- 1School of Life Sciences, Ludong University, Yantai, China
- 2Collaborative Innovation Center for the Pet Infectious Diseases and Public Health in the Middle and Lower Stream Regions of the Yellow River, Yantai, China
- 3Provincial Engineering Research Center for Pet Animal Vaccines, Yantai, China
Introduction: Feline parvovirus (FPV), a single-stranded DNA virus, is accountable for causing feline panleukopenia, a highly contagious and often lethal disease that primarily affects cats. The epidemiology prevalence and pathogenicity of FPV in certain regions of China, however, remains unclear. The aim of this research was to investigate the epidemiology of FPV in different regions of China in 2021 and compare its infectivity and pathogenicity.
Methods: In this research, a total of 36 FPV strains were obtained from diverse regions across China. Phylogenetic analysis was performed based on the VP2 and NS1 sequences, and two representative strains, FPV027 and FPV072, which belonged to different branches, were selected for comparative assessment of infectivity and pathogenicity.
Results and discussion: The results revealed that all strains were phylogenetically classified into two groups, G1 and G2, with a higher prevalence of G1 strains in China. Both in vitro and in vivo experiments demonstrated that FPV072 (G1 group) exhibited enhanced infectivity and pathogenicity compared to FPV027 (G2 Group). The structural alignment of the VP2 protein between the two viruses revealed mutations in residues 91, 232, and 300 that may contribute to differences in infectivity and pathogenicity. The findings from these observations will contribute significantly to the overall understanding of the molecular epidemiology of FPV in China and facilitate the development of an effective FPV vaccine.
1 Introduction
Feline parvovirus (FPV), alternatively termed feline panleukopenia virus (FPLV), is the etiological agent for feline panleukopenia (FPL) in cats, exhibiting a predilection for kittens. This virus demonstrates cross-species infectivity, capable of infecting various members of the felid family, encompassing both wild and domestic species (1–3). Moreover, it has the potential to affect certain species belonging to the Mustelidae, Procyonidae, and Viverridae families, such as raccoons, ring-tailed cats, foxes, and minks (4). FPL is a highly infectious and frequently lethal disease that is distinguished by pancytopenia, acute and severe inflammation of the intestines, dehydration, and sepsis caused by lymphoid depletion (5). FPV has a genome length of about 5.1 kilobases and contains two open reading frames (ORFs), namely ORF1 and ORF2. The ORF1 gene contains two non-structural proteins, specifically NS1 and NS2. The NS1 protein includes a domain that encodes a polyprotein, a domain that binds to the origin of replication (ORI), a helicase domain, and functional domains for transactivation. These domains are crucial for controlling replication, DNA packaging, cytotoxicity, and pathogenicity. On the other hand, the NS2 protein is encoded by combining a 260-nt fragment from the left side with a 238-nt fragment from the right side, both originating from the NS1 open reading frame (6–8). Two capsid proteins, VP1 and VP2, are encoded by the ORF2 region. Encoded by alternative splicing, the VP1 protein includes the entire VP2 protein sequence and possesses a distinct N-terminal sequence of 143 residues, which impacts the transportation of capsids to the nucleus and the infection of cells (9). The VP2 protein constitutes approximately 90% of the viral capsid, making it the predominant capsid protein. It serves as the primary target for neutralizing antibodies against parvovirus and plays a crucial role in determining hemagglutination, cellular tropism, and host range (10). Specific amino acids within the VP2 protein are essential for antigenicity, pathogenicity, and host range determination, and subsequently influencing the viral surface structure (11). The amino acids 80K, 564N, and 568A are critical for FPV replication in cats, and the key amino acids residues (80K, 93K, 103V, 323D, 564N, and 568A) has been widely used to distinguish canine parvovirus type 2 (CPV-2) from FPV (12).
The majority of FPV vaccines are modified live virus vaccines (MLV) that have been passaged numerous times on feline or mink cell lines. Inactivated vaccines were used extensively in the past but provide limited amounts of antibodies that protect for a short time, and have been largely replaced by MLVs (13). Although, commercial FPV vaccines have been extensively employed on a global scale, exhibiting promising outcomes in curtailing the transmission of the viral ailment and yielding substantial reductions in both morbidity and mortality rates, FPV is still prevalent in various countries globally (14–18). There are few studies on the molecular epidemiology of FPV in China, and the epidemic situation and molecular characteristics of epidemic strains are still unclear (6, 12, 19). The objectives of the study were to clarify molecular characterization of the epidemic FPV isolated from the domestic cats in parts of China, and to investigate the pathogenicity of the isolates in cats.
2 Materials and methods
2.1 Sample collection
A total of 167 clinical samples (feces or rectal swabs) were collected from pet cats exhibiting one or more clinical signs of parvovirus infection (lethargy, anorexia, vomiting, retching, and/or diarrhea). The cats were subsequently confirmed to be infected with FPV in veterinary hospitals which were located in different regions in China: Linyi (n = 25), Weifang (n = 15), Rizhao (n = 21), Shanghai (n = 35), Changsha (n = 20) and unidentified areas (n = 51). The presence of FPV in these samples had been confirmed to be positive for FPV in veterinary hospital and rechecked using antigen rapid test kit (Colloidal Gold-Based) (Quicking Biotech, Shanghai, China) in the laboratory. After being mixed in 1 mL of 0.1 M PBS (pH 7.4), the samples were then subjected to centrifugation at 10,000 g for 10 min at a temperature of 4°C, and stored at −80°C until use. Four to six FPV isolates from each area and 10 isolates from unidentified areas (36 strains in total) were chosen for subsequent analysis. The details are summarized in Table 1.
2.2 PCR and DNA sequencing assays
For each sample, the viral DNA was extracted by the EasyPure Viral DNA/RNA Kit (TransGen Biotech, Beijing, China) according to the manufacturer’s instructions. Presence of FPV in extracted viral DNA was further confirmed by PCR, targeting the 681 bp VP2 fragments of FPV (Supplementary Table S1). In accordance with the guidelines provided by the manufacturer, PCR was performed utilizing PrimeSTAR® Max DNA Polymerase (Takara Biotech, Dalian, China) in a total volume of 20 μL, which consisted of 10 μL of PrimeSTAR Max Premix (2×), 0.8 μL of each primer (10 μM), 1 μL of DNA, and 7.4 μL of sterile distilled water. The PCR condition was set as an initial denaturation step at 98°C for 1 min; 35 cycles at 98°C for 10 s, annealing at 57°C for 5 s, extension at 72°C for 50 s; and a final extension at 72°C for 10 min. Afterwards, the positive samples underwent amplification of the entire NS1 and VP2 genes by utilizing two sets of designed primers, NS1-F/R or VP2-F/R (Supplementary Table S1), in a final volume of 50 μL comprising 25 μL of PrimeSTAR Max Premix (2×), 2 μL of each primer (10 μM), 5 μL of DNA, and 16 μL of sterile distilled water. The PCR condition was set as an initial denaturation step at 98°C for 1 min; 35 cycles at 98°C for 10 s, annealing at 57°C for 15 s, extension at 72°C for 2 min 30 s; and a final extension at 72°C for 10 min. Positive amplicons were sent for direct Sanger sequencing to BGI Biotech in Beijing.
2.3 Sequence and phylogenetic analyses
BioEdit version 7.2.5 was used to build sequences in accordance with an overlapping technique. The sequences generated were aligned and compared with FPV sequences from the NCBI database using version 7.0 of the MEGA software package. With MEGA7.0, the phylogenetic relationships were assessed. Maximum-likelihood (ML) methods were utilized to construct the phylogenetic trees. The bootstrap values were produced using a total of 1,000 replicates. Thirty-six NS1 gene sequences and 38 VP2 gene sequences were acquired from GenBank as reference sequences in order to conduct a more thorough analysis.
2.4 Virus isolation
Feline kidney cells CRFK (Procell, Wuhan, China) were cultured with DMEM (Vivacell, Shanghai, China) containing 10% fetal bovine serum (FBS) (ZETA, San Francisco, United States) and 1% penicillin-streptomycin. When cultures reached 80%–90% confluence, subculture was performed. Before being infected with virus, CRFK were digested and suspended in serum-free DMEM at a concentration of 5 × 105/mL. The virus sample supernatants were sterilized by filtration with a 0.22 μm microporous filter, synchronously inoculated into cells at a volume ratio of 1:100, gently mixed and incubated at 37°C with 5% CO2 concentration. After 1 h, the virus supernatant was discarded and substituted with the complete cell culture medium. Following a 72 h culture period, the cells were injected with the freeze-thawed culture media and refrozen twice more in a refrigerator at −80°C. After 3–4 generations of continuous blind passage, the observation of cytopathic effect (CPE) was noted. The CPE which is typical of FPV infection often involves total destruction of the cell monolayer. Earlier CPE evidence includes cell rounding and disassociation of the cells from the flask surface (20).
