- 1College of Animal Science, South China Agricultural University, Guangzhou, China
- 2Yunfu Branch, Guangdong Laboratory for Lingnan Modern Agriculture, Yunfu, China
Mycoplasma synoviae (MS) is an economically important pathogen in the poultry industry. Vaccination is an effective method to prevent and control MS infections. Currently two live attenuated MS vaccines are commercially available, the temperature-sensitive MS-H vaccine strain and the NAD-independent MS1 vaccine strain. Differentiation of vaccine strains from wild-type (WT) strains is crucial for monitoring MS infection, especially after vaccination. In this study, we developed a Taqman duplex real-time polymerase chain reaction (PCR) method to identify MS1 vaccine strains from WT strains. The method was specific and did not cross-react with other avian pathogens. The sensitivity assay indicated that no inhibition occurred between probes or between mixed and pure templates in duplex real-time PCR. Compared with the melt-based mismatch amplification mutation assay (MAMA), our method was more sensitive and rapid. In conclusion, the Taqman duplex real-time PCR method is a useful method for the diagnosis and differentiation of WT-MS and MS1 vaccine strains in a single reaction.
1 Introduction
Mycoplasma synoviae (MS) has been described as an important pathogen causing air sacculitis, infection synovitis and eggshell apex abnormalities (1–3), and is listed as a notifiable mycoplasma by the World Organization for Animal Health (WOAH) (4). MS infection can cause subclinical symptoms and lead to co-infection with Mycoplasma gallisepticum (MG), Newcastle disease virus (NDV), Infectious bronchitis virus (IBV), and other avian pathogens (5–8). Rapid and accurate diagnosis is necessary to monitor MS infection especially after vaccination. Diagnostic methods for MS include bacteriological isolation, serological assays and molecular detection (9). Mycoplasma isolation is inefficient and expensive, as in vitro growth requires a rich medium and is time-consuming (10, 11). The serological assay only provides a history of infection (12). Molecular analysis, such as polymerase chain reaction (PCR) or real-time PCR (qPCR), guarantees earlier detection, is more rapid, more sensitive, and more specific than the others, and is widely used (9).
Measures to prevent and control MS include vaccines and antibiotics. However, the emergence of drug resistance in MS strains has made the use of antibiotics more cautious (13–16). Vaccination is another option to control the disease. At present, in addition to the inactivated vaccine, only two live attenuated vaccines are commercially available: the temperature-sensitive (ts+) MS-H vaccine strain (Vaxsafe® MS, Bioproperties Pty Ltd.) and the NAD-independent MS1 vaccine strain (Nobilis® MS Live, MSD Animal Health Inc.). The MS-H strain was developed by chemical mutagenesis of an Australian strain (86079/7NS), while the MS1 strain is a spontaneous attenuation of the wild-type pathogenic isolate WVU1853. After live vaccine inoculation, the differentiation of vaccine strains from wild-type strains is crucial for monitoring MS infection. Moreover, it is important to determine whether the vaccine strains have successfully colonized the respiratory mucosa to provide effective protection against wild-type (WT) strains (17, 18). Several genotyping techniques have been developed to differentiate MS-H strains from WT strains, including real-time PCR (19), melting curve analysis, agarose gel-based mismatch amplification mutation assay (MAMA) (20), and high-resolution melting curve assays (21, 22). However, only one study provided a way to distinguish the MS1 strain from WT strains, using melt-based MAMA PCR or agarose-MAMA PCR (23).
In this study, we developed a Taqman duplex real-time PCR method that was sensitive, specific and more rapid than melt-based MAMA. The developed method is applicable both in laboratory and clinical testing, and promotes an easier method to differentiate WT-MS strains and MS1 vaccine strains in a single reaction.
