui, N, Su, S, Chen, Z, Zhao, X, and Cui, Z. Genomic sequence analysis and biological characteristics of a rescued clone of avian leukosis virus strain JS11C1, isolated from indigenous chickens. J Gen Virol. (2014) 95:2512–22. doi: 10.1099/vir.0.067264-0
7. Payne, LN, and Nair, V. The long view: 40 years of avian leukosis research. Avian Pathol. (2012) 41:11–9. doi: 10.1080/03079457.2011.646237
8. Fan, H, and Johnson, C. Insertional oncogenesis by non-acute retroviruses: implications for gene therapy. Viruses. (2011) 3:398–422. doi: 10.3390/v3040398
9. Yan, YCS, Liao, LGS, Pang, YZX, and Zhang, HXQ. ALV-miRNA-p19-01 promotes viral replication via targeting dual specificity phosphatase 6. Viruses. (2022) 14:805. doi: 10.3390/v14040805
10. Luo, HHX, Wu, HZG, and Chai, WCH. Activation of lnc-ALVE1-AS1 inhibited ALV-J replication through triggering the TLR3 pathway in chicken macrophage like cell line. Vet Res Commun. (2023) 47:431–43. doi: 10.1007/s11259-022-09960-1
11. Xue, JZD, Zhou, JDX, Zhang, XLX, and Ding, LCZ. miR-155 facilitates the synergistic replication between avian leukosis virus subgroup J and reticuloendotheliosis virus by targeting a dual pathway. J Virol. (2023) 97:e0093723. doi: 10.1128/jvi.00937-23
12. Yang, TQL, Chen, SWZ, Jiang, YBH, Bi, YCG, and Chang, G. Circ_PIAS1 promotes the apoptosis of ALV-J infected DF1 cells by up-regulating miR-183. Genes. (2023) 14:1260. doi: 10.3390/genes14061260
13. Roundtree, IA, Evans, ME, Pan, T, and He, C. Dynamic RNA modifications in gene expression regulation. Cell. (2017) 169:1187–200. doi: 10.1016/j.cell.2017.05.045
14. Warminski, M, Mamot, A, Depaix, A, Kowalska, J, and Jemielity, J. Chemical modifications of mRNA ends for therapeutic applications. Acc Chem Res. (2023) 56:2814–26. doi: 10.1021/acs.accounts.3c00442
15. Gilbert, WV, and Nachtergaele, S. mRNA regulation by RNA modifications. Annu Rev Biochem. (2023) 92:175–98. doi: 10.1146/annurev-biochem-052521-035949
16. Yue, Y, Liu, J, and He, C. RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev. (2015) 29:1343–55. doi: 10.1101/gad.262766.115
17. Zhang, X, Peng, Q, and Wang, L. N6-methyladenosine modification-a key player in viral infection. Cell Mol Biol Lett. (2023) 28:78. doi: 10.1186/s11658-023-00490-5
18. Chen, KLZ, Wang, XFY, Luo, GZLN, Han, DDD, Dai, QPT, and He, C. High-resolution N(6) -methyladenosine (m(6) a) map using photo-crosslinking-assisted m(6) a sequencing. Angew Chem. (2015) 54:1587–90. doi: 10.1002/anie.201410647
19. Meyer, KDSY, Zumbo, PEO, and Mason, CEJSR. Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell. (2012) 149:1635–46. doi: 10.1016/j.cell.2012.05.003
20. Linder, BGAV, Olarerin-George, AOMC, and Mason, CEJSR. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat Methods. (2015) 12:767–72. doi: 10.1038/nmeth.3453
21. Bayoumi, M, Rohaim, MA, and Munir, M. Structural and virus regulatory insights into avian N6-Methyladenosine (m6A) machinery. Front Cell Dev. Biol. (2020) 8:543. doi: 10.3389/fcell.2020.00543
22. Zhao, Q, Yao, Z, Chen, L, He, Y, Xie, Z, Zhang, H, et al. Transcriptome-wide dynamics of m6A methylation in tumor livers induced by ALV-J infection in chickens. Front Immunol. (2022) 13:868892. doi: 10.3389/fimmu.2022.868892
23. Zhou, Y, Zeng, P, Li, YH, Zhang, Z, and Cui, Q. SRAMP: prediction of mammalian N6-methyladenosine (m6A) sites based on sequence-derived features. Nucleic Acids Res. (2016) 44:e91. doi: 10.1093/nar/gkw104