2.5 Virus titration and growth kinetics
Viral titration was assessed using the 50% cell culture infectious dose (TCID50) assay. Briefly, 100 μL CRFK cell suspension, containing 3 × 104 cells in DMEM with 10% FBS and 1% penicillin-streptomycin, was seeded into each well of a 96-well plate and kept at 37°C for incubation. After 24 h incubation, CRFK cells were inoculated with 10-fold dilutions of the virus for 1 h, followed by washing and then incubation with DMEM with 2% FBS and 1% penicillin-streptomycin for 3–4 days and checked daily for CPE. TCID50 end-point titers were calculated using the Reed and Muench method. Virus growth kinetics were determined by testing viral titers after three consecutive freeze-thaw cycles.
2.6 Western blot assay
The FPV-infected and mock cells were lysed in Laemmli sample buffer, boiled for 10 min. Proteins were transferred utilizing the Trans-Blot® TurboTM Transfer System (Bio-Rad Laboratories, Hercules, CA, United States) from gels onto PVDF membranes. Membranes were blocked with PBS, 5% (w/v) skim milk, and 0.5% (v/v) Tween 20 for 2 h at room temperature and incubated with mouse monoclonal antibody recognizing CPV VP2 protein (Abcam, Cambridge, United Kingdom) (9, 21) at 4°C overnight. Following multiple rinses with PBS-0.05% Tween 20, the membranes were then incubated at 37°C for 1 h with HRP-conjugated goat anti-mouse IgG (Abcam, Cambridge, United Kingdom). Afterwards, the membranes were thoroughly rinsed with PBS-0.05% Tween-20 and treated with Pierce ECL Plus Western Blotting (Thermo Fisher Scientific, MA, United States) for visualization.
2.7 Transmission electron microscope
The harvested cell cultures were partially purified by ultracentrifugation through a 15% (w/v) sucrose cushion at a speed of 45,000 g for 3 h at 4°C. After purification, the sample was treated with a solution of alcohol saturated with 2% uranyl acetate and a solution of 2.6% lead citrate, and observed using a transmission electron microscope (H-7800, Hitachi, Japan).
2.8 Immunofluorescence assay
CRFK cells infected with FPV isolate (MOI = 0.1) in 96-well plates were treated with cold 80% (v/v) acetone for 15 min, washed with PBS three times, exposed to a mouse anti-CPV antibody (Abcam, Cambridge, United Kingdom) at 37°C for 1 h, and then treated with an Alexa Fluor 488-conjugated goat anti-mouse IgG H&L antibody (Abcam, Cambridge, United Kingdom). Then, diamidino-2-phenylindole (DAPI) (Solarbio, Beijing, China) dye was added for fluorescent staining of cell nuclei. FPV infected cells were observed under a fluorescence microscope (ZEISS, Oberkochen, Germany) after being washed with PBS; cells evidencing intranuclear fluorescence were considered infected with FPV.
2.9 Animal experiments
Fifteen conventional British Shorthair pet kittens used in the challenge experiments were purchased from Jianyouda Biomedical Technology Co., Ltd. The kittens were brought to the facility when they were between 8 and 10 weeks old. They were acclimated and used in the studies at the age of 12–14 weeks. The animals were kept in separate groups in a closed facility. PCR and/or ELISA tests confirmed that the cats were free from feline herpesvirus (FHV), feline calicivirus (FCV), feline immunodeficiency virus (FIV), and feline leukaemia virus (FELV). Kittens were intraperitoneally challenged with 104.7 TCID50 of the viruses FPV027 (Group 1, n = 5) and FPV072 (Group 2, n = 5), respectively. The kittens in the mock control group were given DMEM intraperitoneally (Group 3, n = 5). The duration of the clinical observation period amounted to 14 days. Throughout this period, daily clinical examinations were conducted on the kittens (22). Briefly, scoring was based on attitude (0–3 points), appetite (0–3 points), vomiting (0–3 points), and fecal appearance (0–4 points). The clinical score was given by the sum of the 4 assigned values. The severity of the values is reported in ascending order. In addition, the white blood cell (WBC) count was recorded every 2 days. Kittens showing signs of terminal FPV were euthanized in order to avoid unnecessary suffering. All animal experiments were approved and supervised by the Institutional Animal Care and Use Committee (IACUC) of Ludong University.
2.10 Detection of virus shedding
In order to ascertain the shedding of the virus, samples of feces were collected and immersed in 500 mL of phosphate buffered saline (PBS). After vortexing and 30 min incubation, the swabs were removed, and the extract was centrifuged at 1,000 g for 10 min to get rid of debris. PCR was performed using the extracted viral DNA from the supernatant. The genome copy numbers of FPV were quantified using the Feline Parvovirus Real Time PCR detection kit (TIANDZ, Beijing, China).
2.11 Pathology, histopathology and immunohistochemical staining
Pathological examination was conducted on all animals that perished or were euthanized during the experiment. Carcasses were dissected within a span of 2 h after the demise. Samples extracted from the intestinal tract and spleen were embedded in paraffin wax after being fixed in 8% formaldehyde. The blocks were sectioned at 4–6 mm and the sections were stained with haematoxylin-eosin and examined under light microscope. Intestinal immunohistochemistry was then scored according to the description in the previous literature (23). In brief, the ratio of apical denudation of villi between 0% and 10%, scored 3; between 11% and 40%, scored 2; between 41% and 70%, scored 1; between 71% and 100%, scored 0. The histopathologically examined organs were subsequently subjected to immunohistochemical analysis. A primary antibody specific to parvovirus (Abcam, Cambridge, United Kingdom) was utilized, followed by a secondary antibody conjugated with HRP and specific for mouse IgG (H + L) (Thermo Fisher Scientific, MA, United States). The binding events were visualized using the peroxidase anti-peroxidase method with diaminobenzidine as the chromogen.
2.12 Molecular dynamics simulation
The initial structure of the VP2 of FPV072 and FPV027 were predicted with trRosetta online server (24).1 Using H++ server, the protonation states of ionizable residues were determined at pH = 7.0 (25). In FPV072 VP2, His102 and His are protonated at Nδ atom; His70, His234, and His483 are deprotonated; the carboxyl groups in Asp373, Asp375, and Glu368 are all protonated. In FPV027 VP2, the protonation state of titrable amino acid residues is almost the same as FPV072 VP2, except for the Nδ protonation of His384. Hydrogen atoms and missing atoms were subsequently added to the complex structures using the t-Leap procedure of AMBER22. To keep the whole system electric neutral state, five Na+ ions were introduced to the systems built above. Then, each system was solvated in an octahedron box with a 10.0 Å distance around the solute.
The ff14SB force field was adopted for the proteins (26). AMBER22 software package was utilized to conduct the molecular dynamics simulations (MD simulations). To minimize water molecules, a total of 5,000 cycles of steepest descent followed by 5,000 cycles of conjugated gradient minimizations were conducted. And then, the minimizations were repeated to minimize the whole system. Thirdly, each system was gradually heated from 0 K to its physiological temperature over a period of 300 ps. After that, 500 ps equilibrium simulation was performed to balance the density. Finally, 200 ns MD simulation was performed for each system under NPT condition to produce trajectory. The particle mesh Ewald (PME) method (27) was used to evaluate long-range electrostatic interactions, while the short range interactions were cut off at 10.0 Å. The integration step was set to 2 fs in all the simulations.
2.13 Statistical analysis
The analysis of the data was conducted using GraphPad Prism Software 9.5 (GraphPad Software, SanDiego, CA, United States). Results were represented as mean ± SEM, and one-way analysis of variance (ANOVA) was used to compare data among three or more groups; p < 0.05 was considered as statistically significant, *p < 0.05, **p < 0.01.
3 Results
3.1 Isolation and identification of FPV strains
CRFK cells were inoculated with FPV-positive samples at a 100-fold dilution, and visible CPE was observed 48 h after inoculation (Figure 1A). Viral supernatants and cell lysates were collected and subjected to both PCR and immunoblotting to test for virus production. As expected, PCR amplification of the viral genomic DNA revealed a specific band of 681 bp corresponding to the partial amplification of the VP2 gene (Figure 1B). Representative WB bands for FPV were detected in infected cells (Figure 1C). Furthermore, FPV antigen was detected by IFA using a parvovirus-specific mouse monoclonal antibody and cell nuclei was detected by DAPI. Cells expressing FPV antigens were identified by the specific green cytoplasmic signals. No positive staining was observed in uninfected cells (Figure 1D). FPV particles in culture media and infected cells were imaged with a transmission electron microscope (TEM). Typical non-enveloped round virus particles were observed with diameters of 20–25 nm (Figure 1E). The 36 strains of FPV (Table 1) were isolated and identified according to the above methods.
