
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci. , 04 December 2023
Sec. Veterinary Epidemiology and Economics
Volume 10 - 2023 | https://doi.org/10.3389/fvets.2023.1298434
 Wenchao Gao1,2,3,4†
Wenchao Gao1,2,3,4† Xiaoxue Jiang1,2,3,4†
Xiaoxue Jiang1,2,3,4† Zhiqiang Hu1,2,3,4
Zhiqiang Hu1,2,3,4 Qing Wang1,2,3,4
Qing Wang1,2,3,4 Yuntong Shi1,2,3,4
Yuntong Shi1,2,3,4 Xiaogang Tian1,2,3,4
Xiaogang Tian1,2,3,4 Mengli Qiao3,4,5
Mengli Qiao3,4,5 Jinyong Zhang3,4,5
Jinyong Zhang3,4,5 Yang Li1,2,3,4,5
Yang Li1,2,3,4,5 Xiaowen Li1,2,3,4,5,6*
Xiaowen Li1,2,3,4,5,6*Introduction: Pseudorabies virus (PRV) is a linear DNA virus with a double-stranded structure, capable of infecting a diverse array of animal species, including humans. This study sought to ascertain the seroprevalence of Pseudorabies Virus (PRV) in China by conducting a comprehensive collection of blood samples from 16 provinces over the course of 2022.
Methods: The presence of PRV gE antibodies was detected through the utilization of an enzyme-linked immunosorbent assay (ELISA) technique. Logistic regression analysis was conducted to identify potential related factors associated with the serologic status of PRV gE at the animal level. Additionally, the SaTScan 10.1 software was used to analyze the spatial and temporal clusters of PRV gE seroprevalence.
Results: A comprehensive collection of 161,880 samples was conducted, encompassing 556 swine farms throughout the country. The analysis revealed that the seroprevalence of PRV gE antibodies was 12.36% (95% confidence interval [CI], 12.20% to 12.52%) at the individual animal level. However, at the swine farm level, the seroprevalence was considerably higher, reaching 46.22% (95% CI, 42.08% to 50.37%). Related factors for PRV infection at the farm level included the geographic distribution of farms and seasonal variables. Moreover, five distinct high seroprevalence clusters of PRV gE were identified across China, with the peak prevalence observed during the months of April through June 2022.
Conclusion: Our findings serve as a valuable addition to existing research on the seroprevalence, related factors, and temporal clustering of PRV gE in China. Furthermore, our study provides a reference point for the development of effective strategies for the prevention and control of pseudorabies and wild virus outbreaks.
Pseudorabies virus (PRV) belongs to the herpesviral subfamily A, which also includes the varicella virus. PRV is the causative agent of pseudorabies (PR), which is more commonly known as Aujeszky's Disease (1). While PRV has the ability to infect various animal species, only pigs serve as the reservoir hosts for this virus (2–7). Upon PRV infection, pigs can exhibit varying clinical symptoms depending on their age at the time of infection. Newborn piglets primarily display neurological symptoms and have high mortality rates, while infected adult sows exhibit reproductive and respiratory disorders (8–10).
Since 2011, there has been a resurgence of PRV in multiple swine farms throughout China. This outbreak is mainly characterized by sow abortion, stillbirth, and increased piglet mortality (11). This resurgence can be attributed to the emergence of PRV variants, such as HN1201, TJ strains, and SDYC-2014 (12–14). Multiple studies have demonstrated that Bartha-K61 deletion strain vaccines are insufficient in providing comprehensive protection against these variants (13, 15).
Despite successful control or eradication of pseudorabies (PR) in many countries through vaccination, the epidemic remains prevalent in Chinese pigs (16). Despite immunization with the PR Bartha-K61 deficiency vaccine, numerous outbreaks of PRV have occurred (13, 17–21). In 2018, there was an outbreak of African swine fever in China, which had a significant impact on the country's swine farming industry. It has led to significant changes in herd distribution, farm biosecurity levels, and herd circulation strategies within the swine farming industry. Zhao et al. found that pig farms sampled after the ASF outbreak demonstrated a lower likelihood of PRV infection compared to those sampled before the outbreak (22). This could be attributed to enhanced biosafety management practices. As a result, the prevalence of PR has been affected (23, 24). There are reports on the serum prevalence of PRV in China before 2021, as well as related factors and spatiotemporal analysis, without relevant data for 2022. Therefore, in this study, more than 160,000 serum samples were collected in China in 2022, its aimed to analyze the current prevalence of pseudorabies and explore the spatiotemporal patterns. Additionally, conducting spatial-temporal analysis of PRV infection can help in identifying clusters with high PRV prevalence and understanding the trends of variation in PRV infection. This information can assist policymakers in designing more precise and cost-effective intervention policies for future PRV control in China.
