
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci., 02 May 2024
Sec. Animal Behavior and Welfare
Volume 11 - 2024 | https://doi.org/10.3389/fvets.2024.1369655
This article is part of the Research TopicHealth And Welfare of Rabbits (On a Farm, in the Laboratory or as a Pet)View all 4 articles
Antimicrobial resistance (AMR) is commonly associated with the inappropriate use of antibiotics during meat-rabbit production, posing unpredictable risks to rabbit welfare and public health. However, there is limited research on the epidemiological dynamics of antibiotic resistance among bacteria indicators derived from local healthy meat-rabbits. To bridge the knowledge gap between antibiotic use and AMR distribution, a total of 75 Escherichia coli (E. coli) and 210 Enterococcus spp. strains were successfully recovered from fecal samples of healthy meat-rabbits. The results revealed that diverse AMR phenotypes against seven commonly used antimicrobials, including ampicillin (AMP), amoxicillin-clavulanic acid (A/C), doxycycline (DOX), enrofloxacin (ENR), florfenicol (FFC), gentamicin (GEN), and polymycin B (PMB), were observed among most strains of E. coli and Enterococcus spp. in two rabbit farms, although the distribution pattern of antibiotic resistance between young and adult rabbits was similar. Among them, 66 E. coli strains showed resistance against 6 antimicrobials except for PMB. However, 164 Enterococcus spp. strains only exhibited acquired resistance against DOX and GEN. Notably, the DOX-based AMR phenotypes for E. coli and Enterococcus spp. strains were predominant, indicating the existing environmental stress conferred by DOX exposure. The MICs tests suggested elevated level of antibiotic resistance for resistant bacteria. Unexpectedly, all GEN-resistant Enterococcus spp. strains resistant high-level gentamicin (HLGR). By comparison, the blaTEM, tetA, qnrS and floR were highly detected among 35 multi-resistant E. coli strains, and aac[6']-Ie-aph[2']-Ia genes widely spread among the 40 double-resistant Enterococcus spp. strains. Nevertheless, the presence of ARGs were not concordant with the resistant phenotypes for a portion of resistant bacteria. In conclusion, the distribution of AMR and ARGs are prevalent in healthy meat-rabbits, and the therapeutic antimicrobials use in farming practice may promote the antibiotic resistance transmission among indicator bacteria. Therefore, periodic surveillance of antibiotic resistance in geographic locations and supervisory measures for rational antibiotic use are imperative strategies for combating the rising threats posed by antibiotic resistance, as well as maintaining rabbit welfare and public health.
In recent years, the escalating public concerns over the evolution and spread of bacteria resistance between livestock and humans have underscored the need for surveillance on the judicious use of antibiotics in food-producing animals (1–3). However, the aggressive use of prophylactic and therapeutic antimicrobials during intensive rabbit production may facilitate population-level transfer of antibiotic resistance in farming environments (4, 5). These inadvertent consequences could potentially expedite the acquisition of antibiotic resistance and virulence among commensal bacteria in gastrointestinal tract (6), such as Enterobacter spp. and Enterococcus spp. (7), thereby contributing to gut microbiota dysbiosis and intestinal barrier impairments (8). Several genotypes of opportunistic E. coli or Enterococci strains have been confirmed as causative agents in newborn or weaning rabbits with diarrhea (9, 10), which urge us to explore the emergence and distribution of antibiotic resistance among the bioindicators derived from healthy rabbit population.
Although the restriction on the use of antibiotic feed additives as growth promoter in food-producing animals has significantly reduced the burden of bacterial antimicrobial resistance (AMR) on food-chains and ecosystems (11, 12), the inappropriate use of therapeutic antibiotics, such as tetracyclines, chloramphenicols and β-lactams (4, 5), remains an overlooked attention to the transferable antibiotic resistance resulting from antibiotics stress or bacteria contamination in food animal production systems. Since the emerging “superbugs” that exhibit resistance to critically important antimicrobials (CIAs), such as extended spectrum beta-lactamases (ESBL) (like blaTEM, blaCTX-M, blaSHV), as well as phosphoethanolamine (pEtN) transferase (like mcr-1and mcr-2) producing E. coli strains, high-level gentamicin-resistant (HLGR) and vancomycin-resistant Enterococcus spp. strains, have been frequently reported in domestic animals (13–17), the relationship between antibiotic use and antibiotic resistance distribution should be continously monitored, particularly in the case of neglected healthy farmed rabbits.
Limited epidemiological studies have reported the presence of antibiotic-resistant E. coli and Enterococcus spp. isolates in domestic or wild rabbit populations (18–21), exhibiting diverse AMR spectrums and distinctive ARG distribution across different geographical areas. However, there is a lack of knowledge regarding updated epidemiological data in Chengdu City, which is one of largest markets for meat-rabbit production and consumption in Southwest China. Therefore, this investigation aims to characterize the antibiotic resistance in indicator E. coli and Enterococcus spp. strains derived from healthy meat-rabbits. The findings will enhance our understanding of the emergence of antibiotic resistance conferred by routinely used therapeutic antibiotics in a context of reduced antibiotic use and residues in food animals, thereby raising awareness about rational antibiotic use to combat the rising threats posed by antibiotic resistance in rabbit farming practices.
All methods involving the animal sampling and handling followed the criteria of animal welfare regulation. Informed consent was obtained from commercial rabbit farm owners, and the research protocol was approved by the Research Ethics Committee of Southwest Minzu University (approval number no. 2021-MDLS-037).
Two hundred and forty-seven freshly voided fecal samples were randomly collected from 118 young (< 8 weeks) and 129 adult healthy rabbits (> 24 weeks) raised in two large-scale rabbit farms situated in suburbs of Chengdu City, Southwest China, from April to June, 2021. The samples were sterilely pooled in the plastic bags and stored at −80°C pending processing. Fecal samples were homogenized in 0.9% saline with a 1:2 ratio, then the diluted samples were inoculated into Luria-Bertani (LB) and Tryptic soy agar (TSA) (Hopebiol, Qingdao, China), respectively, and incubated in thermostatic incubator at 37°C for 24 h in aerobic conditions. The non-repetitive single colony per sample was cultured on Eosin methylene blue (EMB) agar and Enterococcus agar (Hopebiol, Qingdao, China) to preliminarily differentiate the E. coli or Enterococcus spp. isolates, respectively. After that, the presumptive bacteria isolates were further purified for later molecular identification.