2 Materials and methods
2.1 Samples
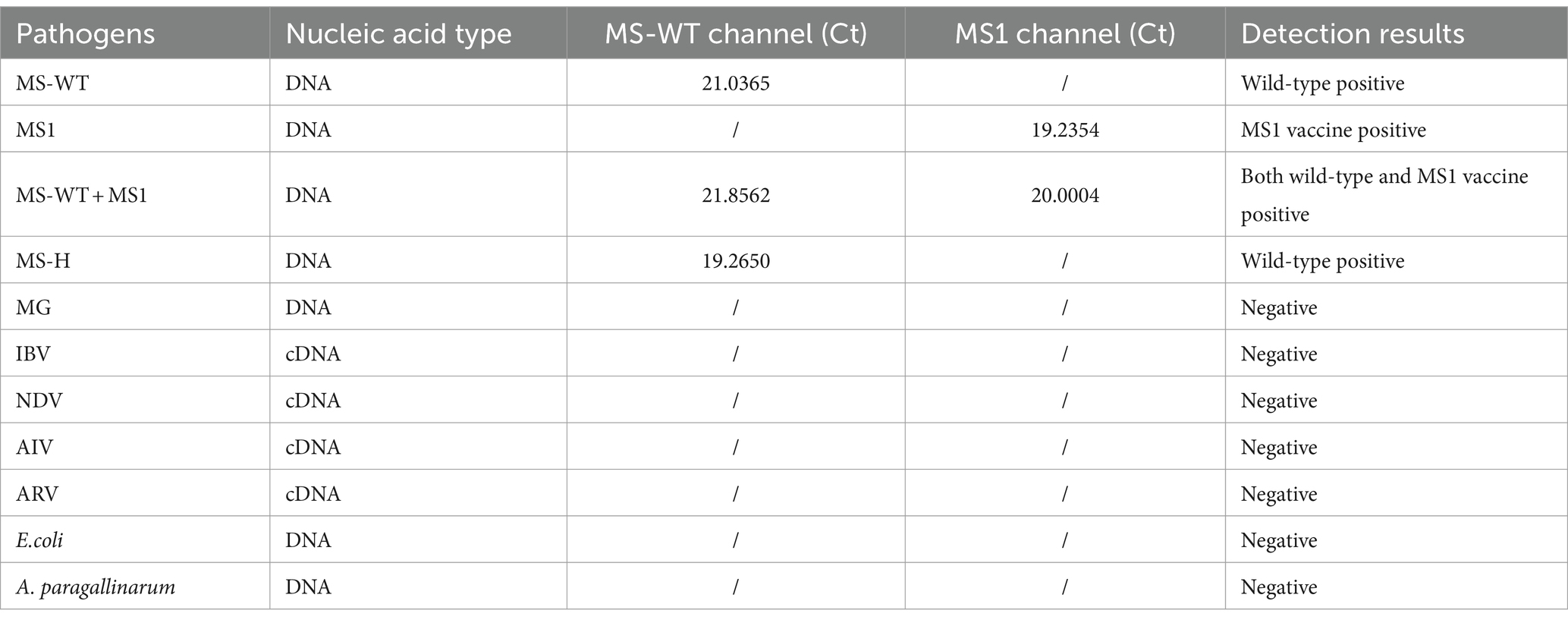
The MS1 (Nobilis® MS Live, MSD) and MS-H (Vaxsafe® MS-H, SINDER) vaccine strains used in this study were obtained from commercial distributors. The WT- MS strains and DNA samples extracted from tracheal swab samples were isolated by the authors (Supplementary Table S1). The genomes of MG, IBV, NDV, Avian influenza virus (AIV), Avian reovirus (ARV), Escherichia coli (E. coli) and Avibacterium paragallinarum (A. paragallinarum) were used for the specific detection of the method. The standard nucleic acid (plasmid) of MS1 and WT- MS used in this study was constructed with pMD-18 T (Takara, China).
2.2 Nucleic acid extraction
The nucleic acids of MS, MG, E.coli and A. paragallinarum were extracted using the Bioer Total DNA Extraction Kit (Bioer Tec., China). The nucleic acids of IBV, NDV, AIV and ARV were extracted using the Bioer Total RNA Extraction Kit (Bioer Tec., China) and then the extracted RNAs were used to synthesize cDNAs using the PrimeScript™ II 1st Strand cDNA Synthesis Kit (Takara, China) according to the manufacturer’s instructions.
