
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci. , 14 May 2024
Sec. Veterinary Epidemiology and Economics
Volume 11 - 2024 | https://doi.org/10.3389/fvets.2024.1366904
 Hieu Van Dong1
Hieu Van Dong1 Giang Thi Huong Tran1
Giang Thi Huong Tran1 Tra Thi Thu Vu1
Tra Thi Thu Vu1 Ngan Hong Thi Le2
Ngan Hong Thi Le2 Yen Thi Hoang Nguyen1
Yen Thi Hoang Nguyen1 Witsanu Rapichai3,4
Witsanu Rapichai3,4 Amonpun Rattanasrisomporn5
Amonpun Rattanasrisomporn5 Chaiwat Boonkaewwan6
Chaiwat Boonkaewwan6 Dao Anh Tran Bui1
Dao Anh Tran Bui1 Jatuporn Rattanasrisomporn3*
Jatuporn Rattanasrisomporn3*Tembusu virus (TMUV) is an important infectious disease, causing economic losses in duck production. Since the first report of TMUV infection in Vietnam in 2020, the disease has persisted and affected poultry production in the country. This study conducted epidemiological and genetic characterization of the viral strains circulating in north Vietnam based on 130 pooled tissue samples collected in six provinces/cities during 2021. The TMUV genome was examined using conventional PCR. The results indicated that 21 (16.15%) samples and 9 (23.68%) farms were positive for the viral genome. The positive rate was 59.26% for ducks at ages 2–4 weeks, which was significantly higher than for ducks at ages >4 weeks and < 2 weeks. Genetic analysis of the partial envelope gene (891 bp) sequences indicated that the five Vietnamese TMUVs shared 99.55–100% nucleotide identity, while the rates were in the range 99.59–100% based on the pre-membrane gene sequences (498 bp). The five Vietnamese TMUV strains obtained formed a novel single subcluster. These strains were closely related to Chinese strains and differed from the vaccine strain, suggesting that Vietnamese TMUV strains were field viruses. It needs to be further studied on vaccine development to prevent effects of TMUV infection on poultry production across Vietnam.
Duck farming is a long-standing agricultural venture in Asia. However, this sector faces challenges from various infectious agents, including duck Circovirus, Sitiawan virus, and duck Tembusu virus (TMUV) (1–3). TMUV, classified within the genus Flavivirus of the Flaviviridae family, is an arthropod-borne virus characterized by a single-stranded, positive-sense RNA genome. It was originally isolated in 1955 from Culex tritaeniorhynchus mosquitoes in Kuala Lumpur, Malaysia (3). Nevertheless, its potential implications for human and animal health remain incompletely understood. Within the Flavivirus genus, several members, such as West Nile virus (WNV), dengue virus (DENV), yellow fever virus (YFV), Japanese encephalitis virus (JEV), and Zika virus, exhibit zoonotic properties and serve as major vector-borne pathogens responsible for millions of annual infections. These infections can manifest with a spectrum of clinical presentations, ranging from mild febrile symptoms to severe and potentially fatal hemorrhagic or neurologic diseases. This viral genus is characterized by its 30–60 nm icosahedral envelope capsid containing a single-stranded positive-sense RNA genome approximately 11 Kb in length. The genomic organization encoding the singular polyprotein is three structural proteins of capsid (C)-premembrane (prM)-envelope (E) and seven non-structural (NS) proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, NS5) that are produced through the actions of viral and cellular proteases. Surrounding the coding region are untranslated regions at both the 5′ and 3′ ends, which form conserved stem-loop structures (3, 4). Primarily, the composition of the viral particle is shaped by the structural proteins, which play a pivotal role in mediating viral fusion with host cells. These proteins are essential in various processes, including binding to virus receptors, facilitating entry, and associating fusion events. In contrast, the non-structural (NS) proteins are primarily engaged in activities, such as viral RNA replication, the assembly of virions, and the evasion of innate immune responses (5).
Within natural ecosystems, avian species are reservoir hosts for a multitude of flaviviruses, such as West Nile virus (WNV) (6), Sitiawan virus (3), Usutu virus (7, 8), and Bagaza virus (9). In April 2010, the causative agent of duck egg-drop disease in China was first recognized as duck TMUV. This ailment is distinguished by a decrease in egg yield, an abrupt reduction in feed intake, and the emergence of neurological manifestations among afflicted egg-laying and breeder ducks (4). Egg-drop disease impacts a diverse array of duck breeds, encompassing both meat-producing and egg-laying categories. This spectrum includes Pekin ducks, Cherry Valley ducks, Shaoxing ducks, Jinyun ducks, Longyan ducks, Jinding ducks, Khaki-Campbell ducks, Muscovy ducks, and domesticated mallards (10). The virus is still circulating and affecting duck production and many countries. It would be further studied on epidemiological and genetic characterization of the viral strains, which gives important information for developing vaccine and preventive strategies.
In Vietnam, the first study on TMUV infection was reported by Dang et al. Those authors used a PCR method to detect the TMUV genome in clinically suspected ducks in Hanoi city. Phylogenetic analysis of the partial NS5B gene sequences suggested that the three Vietnamese TMUV strains were genetically related to the DK/TH/CU-1 strain (KR061333) detected in Thailand (11). The aim of the current study was to carry out further epidemiological and genetic characterization of the TMUV strains circulating in several cities and provinces in north Vietnam based on their partial E and prM gene sequences.
The study did not consist of any studies involving human participants. Samples were collected from ducks farmed in north Vietnam under the auspices of the Vietnam National University of Agriculture and the protocol for sampling purposes was submitted and approved by the Committee on Animal Research and Ethics of the University (CARE-2021/04). Permission was obtained from the duck farm owners before sampling.
In total, 130 tissue samples (brain, lung, liver, kidney and bursa of Fabricius) of broiler ducks aged 2–7 weeks were collected from Bacgiang, Haiduong, Hanoi, Hungyen, Thaibinh and Thainguyen in 2021 (Figure 1). In each flock, 2–6 diseased ducks were collected by local veterinarians, necropsied, packed with dry ice, and transported to the Vietnam National University of Agriculture for laboratory analysis. Tissue samples of each duck were pooled for further testing. A 10% homogenization of pooled samples was prepared in phosphate-buffered saline.