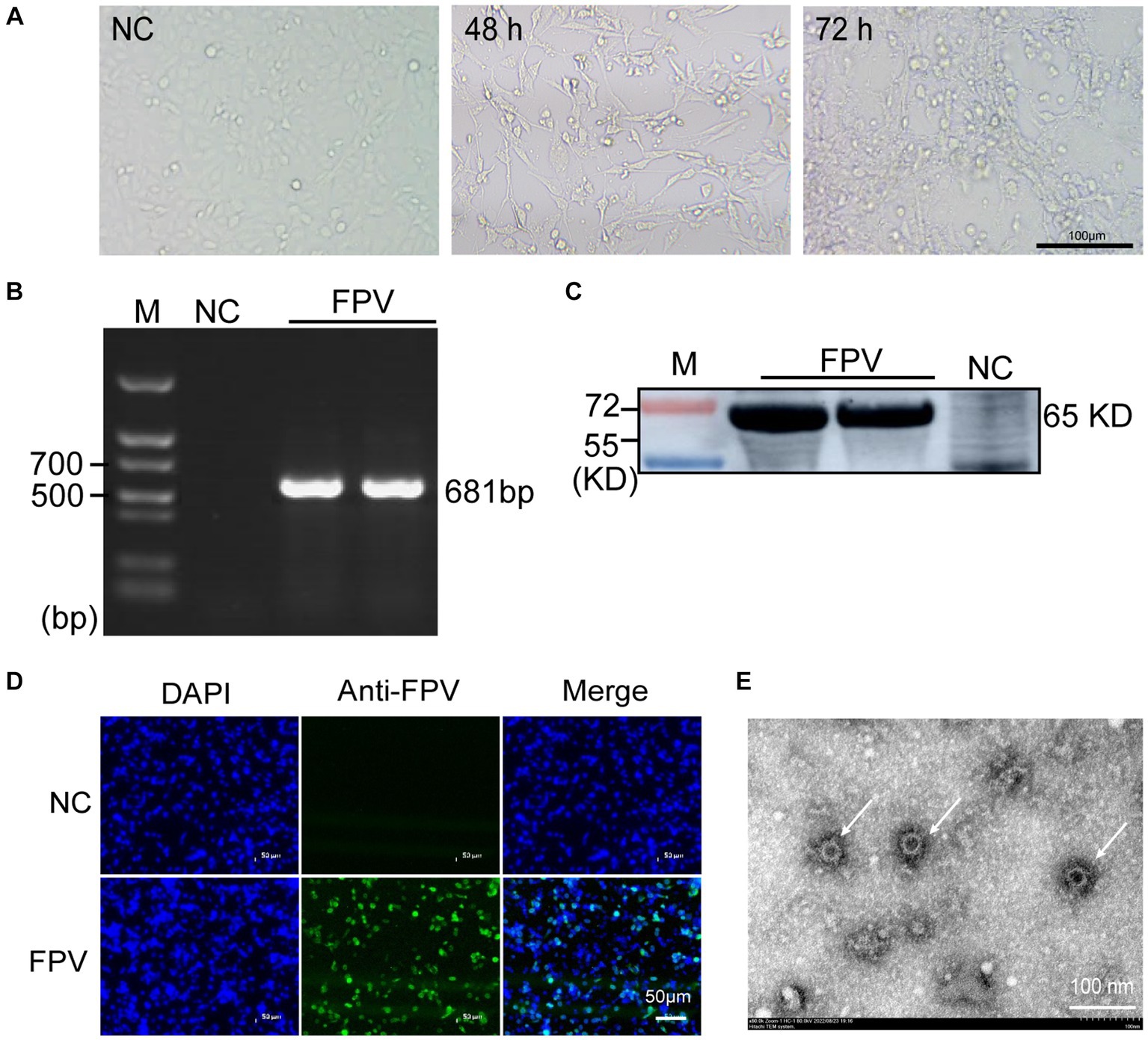
Figure 1. Isolation and identification of FPV in CRFK cells. (A) Cytopathic effect (CPE) of CRFK cells infected with FPV at 48 hpi and 72 hpi (×100). The negative control cells were mock infected with FPV (×100). Scale bar = 100 μm. (B) PCR analysis of the total cell DNA from the third passages of virus-infected cells or mock-infected cells. (C) Western blot analysis of the total cell lysates from virus-infected cells using anti-CPV polyclonal antibodies. (D) Immunofluorescence detection results at 24 h after inoculation. Scale bar = 50 μm. (E) Electron micrograph of FPV virions (arrow heads) in cell culture media of infected CRFK cells. Scale bar = 100 nm.
3.2 Genetic evolution and amino acid mutations of VP2
To further examine the relationship between the 36 strains with FPV strains in other regions of the world, we reconstructed a phylogenetic tree containing 38 FPV reference sequences and the 36 cat-derived parvovirus sequences based on the complete VP2 gene sequence (Figure 2). The phylogenetic tree was divided into two groups, G1 and G2. Strains in G1 mostly comprised FPV isolates from Asia (China and Korea) and a few from Europe (Italy and Portugal). Strains in G2 consisted almost exclusively of Chinese isolates, and included two vaccine strains, indicating a close relationship between G2 strains and vaccine strains. A total of 24 of the 36 FPV strains (66.7%) were distributed in G1, suggesting that G1 strains were the predominant strains circulating in China.
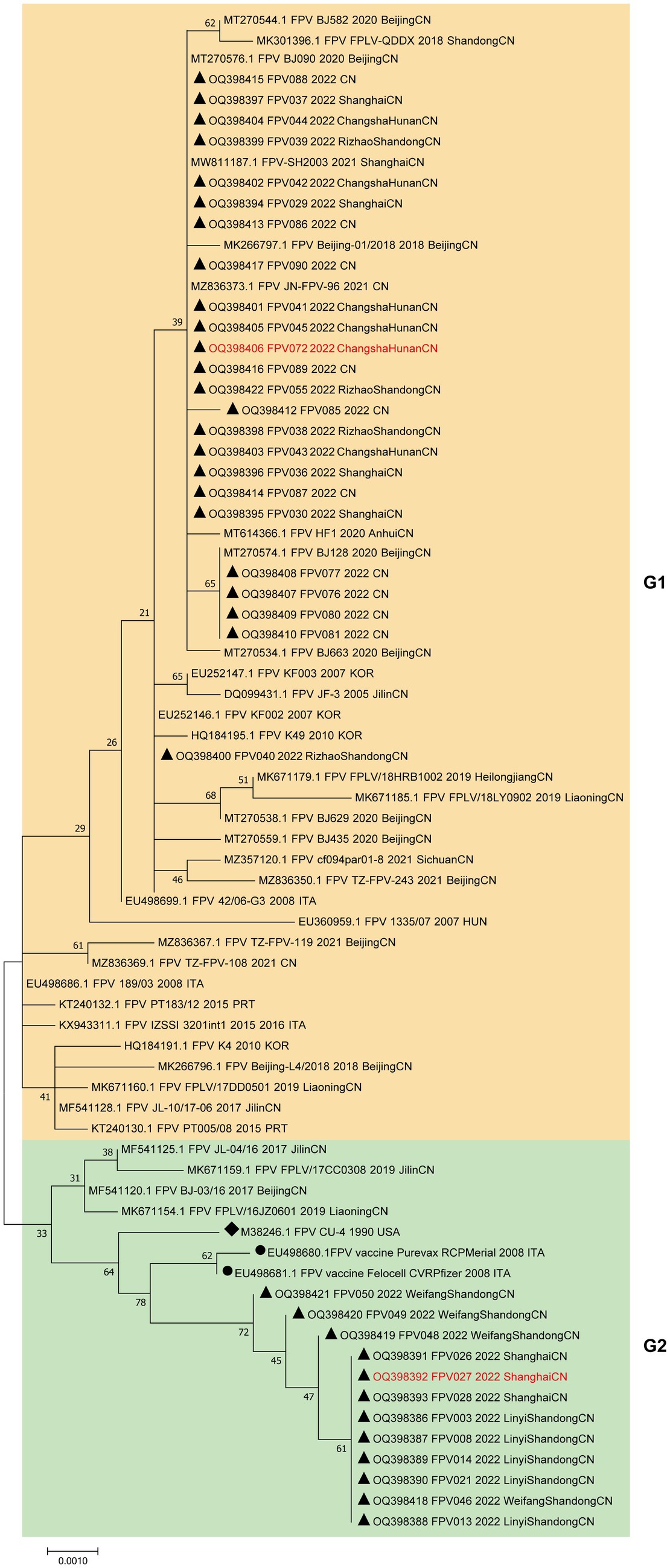
Figure 2. Phylogenetic tree analysis of the VP2 nucleotide sequences of the FPV strains. A phylogenetic tree based on 36 VP2 gene sequences from domestic cats and 38 reference FPV strains was constructed by the maximum likelihood method using the MEGA software, version 7.0. ▲ Indicates the FPV strains in this study. ● Indicates the FPV vaccine strains. ◆ Indicates the FPV standard strain. Horizontal branch lengths are proportional to genetic distances. Scale bars indicate nucleotide substitutions per site. Bootstrap values were calculated based on 1,000 replicates.