In total, 161,880 serum samples were collected from 556 swine farms belonging to 106 companies across 16 provinces in China in 2022, covering six regions including Northeast China (Heilongjiang Province and Liaoning Province), Central South China (Henan, Hubei, Hunan, and Guangxi), East China (Jiangsu and Shandong), Northwest China (Gansu and Shaanxi), North China (Hebei, Inner Mongolia, Shanxi, and Tianjin), and Southwest China (Guizhou and Sichuan). Furthermore, the location coordinates of pig farms were obtained from Baidu Map (https://map.baidu.com/). The breeding farms, which had a breeding stock ranging from 750 to 3,000 sows, maintained a consistent herd composition. The consistent herd composition of the breeding farms included pre-weaning piglets (piglets that are 0–21 days old), gilts (female pigs that are between 90 and 230 days old), adult sows (over 230 days old, either in gestation or with a history of gestation), and boars (male pigs that are over 300 days old). The pigs in the fattening farm are divided into two categories: nursery piglets and growing-finishing pigs. Nursery piglets were between 21 and 70 days old, while growing-finishing pigs were between 70 and 180 days old. These farms had a production scale of over 6,000 pigs. All adult sows and boars were vaccinated three times a year with live PR vaccines (Bartha-K61 strain) and an inactivated vaccine (Bartha-K61 strain). All fattening pigs on the farms received vaccinations as part of their healthcare routine. At the age of 56 days, they were vaccinated with live PR vaccines. Then, at the age of 84 days, they received vaccinations using inactivated vaccines. For all gilts on the farms, at the age of 119 days, they received vaccinations with live PR vaccines. Subsequently, at the age of 147 days, they were vaccinated with inactivated vaccines. The sampling of pigs was conducted based on the scale and model of breeding. An approximately equal number of samples were collected from various growth stages, including suckling piglets, nursery pigs, fattening pigs, sows, and gilts. This approach ensured a representative distribution of samples across various age groups and production stages, enabling a thorough analysis of the swine being studied. In each season, the sampling frequency varied depending on the size of the farms. Approximately 50–60 serum samples were collected from each small farm (with fewer than 1,000 pigs), 70–100 serum samples from each medium-sized farm (with 1,000–2,000 pigs), and 100–150 serum samples from each large farm (with more than 3,000 pigs). This sampling approach aimed to ensure adequate representation and provide sufficient data for analysis across various farm sizes. All serum samples were collected and stored at −20°C.
Anti-gE antibody levels in serum were quantified using commercially available blocking ELISA Kits (Cat: CP144, IDEXX Laboratories, Westbrook, ME) in accordance with the manufacturer's instructions. Commercial blocking ELISA kits have been specifically developed to distinguish between the vaccine strain and wild strains of PR. In the study, the findings of the blocking ELISA test were reported in terms of sample/negative (S/N) values. A S/N value below 0.60 was deemed positive, suggesting the existence of anti-gE antibodies. Conversely, a S/N value exceeding 0.70 was considered to be indicative of an negative result, suggesting the lack of anti-gE antibodies. S/N values ranging from 0.60 to 0.70 were deemed questionable, necessitating additional testing or repeated testing over a period of time to ascertain the presence or absence of antibodies.
In this study, all the gathered data were entered and organized utilizing Microsoft Excel 2021, a widely used spreadsheet software developed by Microsoft in the United States. The associations between the seroprevalence of PRV-gE antibodies and various factors, such as regions, quarters, and pig herds, were analyzed using the logistic regression model in SPSS 26.0 software (IBM, Chicago, IL, USA). The study examined the 95% confidence intervals (CIs) of the results. In this study, statistical significance was determined by a p-value of <0.05.
A trend analysis of seroprevalence for PRV gE was conducted by following the steps outlined below. A pig farm was deemed to be positive for wild-type PRV if at least one sample tested positive for gE antibodies. The serological status of a swine farm with regards to PRV was considered as a dichotomous variable, indicating that it was categorized as either PRV-positive or PRV-negative. Spatio-temporal scanning was performed utilizing the Poisson distribution model. The researchers employed SaTScan version 10.1 software to forecast the spatial-temporal distribution of high PRV gE seroprevalence aggregation using the Bernoulli model. The samples obtained from each pig farm were categorized into two groups by the researchers. The first group, referred to as the experimental group, comprised of samples that tested positive for PRV gE antibodies. The second group, known as the control group, consisted of samples that tested negative for PRV gE antibodies. Time aggregation was conducted on a monthly basis, encompassing the entire duration of the experiment, which spanned from January 1, 2022, to December 31, 2022. When the P-value of the log-likelihood ratio (LLR) test is <0.05, the region is deemed to demonstrate aggregation. In addition, the creation of maps was facilitated through the utilization of ArcGIS 10.7 software, developed by ESRI, USA.
Between the period of January 2022 and December 2022, an extensive collection of 161,880 blood samples was conducted from various sources within China's swine farms. Specifically, the samples were collected from a total of 556 swine farms, which are owned by 106 different companies across 16 provinces (see Figure 1). Out of the total of 556 swine farms surveyed, a significant proportion of 257 farms were found to have tested positive for PRV gE antibodies. According to the data collected in this study, the prevalence of PRV gE seropositivity at the animal level demonstrated significant variation, with a range of 0–100% observed within each farm (Figure 2). The overall prevalence of PRV-gE antibodies in serum samples was found to be 12.36% (20,009/161,880, 95% CI 12.20–12.52%), with a farm positivity rate of 46.22% (257/556, 95% CI 42.08–50.37%) (Table 1).