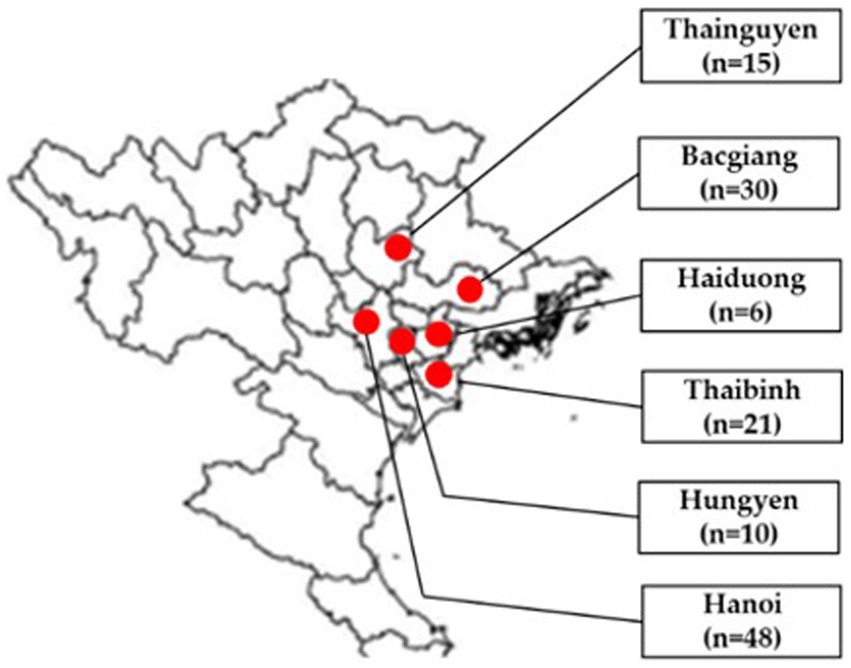
Figure 1. Location of sampling areas in north Vietnam in provinces/cities (red circles): Thainguyen (n = 15), Bacgiang (n = 30), Haiduong (n = 6), Thaibinh (n = 21), Hungyen (n = 10), and Hanoi (n = 48). Adapted from Dong et al. (12); licensed under CC BY 4.0.
A Viral Gene-spin™ Viral DNA/RNA Extraction Kit (iNtRON Biotechnology; Seoul, Korea) was used for the RNA extraction from homogenized samples, following the manufacturer’s instruction. M-MLV enzyme (Invitrogen; Carlsbad, CA, United States) was used to synthesize cDNA. In total, 20 μL of reagents, consisting of 4 μL of 5X M-MLV buffer, 1 μL of dNTP, 1 μL of random primer (Invitrogen; Carlsbad, CA, United States), 1 μL of MMLV reverse transcriptase (Invitrogen; Carlsbad, CA, United States), and 9 μL of distilled water, were mixed with 4 μL of RNA. Then, the mixture was placed in the thermal machine at 25°C for 10 min, 37°C for 1 h, and 65°C for 10 min.
Identification of the TMUV genome was carried out using PCR to detect the target 400 bp gene, with the TV-3F and TV-3R primers (Table 1), as described elsewhere (13). PCR was performed using GoTag™ Green Master Mix (Promega) at 94°C for 5 min, 40 cycles of 94°C for 30 s, then 55°C for 30 s and 72°C for 30 s, with an extension step at 72°C for 10 min. The PCR products were loaded in 1.5% agarose gels for electrophoresis and then the gel was photographed under UV light.
Two pairs of primers (E-F/R and prM-F/R; Table 1) were used for amplification of the partial prM and E gene sequences of the TMUV strains. The thermal conditions were 94°C for 4 min, followed by 35 cycles of 94°C for 60 s, then 50°C for 60 s and 72°C for 60 s, with a final extension step at 72°C for 5 min. The PCR products were loaded in 1.5% agarose gels for electrophoresis. The PCR products were purified using GeneClean® II Kits (MP Biomedicals; Santa Ana, CA, United States). Sequencing of the TMUV strains was performed by 1st BASE (Malaysia).
The nucleotide sequences from the Vietnamese TMUV strains identified in this study were aligned using the Bioedit software supplemented with Clustal W (14, 15). Nucleotide identity among the Vietnamese and other sequences downloaded in GenBank were identified using the Basic Local Alignment Search Tool (BLAST)1 and the GENETYX version 10.0 software (GENETYX Corp.; Tokyo, Japan). In total, 40 and 38 sequences of the E and prM genes, respectively, from different TMUV strains from GenBank (Table 2) were used to construct phylogenetic trees and further conduct genetic characterization of the viral strains. Maximum likelihood methods, based on the Tamura 2-parameter model, were used to established phylogenetic trees. The MEGA X2 software was used with bootstrapping of 1,000 replicates to determine the confidence values of tree branches. The partial E and partial prM gene sequences obtained were deposited into GenBank, with the accession numbers OR727885 to OR727894, respectively.
The selected TMUV strains obtained in this study and other sequences from GenBank were used to examine recombination events based on the Recombination Detection Program (RDP; version Beta 4.97), containing the RDP, GENECONV, BootScan, MaxChi, Chimera, SiScan, Phyl-Pro, LARD, and 3Seq methods, with default settings (13). Evaluation of natural selection profiles was performed using the FUBAR (a Fast Unconstrained Bayesian AppRoximation) method (16).3
Significant differences in the rate of the TMUV genome between geographical regions, ages, or flock size groups were detected using Fisher’s exact test. A value of p < 0.05 was used to determine significant differences.
In this study, 21 (16.15%) out of 130 samples were detected as positive for the TMUV genome based on the PCR method. Detailed information of positive samples was indicated in Supplementary Table 1. The TMUV-positive rates were 30 and 26.67% in Bac Giang and Thai Nguyen, respectively, which were significantly greater than for Haiduong (16.67%), Hanoi (10.42%), Hungyen (10%), and Thaibinh (4.76%). Of the 38 tested duck farms, 9 (23.68%) were positive for the viral genome. Among the cities/provinces, the highest positive rate was 50% in Hai Duong, which was significantly higher than for Thainguyen (33.33%), Bacgiang (27.27%), Hungyen (25%), Hanoi (18.18%), and Thaibinh (14.29%), as shown in Table 3.