Moreover, an analysis was conducted to compare the amino acid sequences of VP2. The amino acid sites 80/93/103/323/568 in all 36 sample sequences were found to be entirely in agreement with the characteristic amino acid sites of FPV, as depicted in Figure 3 (28). In comparison to FPV prototype CU-4 (GenBank accession number M38246), mutations were mainly concentrated in 6 positions (A91S, I101T, I145L, V232I, A300P, and V562L), where the most common mutations were A91S (67.6%) and I101T (73.0%).
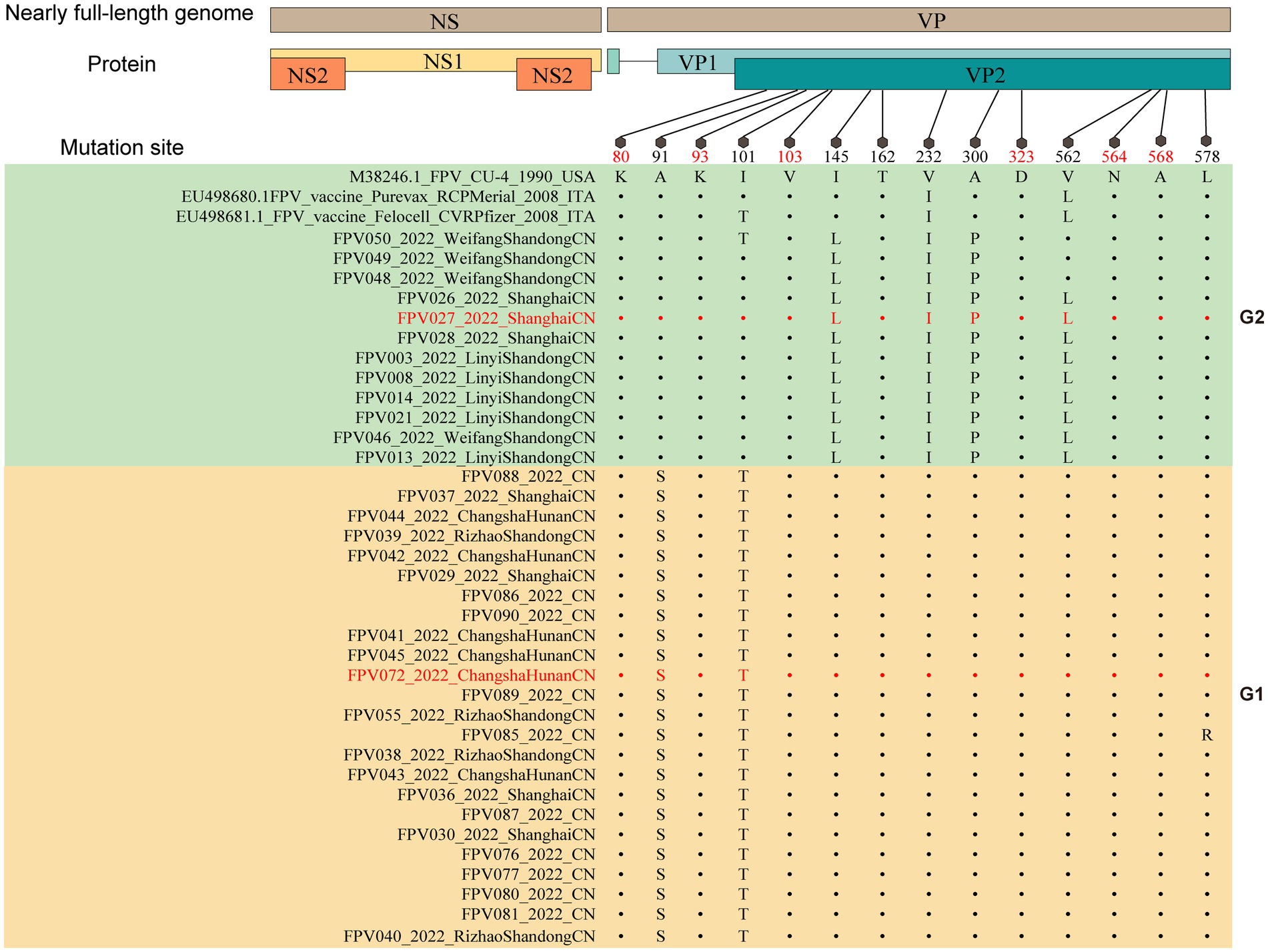
Figure 3. Molecular schematic representation showing the organization of FPV and amino acid mutations in the VP2 protein. The FPV strains explored herein are colored with red. Capital letters represent amino acids.
3.3 Genetic evolution and amino acid mutations of NS1
Since non-structural proteins are important in virus replication and pathogenicity, their phylogeny and amino acid mutations were investigated as well. Phylogenetically, 36 NS1 nucleotide sequences from this study and 38 reference sequences from GenBank formed three different groups, G1, G2 and G3 (Supplementary Figure S1). The G1 group mainly comprised strains from China and Japan, and contained 18 (50%) of the isolated strains in this study. The G2 group consisted exclusively of Chinese isolates, with 18 (50%) of the isolated strains in this study. G1 and G2 branches originated from a single node, suggesting the NS1 sequences of the strains isolated in this study is limited (Supplementary Figure S1).
Compared to FPV prototype CU-4, amino acid sequence analysis demonstrated that the mutation sites were concentrated in 12 sites (N23D, E72K, E73K, S136R, R148H, K201R, H247Q, T248I, I574V, H595Q, V596L and D616N). Among these, the most common mutations detected were H247Q (45.9%), T248I (48.6%), H595Q (91.9%) and V596L (91.9%) (Supplementary Figure S2). The biological significance of these mutation sites needs to be investigated further.
3.4 Viral titers and growth characteristics of FPV strains
To analyze the replication characteristics of FPV strains, the virus titers were calculated in a defined time. CRFK cells were infected with the FPV strain at an MOI of 0.1. Then, the supernatants were harvested at 80 h after induction, and the virus titers in the samples were determined. As shown in Table 2, the viral titers ranged from 2.7 ± 0.6 log10 TCID50/mL to 7.2 ± 0.3 log10 TCID50/mL. Among them, the titers of FPV072 were highest with 7.2 ± 0.3 log10 TCID50/mL, followed by FPV027, with 6.9 ± 0.5 log10 TCID50/mL. In addition, the growth characteristics of FPV072 and FPV027 were studied using one-step growth experiments. CRFK cells were inoculated with FPV strains and the supernatants were collected every 8 h post-infection to determine viral titers. As shown in Figure 4, viral growth of FPV027 and FPV072 reached peak titer 80–88 h post-infection and declined thereafter. No significant difference was found between the two strains.
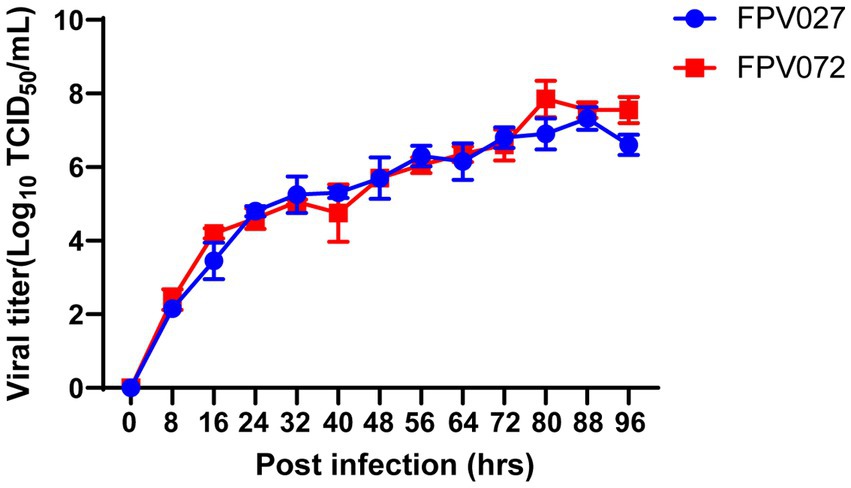
Figure 4. Single growth curve analysis of CRFK cells infected with FPV027 and FPV072 strains over 96 h.