2.3 Monoplex and duplex real-time PCR
By sequencing and comparing the whole genomes of the vaccine strain and wild-type strains (24), we found a single nucleotide mutation site, and designed probes and primers that could be used to distinguish the vaccine strain (MS1) from wild-type strains (Supplementary Table S2). All real-time PCR reactions were carried out on an ABI 7500fast Real-time PCR Detection System. A volume of 20 μL reaction mixture contained 10 μL 2x THUNDERBIRD Probe qPCR Mix (TOYOBO, China), 200 nM each primer, 100 nM each probe, and 2 μL templates. The reaction conditions involved incubation at 95°C for 3 min, followed by 40 cycles of denaturation at 95°C for 15 s and a combined annealing and extension step at 60°C for 30s.
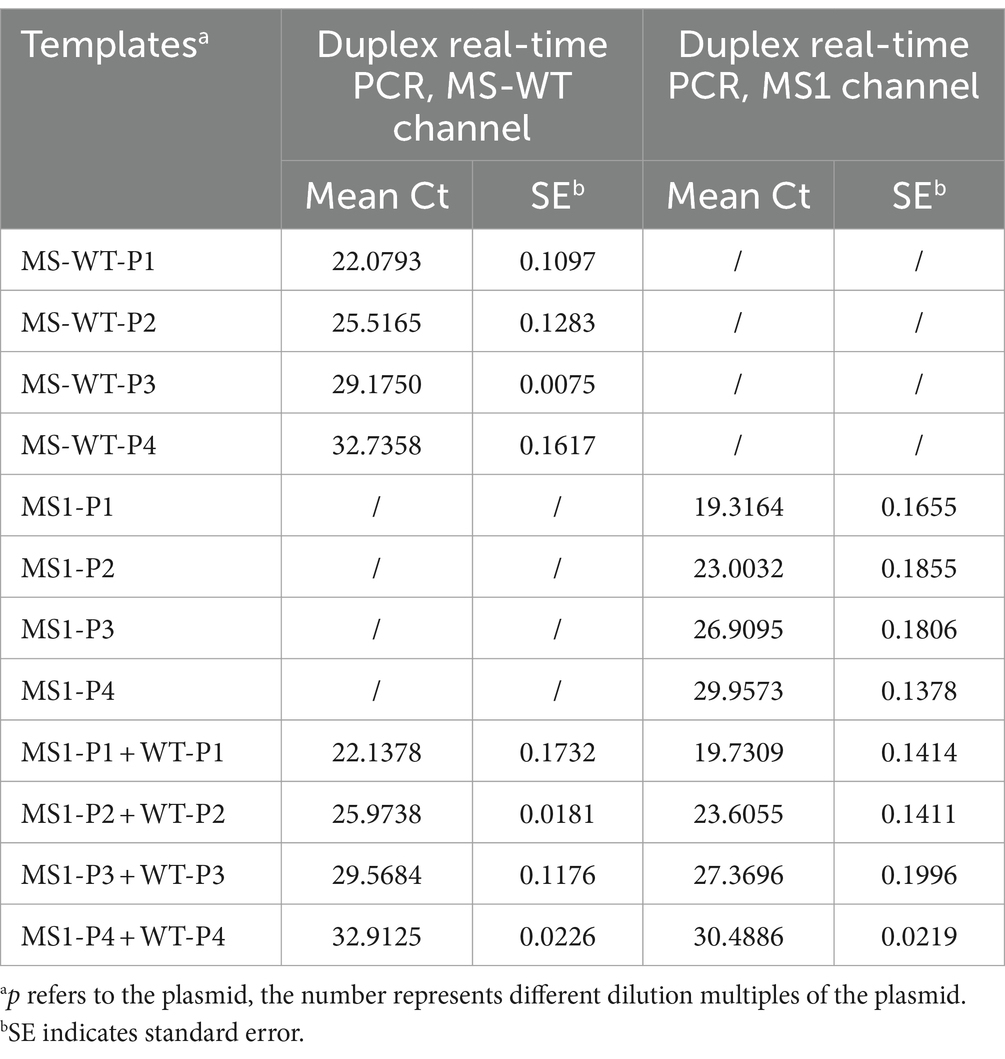
For the sensitivity of real-time PCR assays, MS1 and MS-WT standard plasmids were constructed. Briefly, qPCR amplification products were collected, purified by gel, and then connected to the pMT-18 T vector. The recombinant plasmids with correct sequencing were used as the standard plasmids for subsequent experiments. Each standard plasmid was serially diluted tenfold to achieve concentrations of 101 to 107 copies/μL. The serially diluted plasmids were used to establish a standard curve for each target after three technical replications. For duplex real-time PCR, two plasmids were equally mixed and then serially diluted as described above. The single and mixed plasmids were used to compare detection sensitivities between the duplex reaction and the individual singular reactions.