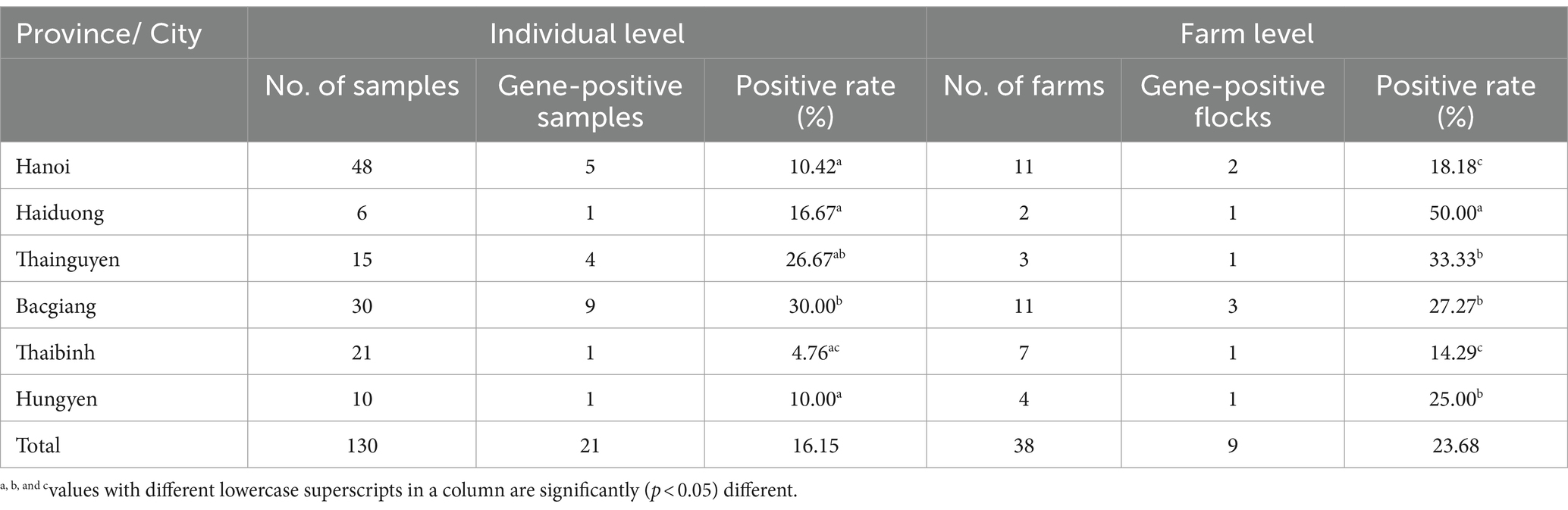
Table 3. Identification of Tembusu virus in individual ducks and farms from different locations in north Vietnam.
The percentage of TMUV-positive ducklings at age 2–4 weeks was 59.26%, which was significantly higher than for those aged >4 weeks (5.43%), while there were no positive samples from ducks aged <2 weeks for the viral genome using the PCR method. In this study, the farm levels were divided into three levels (1 with the number of ducklings <500, 2 with the number of ducks in the range 500–1,000, and 3 with the number of ducks >1,000 ducks). The positive rates were 17.24, 18.75, and 10.81% for levels 1, 2, and 3 flocks; however, these rates were not significantly different (Table 4).
Among the TMUV-positive samples, five were randomly selected for sequencing of the partial E and prM genes of the viral strains. The Vietnamese TMUV strains obtained in this study were designated as Vietnam/VNUA-28, −31, −101, −102, and 117/2021. Regarding the partial E gene (891 bp) sequences, the nucleotide identity ranged from 99.55% (VNUA-101/2021 vs. VNUA-102/2021) to 100% (VNUA-28/2021 vs. VNUA-101/2021). The Vietnamese nucleotide identities were from 94.51 to 94.73% compared to the vaccine strain (China/JXSP-310/2017), as shown in Table 5.
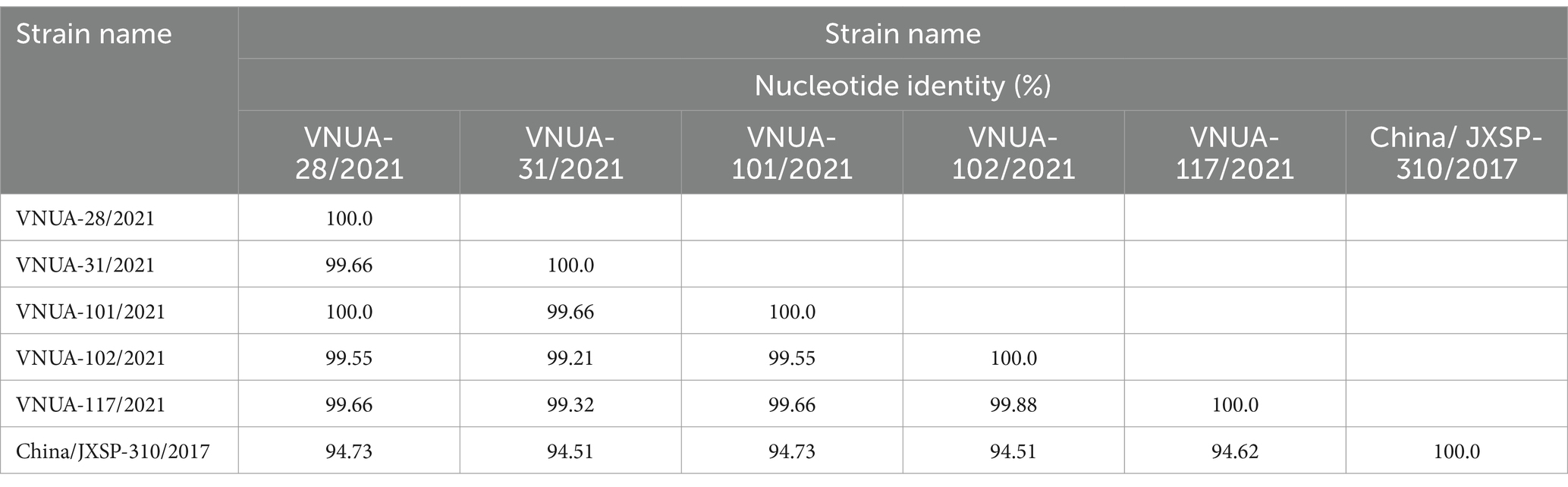
Table 5. Nucleotide identities of partial E (891 bp) gene sequences among Vietnamese and vaccine TMUV strains.