3.5 Pathogenicity of the FPV 027 and FPV072 strains
As previously mentioned, the phylogenetic tree, which was constructed using VP2 sequences of these 36 FPV strains, was categorized into two groups, namely G1 and G2. However, it remains unknown if there are differences in pathogenicity between viruses in the two groups. Then, two representative strains with high titers and from different evolutionary branches and regions (FPV072: 7.2 ± 0.3 log10 TCID50/mL, Hunan; FPV027: 6.9 ± 0.5 log10 TCID50/mL, Shanghai) were chosen to conduct further studies on pathogenicity comparison. Cats were challenged with 104.7 TCID50 FPV strains and the clinical signs were monitored and compared daily. As shown in Table 3, the number of total WBCs significantly declined 4–6 days post-inoculation, and death occurred when it dropped to below 2 × 109/L. In addition, WBCs were significantly lower in FPV072 infected cats than in FPV027 infected cats. According to the fecal scoring system (Supplementary Table S2), cats infected with the FPV072 strain developed higher fecal scores on days 2, 4, 10 and 12, indicating more severe leukopenia diarrhea in FPV072-infected cats (Figure 5A). There was no significant difference in FPV shedding in feces between two groups (Figure 5B). Compared with those infected with FPV027, the weight loss of mice infected with FPV072 was more obvious (Figure 5C).
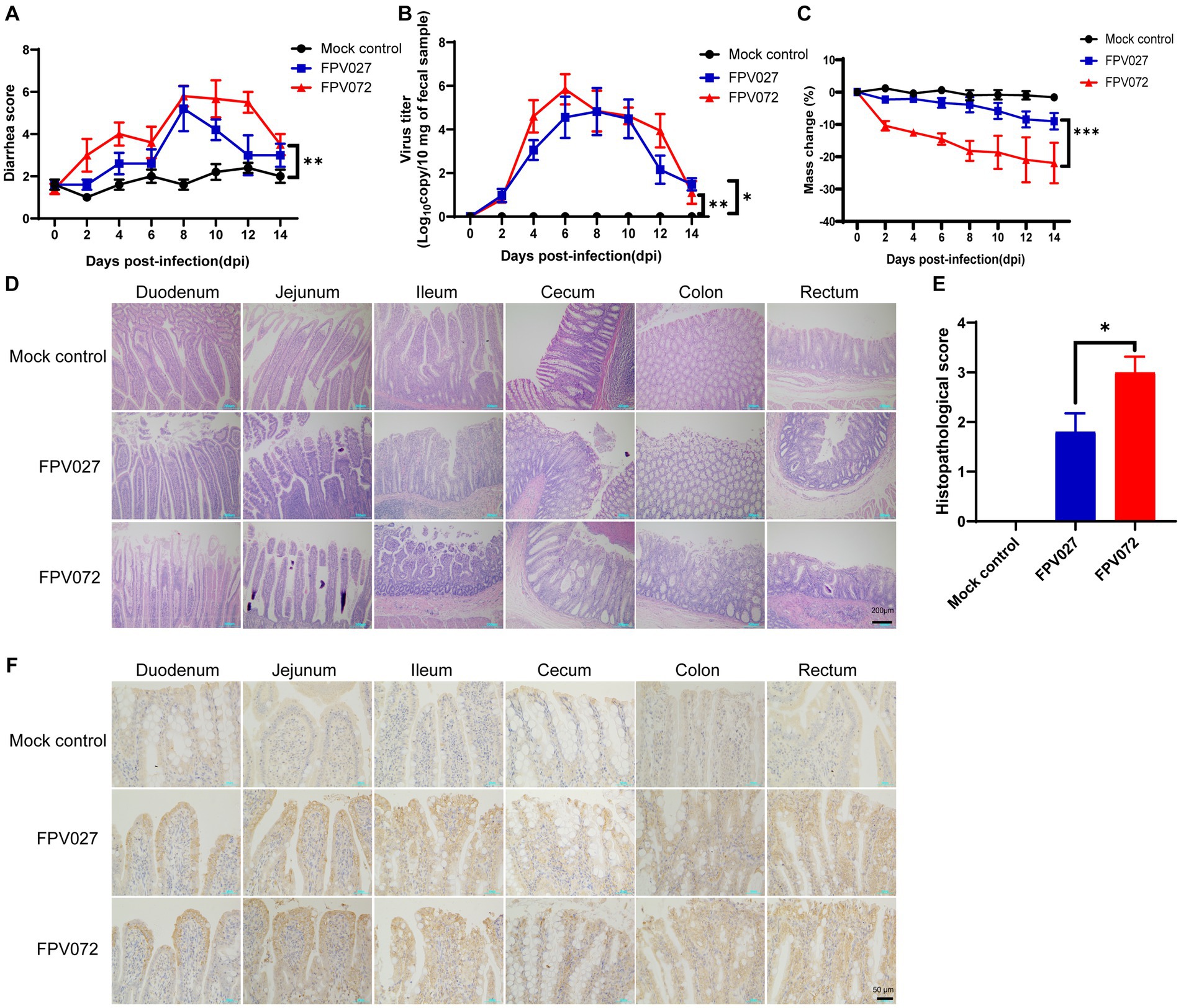
Figure 5. Diarrhea score, virus shedding in feces, weight loss, histopathology and immunohistochemistry (IHC) of cats inoculated with FPV. (A) Diarrhea score in cats after FPV inoculation. (B) Virus shedding in feces. (C) Weight loss of cats. (D,E) Intestinal histopathology and score in FPV-injected and negative control cats. (F) Detection of FPV antigen by IHC analysis of intestinal tissue sections from FPV infected and mock control (400× magnification). Data was processed based on the results of each group (n = 5).
Histopathological examination showed prominent changes, including villous necrosis of the intestine, mucosal erosion and ulceration of the cecum colon and rectum. A higher intestinal histopathological score in the FPV072 group indicated more severe intestinal damage (Figures 5D,E). Furthermore, immunohistochemical examinations revealed that FPV antigens were predominant in the cytoplasm of the epithelial cells of atrophied villi in the small intestines (Figure 5F). Thus, cats in the FPV072 group showed more severe pathological changes. In addition, enlarged and mottled spleens were also found in a minority of infected cats (Supplementary Figure S3A). On 14 dpi, spleens of infected mice exhibited significant congestion and an increase in size of the red and white pulp (Supplementary Figure S3B).
3.6 Comparison of VP2 and NS1 protein structures between FPV027 and FPV072 strains
As previously mentioned, there exists a disparity in the pathogenic potential between the FPV072 and FPV027 virus strains, which cannot be attributed to host-specific variations. Hence, we tried to elucidate this distinction from the perspective of viral molecular protein structures. A 200 ns MD simulation was performed for the VP2 protein of each virus. The representative conformations obtained from the cluster analysis of the two systems are superimposed. As shown in Figure 6, the mutation of residues at 101, 145, and 562 site have no significant effect on the structure. It’s worth noting that Ser91 is part of the alpha helix in FPV072 VP2, whereas Ala91 has no stable secondary structure in FPV027 VP2. This change also induces a certain bend in the β-sheet where residue 232 located. Besides, due to the mutation of residue 300, a large conformational shift occurs near it. The above conformation changes may affect the virulence of the virus.