The bacterial DNA was extracted using phenol-chloroform protocol (Tiangen, Beijing, China) as instruction described. The PCR assay targeting highly conserved 16S rRNA gene fragment was applied to test all suspected isolates. The primers 27F-ATGGCTCAGATTGAACG and 1492R-CAGGTTCCCCTACGGTTA were used for batcerial identification as described in the literature (22). Reaction system: 2 × Taq PCR MasterMix II (10 μM) (Tiangen, Beijing, China), 12.5 μL, 27F and 1492R primers (10 μM) 0.5 μL each, 1 μL of DNA template, and ddH2O replenished the system to 25 μL. PCR reaction conditions: pre-denaturation temperature of 94 °C for 3 min, denaturation temperature of 94 °C for 30 s, annealing and restitution temperature of 55 °C, PCR reaction conditions: pre-denaturation at 94°C for 3 min, denaturation at 94°C for 30 s, annealing at 55°C for 30 s, extension at 72°C for 1 min and 30 s for 30 cycles, and final extension at 72°C for 5 min on veriti 96-well thermal cycler (Thermo Fisher Scientific, USA). The 5 μL of each reaction were analyzed on 1% (w/v) agarose gel and submitted for DNA sequencing (Sangon Biotech, Shanghai, China). The species identification of E. coli and Enterococcus spp. isolates was accomplished by nucleotide sequence alignments on BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
The availablility of therapeutic antibiotics were confirmed by the farm owners, and 7 commonly used antimicrbiobials including ampicillin (AMP), amoxicillin-clavulanate (A/C), doxycycline (DOX), enrofloxacin (ENR), gentamicin (GEN), florfenicol (FFC) and polymyxin B (PMB), were prepared in injectable or oral formulations. These antimicrobials are routinely or sporadically used in the two rabbit farms for suspected bacterial infection. Herein, the antibiotic susceptibility of bacteria isolates were tested for AMP, A/C, DOX, ENR, GEN, FFC and PMB using Kirby-Bauer (K-B) agar disk diffusion method (Hangzhou microbial reagent CO., LTD, Hangzhou, China). Then, the minimal inhibitory concentrations (MICs) of these antimirobials (Meilunbio, Dalian, China) for antibiotic-resistant E.coli and Enterococcus spp. strains were further analyzed by microbroth dilution method according to the recommendation of Clinical and Laboratory Standards Institute (CLSI, 2018) (23). In addition, high-level GEN-resistant (HLGR) Entercococcus spp. strains (MIC ≥ 500 μg/mL) were determined according to the recommended screen test (CLSI, 2014) (24). E. coli ATCC 25922 and Enterococcus faecalis ATCC29212 were used as quality control strains.
Based on the AMR phenotypes of multi-resistant E.coli or double-resistant Enterococcus spp. strains, representative antibiotic resistance genes (ARGs) that belonged to β-lactams (blaTEM), tetracyclines (tetA, tetB, tetM), fluroquinoloncs (qnrD, qnrS), chloramphenicols (fexA, floR), and aminoglycosides (aac[6']-Ib for E. coli strains, aac[6']-Ie-aph[2”]-Ia for HLGR Enterococcsu spp. strains) were detected using established PCR methods for these target genes (9, 25–27) (Supplementary Table 1.).
The frequencies and percentages of all categorical variables were tabulated. The statistical significance (P < 0.05) was determined using Fisher's exact test or chi-squared tests in SPSS 18.0 (IBM Armonk Corp., NY, United States). The descriptive histograms were achieved using Graphpad Prism 8.0 (Graphpad Software Inc, San Diego, United States).
Totally, 75 suspected isolates of E. coli and 210 Enterococcus spp. were preliminary identified from 247 fecal samples by differential culture. The Gram-negative E. coli strains appeared to be rod-like bacillus and formed deep purplish-black colonies on EMB agar (Supplementary Figures 1A, C). On the other hand, the Gram-positive Enterococcus spp. strains appeared to be a spherical morphology and formed black colonies on Enterococci differential medium (Supplementary Figures 1B, D). DNA sequencing and molecular alignment further confirmed all suspected E. coli or Enterococcus spp. strains. In addition, the Enterococcus spp. isolates consisted of 142 Enterococcus faecalis (E. faecalis), 47 Enterococcus faecium (E. faecium), 15 Enterococcus gallinarum (E. gallinarum), and 6 Enterococcus hirae (E.hirae) strains. Among them, 51 E. coli strains (27 from young rabbits and 24 from adult rabbits) and 82 Enteroccocus spp. strains (45 from young rabbits and 37 from adult rabbits) were recovered from farm A, while 24 E. coli strains (9 from young rabbits and 15 from adult rabbits) and 128 Enteroccocus spp. strains (54 from young rabbits and 74 from adult rabbits) were recovered from farm B (Table 1).