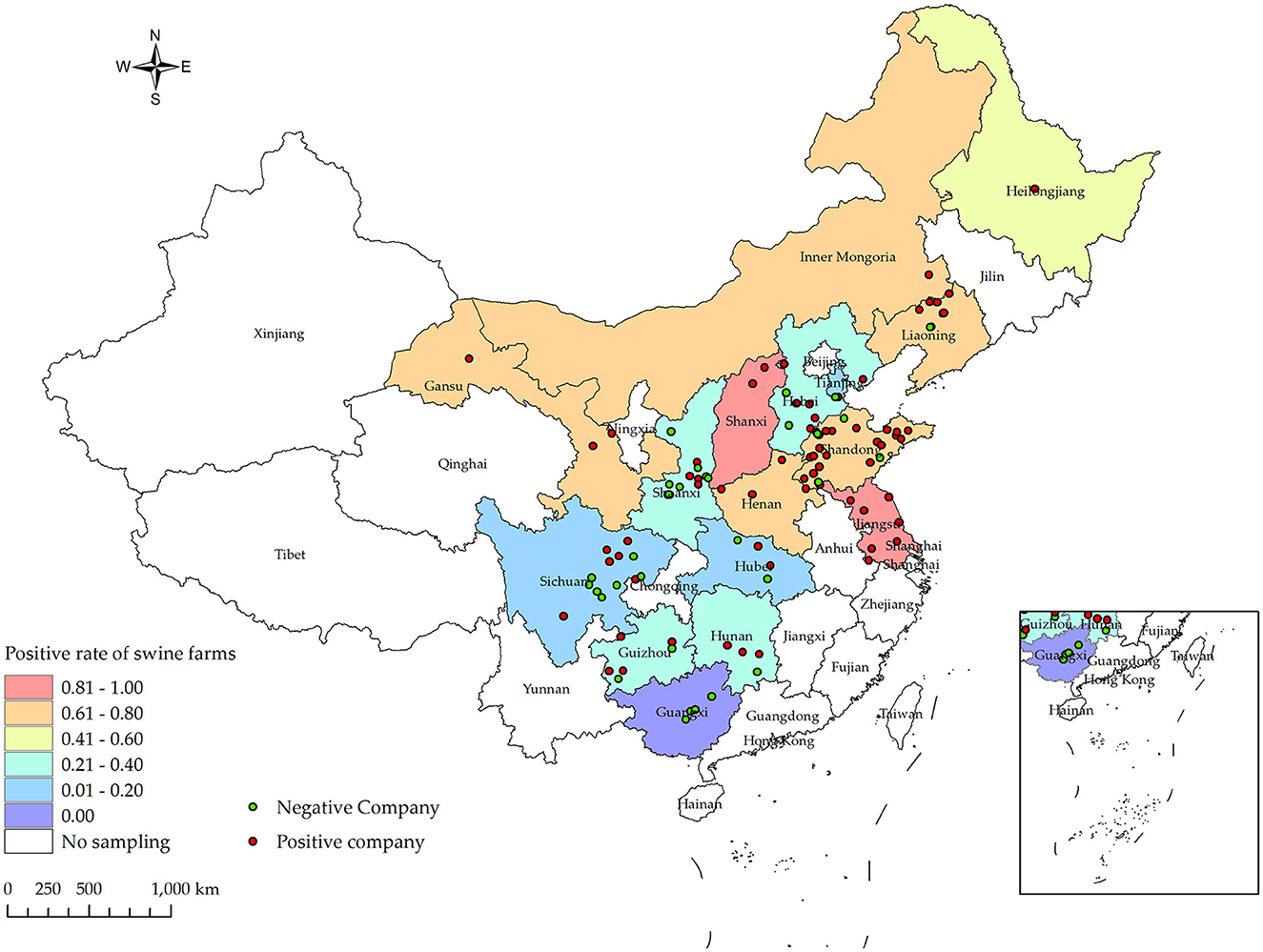
Figure 1. Distribution of PRV gE infection rates at the animal level for samples collected from January 2022 to December 2022 in various provinces and cities in China, along with their respective collection locations. Different shades of box colors represent infection rates, green dots indicate PRV gE antibody-negative companies, and red dots indicate PRV gE antibody-positive companies.
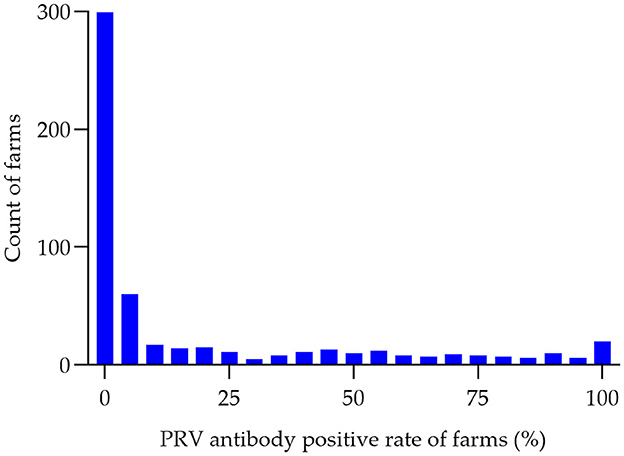
Figure 2. Histogram of PRV antibody-positive rates (%) in pig farms. The seropositivity rate of PRV was determined for each of the 556 swine farms in 2022. The number of swine farms falling within each positivity range was then counted in 5% increments, ranging from 0 to 100%. This data was used to create a histogram, with the horizontal axis representing the positivity range (in groups of 5%) and the vertical axis representing the number of swine farms.