For the specificity assay, potential cross-reactions with other avian pathogens were measured to ensure the specificity of our method. The templates used in this assay included DNA from MS1, MS-H, MS-WT, MG, E.coli and A. paragallinarum, and cDNA from IBV, NDV, AIV, and ARV.
2.4 Melt-based mismatch amplification mutation assays
As described by Kreizinger et al. (23), MAMA is based on allele-specific competing primers and is widely used for SNP detection. One volume of Melt-MAMA PCR reaction was performed in 20 μL, containing 2 μL templates, 150 nM each primer, 4 μL 5x Colorless GoTaq Flexi Buffer (Promega), 2 μL MgCl2 (25 mM), 0.6 μL dNTP (10 mM, Takara), 1 μL EvaGreen (Biotium Inc.) and 0.16 μL GoTaq DNA polymerase (5 U/μL, Promega). Melt-based MAMA PCR reactions were carried out on an ABI 7500fast Real-time PCR Detection System with High Resolution Melting (HRM) Software (v3.2, Thermo Fisher). The thermocycling parameters were 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 58°C for 1 min and a dissociation protocol comprising 95°C for 15 s, followed by an incremental temperature ramp (0.2°C) from 58°C to 95°C.
2.5 Image and statistical analyses
All graphs and statistics in this study were created with GraphPad Prism 8 software (v8.0.2). All data are presented as standard errors (SEs) of at least three independent experiments.
3 Results
3.1 Duplex real-time PCR specificity analysis
To determine the specificity of the method, genomes extracted from different chicken pathogens were used as templates. As shown in Table 1, no specific amplifications occurred between the reaction systems of other avian pathogens. As expected, in the duplex real-time PCR reaction system, the nucleic acids of WT-MS were positive in the MS-WT measurement channel and negative in the MS1 measurement channel and vice versa. The MS-H strain was identified as the WT strain in the duplex real-time PCR system.
3.2 Duplex real-time PCR sensitivity analysis
The sensitivity of the duplex real-time PCR was investigated from two perspectives. One was to compare the detection limits of monoplex qPCR with duplex qPCR. The other was carried out with templates containing a single target or mixed targets.
To make the comparison more intuitive, we constructed the standard plasmids of MS-WT and MS1 respectively, and plotted the standard curves. As shown in Figure 1, targeting the same plasmids, the curves shown for duplex and monoplex real-time PCRs, respectively, were in practical agreement, and the minimum detection limits of all qPCRs were between 101 and 102 copies/μL (Figure 1). No evidence of inhibition between probes was observed in the duplex reaction.
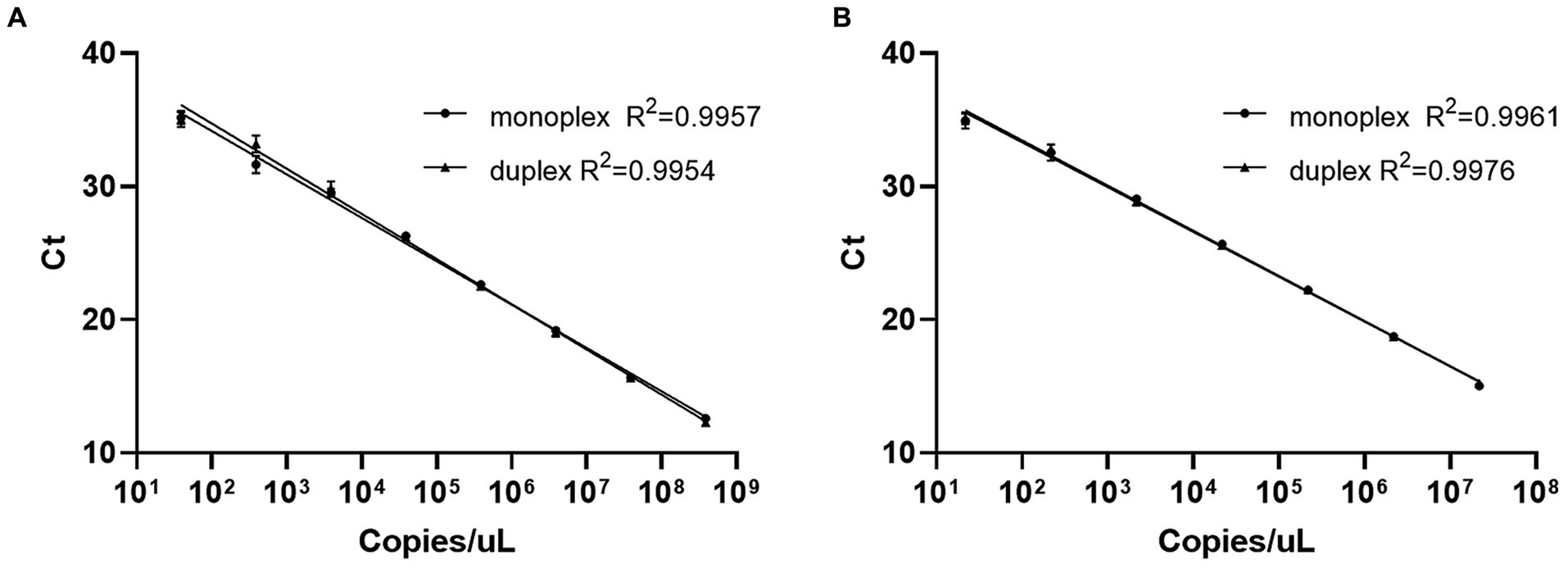
Figure 1. Standard curves of monoplex and duplex real-time PCRs with the same targets. (A) Standard curves of monoplex and duplex real-time PCRs using the MS1 standard plasmid template. (B) Standard curves of monoplex and duplex real-time PCRs using the MS-WT standard plasmid template.