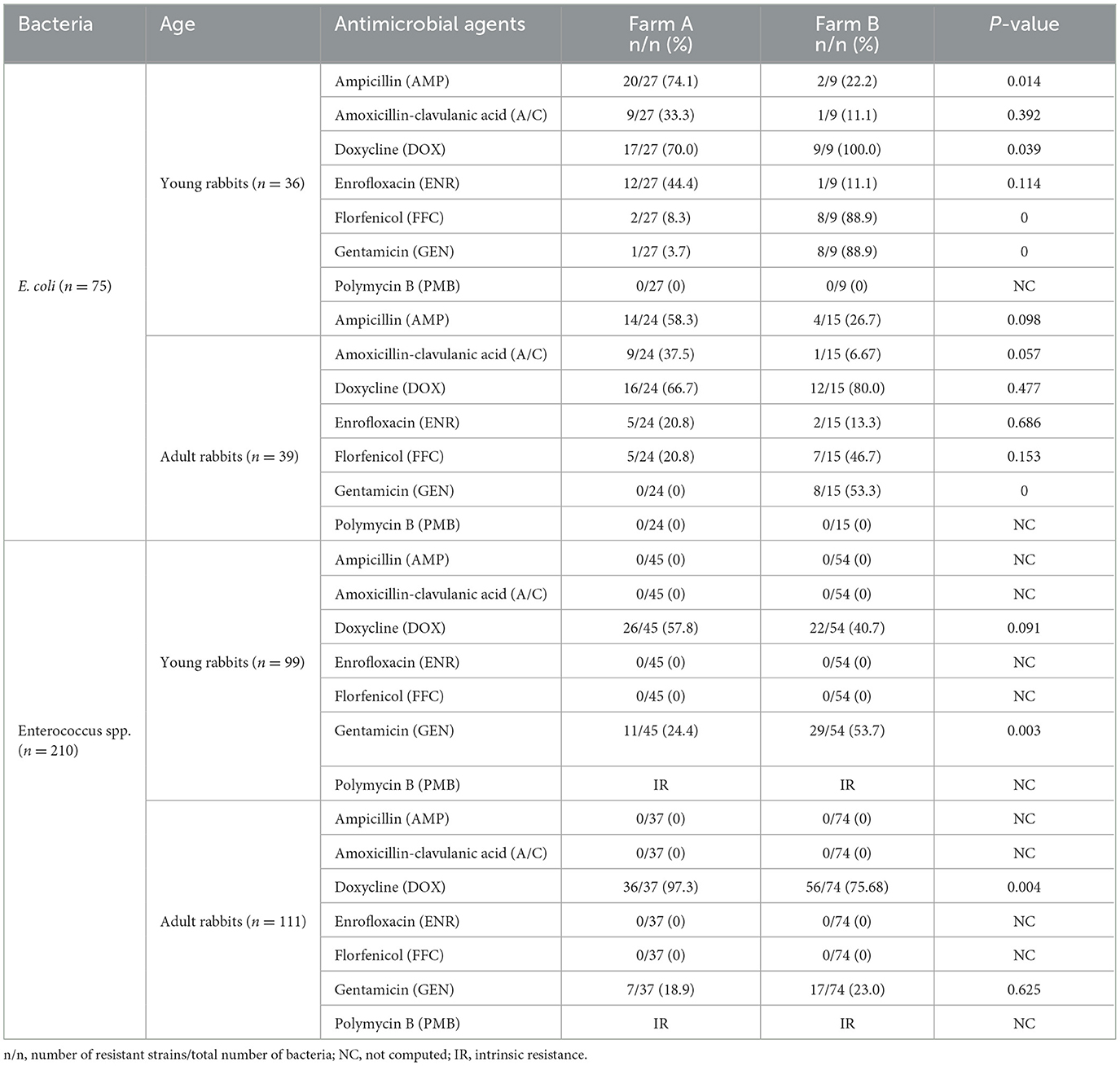
Table 1. AMR frequencies of E. coli and Enterococcus spp. isolates from farming rabbits against commonly used antibiotics using the K-B method.
All bacteria strains were tested for their antibiotic resistance phenotype against seven commonly used antimicrobials using the K-B method. Among them, majority of E. coli strains exhibited resistance to DOX (54/75, 72.0%), AMP (40/75, 53.3%), FFC (22/75, 29.3%), A/C (20/75, 26.7%), ENR (20/75, 26.7%), and GEN (17/75, 22.7%), respectively. However, all E. coli strains were susceptible to PMB. Furthermore, all Enterococcus spp. strains showed sensitivity to A/C, AMP, FFC, and ENR while possessing intrinsic resistance (IR) to PMB. Notably, a significant number of resistant Enterococcus spp. strains against DOX (140/210, 66.7%) and GEN (64/210, 30.5%) were detected (Table 1).
In contrast, the prevalence of resistant E. coli strains against AMP (34/51, 66.7%), A/C (18/51, 35.3%), and ENR (17/51, 33.3%) from farm A were higher compared to farm B (AMP, 6/24, 25.0%; A/C, 2/24, 8.3%; ENR, 3/25, 12.0%), respectively. Conversely, the DOX-, FFC-, and GEN-resistant E. coli strains were more frequently observed in farm B (DOX, 21/24, 87.5%; FFC, 15/24, 62.5%; GEN, 16/24, 66.7 %) than farm A (DOX, 33/51, 64.7%; FFC, 7/51, 13.7%; GEN, 1/51, 2.0 %). Moreover, the proportion of DOX-resistant Enterococcus spp. strains from farm A (62/82, 75.6%) was higher than that from farm B (78/128, 60.9%), while the occurrence of GEN-resistant strains was lower in farm A (18/82, 22.0%) compared to farm B (46/128, 35.9%). The distribution pattern of antibiotic resistance among E. coli or Enterococcus spp. strains between young and adult rabbits was similar in two farms, but the statistical differences for the ratios of resistant bacteria against certain antimicrobials were not always concordant (Figure 1 and Table 1).
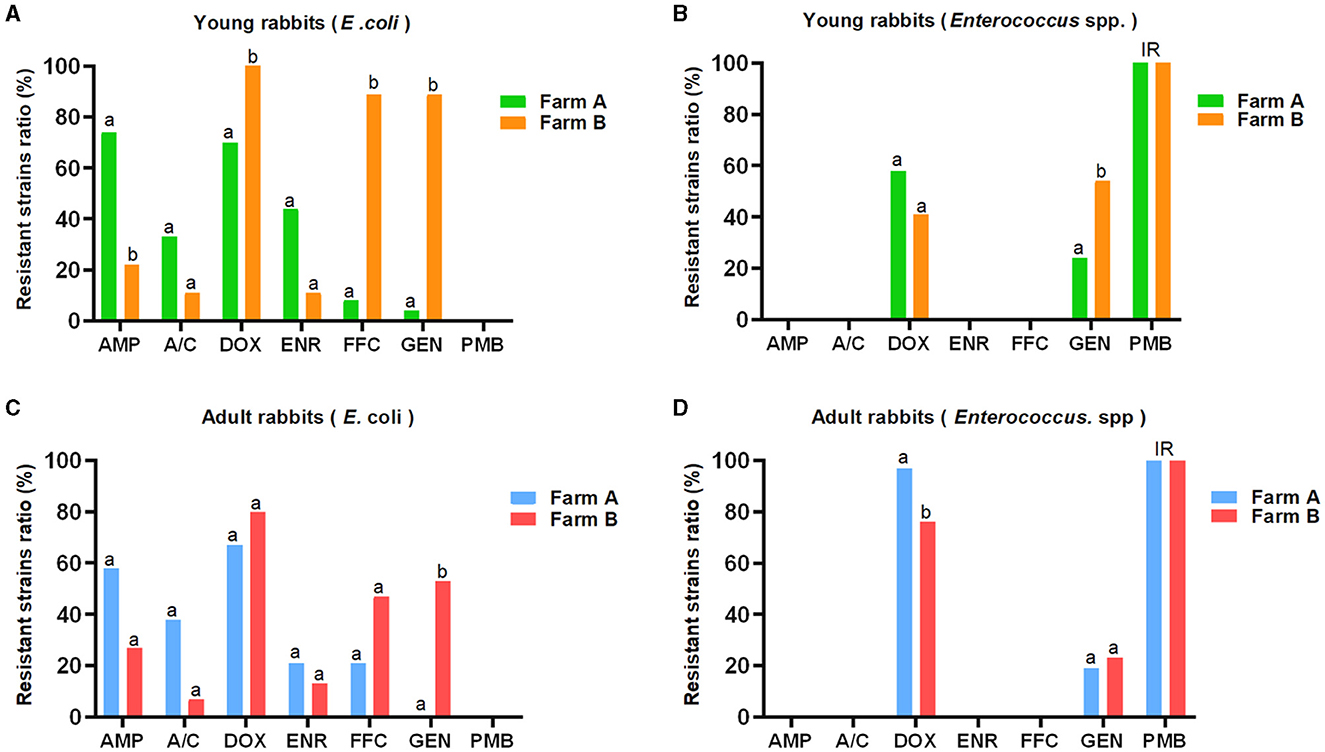