In terms of the prM gene (498 bp), the nucleotide identity ranged from 99.59% (VNUA-28/2021 vs. VNUA-31/2021) to 100% (VNUA-102/2021 vs. VNUA-117/2021). Comparison with the vaccine strain indicated that the nucleotide identities were from 93.57 to 93.97% (Table 6).
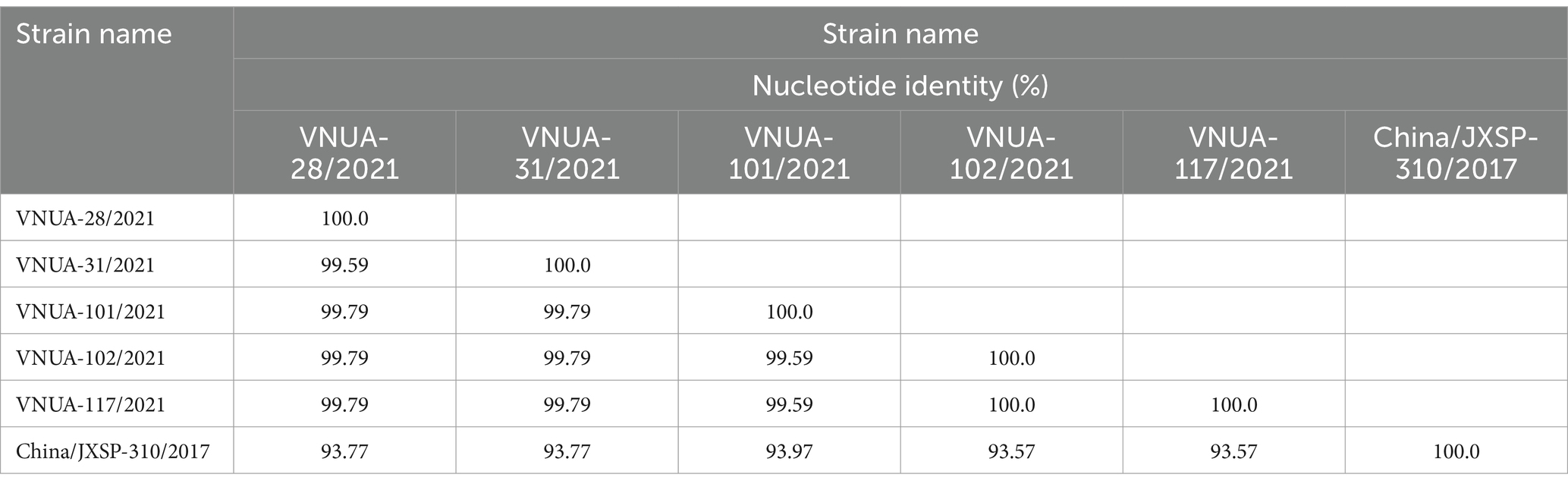
Table 6. Nucleotide identities of partial prM gene sequences (498 bp) among Vietnamese and vaccine TMUV strains.
In addition, the five Vietnamese TMUV strains were compared with sequences from abroad downloaded from GenBank. These results indicated that the highest nucleotide identities for the partial E gene sequences were 96.52% (China/GX2012 vs. VNUA-31/2021), 96.64% (China/GX2013H VNUA-102; China/GX2012 vs. VNUA-117/2021), and 96.75% (China/GX2012 vs. VNUA-28 and -101/2021) for the partial E gene sequences, while the highest nucleotide identities for the partial prM gene sequences were 96.38% (China/SD201120 vs. VNUA-28/2021), 96.58% (China/SD201120 vs. VNUA-101, −102, −117/2021), and 96.78% (China/SD201120 vs. VNUA-31/2021), as shown in Table 7.
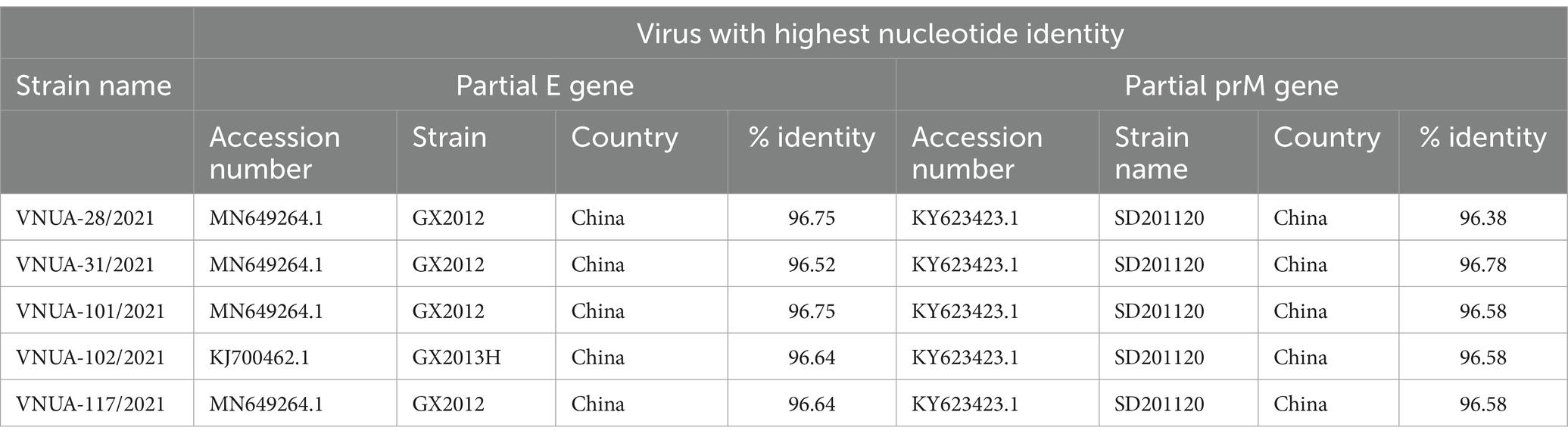
Table 7. Nucleotide identities of partial E (891 bp) and prM (498 bp) gene sequences between Vietnamese TMUV strains and viruses retrieved from GenBank.