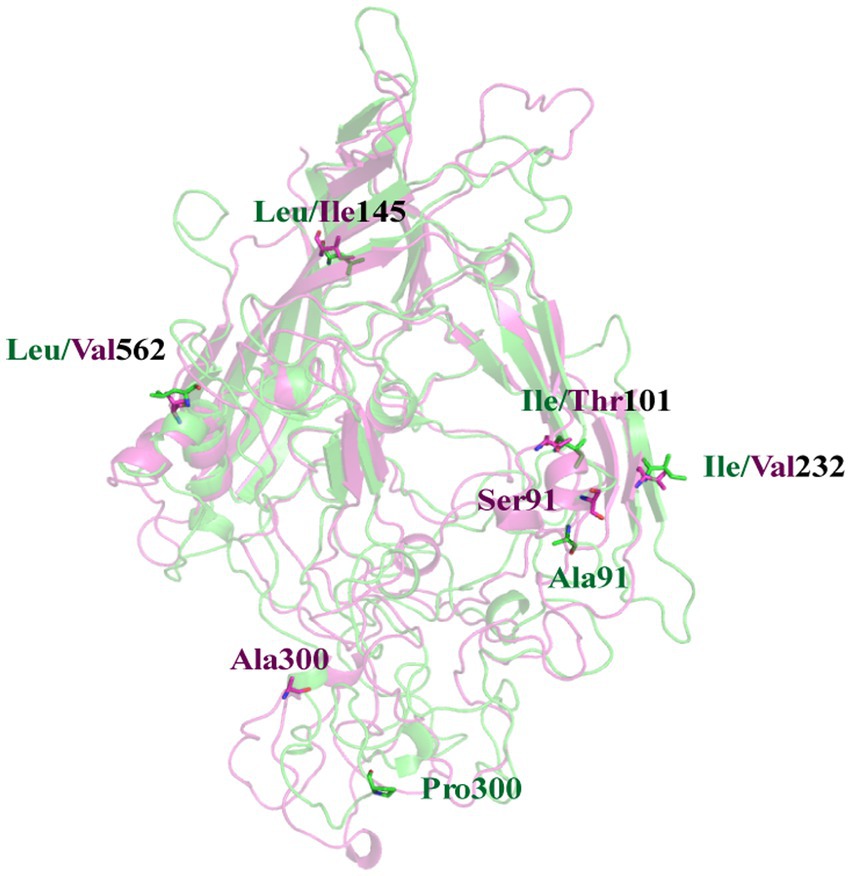
Figure 6. A superposition of FPV027 VP2 (green) and FPV072 VP2 (magentas). Mutated residues are shown in sticks. The residue numbers are labeled nearby with the corresponding color.
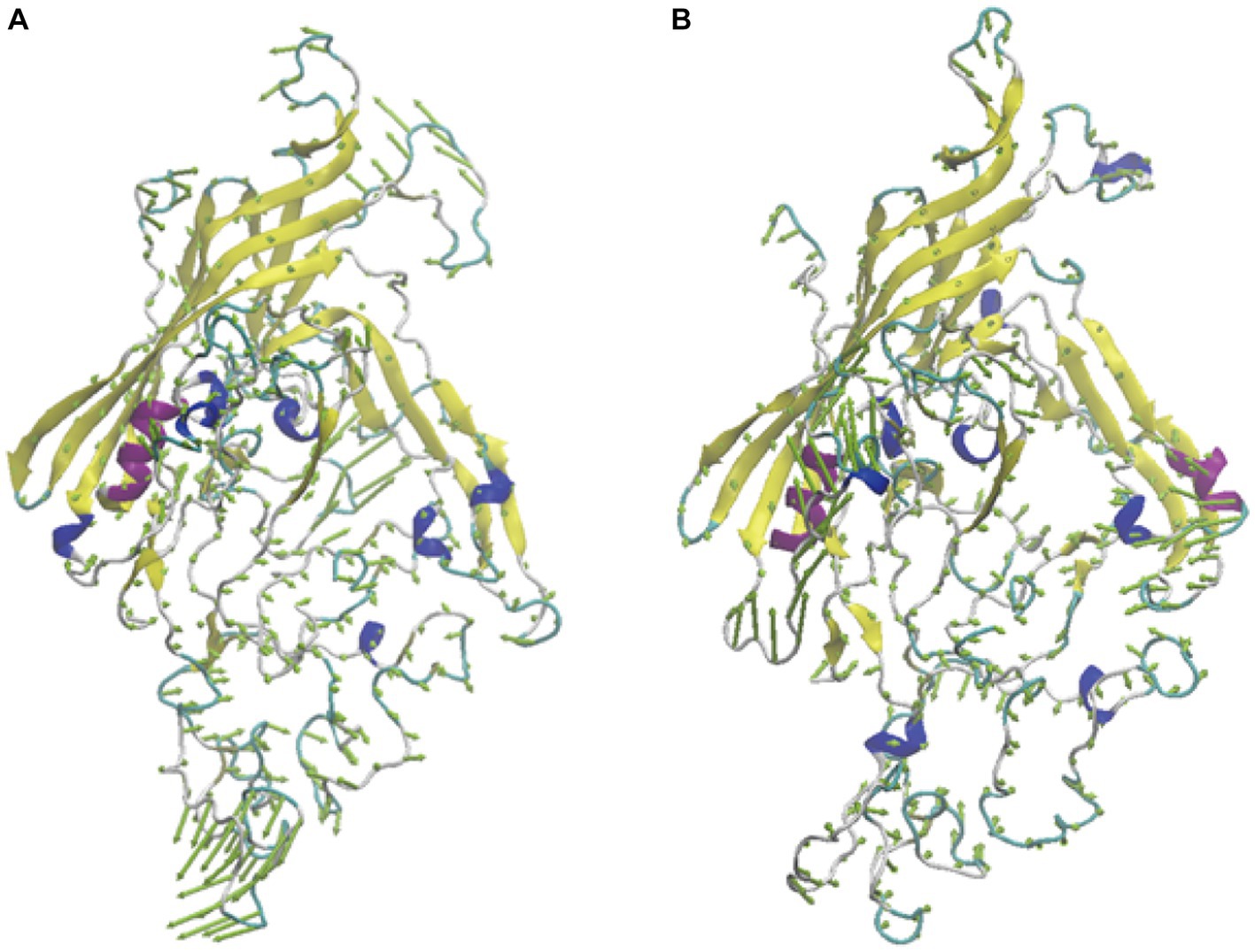
To further analyze the effect of mutations on protein movement patterns, we performed principal component analysis of these two proteins and presented the results in the form of porcupine diagrams. Our main emphasis in this section will be on the principal modes derived from the highest eigenvalue (PC1) and its corresponding eigenvector. In Figure 7, the spine direction represents the eigenvector, and the length of the spine represents the eigenvalue. There are different motion modes in the two different systems, which could be the reason for the difference in infectivity and pathogenicity.
4 Discussion
The feline parvovirus infection is a prevalent ailment that induces diarrhea in cats and can be life-threatening in severe cases. In recent years, the prevalence of FPV infection remains high in different countries and regions. A findings of a study conducted in central and eastern China from 2018 to 2022 revealed that 27.6% of cats with clinical gastroenteritis were infected with FPV (29). While, the prevalence of FPV in southwest China from 2019 to 2021 was 56.7% (30). Here, a total of 167 FPV positive specimens were obtained during a single quarterly period from 8 veterinary hospitals in different aeras, indicating the FPV positive cats in China was considerable. Furthermore, the prevalence of FPV exhibits regional variations. It was reported that the prevalence positivity of FPV in Italy, Egypt, and Spain are 73.5%, 43%, and 18%, respectively (31–33). The investigation into the epidemiological characteristics, pathogenic features, and underlying mechanisms of FPV will establish a fundamental framework for the prevention and control strategies against FPV.
In this work, 36 FPV strains were isolated from different parts of China in 2021, and their molecular prevalence was investigated. The phylogenetic tree based on VP2 or NS1 genes showed that all the isolated strains were distributed on two branches. Two genetically different FPV strains (027 and 072) from each branch were chosen to compare pathogenicity in a domestic cat infection model, showing differences in degree of infectivity and pathogenicity. While individual variations in feline characteristics may account for these disparities, the pathogenicity is also influenced by mutations occurring in viral proteins. Comparison of sequences in the two strains showed 6 amino acid substitutions in VP2, at positions 91, 101, 145, 232, 300 and 562 (Figure 6). Residue 91 is located within the LOOP1 region, contradicting the assertion that the LOOP1 segment is highly conserved (34). The residue is situated close to residue 93, positioned at the apex of the capsid’s 3-fold spike. It is among the residues that play a role in determining the range of hosts, although the precise phenotypic impact of these mutations remains uncertain. It has been suggested that mutation at residue 91 increases strain virulence, in agreement with our pathogenicity test results (35). Additional research has suggested that residue 101 is situated not only within the central region of the receptor-binding area (36) but also within the site where antibody bind (37). Structural data has also shown that a Thr at position 101 leads to the formation of a polar contact with Asp99. The modification alters the outer layer of the central area responsible for binding to receptors, thereby affecting the strength of the bond between the transferrin receptor and the virus (11). Moreover, a mutation at position 101 may cause immune failure (38), which may also explain why the current vaccine is only partially effective. Since residues 299–301 on LOOP3 are part of neutralizing site B, a mutation at residue 300 may affect antigenicity (39).