Figure 1. AMR distribution among E. coli and Enterococcus spp. strains derived from young and adult rabbits against seven commonly used antibiotics (K-B method). The E. coli (A, C) and Enterococcus spp. (B, D) strains showed distinct resistant rates against seven commonly used antibiotics. The prevalence of resistant E. coli strains from farm A was higher than that from farm B with respect to AMP, A/C, and ENR. However, it was lower for DOX, FFC, and GEN, respectively (A, C). Similarly, the rate of DOX-resistant Enterococcus spp. strains from farm A exceeded that from farm B, but was lower for GEN (B, D). Although the distribution patterns of antimicrobial resistance between young and adult rabbits were similar, there were no consistent statistical differences observed in terms of resistant E. coli or Enterococcus spp. strains for most antimicrobials. IR: intrinsic resistance. a, b: different letters mark statistical significance between variables.
To investigate the AMR profiles of E. coli and Enterococcus spp. strains we further characterized the antibiotic resistance phenotypes of each bacteria (Table 2). Diverse AMR profiles were shown among resistant E. coli strains with single, double, and multiple resistance phenotypes against six commonly used antimicrobials, except for PMB. However, only single and double resistance phenotypes against DOX and GEN were found among Enterococcus spp. strains. Notably, the DOX-based AMR phenotypes in E. coli and Enterococcus spp. strains were most predominant. Additionally, a subset of multi-resistant E. coli (35/66, 53.0%) and double-resistant Enterococcus spp. strains (40/164, 24.4%) contributed to the diversity of AMR phenotypes.
The MICs of resistant bacteria were determined using the microbroth dilution method, which yielded consistent results with the K-B method. Resistant strains of E. coli and Enterococcus spp. exhibited elevated levels of antibiotic resistance, as indicated by increased MIC values. Furthermore, all GEN-resistant Enterococcus spp. strains (64/64, 100%) demonstrated a high-level gentamicin resistance, whereas this phenomenon was not observed among GEN-resistant E. coli strains (0/17, 0%) (Table 3).
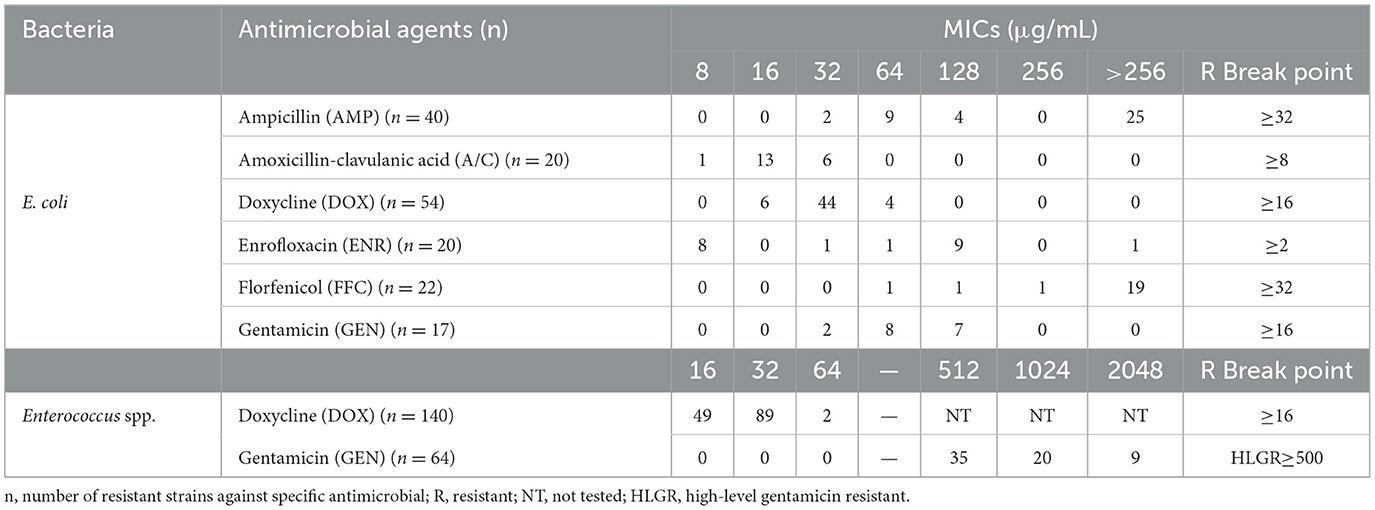
Table 3. Minimal inhibitory concentrations (MICs) of antibiotic-resistant E. coli and Enterococcus spp. strains.