Phylogenetic trees were established based on the partial E (891 bp) and prM (498 bp) sequences of the five TMUVs and other viruses from GenBank. The results indicated that the five Vietnamese TMUV strains obtained in this study formed a novel single cluster (2.1b). The five Vietnamese TMUV strains were genetically related to Chinese strains and differed from the vaccine strain China/JXSP-310/2017 (MZ031023.1), as shown in Figures 2, 3.

Figure 2. A Maximum likelihood phylogenetic tree of partial E gene (891 bp) sequences of Vietnamese Tembusu virus strains compared with those available in GenBank. GenBank sequences are indicated by country name/accession number. The maximum likelihood method in the MEGA X software was used to establish phylogenetic trees (1,000 bootstrap replicates). Numbers at each branch point indicated bootstrap values ≥50% in the bootstrap interior branch test. The current Vietnamese strains are indicated by black-filled circles, while the vaccine strain is marked by a black-filled square.
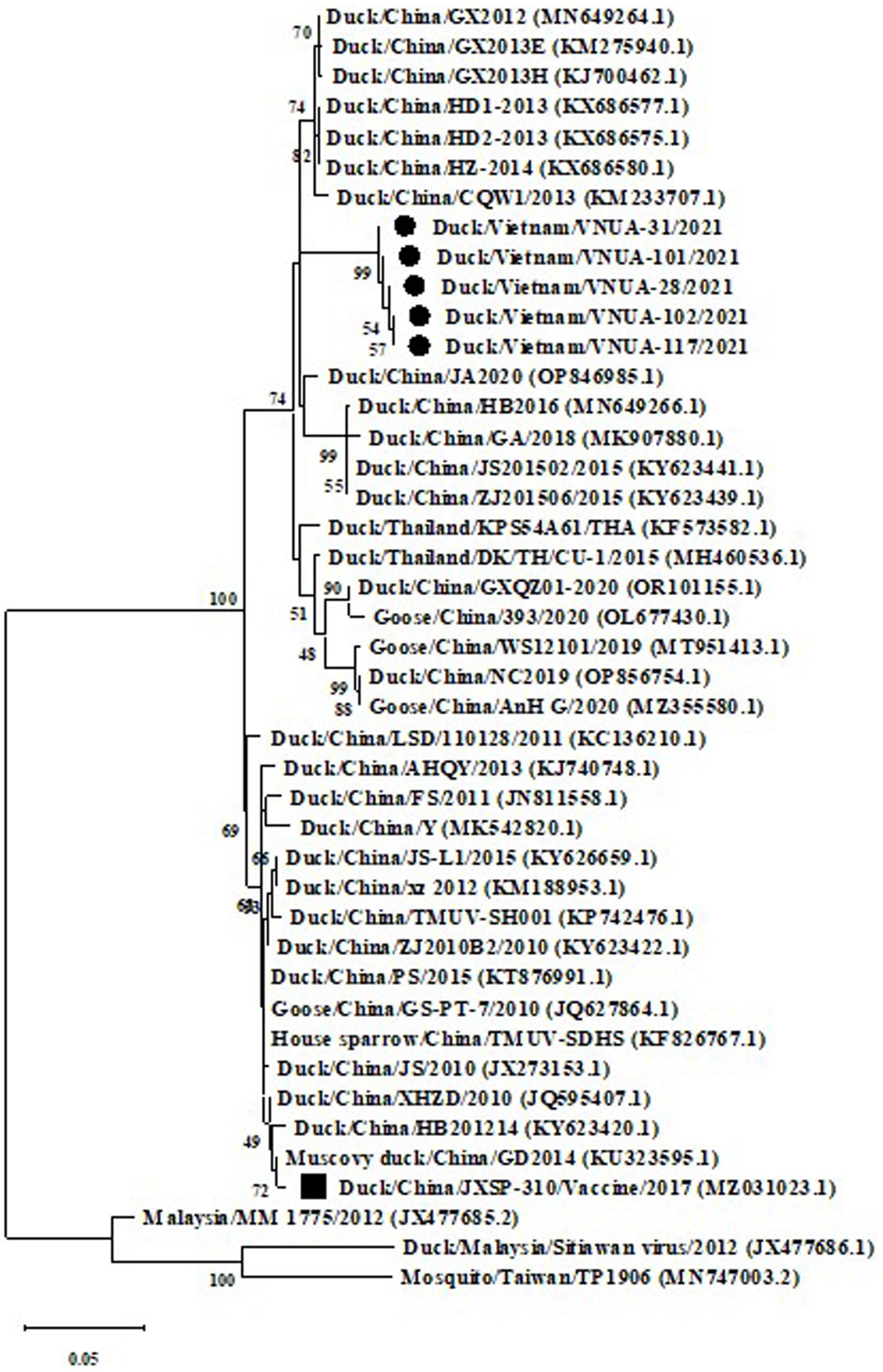
Figure 3. A Maximum likelihood phylogenetic trees of partial prM gene (498 bp) sequences of Vietnamese Tembusu virus strains compared with those available in GenBank. GenBank sequences are indicated by country name/accession number. The maximum likelihood method in the MEGA X software was used to establish phylogenetic trees (1,000 bootstrap replicates). Numbers at each branch point indicated bootstrap values ≥50% in the bootstrap interior branch test. The current Vietnamese strains are indicated by black-filled circles, while the vaccine strain is marked by a black-filled square.