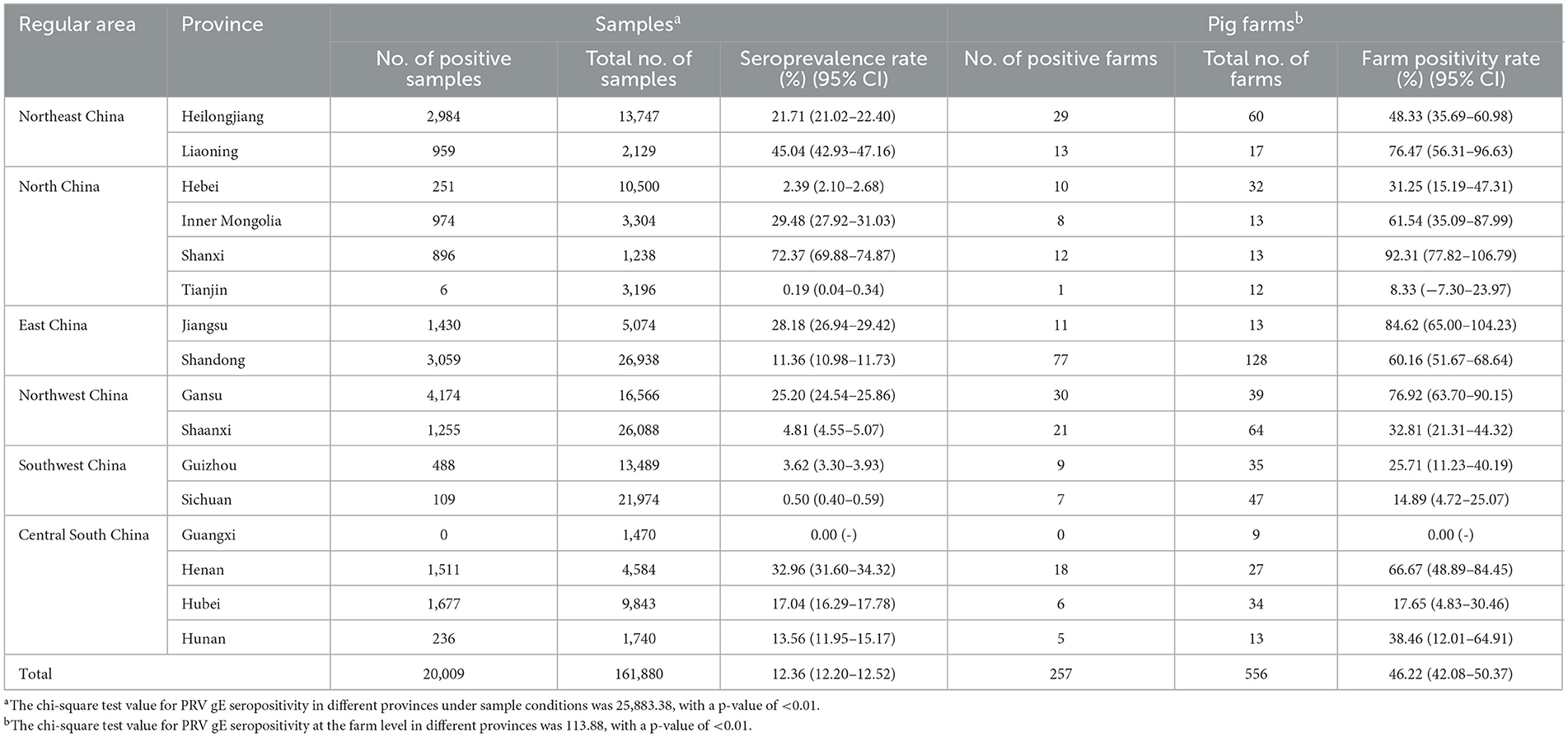
Table 1. Prevalence of PRV gE antibody determined by the Pearson chi-square test in the provinces of China.
At the animal level, the provinces exhibiting higher rates of PRV gE antibodies are Shanxi, Liaoning, and Henan, with prevalence rates of 72.37% (95% CI, 69.88–74.87%), 45.04% (95% CI, 42.93–47.16%), and 32.96% (95% CI, 31.60–34.32%), respectively. In contrast, the prevalence of PRV gE antibodies in three provinces (Sichuan Province, Tianjin City, and Guangxi Autonomous Region) is found to be <1%. The chi-square test for PRV gE serum numbers indicated a significant difference between the provinces (ranging from 0.00 to 72.37%), with a P-value of <0.01 (Table 1). At the farm level, the positivity rates of PRV gE antibodies were highest in Shanxi and Jiangsu provinces, at 92.31% (95% CI, 77.82–106.79%), 84.62% (95% CI, 65.00–104.23%), respectively. In contrast, the lowest rates of PRV gE antibody positivity are 0.00% in the Guangxi provinces. The chi-square test for PRV gE farm numbers indicated a significant difference between the provinces (ranging from 0.00 to 92.31%), with a P-value of <0.01 (Table 1).