The NS1 protein has crucial functions in regulating replication, DNA packaging, cytotoxicity, and pathogenicity. Three substitutions in the NS1 coding region, at positions 247, 248 and 574, were also found (data not shown). Mutations at positions 247 and 248 were also found in mink enteritis virus (40). Mutation at position 574 of the NS1 protein has not been reported previously in FPV. Additional research is necessary to ascertain its importance in the contagion of cats.
Our phylogenetic tree and homology analysis revealed that all the 36 FPV strains isolated clustered into two branches, where genogroup G1 contains most (24/36, 66.7%) of the isolates and G2 contains a smaller fraction (12/36, 33.3%). Notably, two vaccine strains were clustered within the G2 group, and they were distantly related to the currently prevalent strains, which may explain the low protective effect of the current vaccine. The development of vaccines utilizing newly circulating strains is imperative.
The incubation period of FPV is generally less than 14 days (5), and the median time from shelter admission to FPL diagnosis is 9 days (41). The risk of acquiring FPV infection is even higher in the stressful environment of shelters (42). In our study, domestic cats were maintained under well-controlled and standardized barrier conditions for 14 days followed by antigen and serology tests to screen cats that had been previously infected with FPV or in the incubation period. Nonetheless, after the observation period, a minority of non-inoculated cats showed clinical symptoms of FPV infection, indicating possible infection by direct fecal-oral contact during the observation period. Since the experimental cats used in this study were 8–12 weeks of age, the balance period could not be extended and this may add some uncertainty to our data.
It has been reported that FPV mainly replicates in small intestinal crypt cells and lymphoid cells (43). Replication often leads to bleeding in the small intestine and stomach, which is the main cause of hemorrhagic diarrhea in cats (41). However, in the present study, no hemorrhagic diarrhea was observed. This discrepancy may be caused by different doses and strains of FPV. Other lesions were also detected, such as enlarged spleen and lymph nodes in the mesentery. Despite splenomegaly is rarely described in clinical symptoms of FPV infection, about one-third of cats exhibited splenomegaly with rough spleen surfaces, possibly due to antiviral host responses, such as antibody synthesis and lymphocyte proliferation (44).
5 Conclusion
Based on phylogenetic analysis, the 36 strains isolated in China in 2021 were classified into two groups, G1 and G2, with most strains clustering in the G1. G1 group isolate FPV072 exhibits higher infectivity and pathogenicity than G2 group isolate FPV027. Structural alignment of the VP2 protein of the two viruses showed that there were mutations of residues at 91, 232, and 300, which might be related to the difference in infectivity and pathogenicity between the two groups. These observations will be helpful for better understanding the prevalence of FPV infection in China and the development of an FPV vaccine.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Ethics statement
The animal study was approved by Institutional Animal Care and Use Committee (IACUC) of Ludong University. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
QX: Methodology, Writing – original draft. ZS: Methodology. XX: Validation. YP: Methodology. SZ: Methodology. YL: Investigation. JiuZ: Formal Analysis. LJ: Visualization. JiaZ: Resources. HZ: Resources. XY: Project administration, Writing – review & editing. XZ: Funding acquisition.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by a Universities-Industry Collaboration Program funded by the Bureau of Education of Yantai City, China (Development of Next-Generation Vaccines Platform for Pet Animals).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2024.1328244/full#supplementary-material
Footnotes
References
1. Pacini, MI, Forzan, M, Franzo, G, Tucciarone, CM, Fornai, M, Bertelloni, F, et al. Feline parvovirus lethal outbreak in a group of adult cohabiting domestic cats. Pathogens. (2023) 12:822. doi: 10.3390/pathogens12060822
2. Kim, YJ, Yoon, S-W, Jang, JH, Jeong, DG, Lee, BJ, and Kim, HK. Genetic characterization of feline parvovirus isolate Fe-P2 in Korean cat and serological evidence on its infection in wild leopard cat and Asian badger. Front Vet Sci. (2021) 8:650866. doi: 10.3389/fvets.2021.650866
3. Duarte, MD, Henriques, AM, Barros, SC, Fagulha, T, Mendonça, P, Carvalho, P, et al. Snapshot of viral infections in wild carnivores reveals ubiquity of parvovirus and susceptibility of Egyptian mongoose to feline panleukopenia virus. PLoS One. (2013) 8:e59399. doi: 10.1371/journal.pone.0059399
4. Stuetzer, B, and Hartmann, K. Feline parvovirus infection and associated diseases. Vet J. (2014) 201:150–5. doi: 10.1016/j.tvjl.2014.05.027
5. Rehme, T, Hartmann, K, Truyen, U, Zablotski, Y, and Bergmann, M. Feline panleukopenia outbreaks and risk factors in cats in animal shelters. Viruses. (2022) 14:1248. doi: 10.3390/v14061248
6. Li, S, Chen, X, Hao, Y, Zhang, G, Lyu, Y, Wang, J, et al. Characterization of the VP2 and NS1 genes from canine parvovirus type 2 (CPV-2) and feline panleukopenia virus (FPV) in northern China. Front Vet Sci. (2022) 9:934849. doi: 10.3389/fvets.2022.934849
7. Allison, AB, Organtini, LJ, Zhang, S, Hafenstein, SL, Holmes, EC, and Parrish, CR. Single mutations in the VP2 300 loop region of the three-fold spike of the carnivore parvovirus capsid can determine host range. J Virol. (2016) 90:753–67. doi: 10.1128/JVI.02636-15
8. Fei-Fei, D, Yong-Feng, Z, Jian-Li, W, Xue-Hua, W, Kai, C, Chuan-Yi, L, et al. Molecular characterization of feline panleukopenia virus isolated from mink and its pathogenesis in mink. Vet Microbiol. (2017) 205:92–8. doi: 10.1016/j.vetmic.2017.05.017
9. Chung, HC, Kim, SJ, Nguyen, VG, Shin, S, Kim, JY, Lim, SK, et al. New genotype classification and molecular characterization of canine and feline parvoviruses. J Vet Sci. (2020) 21:e43. doi: 10.4142/jvs.2020.21.e43
10. Yi, S, Liu, S, Meng, X, Huang, P, Cao, Z, Jin, H, et al. Feline panleukopenia virus with G299E substitution in the VP2 protein first identified from a captive giant panda in China. Front Cell Infect Microbiol. (2021) 11:820144. doi: 10.3389/fcimb.2021.820144
11. Hoang, M, Wu, CN, Lin, CF, Nguyen, HTT, Le, VP, Chiou, MT, et al. Genetic characterization of feline panleukopenia virus from dogs in Vietnam reveals a unique Thr101 mutation in VP2. PeerJ. (2020) 8:e9752. doi: 10.7717/peerj.9752
12. Chen, Y, Wang, J, Bi, Z, Tan, Y, Lv, L, Zhao, H, et al. Molecular epidemiology and genetic evolution of canine parvovirus in east China, during 2018–2020. Infect Genet Evol. (2021) 90:104780. doi: 10.1016/j.meegid.2021.104780
13. Truyen, U, and Parrish, CR. Feline panleukopenia virus: its interesting evolution and current problems in immunoprophylaxis against a serious pathogen. Vet Microbiol. (2013) 165:29–32. doi: 10.1016/j.vetmic.2013.02.005
14. Jenkins, E, Davis, C, Carrai, M, Ward, MP, O’Keeffe, S, van Boeijen, M, et al. Feline parvovirus seroprevalence is high in domestic cats from disease outbreak and non-outbreak regions in Australia. Viruses. (2020) 12:320. doi: 10.3390/v12030320