Among the five Vietnamese TMUV strains, the deduced amino acid (aa) of the partial E protein indicated that there were three major substitutions, consisting of 110 K/E, 157A/V, and 345D/N. Substitution at the 345 residue was unique, in the Vietnam/VNUA-102 and -117/2021 strains, D → N. For the vaccine strain (China/JXSP-310/2017), there were 11 aa substitutions detected at residues 110, 122, 157, 176, 196, 277, 314, 345, 367, 391, and 394 (Table 8).
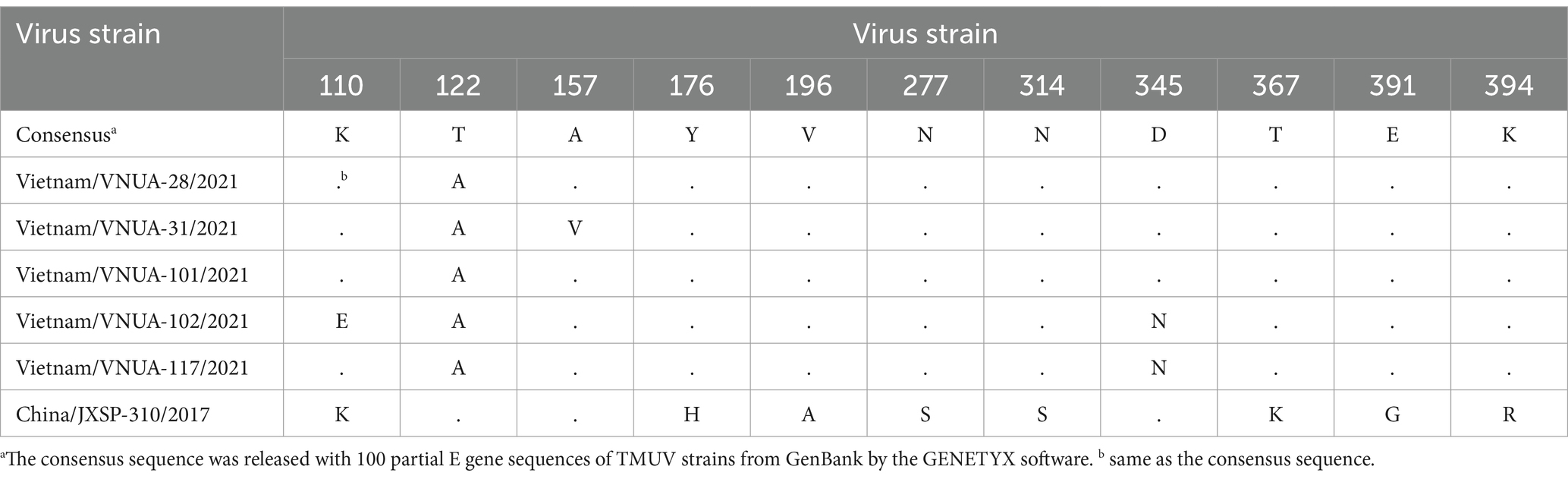
Table 8. Amino acid substitutions of E protein among Vietnamese TMUV strains compared with vaccine strain.
No recombination event was found among the Vietnamese TMUVs obtained in this study based on the partial E gene sequences. The selection profiles of the five obtained TMUV strains and other strains from GenBank were analyzed based on the partial E protein (297 amino acids). These results indicated that 169 sites had negative selection (Supplementary Table 2), whereas no positive selection was found on the partial E protein of the Vietnamese TMUV strains.
It has been reported that TMUV was successfully isolated from mosquitoes in Malaysia in 1950s (17). Subsequently, the virus spread and has been reported in many countries globally (1, 2, 4, 7, 11, 18). Understanding infection is important in creating strategies to control the disease. In Vietnam, the first report of TMUV infection was conducted in 2019 (11). The current study has continued to describe viral infection among ducks raised in some provinces in north Vietnam. Epidemiological analysis was first conducted in this study, with the five novel Vietnamese TMUV strains obtained in this study genetically forming a single cluster (2b), separated from the vaccine strain based on the partial E gene sequences (891 bp). This finding suggested that these strains were field strains.
Another study noted that TMUV infection could reach 90% among ducks within a farm, while the mortality rate varied from 5 to 30% (19). In the current study, The rates for TMUV-positive samples and farms were 16.15 and 23.68%, respectively, based on PCR. These rates were lower than the rate of 46.59% (41/88) reported in China during 2010–2016 (20) due to the fact that the sampling number, location, and time parameters were different, resulting in differences in the infection rates. Li et al. conducted a study to access pathogenicity of TMUV strains isolated in China (21). One-, 3-, and 7-weeks-old ducks were inoculated with TMUV strains. The study pointed out that ducks at one-week-old indicated severe symptoms, while other groups showed milder pathological lesions (21). This finding agreed with a previous study revealed that young ducks were more sensitive to TMUV strains than older ducks, suggesting a relationship between viral susceptibility and age (21, 22). Later researchers found that ducks aged 18–21 weeks and 55 weeks were susceptible to the disease caused by TMUV (23, 24). Changes in age-related susceptibility occurred among ducks infected with TMUV strains. It is due to the fact that host immune responses, different viral strains, or evolution among viral strains may contribute to age-related susceptibility. Further studies should be conducted to elucidate these points. The current study indicated that the highest rate of viral infection was in young ducks aged 2–4 weeks (59.26%), compared to older ducks aged >4 weeks (5.43%), with no positive samples being detected in ducks aged <2 weeks. Older ducks may have a stronger immune response, leading to lower positive rates (21). Further studies should be conducted to explain the relationship between age and infection rates in north Vietnam.