Do mixed targets have any effect on duplex real-time PCR? The assay was performed with single or mixed plasmids of MS-WT and MS1. As shown in Table 2, similar Ct values were obtained from the same target in mixed and single templates. There was no evidence of inhibition as both targets only reacted with their specific probe.
3.3 Comparison of duplex real-time PCR with melt-based MAMA
To confirm the practicality of duplex real-time PCR, we detected the same templates by real-time PCR and melt-based MAMA methods, respectively. The template information is shown in Table 1. Results showed that both methods could distinguish between WT strains and the MS1 vaccine strain (Table 3). Test results of clinical swab samples showed a higher detection rate of duplex real-time PCR than melt-based MAMA, especially when the nucleic acid content of the samples was low (Supplementary Table S3). It should be noted that the melt-based MAMA method was more suitable for qualitative analysis because the instrumentation system only showed the Ct values of the higher peak when there were two detection targets in the same sample (Table 3). All of the results indicated that the developed duplex real-time PCR method was more sensitive and more suitable for quantitative analysis than the existing method.

Table 3. Ct values of samples detected by duplex real-time polymerase chain reaction (PCR) and melt-based mismatch amplification mutation assay (MAMA) method.
In addition, the duplex real-time PCR took less time for detection than the existing method because there was no slow warming step (0.2°C/s).
4 Discussion
MS is distributed worldwide and has become one of the most important pathogens threatening the global poultry industry (24, 25). Furthermore, MS co-infections with other infectious agents such as NDV, IBV, E. coli, and MG increase economic losses (26–28). Research on MS can lay the foundation for the prevention and treatment of MS-related diseases. With the increase in positive and incidence rates, prevention and control of MS have become the focus of the poultry industry in China (29–31).
To increase the knowledge of MS epidemiology and to improve control and eradication programs, it is important to monitor MS infection and identify sources of infection and modes of transmission. In order to avoid economic losses due to disease outbreaks, vaccination has become the primary prevention and control measure in the poultry industry. For MS, the live vaccine stands out among other types of vaccines because it prevents infection with wild-type strains by colonizing the trachea and continuously stimulating the immune response (17, 18). To date, only two commercial live vaccines are available in the world. After immunization with a live vaccine, differentiation between wild-type strains and vaccine strains is imperative. There have been several reports on distinguishing the MS-H vaccine strain from wild-type strains but only one report on the MS1 vaccine strain (23). In this study, we developed a quantitative and rapid Taqman-based duplex real-time PCR method to differentiate and quantify the MS1vaccine strain and wild-type strains simultaneously.