24. Tamura, K, Stecher, G, and Kumar, S. MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol. (2021) 38:3022–7. doi: 10.1093/molbev/msab120
25. Chen, C, Wu, Y, Li, J, Wang, X, Zeng, Z, Xu, J, et al. TBtools-II: a "one for all, all for one" bioinformatics platform for biological big-data mining. Mol Plant. (2023) 16:1733–42. doi: 10.1016/j.molp.2023.09.010
26. Xie, Y, Li, H, Luo, X, Li, H, Gao, Q, Zhang, L, et al. IBS 2.0: an upgraded illustrator for the visualization of biological sequences. Nucleic Acids Res. (2022) 50:W420–6. doi: 10.1093/nar/gkac373
27. Kierzek, E, Zhang, X, Watson, RM, Kennedy, SD, Szabat, M, Kierzek, R, et al. Secondary structure prediction for RNA sequences including N6-methyladenosine. Nat Commun. (2022) 13:1271. doi: 10.1038/s41467-022-28817-4
28. Song, J, and Yi, C. Chemical modifications to RNA: a new layer of gene expression regulation. ACS Chem Biol. (2017) 12:316–25. doi: 10.1021/acschembio.6b00960
29. Cheng, B, Leng, L, Li, Z, Wang, W, Jing, Y, Li, Y, et al. Profiling of RNA N6-Methyladenosine methylation reveals the critical role of m6A in chicken adipose deposition. Front Cell Dev Biol. (2021) 9:590468. doi: 10.3389/fcell.2021.590468
30. Yu, B, Liu, J, Cai, Z, Wang, H, Feng, X, Zhang, T, et al. RNA N6-methyladenosine profiling reveals differentially methylated genes associated with intramuscular fat metabolism during breast muscle development in chicken. Poult Sci. (2023) 102:102793. doi: 10.1016/j.psj.2023.102793
31. Chiu, R, and Grandgenett, DP. Avian retrovirus DNA internal attachment site requirements for full-site integration in vitro. J Virol. (2000) 74:8292–8. doi: 10.1128/jvi.74.18.8292-8298.2000
32. Scheifele, LZ, Ryan, EP, and Parent, LJ. Detailed mapping of the nuclear export signal in the Rous sarcoma virus gag protein. J Virol. (2005) 79:8732–41. doi: 10.1128/JVI.79.14.8732-8741.2005
33. Gallo, RC. Reverse transcriptase, the DNA polymerase of oncogenic RNA viruses. Nature. (1971) 234:194–8. doi: 10.1038/234194a0
34. Eisenman, RN, Mason, WS, and Linial, M. Synthesis and processing of polymerase proteins of wild-type and mutant avian retroviruses. J Virol. (1980) 36:62–78. doi: 10.1128/JVI.36.1.62-78.1980
35. Pandiri, ARRWM, and Mays, JKFAM. Influence of strain, dose of virus, and age at inoculation on subgroup J avian leukosis virus persistence, antibody response, and oncogenicity in commercial meat-type chickens. Avian Disease. (2007) 51:725–32. doi: 10.1637/0005-2086(2007)51[725:IOSDOV]2.0.CO;2
Keywords: avian leukosis virus subgroup J, m6A, DRACH motif, RNA secondary structure, comparative analysis
Citation: Ji J, Mu X, Xu S, Xu X, Zhang Z, Yao L, Xie Q and Bi Y (2024) Conservation and distribution of the DRACH motif for potential m6A sites in avian leukosis virus subgroup J. Front. Vet. Sci. 11:1374430. doi: 10.3389/fvets.2024.1374430
Received: 22 January 2024; Accepted: 03 April 2024;
Published: 12 April 2024.
Edited by:
Dirk Werling, Royal Veterinary College (RVC), United KingdomReviewed by:
Huansheng Wu, Jiangxi Agricultural University, ChinaCopyright © 2024 Ji, Mu, Xu, Xu, Zhang, Yao, Xie and Bi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jun Ji, amlqdW4wMjBAMTI2LmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.