Genetic and phylogenetic analyses based on the partial E and prM gene sequences revealed that the current five Vietnamese TMUV strains formed a novel subcluster, which was closely related to Chinese strains. The results suggested that the TMUV strains obtained in this study might have a similar origin to Chinese strains. N-linked glycosylation, which contributes to the entry of the virus into host cells, was predicted in the E protein of TMUV at the 103, 154, and 314 residues (25, 26). However, in the current study, substitutions were not found at these three residues, suggesting the conservation of the glycosylation sites of E protein among Vietnamese TMUV strains, which was similar to the findings of Huang et al. Fritz et al. suggested that protonation of histidine’s plays an important role in the membrane fusion of flaviviruses (27). No substitutions of histidine was observed at the 144, 153, 163, 219, 246, 263, 285, 320, and 398 residues of the E protein of the five Vietnamese TMUVs in the current study, nor were substitutions found in the epitopes 220–226 and the 374–380 residues, located in domains DII and DIII in the E protein of the Vietnamese TMUV strains. In the current study, three substitutions were found among Vietnamese TMUV strains: residues 110 (K → E), 157 (A → V), and 345 (D → N). To our knowledge, no evidence on the contribution of these substitutions has been reported; other studies will be conducted to evaluate this point.
One of the evolutionary processes is recombination, was first found among TMUV strains in China (28). The current study did not find any recombination events among the Vietnamese TMUV strains based on analysis of the partial S gene. Expanding analysis of the complete genome and increasing the number of sequences are necessary to examine possible recombination events among Vietnamese TMUV strains. Dai et al. (16) reported that negative selection was strong among Chinese TMUV strains. A few positive selection sites were found among the TMUV strains as a result of genetic drifts, similar to other flaviviruses (29, 30). In the current study, 169 residues of Vietnamese TMUV E protein had negative selection.
In this study, TMUV infection in samples and on duck farms were 16.15 and 23.68%, respectively, in six provinces/city in north Vietnam in 2021. The infection was most commonly detected in young ducks aged 2–4 weeks (59.26%), at a significantly higher level than in ducks aged <2 weeks and > 4 weeks. Genetic and phylogenetic analyses of the five Vietnamese TMUV strains based on the partial E and prM gene sequences supported that the current Vietnamese TMUV strains belonged to a novel subcluster, which was closely related to the Chinese strains and differed from the vaccine strain. No putative recombination event was detected among the Vietnamese TMUV strains. Strong negative selection was found among the Vietnamese TMUV strains, based on the analysis of the partial E protein. Further studies should be conducted to better understand the evaluation of TMUV strains across the country.
The original contributions presented in the study are publicly available. This data can be found here: https://www.ncbi.nlm.nih.gov/nuccore/; OR727885-OR727894.
The animal studies were approved by the Committee on Animal Research and Ethics of the University (CARE-2021/04). The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent was obtained from the owners for the participation of their animals in this study.
HD: Conceptualization, Methodology, Software, Writing – original draft, Data curation, Formal analysis, Investigation, Visualization. GT: Conceptualization, Writing – original draft, Data curation, Formal analysis, Investigation, Visualization. TV: Writing – original draft. NL: Writing – original draft. YN: Writing – original draft. WR: Writing – original draft. AR: Writing – original draft. CB: Writing – original draft. DB: Writing – original draft, Conceptualization, Software, Supervision, Writing – review & editing. JR: Conceptualization, Funding acquisition, Methodology, Project administration, Resources, Software, Supervision, Validation, Writing – original draft, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. The research was partly supported by the Faculty of Veterinary Medicine, Kasetsart University, Bangkok, Thailand and the National Research Council of Thailand (NRCT) through R. Thanawongnuwech NRCT Senior Scholar 2022 #N42A650553.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2024.1366904/full#supplementary-material
1. Fang, Y, Hang, T, Yang, LM, Xue, JB, Fujita, R, Feng, XS, et al. Long-distance spread of Tembusu virus, and its dispersal in local mosquitoes and domestic poultry in Chongming Island, China. Infect Dis Poverty. (2023) 12:52. doi: 10.1186/s40249-023-01098-9
2. Zhang, X, Jiang, S, Wu, J, Zhao, Q, Sun, Y, Kong, Y, et al. An investigation of duck circovirus and co-infection in Cherry Valley ducks in Shandong Province. China Vet Microbiol. (2009) 133:252–6. doi: 10.1016/j.vetmic.2008.07.005
3. Kono, Y, Tsukamoto, K, Abd Hamid, M, Darus, A, Lian, TC, Sam, LS, et al. Encephalitis and retarded growth of chicks caused by Sitiawan virus, a new isolate belonging to the genus Flavivirus. Am J Trop Med Hyg. (2000) 63:94–101. doi: 10.4269/ajtmh.2000.63.94
4. Su, J, Li, S, Hu, X, Yu, X, Wang, Y, Liu, P, et al. Duck egg-drop syndrome caused by BYD virus, a new Tembusu-related flavivirus. PLoS One. (2011) 6:e18106. doi: 10.1371/journal.pone.0018106
5. Baltimore, D. Expression of animal virus genomes. Bacteriol Rev. (1971) 35:235–41. doi: 10.1128/br.35.3.235-241.1971
6. Swayne, DE, Beck, JR, Smith, CS, Shieh, WJ, and Zaki, SR. Fatal encephalitis and myocarditis in young domestic geese (Anser anser domesticus) caused by West Nile virus. Emerg Infect Dis. (2001) 7:751–3. doi: 10.3201/eid0704.017429
7. Becker, N, Jost, H, Ziegler, U, Eiden, M, Hoper, D, Emmerich, P, et al. Epizootic emergence of Usutu virus in wild and captive birds in Germany. PLoS One. (2012) 7:e32604. doi: 10.1371/journal.pone.0032604
8. Weissenbock, H, Kolodziejek, J, Url, A, Lussy, H, Rebel-Bauder, B, and Nowotny, N. Emergence of Usutu virus, an African mosquito-borne flavivirus of the Japanese encephalitis virus group, Central Europe. Emerg Infect Dis. (2002) 8:652–6. doi: 10.3201/eid0807.020094
9. Aguero, M, Fernandez-Pinero, J, Buitrago, D, Sanchez, A, Elizalde, M, San Miguel, E, et al. Bagaza virus in partridges and pheasants, Spain, 2010. Emerg Infect Dis. (2011) 17:1498–501. doi: 10.3201/eid1708.110077
10. Petz, LN, Turell, MJ, Padilla, S, Long, LS, Reinbold-Wasson, DD, Smith, DR, et al. Development of conventional and real-time reverse transcription polymerase chain reaction assays to detect Tembusu virus in Culex tarsalis mosquitoes. Am J Trop Med Hyg. (2014) 91:666–71. doi: 10.4269/ajtmh.13-0218
11. Dang, HA, Nguyen, VG, and Huynh, TML. Some initial result of study on Tembusu virus in the infected ducks in ha Noi. J Vet Tech. (2020) 27:22–8.