The Northeast region had the highest seroprevalence in serum samples at 24.84% (3,943/15,876, 95% CI 24.16–25.51%). On the other hand, the lowest seroprevalence was observed in the Southwest region at 1.68% (597/35,463, 95% CI 1.55–1.82%). The East region had the highest positive rate of pig farms at 62.41% (88/141, 95% CI 54.42–70.41%). On the other hand, the Southwest region had the lowest positive rate at 19.51% (16/82, 95% CI 10.93–28.09%). Furthermore, significant variations in the positive rates of sera were observed across different regions (chi-square test, p < 0.01) (Table 2).
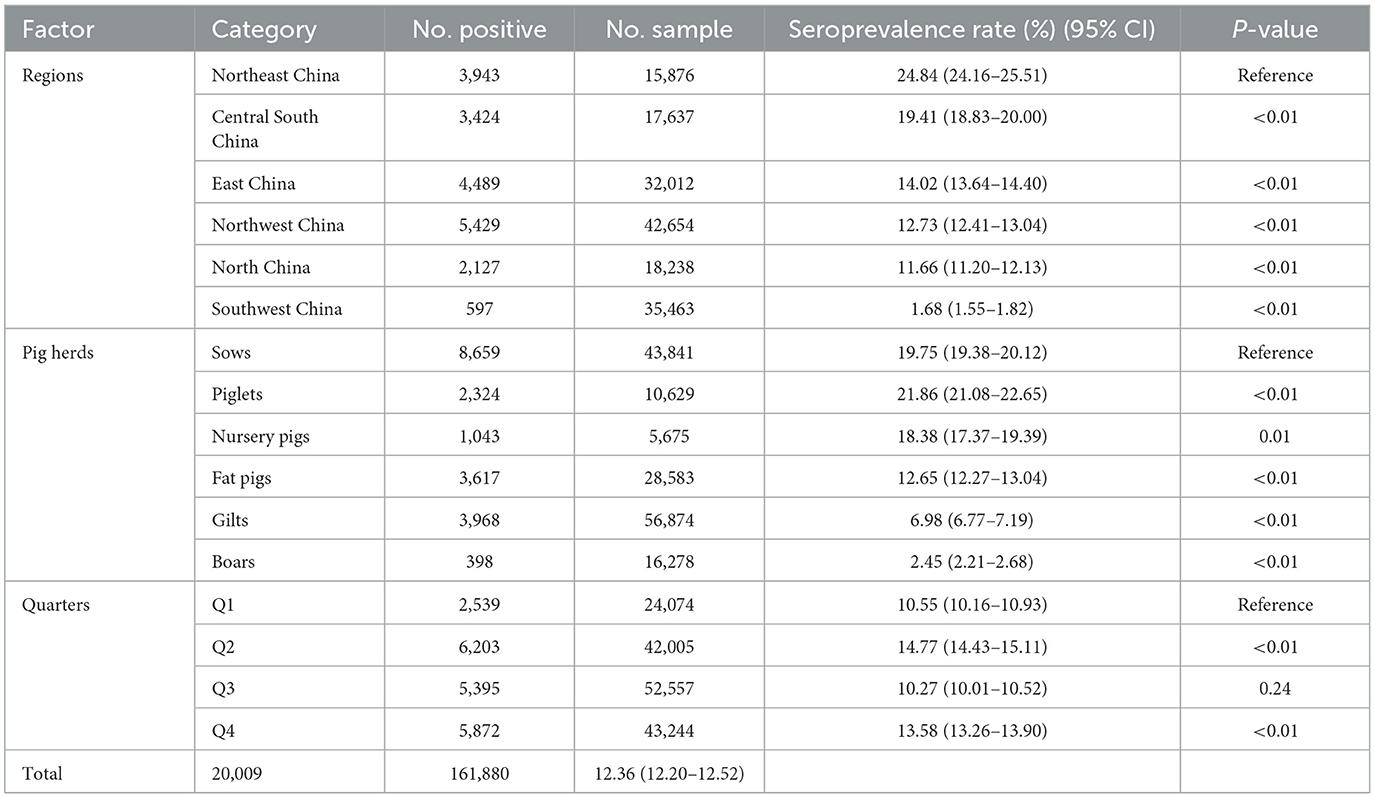
Table 2. The Chi-square test of related factors associated with PRV serological status at the sample level.
In breeding farms, pigs are categorized into piglets, gilts, sows, and boars based on their growth stages. Fattening farms, on the other hand, house nursery pigs and fattening pigs. As indicated in Table 2, the seroprevalence of serum samples was highest in piglet herds at 21.86% (2,324/10,629, 95% CI 21.08–22.66%), and lowest in boar herds at 2.45% (398/16,278, 95% CI 2.21–2.68%). Furthermore, significant differences were observed between sows and other herds (chi-square test, p < 0.05). Interestingly, there was a linear downward trend in seroprevalence from piglets to gilts (Figure 3).