The specificity assay indicated that no fluorescent signal was detected among the nucleic acids of MG, IBV, NDV, AIV, ARV, E. coli and A. paragallinarum in our reaction system. Since the purpose of this study is to distinguish the MS1 strain from the wild-type strains, we did not take the MS-H strain into account. According to the results, the MS-H strain was identified as wild-type as expected. The quantification method requires knowledge of the detection limit. Therefore, we determined the limit of our method by 10-fold serial dilution of the standard plasmids. The lowest detection range was between 101 and 102 copies/μL, regardless of whether it was an MS1 plasmid or MS-WT plasmid. We also found that duplex reactions or mixed targets did not significantly influence the detection results.
Duplex real-time PCR has distinct advantages over melt-based MAMA, because it allows quantification and differentiation at the same time. In addition, the detection limit of duplex real-time PCR is more sensitive than that of melt-based MAMA considering the lower Ct values.
5 Conclusion
In conclusion, a duplex real-time PCR method was developed to distinguish between wild-type MS strains and MS1 vaccine strains. This method was highly specific and sensitive, and allowed the simultaneous quantification of MS1 and MS-WT. Based on the above, duplex real-time PCR can be used as a diagnostic tool for the detection and quantification of MS strains after inoculation with the MS1 live vaccine.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
Author contributions
CL: Writing – original draft. YC: Writing – review & editing. ZY: Investigation, Writing – review & editing. YS: Methodology, Writing – review & editing. QZ: Writing – review & editing, Software. PZ: Writing – review & editing, Data curation. XH: Writing – review & editing, Formal analysis. WL: Writing – review & editing, Resources. FC: Writing – review & editing, Investigation.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the China Postdoctoral Science Foundation [grant number 2021M692455].
Acknowledgments
Thanks to Professor Xiaona Wei of Zhengzhou University for providing technical guidance.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2024.1354548/full#supplementary-material
References
1. Kleven, SH , King, DD , and Anderson, DP . Air sacculitis in broilers from Mycoplasma synoviae: effect on air-sac lesions of vaccinating with infectious bronchitis and Newcastle virus. Avian Dis. (1972) 16:915–24. doi: 10.2307/1588772
2. Kang, MS , Gazdzinski, P , and Kleven, SH . Virulence of recent isolates of Mycoplasma synoviae in turkeys. Avian Dis. (2002) 46:102–10. doi: 10.1637/0005-2086(2002)046[0102:VORIOM]2.0.CO;2
3. Feberwee, A , and Landman, WJ . Induction of eggshell apex abnormalities in broiler breeder hens. Avian Pathol. (2010) 392:133–7. doi: 10.1080/03079451003657637
4. OIE . Avian mycoplasmosis (Mycoplasma gallisepticum, M. synoviae) in manual of diagnostic tests and vaccines for terrestrial animals. Paris, France: World Organization for Animal Health (2015).
5. Ball, C , Forrester, A , and Ganapathy, K . Co-circulation of genetically diverse population of vaccine related and unrelated respiratory mycoplasmas and viruses in UK poultry flocks with health or production problems. Vet Microbiol. (2018) 225:132–8. doi: 10.1016/j.vetmic.2018.09.009
6. Khatoon, A , Abidin, ZU , Gul, ST , Khan, AR , Naeem, M , Qureshi, MA, et al. Lyophilization as a possible way to enhance the viability of live Newcastle disease (LaSota) vaccine: suggesting the optimized method by comparing five different protocols. Pak Vet J. (2022) 423:404–8. doi: 10.29261/pakvetj/2022.047