Based on the AMR phenotypes, a total of 35 multi-resistant E. coli strains and 40 double-resistant Enterococcus spp. strains (n = 40) were selected for further screening of relevant ARGs. Among the multi-resistant E.coli strains, blaTEM (97.1%, 34/35), tetA (94.3%, 33/35), tetB (25.7%, 9/35), tetM (2.9%, 1/35), qnrS (71.43%, 25/35), floR (57.14%, 20/35), and aac(6')-lb (11.4%, 4/35) genes were widely detected, respectively. Accordingly, infrequent distributions of tetM (25.0%, 10/40), fexA (10.0%, 4/40), and floR (7.5%, 3/40) were also observed among double-resistant Enterococcus spp. strains, while the aac[6']-Ie-aph[2']-Ia (77.5%, 31/40) gene was highly prevalent. Comparatively, blaTEM, tetA, qnrS, and floR genes for multi-resistant E. coli strains and the aac[6']-Ie-aph[2”]-Ia gene for double-resistant Enterococcus spp. strains were overrepresented (over 50%). However, the presence of ARGs were not consistent with the AMR phenotypes for a portion of resistant strains (e.g., fexA and floR genes were detected in DOX/GEN-resistant Enterococcus spp. strains) (Table 4).
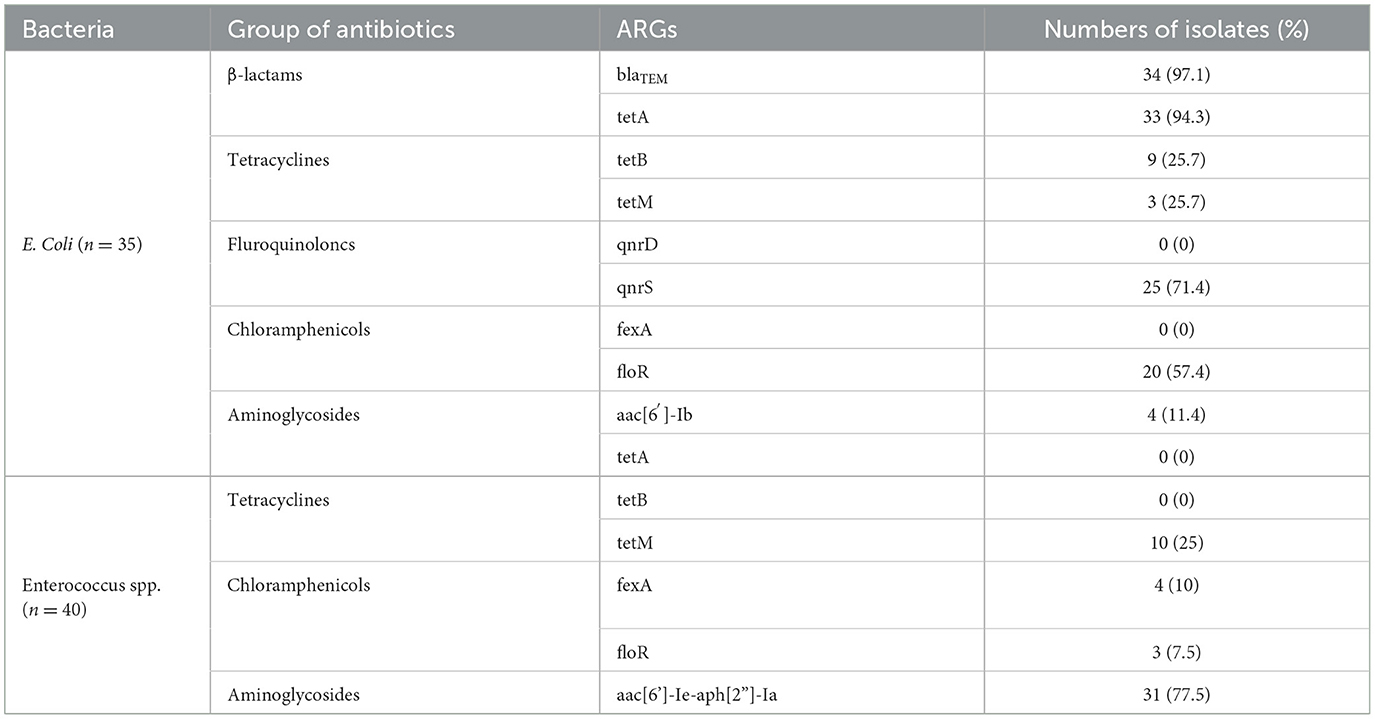
Table 4. Antibiotic resistance genes (ARGs) of multi-resistance E. coli and double-resistant Enterococcus spp. isolates from healthy rabbits.
It is widely recognized that symbiotic E. coli and Enterococcus spp. play a crucial role in maintaining intestinal homeostasis and integrity (28). However, they are also prone to acquiring antibiotic resistance and harboring antimicrobial resistance genes (ARGs) when repeatedly exposed to antibiotics, particularly in intensive food-animal husbandry (6, 7). Moreover, these antibiotic-resistant commensal bacteria have been reported as opportunistic pathogens causing diarrhea among rabbits (9, 10), and they even act as negative mediators to shield the antibiotic-sensitive pathogens during antibiotic treatment (29). Therefore, it is imperative to conduct surveillance on these bioindicators to assess the potential challenge posed by transferable antibiotic resistance for animal health.
Considering that commercial meat-rabbits may receive prophylactic or therapeutic antimicrobials to address suspected bacterial infection, such as respiratory or gastrointestinal infectious diseases, either through direct contact or environmental contamination (5, 30), which may significantly changes the composition of gastrointestinal microbiota (31) and antimicrobial resistance (AMR) phenotypes of commensal E. coli and Enterocuccus spp. strains (32). In this study, we successfully isolated two bacteria species from rabbit feces with a higher identification rate for Enterococcus spp. strains (210/247, 85%) compared to E. coli strains (75/247, 30.4%), indicating the dominant presence of Enterococcus spp. in the fecal flora under local farming conditions. However, this lower recovery rate, possibly attributed to the reduced bioactivity of E. coli during feces processing and cell culture, should also be considered when compared with high isolation rates (57.1~100%) observed in other rabbit populations (19–21). Despite this discrepancy, an adequate number of E. coli and Enterococcus spp. strains were obtained for subsequent analysis.