15. Miranda, C, Vieira, MJ, Silva, E, Carvalheira, J, Parrish, CR, and Thompson, G. Genetic analysis of feline panleukopenia virus full-length VP2 gene in domestic cats between 2006–2008 and 2012–2014, Portugal. Transbound Emerg Dis. (2017) 64:1178–83. doi: 10.1111/tbed.12483
16. Soma, T, Ogata, M, Ohta, K, Yamashita, R, and Sasai, K. Prevalence of astrovirus and parvovirus in Japanese domestic cats. J Vet Med Sci. (2020) 82:1243–6. doi: 10.1292/jvms.20-0205
17. Dall’Ara, P, Labriola, C, Sala, E, Spada, E, Magistrelli, S, and Lauzi, S. Prevalence of serum antibody titres against feline panleukopenia, herpesvirus and calicivirus infections in stray cats of Milan, Italy. Prev Vet Med. (2019) 167:32–8. doi: 10.1016/j.prevetmed.2019.03.010
18. Barrs, VR. Feline panleukopenia: a re-emergent disease. Vet Clin North Am Small Anim Pract. (2019) 49:651–70. doi: 10.1016/j.cvsm.2019.02.006
19. Tang, Y, Tang, N, Zhu, J, Wang, M, Liu, Y, and Lyu, Y. Molecular characteristics and genetic evolutionary analyses of circulating parvoviruses derived from cats in Beijing. BMC Vet Res. (2022) 18:195. doi: 10.1186/s12917-022-03281-w
20. Mochizuki, M, and Hashimoto, T. Growth of feline panleukopenia virus and canine parvovirus in vitro. Nihon Juigaku Zasshi. (1986) 48:841–4. doi: 10.1292/jvms1939.48.841
21. Jiao, C, Zhang, H, Liu, W, Jin, H, Liu, D, Zhao, J, et al. Construction and immunogenicity of virus-like particles of feline parvovirus from the tiger. Viruses. (2020) 12:315. doi: 10.3390/v12030315
22. Ferri, F, Porporato, F, Rossi, F, Enache, D, Callegari, C, Gerardi, G, et al. Treatment with class a CpG oligodeoxynucleotides in cats with naturally occurring feline parvovirus infection: a prospective study. Viruses. (2020) 12:640. doi: 10.3390/v12060640
23. Cheat, S, Gerez, J, Cognié, J, Alassane-Kpembi, I, Bracarense, A, Raymond-Letron, I, et al. Nivalenol has a greater impact than deoxynivalenol on pig jejunum mucosa in vitro on explants and in vivo on intestinal loops. Toxins. (2015) 7:1945–61. doi: 10.3390/toxins7061945
24. Yang, J, Anishchenko, I, Park, H, Peng, Z, Ovchinnikov, S, and Baker, D. Improved protein structure prediction using predicted interresidue orientations. Proc Natl Acad Sci U S A. (2020) 117:1496–503. doi: 10.1073/pnas.1914677117
25. Gordon, JC, Myers, JB, Folta, T, Shoja, V, Heath, LS, and Onufriev, A. H++: a server for estimating pKas and adding missing hydrogens to macromolecules. Nucleic Acids Res. (2005) 33:W368–71. doi: 10.1093/nar/gki464
26. Maier, JA, Martinez, C, Kasavajhala, K, Wickstrom, L, Hauser, KE, and Simmerling, C. ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J Chem Theory Comput. (2015) 11:3696–713. doi: 10.1021/acs.jctc.5b00255
27. Harris, JA, Liu, R, Martins de Oliveira, V, Vazquez-Montelongo, EA, Henderson, JA, and Shen, J. GPU-accelerated all-atom particle-mesh Ewald continuous constant pH molecular dynamics in amber. J Chem Theory Comput. (2022) 18:7510–27. doi: 10.1021/acs.jctc.2c00586
28. Jing, Z, Ji, P, Wei, Y, Hao, F, and Wei, Y. Isolation and identification of a novel canine parvovirus type 2c strain in domestic cats in Dalian, China. Front Vet Sci. (2022) 9:1001604. doi: 10.3389/fvets.2022.1001604
29. Pan, S, Jiao, R, Xu, X, Ji, J, Guo, G, Yao, L, et al. Molecular characterization and genetic diversity of parvoviruses prevalent in cats in central and eastern China from 2018 to 2022. Front Vet Sci. (2023) 10:1218810. doi: 10.3389/fvets.2023.1218810
30. Chen, X, Wang, J, Zhou, Y, Yue, H, Zhou, N, and Tang, C. Circulation of heterogeneous carnivore protoparvovirus 1 in diarrheal cats and prevalence of an A91s feline panleukopenia virus variant in China. Transbound Emerg Dis. (2022) 69:e2913–25. doi: 10.1111/tbed.14641
31. Amoroso, MG, Serra, F, Miletti, G, Cardillo, L, de Martinis, C, Marati, L, et al. A retrospective study of viral molecular prevalences in cats in southern Italy (Campania region). Viruses. (2022) 14:2583. doi: 10.3390/v14112583
32. Abdel-Baky, MMM, El-Khabaz, KAS, Abdelbaset, AE, and Hamed, MI. Clinico-epidemiological survey of feline parvovirus circulating in three Egyptian provinces from 2020 to 2021. Arch Virol. (2023) 168:126. doi: 10.1007/s00705-023-05751-4
33. Millan, J, and Rodriguez, A. A serological survey of common feline pathogens in free-living European wildcats (Felis silvestris) in central Spain. Eur J Wildl Res. (2009) 55:285–91. doi: 10.1007/s10344-008-0246-z
34. Parker, JS, and Parrish, CR. Canine parvovirus host range is determined by the specific conformation of an additional region of the capsid. J Virol. (1997) 71:9214–22. doi: 10.1128/JVI.71.12.9214-9222.1997
35. Garcia Rde, C, de Castro, TX, de Miranda, SC, Lopes, G Jr, de Lima, M, Labarthe, NV, et al. Characterization of parvoviruses from domestic cats in Brazil. J Vet Diagn Invest. (2011) 23:951–5. doi: 10.1177/1040638711417140
36. Lee, H, Callaway, HM, Cifuente, JO, Bator, CM, Parrish, CR, and Hafenstein, SL. Transferrin receptor binds virus capsid with dynamic motion. Proc Natl Acad Sci U S A. (2019) 116:20462–71. doi: 10.1073/pnas.1904918116
37. Organtini, LJ, Lee, H, Iketani, S, Huang, K, Ashley, RE, Makhov, AM, et al. Near-atomic resolution structure of a highly neutralizing fab bound to canine parvovirus. J Virol. (2016) 90:9733–42. doi: 10.1128/JVI.01112-16
38. Wen, JWZ, and Ren, J. Analysis of genetic variation of VP2 gene in 6 FPV strains in China. Adv Microbiol. (2021) 11:191–8. doi: 10.4236/aim.2021.114014
39. Mietzsch, M, Penzes, JJ, and Agbandje-McKenna, M. Twenty-five years of structural parvovirology. Viruses. (2019) 11:4. doi: 10.3390/v11040362
40. Wang, J, Cheng, S, Yi, L, Cheng, Y, Yang, S, Xu, H, et al. Evidence for natural recombination between mink enteritis virus and canine parvovirus. Virol J. (2012) 9:252. doi: 10.1186/1743-422x-9-252
41. Van Brussel, K, Carrai, M, Lin, C, Kelman, M, Setyo, L, Aberdein, D, et al. Distinct lineages of feline parvovirus associated with epizootic outbreaks in Australia, New Zealand and the United Arab Emirates. Viruses. (2019) 11:1155. doi: 10.3390/v11121155
42. Jacobson, LS, Janke, KJ, Ha, K, Giacinti, JA, and Weese, JS. Feline panleukopenia virus DNA shedding following modified live virus vaccination in a shelter setting. Vet J. (2022) 279:105783. doi: 10.1016/j.tvjl.2021.105783
43. Isaya, R, Ciccarelli, S, Enache, D, Specchi, S, Pesaresi, M, Ferri, F, et al. Gastrointestinal ultrasonographic findings in cats with feline panleukopenia: a case series. BMC Vet Res. (2021) 17:20. doi: 10.1186/s12917-020-02720-w
Keywords: feline parvovirus (FPV), VP2, infectivity, pathogenicity, China
Citation: Xie Q, Sun Z, Xue X, Pan Y, Zhen S, Liu Y, Zhan J, Jiang L, Zhang J, Zhu H, Yu X and Zhang X (2024) China-origin G1 group isolate FPV072 exhibits higher infectivity and pathogenicity than G2 group isolate FPV027. Front. Vet. Sci. 11:1328244. doi: 10.3389/fvets.2024.1328244
Edited by:
Lester J. Perez, Abbott, United StatesReviewed by:
Gianmarco Ferrara, University of Naples Federico II, ItalyGuixue Hu, Jilin Agriculture University, China
Feng Na, Chinese Academy of Agricultural Sciences (CAAS), China
Copyright © 2024 Xie, Sun, Xue, Pan, Zhen, Liu, Zhan, Jiang, Zhang, Zhu, Yu and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongwei Zhu, aG5nd3podUBvdXRsb29rLmNvbQ==; Xin Yu, eXV4aW56Z2huQGxkdS5lZHUuY24=; Xingxiao Zhang, emhhbmd4aW5neGlhb0BsZHUuZWR1LmNu
 Qiaoqiao Xie1,2,3
Qiaoqiao Xie1,2,3 Yang Liu
Yang Liu Jianlong Zhang
Jianlong Zhang Hongwei Zhu
Hongwei Zhu Xin Yu
Xin Yu Xingxiao Zhang
Xingxiao Zhang