7. Derksen, T , Lampron, R , Hauck, R , Pitesky, M , and Gallardo, RA . Biosecurity assessment and Seroprevalence of respiratory diseases in backyard poultry flocks located close to and far from commercial premises. Avian Dis. (2018) 62:1–5. doi: 10.1637/11672-050917-Reg.1
8. Rehman, AU , Shah, AH , Rahman, SU , Sajid, S , Khan, IU , Ullah, Q, et al. Molecular confirmation and immunological cross reactivity among Mycoplasma gallisepticum isolates recovered from broiler chicken in Khyber Pakhtunkhwa, Pakistan. Pak Vet J. (2022) 424:487–92. doi: 10.29261/pakvetj/2021.045
9. Feberwee, A , Mekkes, DR , de Wit, JJ , Hartman, EG , and Pijpers, A . Comparison of culture, PCR, and different serologic tests for detection of Mycoplasma gallisepticum and Mycoplasma synoviae infections. Avian Dis. (2005) 49:260–8. doi: 10.1637/7274-090804R
10. Sun, SK , Lin, X , Chen, F , Wang, DA , Lu, JP , Qin, JP, et al. Epidemiological investigation of Mycoplasma Synoviae in native chicken breeds in China. BMC Vet Res. (2017) 13:115. doi: 10.1186/s12917-017-1029-0
11. Ali, I , Shoukat, T , Parveen, T , Raza, S , Jamil, F , Kanwal, S, et al. Multi epitope based vaccine design and analysis against Mycoplasma bovis using Immunoinformatic approaches. Pak Vet J. (2022) 421:33–40. doi: 10.29261/pakvetj/2021.068
12. Reck, C , Menin, A , Canever, MF , Pilatic, C , and Miletti, LC . Molecular detection of Mycoplasma synoviae and avian reovirus infection in arthritis and tenosynovitis lesions of broiler and breeder chickens in Santa Catarina state. J S Afr Vet Med Assoc. (2019) 90:e1–5. doi: 10.4102/jsava.v90i0.1970
13. Wang, C , Ewing, M , and Aarabi, SY . In vitro susceptibility of avian mycoplasmas to enrofloxacin, sarafloxacin, tylosin, and oxytetracycline. Avian Dis. (2001) 45:456–60. doi: 10.2307/1592988
14. Cerda, RO , Giacoboni, GI , Xavier, JA , Sansalone, PL , and Landoni, MF . In vitro antibiotic susceptibility of field isolates of Mycoplasma synoviae in Argentina. Avian Dis. (2002) 46:215–8. doi: 10.1637/0005-2086(2002)046[0215:IVASOF]2.0.CO;2
15. Le, CJ , Reinhardt, AK , Kempf, I , and Gautier-Bouchardon, AV . Persistence of Mycoplasma synoviae in hens after two enrofloxacin treatments and detection of mutations in the par C gene. Vet Res. (2006) 37:145–54. doi: 10.1051/vetres:2005046
16. Lysnyansky, I , Gerchman, I , Mikula, I , Gobbo, F , Catania, S , and Levisohn, S . Molecular characterization of acquired enrofloxacin resistance in Mycoplasma synoviae field isolates. Antimicrob Agents Chemother. (2013) 57:3072–7. doi: 10.1128/AAC.00203-13
17. Raviv, Z , Callison, SA , Ferguson-Noel, N , and Kleven, SH . Strain differentiating real-time PCR for Mycoplasma gallisepticum live vaccine evaluation studies. Vet Microbiol. (2008) 129:179–87. doi: 10.1016/j.vetmic.2007.11.017
18. Ferguson-Noel, NM , Laibinis, VA , and Kleven, SH . Evaluation of Mycoplasma gallisepticum K-strain as a live vaccine in chickens. Avian Dis. (2012) 56:44–50. doi: 10.1637/9833-061411-Reg.1
19. Dijkman, R , Feberwee, A , and Landman, WJM . Development, validation and field evaluation of a quantitative real-time PCR able to differentiate between field Mycoplasma synoviae and the MS-H-live vaccine strain. Avian Pathol. (2017) 46:403–15. doi: 10.1080/03079457.2017.1296105
20. Kreizinger, Z , Sulyok, KM , Pasztor, A , Erdelyi, K , Felde, O , Povazsán, J, et al. Rapid, simple and cost-effective molecular method to differentiate the temperature sensitive (ts+) MS-H vaccine strain and wild-type Mycoplasma synoviae isolates. PLoS One. (2015) 10:e0133554. doi: 10.1371/journal.pone.0133554