We conducted an investigation on antibiotic resistance and relevant ARG distribution among these bacteria, and evaluated their susceptibility to seven available antimicrobials in two rabbit farms. A significant proportion of E. coli strains exhibited resistance to most antimicrobials (6/7) with diverse AMP spectrums, in contrast to Enterococcus spp. strains (3/7). This observation supports the previous finding that Enterobacter spp. harbor more mobile genetic elements (MGEs), facilitating robustly horizontal transfer of ARGs and acquisition of antibiotic resistance against specific antimicrobials (33). Notably, most strains of E. coli demonstrated reduced susceptibility to these antimicrobials, suggesting the need to consider alternative antibiotic prescription in light of the prevalence of DOX and AMP-resistant phenotypes. In addition, it is worth mentioning that no E. coli strain demonstrated resistance to PMB, which may be associated with less frequent medication. Although oral administration of ampicillin and amoxicillin/clavulanate may pose a risk of antibiotic-associated dysbiosis in certain rabbit populations (5), parenteral administration is still recommended for susceptible pathogens (18, 20). Moreover, antibiotics from the penicillin family appear to be more readily available for veterinary use in local rabbit farming practices, which may also influence the choice of antibiotic prescription and the prevalence of relevant antimicrobial resistance. Similarly, the frequent use of DOX and the sporadical use of PMB as informed by farm owners may influence the antibiotic susceptibility of E. coli strains.
Interestingly, most Enterococcus spp. strains only exhibited resistance to DOX and GEN, while no resistant strains against other antimicrobials were found. This disparity between Enterobacter spp. and Enterococcus spp. may be associated with their distinctive evolutionary pathways in antibiotic resistance against specific antimicrobials (34, 35). In particular, the difference in AMR spectrums between two farms was evident, possibly reflecting the preference of antibiotic prescriptions for veterinary technicians in farming practices, although the detailed medical history of antibiotic use was not comprehensively evaluated. Furthermore, bacteria of young rabbits origin exhibited a higher prevalence of antibiotic resistance compared to their adult counterparts, indicating that early-life exposure to antibiotics may robustly contribute to the emergence of antibiotic resistance among commensal bacteria (36). Therefore, reducing the inappropriate use of antimicrobials and medication overload will be a sustainable meat-rabbit farming practice.
According to the MIC values tested for resistant bacteria, increased levels of antibiotic resistance was prevalent, which may be inferred to the substantial exposure to antibiotics for rabbits in farming environments (30, 37). Furthermore, all GEN-resistant Enterococcus spp. strains showed high-level gentamicin resistance, whereas this characteristic was absent in GEN-resistant E. coli strains. This implies that Enterococcus spp. may serve as an active reservoir for acquired GEN resistance, which has also been detected in European wild rabbits (19). Previous research studies have indicated that patients with HLGR Enterococcus spp. strains of hospital origin could increase the risk of nosocomial infections, particularly the E. faecalis strains that typically harbor the aac[6']-Ie-aph[2”]-Ia gene (38, 39), and even produce cross-resistance with other critical important antibiotics (CIAs), such as vancomycin (40). It is important to note that the vancomycin-resistant Enterococcus spp. have been confirmed in food-producing animals (14, 15). Thus, increasing public attention should be paid toward these opportunistic pathogens of animal origin.
Several surveillance studies have reported a high prevalence of multi-resistant E. coli strains in farmed rabbits (10, 19–21). However, a similar occurrence (35/66, 53.0%) was still observed in our healthy rabbits. Additionally, we also identified a proportion of double-resistant Enterococcus spp. strains (40/164, 24.4%). As expected, most of multi-resistant E. coli strains carried ARGs associated with β-lactams (blaTEM, 34/35, 97.1%), tetracyclines (tetA, 33/35, 94.3%), fluroquinoloncs (qnrS, 25/35, 71.4%), and chloramphenicols (floR, 20/35, 57.4%) resistance. On the other hand, double-resistant Enterococcus spp. strains predominantly harbored the aac[6']-Ie-aph[2”]-Ia gene (31/40, 77.5%). Similar ARG distribution among resistant E. coli or Enterococcus spp. has been also found in farmed or wild rabbits, which may be attributed to environmental antibiotics co-selection (9, 15, 19, 41). Particularly, some resistant bacteria only carried a small number of ARGs (for example, fexA, qnrD, tetB, tetM, and aac[6′]-lb) or none at all, suggesting that certain ARG-independent mechanisms for antimicrobial resistance may be implicated for these bacteria (34, 35). It is well known that co-harboring various ARGs is one of the most important mechanisms to acquire antibiotic resistance (1, 12, 13); thus, the wide spread of antibiotic-specific ARGs among commensal E. coli and Enterococcus spp. strains may confer their resistance against commonly used antimicrobials, although other strain-level mobile genetic elements, including transposons, insertion sequences, and integrons (42), have not been evaluated in current study. In general, the commensal bacteria bearing abundant ARGs may serve as a high-risk reservoir of transferable antibiotic resistance, which pose unintended threats to rabbit welfare, farming biosecurity, and public health in a “One Health” perspective.
One limitation of the current study is the unexpanded AMR spectrum analysis for other groups of antibiotics, such as cephalosporins (e.g., ceftiofur), macrolides (e.g., azithromycin), sulfonamides (e.g., sulfadiazine/trimethoprim), and tricyclic glycopeptides (e.g., vancomycin), although they are rarely or never used in local rabbit farming practices. Second, the influence determinants involving antibiotic usage and consumption, bacteria genetic lineages, and biosecurity measures (15, 30, 31, 37) should be correlated with the emergence and enhancement of antibiotic resistance in further study. Even so, our findings provided a novel perspective on the impact of commonly used antimicrobials in shaping distinctive AMR spectrums and the increasing level of antibiotic resistance among indicator bacteria in meat-rabbit farming practices.