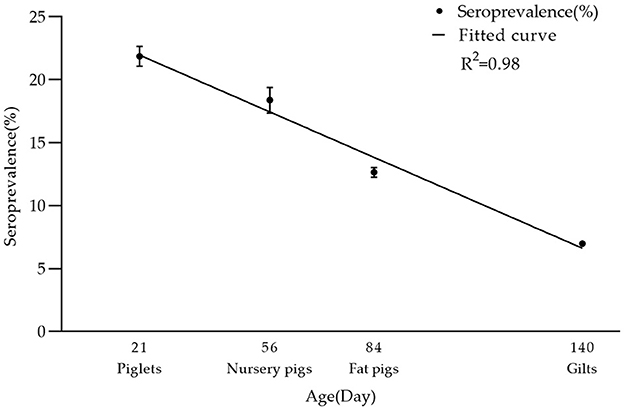
Figure 3. Seroprevalence rate of PRV-gE in different age groups. There was a linear decrease in seropositivity of serum samples from piglets, nursery pigs, fat pigs, to gilts: 21.86% (2,324/10,629, 95% CI 21.08–22.66%), 18.38% (1,043/5,675, 95% CI 17.37–19.39%), 12.65% (3,617/28,583, 95% CI 12.27–13.04%), 6.98% (3,968/56,874, 95% CI 6.77–7.19%). The R2 value of its trendline is 0.98.
In the present investigation, the initial quarter encompasses the months of January, February, and March. The second quarter encompasses the months of April, May, and June. The third quarter encompasses the months of July, August, and September. The fourth quarter, which includes the months of October, November, and December, marks the final period of the year. In terms of quarters, the seropositive rates of the first quarter (Q1) and the third quarter (Q3) were significantly lower than those of the second quarter (Q2) and the fourth quarter (Q4) (chi-square test, p < 0.05). Q2 had the highest seropositive rate of 14.77% (6,203/42,005, 95% CI 14.43–15.11%) (Table 2).
In the chi-square test result (Table 2), it was observed that variables such as regions, pig herds, and quarters had P-values of <0.05. Consequently, these variables were included in the multiple logistic regression model. Three related factors associated with the serological status of PRV were identified through multivariate logistic analysis (Table 3). The population density in North China, East China, Northwest, Southwest, and Central South regions is comparatively lower than that in Northeast China. In comparison to the Northeast region, the Southwest region exhibited a significantly lower probability of PRV infection in pigs, with an odds ratio of 0.05 (95% CI, 0.04–0.05). The probability of PRV infection in boars (OR, 0.09; 95% CI, 0.08–0.10), fattening pigs (OR, 0.09; 95% CI, 0.08–0.10), and gilts (OR, 0.32; 95% CI, 0.30–0.33) was significantly lower than in sows. Additionally, the likelihood of PRV infection was found to be higher in Q2, Q3, and Q4 compared to Q1. The probability of PRV infection in pigs during the second quarter is 1.65 times higher (95% CI, 1.57–1.74) compared to the first quarter.
In Figure 4 and Table 4, the survey results show the high serum prevalence of five clusters of PRV gE found in China from January 2022 to December 2022. The first cluster was located at latitude 42.313505° N, longitude 121.849035° E, with a radius of 92.88 km. It spanned from June 1, 2022, to November 30, 2022, and had a relative risk value of 6.50 and a log-likelihood ratio (LLR) value of 2,037.92 (p < 0.001). The second cluster was located at latitude 32.183738° N, longitude 111.832792° E, with a radius of 247.57 km. It occurred from December 1, 2022, to December 31, 2022, with a relative risk value of 6.54 and an LLR value of 1,675.08 (p < 0.001). The third cluster is the first large area with a radius of 297.86 km, located at latitude 34.040922° N and longitude 118.058937° E. It spans from May 1, 2022, to October 31. The relative risk value is 2.97, and the LLR value is 927.48 (p < 0.001). The fourth cluster is situated at coordinates 39.226308° N and 112.643375° E, covering an area with a radius of 95.62 km. The cluster encompasses the time period from April 1, 2022, to September 30, 2022. The relative risk value was found to be 7.07, while the LLR value was calculated to be 927.34 (p < 0.001). The fifth cluster was identified at geographical coordinates 37.074554° N, 104.895751° E, encompassing an area with a radius of 113.76 km. The time range of the cluster spans from August 1, 2022, to December 31, 2022. The relative risk value was 2.94, and the likelihood ratio value was 898.77 (p < 0.001). Meanwhile, Figure 5 and Table 5 show a high seropositivity rate of PRV gE during the period from April 2022 to June 2022. The relative risk value was 1.27 and the LLR value was 120.88 (p < 0.01).