21. Shahid, MA , Markham, PF , Marenda, MS , Agnew-Crumpton, R , and Noormohammadi, AH . High-resolution melting-curve analysis of obg gene to differentiate the temperature-sensitive Mycoplasma synoviae vaccine strain MS-H from non-temperature-sensitive strains. PLoS One. (2014) 9:e92215. doi: 10.1371/journal.pone.0092215
22. Zhu, L , Konsak, BM , Olaogun, OM , Agnew-Crumptona, R , Kanci, A , Marc, SM, et al. Identification of a new genetic marker in Mycoplasma synoviae vaccine strain MS-H and development of a strategy using polymerase chain reaction and high-resolution melting curve analysis for differentiating MS-H from field strains. Vet Microbiol. (2017) 210:49–55. doi: 10.1016/j.vetmic.2017.08.021
23. Kreizinger, Z , Sulyok, KM , Grozner, D , Beko, K , Dan, A , Szabo, Z, et al. Development of mismatch amplification mutation assays for the differentiation of MS1 vaccine strain from wild-type Mycoplasma synoviae and MS-H vaccine strains. PLoS One. (2017) 12:e0175969. doi: 10.1371/journal.pone.0175969
24. Sun, Q , Wei, X , Chen, W , Zhong, Q , Yan, Z , Zhou, Q, et al. Characterization and evaluation of a novel conserved membrane antigen P 35 of Mycoplasma synoviae. Front Vet Sci. (2022) 9:836110. doi: 10.3389/fvets.2022.836110
25. Sui, C , Cui, H , Ji, J , Xu, X , Kan, Y , Yao, L, et al. Epidemiological investigations and locally determined genotype diversity of Mycoplasma synoviae in Central China from 2017 to (2019). Poult Sci. (2022) 101:101522. doi: 10.1016/j.psj.2021.101522
26. Fayyaz, A , Saleemi, MK , Gul, ST , Gilani, MM , and Irshad, H . Sero-epidemiology and pathology of infectious bronchitis in commercial poultry from Faisalabad division. Pak Vet J. (2023) 431:146–52. doi: 10.29261/pakvetj/2021.065
27. Wei, X , Zhong, Q , Wang, D , Yan, Z , Liang, H , Zhou, Q, et al. Epidemiological investigations and multilocus sequence typing of Mycoplasma gallisepticum collected in China. Poult Sci. (2023) 102:102930. doi: 10.1016/j.psj.2023.102930
28. Telli, AE , Biçer, Y , Biçer, Y , Telli, N , Gungor, C , Turkal, G, et al. Pathogenic Escherichia coli and Salmonella spp. in chicken carcass rinses: isolation and genotyping by ERIC-PCR. Pak Vet J. (2022) 42:493–8. doi: 10.29261/pakvetj/2022.049
29. Wei, X , Chen, W , Sun, Q , Zhong, Q , Yan, Z , Zhou, Q, et al. Epidemiological investigations and multilocus sequence typing of Mycoplasma synoviae isolates from chicken farms in China. Poult Sci. (2023) 102:102006. doi: 10.1016/j.psj.2022.102006
30. Chen, W , Sun, Q , Yan, Z , Zhou, Q , Cao, Y , Chen, F, et al. Transcriptional profiling of the chicken tracheal and splenic response to virulent Mycoplasma synoviae. Poult Sci. (2022) 101:101660. doi: 10.1016/j.psj.2021.101660
Keywords: real-time PCR, differentiation methods, Mycoplasma synoviae, MS attenuated live vaccines, wild-type strains
Citation: Liao C, Chen Y, Yan Z, Song Y, Zhou Q, Zhu P, He X, Li W and Chen F (2024) Development of a rapid quantitative method to differentiate MS1 vaccine strain from wild-type Mycoplasma synoviae. Front. Vet. Sci. 11:1354548. doi: 10.3389/fvets.2024.1354548
Edited by:
Yuefeng Chu, Chinese Academy of Agricultural Sciences, ChinaReviewed by:
Jun Ji, Nanyang Normal University, ChinaLijun Ling, University of Alabama at Birmingham, United States
Copyright © 2024 Liao, Chen, Yan, Song, Zhou, Zhu, He, Li and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhuanqiang Yan, WnF5YW4tMjAxNUB3ZW5zLmNvbS5jbg==; Feng Chen, ZmVuZ2NoQHNjYXUuZWR1LmNu
†These authors have contributed equally to this work and share first authorship
 Changtao Liao1,2†
Changtao Liao1,2† Yiquan Chen
Yiquan Chen Feng Chen
Feng Chen