Overall, the distribution of AMR and ARGs is prevalent in healthy meat-rabbits, and the use of therapeutic antimicrobials in farming practices may promote the antibiotic resistance transmission among indicator bacteria. Therefore, periodic surveillance of antibiotic resistance in different geographic locations and supervisory measures for rational antibiotic use are considered to be imperative strategies for combating the rising threats posed by antibiotic resistance and maintaining rabbit welfare and public health.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
CS: Writing – original draft, Writing – review & editing, Formal analysis. ZW: Investigation, Writing – original draft, Writing – review & editing. YL: Formal analysis, Methodology, Writing – review & editing. JH: Conceptualization, Funding acquisition, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was supported by the Natural Science Foundation of Sichuan Province (Grant no. 24NSFSC0735), the Southwest Minzu University Double World-Class Project (XM2023013), and the Scientific and Technological Innovation Team for Qinghai-Tibetan Plateau Research in Southwest Minzu University (2024CXTD08).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2024.1369655/full#supplementary-material
1. He T, Wang R, Liu D, Walsh TR, Zhang R, Lv Y, et al. Emergence of plasmid-mediated high-level tigecycline resistance genes in animals and humans. Nat Microbiol. (2019) 4:1450–6. doi: 10.1038/s41564-019-0445-2
2. Cao H, Bougouffa S, Park TJ, Lau A, Tong MK, Chow KH, et al. Sharing of antimicrobial resistance genes between humans and food animals. MSystems. (2022) 7:e00775–22. doi: 10.1128/msystems.00775-22
3. Masebo NT, Marliani G, Cavallini D, Accorsi PA, Di Pietro M, Beltrame A, et al. Health and welfare assessment of beef cattle during the adaptation period in a specialized commercial fattening unit. Res Vet Sci. (2023) 158:50–5. doi: 10.1016/j.rvsc.2023.03.008
4. Caneschi A, Bardhi A, Barbarossa A, Zaghini A. The use of antibiotics and antimicrobial resistance in veterinary medicine, a complex phenomenon: a narrative review. Antibiotics. (2023) 12:487. doi: 10.3390/antibiotics12030487
5. Crovato S, Menegon F, Mascarello G, Pinto A, Nadin A, Piovan G, et al. Development of a training strategy aimed at increasing veterinarians' awareness of the proper use of antibiotics on rabbit farms. Animals. (2023) 13:2411. doi: 10.3390/ani13152411
6. Roth N, Käsbohrer A, Mayrhofer S, Zitz U, Hofacre C, Domig KJ. The application of antibiotics in broiler production and the resulting antibiotic resistance in Escherichia coli: a global overview. Poult Sci. (2019) 98:1791–804. doi: 10.3382/ps/pey539
7. Pissetti C, Kich JD, Allen HK, Navarrete C, de Freitas Costa E, Morés N, et al. Antimicrobial resistance in commensal Escherichia coli and Enterococcus spp. isolated from pigs subjected to different antimicrobial administration protocols. Res Vet Sci. (2021) 137:174–85. doi: 10.1016/j.rvsc.2021.05.001
8. Zhang Q, Cheng L, Wang J, Hao M, Che H. Antibiotic-induced gut microbiota dysbiosis damages the intestinal barrier, increasing food allergy in adult mice. Nutrients. (2021) 13:3315. doi: 10.3390/nu13103315
9. Zhao X, Yang J, Ju Z, Chang W, Sun S. Molecular characterization of antimicrobial resistance in Escherichia coli from rabbit farms in Tai'an, China. Biomed Res Int. (2018) 2018:8607647. doi: 10.1155/2018/8607647
10. Solans L, Arnal JL, Sanz C, Benito A, Chacón G, Alzuguren O, et al. Rabbit Enteropathies on commercial farms in the Iberian peninsula: etiological agents identified in 2018–2019. Animals. (2019) 9:1142. doi: 10.3390/ani9121142
11. Tang KL, Caffrey NP, Nóbrega DB, Cork SC, Ronksley PE, Barkema HW, et al. Comparison of different approaches to antibiotic restriction in food-producing animals: stratified results from a systematic review and meta-analysis. BMJ Global Health. (2019) 4:e001710. doi: 10.1136/bmjgh-2019-001710
12. Nobrega DB, Tang KL, Caffrey NP, De Buck J, Cork SC, Ronksley PE, et al. Prevalence of antimicrobial resistance genes and its association with restricted antimicrobial use in food-producing animals: a systematic review and meta-analysis. J Antimicrob Chemother. (2021) 76:561–75. doi: 10.1093/jac/dkaa443
13. Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. (2016) 16:161–8. doi: 10.1016/S1473-3099(15)00424-7
14. Gousia P, Economou V, Bozidis P, Papadopoulou C. Vancomycin-resistance phenotypes, vancomycin-resistance genes, and resistance to antibiotics of enterococci isolated from food of animal origin. Foodborne Pathog Dis. (2015) 12:214–20. doi: 10.1089/fpd.2014.1832
15. Lengliz S, Cheriet S, Raddaoui A, Klibi N, Ben Chehida N, Najar T, et al. Species distribution and genes encoding antimicrobial resistance in Enterococcus spp. isolates from rabbits residing in diverse ecosystems: a new reservoir of linezolid and vancomycin resistance. J Appl Microbiol. (2022) 132:2760–72. doi: 10.1111/jam.15461
16. Tseng CH, Liu CW, Liu PY. Extended-spectrum β-lactamases (ESBL) producing bacteria in animals. Antibiotics. (2023) 12:661. doi: 10.3390/antibiotics12040661
17. Kim YB, Seo KW, Shim JB, Hyun Son S, Noh EB, Lee YJ. Molecular characterization of antimicrobial-resistant Enterococcus faecalis and Enterococcus faecium isolated from layer parent stock. Poultry Sci. (2019) 98:5892–9. doi: 10.3382/ps/pez288
18. Fernández M, Garcias B, Duran I, Molina-López RA, Darwich L. Current situation of bacterial infections and antimicrobial resistance profiles in pet rabbits in Spain. Vet Sci. (2023) 10:352. doi: 10.3390/vetsci10050352
19. Silva N, Igrejas G, Figueiredo N, Gonçalves A, Radhouani H, Rodrigues J, et al. Molecular characterization of antimicrobial resistance in enterococci and Escherichia coli isolates from European wild rabbit (Oryctolagus cuniculus). Sci Total Environ. (2010) 408:4871–6. doi: 10.1016/j.scitotenv.2010.06.046
20. Kylie J, McEwen SA, Boerlin P, Reid-Smith RJ, Weese JS, Turner PV. Prevalence of antimicrobial resistance in fecal Escherichia coli and Salmonella enterica in Canadian commercial meat, companion, laboratory, and shelter rabbits (Oryctolagus cuniculus) and its association with routine antimicrobial use in commercial meat rabbits. Prev Vet Med. (2017) 147:53–7. doi: 10.1016/j.prevetmed.2017.09.004
21. Ben Said L, Jouini A, Fliss I, Torres C, Klibi N. Antimicrobial resistance genes and virulence gene encoding intimin in Escherichia coli and Enterococcus isolated from wild rabbits (Oryctolagus cuniculus) in Tunisia. Acta Vet Hung. (2019) 67:477–88. doi: 10.1556/004.2019.047
22. Dos Santos HRM, Argolo CS, Argôlo-Filho RC, Loguercio LL. A 16S rDNA PCR-based theoretical to actual delta approach on culturable mock communities revealed severe losses of diversity information. BMC Microbiol. (2019) 19:1–14. doi: 10.1186/s12866-019-1446-2
23. CLSI. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated From Animals. 4th Supplement VET08. Wayne, PA: Clinical and Laboratory Standards Institute (2018).