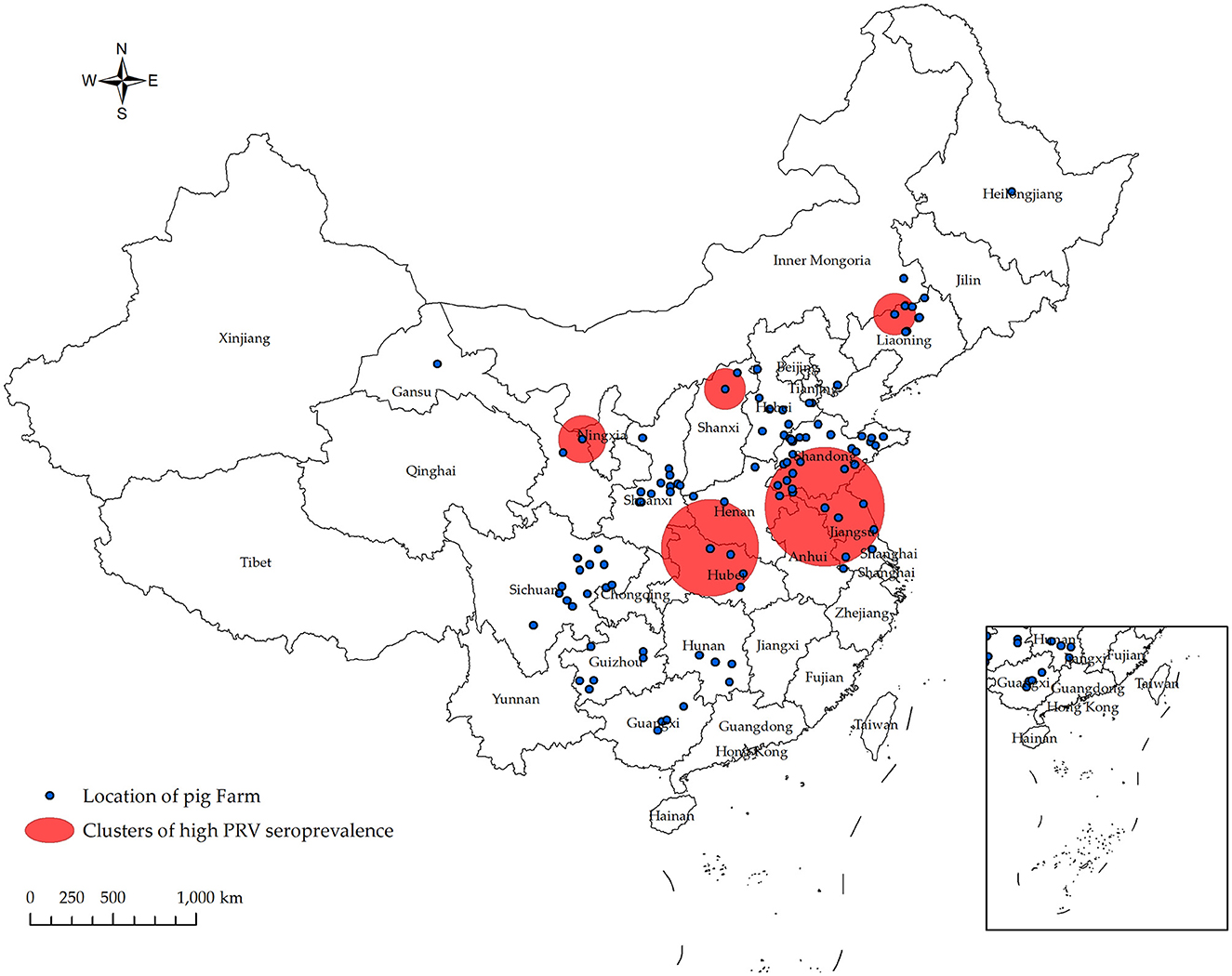
Figure 4. Significant spatial clusters (p < 0.05) of high PRV gE seroprevalence were observed in China from January 2022 to December 2022 with a maximum window size of 50% of the population at risk. Using SaTScan 10.1 software, the PRV gE antibody status of 556 pig farms from 106 companies was analyzed. The Bernoulli model was used to predict the temporal and spatial distribution of PRV gE serum high prevalence aggregation. From January 2022 to December 2022, five populations of PRV gE with a high seroprevalence were identified in China.
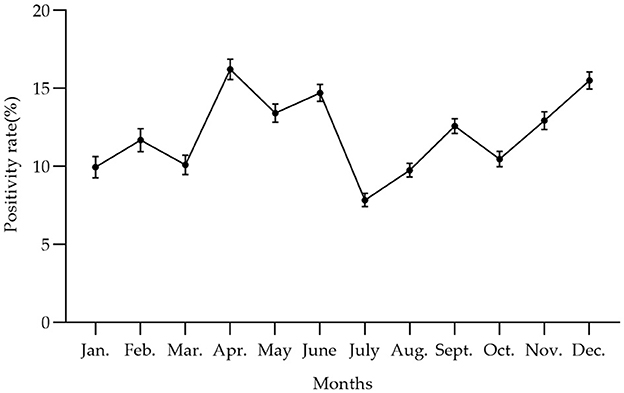
Figure 5. The trend of PRV gE antibody positivity rate at the animal level varied by month from January 2022 to December 2022 in China, with the peak of PRV gE seroprevalence occurring from April to June. The positivity rates of the PRV gE antibody from January to December were 9.96, 11.69, 10.11, 16.23, 13.42, 14.71, 7.84, 9.76, 12.59, 10.48, 12.94, and 15.51%, respectively.