12. Dong, HV, Truong, TH, Tran, GTH, Rapichai, W, Rattanasrisomporn, A, Choowongkomon, K, et al. Porcine sapovirus in northern Vietnam: genetic detection and characterization reveals co-circulation of multiple genotypes. Vet Sci. (2023) 10:430. doi: 10.3390/vetsci10070430
13. Martin, DP, Murrell, B, Golden, M, Khoosal, A, and Muhire, B. RDP4: detection and analysis of recombination patterns in virus genomes. Virus Evol. (2015) 1:vev003. doi: 10.1093/ve/vev003
14. Hall, TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp Ser. (1999) 41:95–8.
15. Thompson, JD, Higgins, DG, and Gibson, TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. (1994) 22:4673–80. doi: 10.1093/nar/22.22.4673
16. Murrell, B, Moola, S, Mabona, A, Weighill, T, Sheward, D, Kosakovsky Pond, SL, et al. FUBAR: a fast, unconstrained bayesian approximation for inferring selection. Mol Biol Evol. (2013) 30:1196–205. doi: 10.1093/molbev/mst030
17. US Army Medical Research Unit (Malaya). Institute for Medical Research, Federation of Malaya, Annual Report. Kuala Lampur, Malaya, 1000–1103. (1957).
18. Mansour, SMG, Mohamed, FF, ElBakrey, RM, Eid, AAM, Mor, SK, and Goyal, SM. Outbreaks of duck hepatitis a virus in Egyptian duckling flocks. Avian Dis. (2019) 63:68–74. doi: 10.1637/11975-092118-Reg.1
19. Cao, Z, Zhang, C, Liu, Y, Liu, Y, Ye, W, Han, J, et al. Tembusu virus in ducks, China. Emerg Infect Dis. (2011) 17:1873–5. doi: 10.3201/eid1710.101890
20. Huang, X, Qiu, H, Peng, X, Zhao, W, Lu, X, Mo, K, et al. Molecular analysis and serological survey of Tembusu virus infection in Zhejiang, China, 2010-2016. Arch Virol. (2018) 163:3225–34. doi: 10.1007/s00705-018-3994-4
21. Li, N, Lv, C, Yue, R, Shi, Y, Wei, L, Chai, T, et al. Effect of age on the pathogenesis of duck tembusu virus in Cherry Valley ducks. Front Microbiol. (2015) 6:581. doi: 10.3389/fmicb.2015.00581
22. Sun, XY, Diao, YX, Wang, J, Liu, X, Lu, AL, Zhang, L, et al. Tembusu virus infection in Cherry Valley ducks: the effect of age at infection. Vet Microbiol. (2014) 168:16–24. doi: 10.1016/j.vetmic.2013.10.003
23. Lv, C, Li, R, Liu, X, Li, N, and Liu, S. Pathogenicity comparison of duck Tembusu virus in different aged Cherry Valley breeding ducks. BMC Vet Res. (2019) 15:282. doi: 10.1186/s12917-019-2020-8
24. Lu, Y, Dou, Y, Ti, J, Wang, A, Cheng, B, Zhang, X, et al. The effect of Tembusu virus infection in different week-old Cherry Valley breeding ducks. Vet Microbiol. (2016) 192:167–74. doi: 10.1016/j.vetmic.2016.07.017
25. Li, C, Liu, J, Shaozhou, W, Bai, X, Zhang, Q, Hua, R, et al. Epitope identification and application for diagnosis of duck tembusu virus infections in ducks. Viruses. (2016) 8:306. doi: 10.3390/v8110306
26. Yu, K, Sheng, ZZ, Huang, B, Ma, X, Li, Y, Yuan, X, et al. Structural, antigenic, and evolutionary characterizations of the envelope protein of newly emerging duck tembusu virus. PLoS One. (2013) 8:e71319. doi: 10.1371/journal.pone.0071319
27. Fritz, R, Stiasny, K, and Heinz, FX. Identification of specific histidines as pH sensors in flavivirus membrane fusion. J Cell Biol. (2008) 183:353–61. doi: 10.1083/jcb.200806081
28. Dai, L, Li, Z, and Tao, P. Evolutionary analysis of Tembusu virus: evidence for the emergence of a dominant genotype. Infect Genet Evol. (2015) 32:124–9. doi: 10.1016/j.meegid.2015.03.004
29. Añez, G, Morales-Betoulle, ME, and Rios, M. Circulation of different lineages of dengue virus type 2 in Central America, their evolutionary time-scale and selection pressure analysis. PLoS One. (2011) 6:e27459. doi: 10.1371/journal.pone.0027459
Keywords: ducks, genetic characterization, PCR, Tembusu virus, Vietnam
Citation: Dong HV, Tran GTH, Vu TTT, Le NHT, Nguyen YTH, Rapichai W, Rattanasrisomporn A, Boonkaewwan C, Bui DAT and Rattanasrisomporn J (2024) Duck Tembusu virus in North Vietnam: epidemiological and genetic analysis reveals novel virus strains. Front. Vet. Sci. 11:1366904. doi: 10.3389/fvets.2024.1366904
Received: 15 January 2024; Accepted: 18 April 2024;
Published: 14 May 2024.
Edited by:
Vuong Bui, National Institute of Veterinary Research (NIVR), VietnamReviewed by:
Kunfeng Sun, University of Pennsylvania, United StatesCopyright © 2024 Dong, Tran, Vu, Le, Nguyen, Rapichai, Rattanasrisomporn, Boonkaewwan, Bui and Rattanasrisomporn. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jatuporn Rattanasrisomporn, ZnZldGpwbkBrdS5hYy50aA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.