24. CLSI. Performance Standards for Antimicrobial Susceptibility Testing: Screening Test for Detection of High-Level Aminoglycoside Resistance (HLAR) in Enterococcus species. 24th Supplement M100-s24. Wayne, PA: Clinical and Laboratory Standards Institute (2014).
25. Chunjing L. Establishment on PCR method for the main drug resistant genes of tetracyclines antibiotics of E.coli (in Chinese). Modern J Anim Husb Vet Med. (2017) 12:7.
26. Ahmed AM, Motoi Y, Sato M, Maruyama A, Watanabe H, Fukumoto Y, et al. Zoo animals as reservoirs of gram-negative bacteria harboring integrons and antimicrobial resistance genes. Appl Environ Microbiol. (2007) 73:6686–90. doi: 10.1128/AEM.01054-07
27. Aishwarya KVL, Geetha PV, Eswaran S, Mariappan S, Sekar U. Spectrum of aminoglycoside modifying enzymes in gram-negative bacteria causing human infections. J Lab Physicians. (2020) 12:27–31. doi: 10.1055/s-0040-1713687
28. Chen Y, Cui W, Li X, Yang H. Interaction between commensal bacteria, immune response and the intestinal barrier in inflammatory bowel disease. Front Immunol. (2021) 12:761981. doi: 10.3389/fimmu.2021.761981
29. Gjonbalaj M, Keith JW, Do MH, Hohl TM, Pamer EG, Becattini S. Antibiotic degradation by commensal microbes shields pathogens. Infect Immun. (2020) 88:10–1128. doi: 10.1128/IAI.00012-20
30. Agnoletti F, Brunetta R, Bano L, Drigo I, Mazzolini E. Longitudinal study on antimicrobial consumption and resistance in rabbit farming. Int J Antimicrob Agents. (2018) 51:197–205. doi: 10.1016/j.ijantimicag.2017.10.007
31. Velasco-Galilea M, Guivernau M, Piles M, Viñas M, Rafel O, Sánchez A, et al. Breeding farm, level of feeding and presence of antibiotics in the feed influence rabbit cecal microbiota. Anim Microbiome. (2020) 2:40. doi: 10.1186/s42523-020-00059-z
32. Zhang L, Guo H, Gu J, Hu T, Wang X, Sun Y, et al. Metagenomic insights into dietary remodeling of gut microbiota and antibiotic resistome in meat rabbits. Sci Total Environ. (2023) 874:162006. doi: 10.1016/j.scitotenv.2023.162006
33. Ellabaan MMH, Munck C, Porse A, Imamovic L, Sommer MOA. Forecasting the dissemination of antibiotic resistance genes across bacterial genomes. Nat Commun. (2021) 12:2435. doi: 10.1038/s41467-021-22757-1
34. Baquero F, Martinez JL, Lanza V, Rodríguez-Beltrán J, Galán JC, San Millán A, Coque TM. Evolutionary pathways and trajectories in antibiotic resistance. Clin Microbiol Rev. (2021) 34:e00050-19. doi: 10.1128/CMR.00050-19
35. Sun Y, Lu H, Zhang X, Wu Q, Bi W, Liu H, et al. Phenotype and genotype alteration during adaptive evolution of Enterococcus faecalis to antimicrobials. Inf Genetics Evol. (2018) 62:80–5. doi: 10.1016/j.meegid.2018.03.029
36. Reyman M, Van Houten MA, Watson RL, Chu MLJ, Arp K, De Waal WJ, et al. Effects of early-life antibiotics on the developing infant gut microbiome and resistome: a randomized trial. Nat Commun. (2022) 13:893. doi: 10.1038/s41467-022-28525-z
37. Mencia-Ares O, Argüello H, Puente H, Gómez-Garcia M, Manzanilla EG, Álvarez-Ordónez A, et al. Antimicrobial resistance in commensal Escherichia coli and Enterococcus spp. is influenced by production system, antimicrobial use, and biosecurity measures on Spanish pig farms. Porcine Health Manag. (2021) 7:27. doi: 10.1186/s40813-021-00206-1
38. Sparo M, Delpech G, García Allende N. Impact on public health of the spread of high-level resistance to gentamicin and vancomycin in enterococci. Front Microbiol. (2018) 9:410861. doi: 10.3389/fmicb.2018.03073
39. Khodabandeh M, Mohammadi M, Abdolsalehi MR, Hasannejad-Bibalan M, Gholami M, Alvandimanesh A, et al. High-level aminoglycoside resistance in Enterococcus faecalis and Enterococcus faecium; as a serious threat in hospitals. Inf Disorders-Drug Targets. (2020) 20:223–8. doi: 10.2174/1871526519666181130095954
40. Woodford N, Jones BL, Baccus Z, Ludlam HA, Brown DFJ. Linkage of vancomycin and high-level gentamicin resistance genes on the same plasmid in a clinical isolate of Enterococcus faecalis. J Antimicrob Chemother. (1995) 35:179–84. doi: 10.1093/jac/35.1.179
41. Baoguang L, Shengli L, Dongdong Y, Caiyu Y, Yihan Z, Ping L, et al. Molecular detection of resistance genes to aminoglycosides, tetracyclines and sulfonamides among Escherichia coli strains iolated from rabbits in He'nan province (in Chinese). J Dom Anim Ecol. (2016) 37:60–3.
Keywords: antimicrobial resistance, antibiotics use, healthy meat-rabbits, epidemiological surveillance, ARG distribution
Citation: Sun C, Wang Z, Li Y and Huang J (2024) Antibiotic resistance spectrums of Escherichia coli and Enterococcus spp. strains against commonly used antimicrobials from commercial meat-rabbit farms in Chengdu City, Southwest China. Front. Vet. Sci. 11:1369655. doi: 10.3389/fvets.2024.1369655
Received: 12 January 2024; Accepted: 12 April 2024;
Published: 02 May 2024.
Edited by:
Michaela Fels, University of Veterinary Medicine Hannover, GermanyReviewed by:
Mehmet Cemal Adiguzel, Atatürk University, TürkiyeCopyright © 2024 Sun, Wang, Li and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jian Huang, aGp2ZXQwM0BzaW5hLmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.