The PRV-gE deletion vaccine has demonstrated efficacy in preventing PRV and has been used in both large-scale and small-scale Chinese farms for an extended period (25). Consequently, the presence of PRV-gE antibodies serves as an indicator of the prevalence of wild-type PRV strains. Previous investigations have indicated that the prevalence of PRV-gE antibodies in pig farms in China has exhibited a gradual increase from 2011 to 2016, followed by a subsequent decline from 2016 to 2021. Notably, the overall positive rate of PRV-gE antibodies has remained at ~20% over the past 2 years (26–35).
In this study, the objective was to evaluate the existing seroprevalence of wild-type PRV gE in China by collecting blood samples from pigs from January 2022 to December 2022. We conducted a series of tests to detect antibodies against PRV gE. This study encompassed an analysis of 161,880 serum samples obtained from 556 pig farms of different scales (large, medium, and small) located in 16 provinces throughout China. We conducted an analysis on the spatiotemporal distribution characteristics of the infection rate of wild-type PRV strains in China, and successfully identified the associated related factors.
In the conducted survey, the collective serum samples exhibited an overall positive rate of PRV gE antibodies at 12.36%, while the positive rate in farms was significantly higher at 46.22%. There are significant variations in the rates of antibody positivity and farm positivity among pig herds in different provinces. This suggests that the prevalence of pseudorabies wild virus infection varies across regions. Chen et al. (33) conducted an analysis on a total of 35,796 serum samples collected in Henan Province between 2019 and 2021. They observed a decline in the prevalence of PRV gE antibodies from 25 to 16.69%. Similarly, Zhou et al. (27) found that the positive rate in Heilongjiang Province from 2013 to 2018 was 16.3%. This study revealed that the prevalence of positivity in Henan and Heilongjiang provinces was 32.96% (1,511/4,584, 95% CI 30.59–35.33%) and 21.71% (2,984/13,747, 95% CI 21.02–22.40%), respectively. These rates indicate a higher prevalence compared to the study conducted by Zhou et al. (27) and Chen et al. (33). This phenomenon could potentially be associated with variables such as the quantity of serum samples collected and the number of pig farms included in the study. The serological survey of PRV in Hunan Province, conducted by Lin et al. from 2016 to 2020, revealed a PRV-gE antibody positivity rate of 23.55% (4,271/18,138, 95% CI 22.9–24.2%) (28). In the present study, the prevalence of PRV gE antibody positivity in Hunan Province was found to be 13.56% (236/1,740, 95% CI 11.95–15.17%), which is significantly lower than the findings reported by Lin et al. (28) in their survey. Similarly, in previous reports, the serum positive rate in Shandong Province from 2013 to 2016 was recorded at 57.8% (2,909 out of 5,033) (26). In the case of Tianjin, the positive rate from 2010 to 2018 was reported as 46.70% (34). Lastly, Hebei Province recorded a positivity rate of 46.27% from 2017 to 2018 (29). In the current investigation, the prevalence rates recorded in Shandong (11.36%), Tianjin (0.19%), and Hebei (17.04%) were determined to be lower compared to previously documented rates. This phenomenon can be ascribed to alterations in the distribution of Chinese pig populations resulting from the African swine fever crisis. The continuous decline in the serum positivity rate of PRV gE antibodies indicates that significant advancements have been made in the country's efforts to manage the transmission of wild pseudorabies virus infection.
Sows infected with PRV have the ability to transmit maternal antibodies to their offspring via colostrum, which can persist in piglets for a duration of 12–14 weeks (36). The present investigation revealed that the seroprevalence was significantly higher in piglets (21.86%) compared to sows (19.75%). However, the odds ratio (OR) value of piglets relative to sow infection was 1.04 (0.98–1.10), and the p-value was >0.05. This difference may be attributed to the presence of positive maternal antibodies against PRV-gE in sows. Notably, the seropositive rate of pigs declined gradually with age, from nurseries to gilts, indicating the effectiveness of current strategies in preventing wild-type PRV strains. While the seropositive rate of sows is still high, there is a need to develop more effective strategies in the future to specifically target sows and pre-wean piglets.
From a geographical perspective, the rates of serum positivity for PRV are higher in the Northeast and Central South regions compared to other regions. The Northeast region has the highest likelihood of PRV infection, while the Southwest region has the lowest likelihood. This result is slightly different from the research findings of Liu et al. (37). A study on the prevalence of PRV in Chinese pig farms from 2013 to 2016 showed that in 2016, the PRV-gE antibody serum positive rate was highest, and the risk was highest in North China, followed by the central and southern regions. It may be related to the variations in pig distribution across different regions and the increased frequency of updates on pig populations following the outbreak of African swine fever diseases. The positive rates of PRV gE antibodies in the six regions studied showed significant differences (p < 0.01), indicating variations in PRV wild-type virus infection rates across the country.
On the spatiotemporal analysis of PRV serum positivity rate, Allepuz et al. (38) conducted a study to analyze the spatial distribution of PRV incidence rate among pig farms in Catalonia, Spain, from 2003 to 2007. The study aimed to determine the clustering of PRV infection in both sows and fattening pig farms. Berke et al. (39) detected the spatial aggregation of two PRV infections in Germany, with radii of 2.6 and 1.7 km, and relative risk values of 2.4 and 3.3, respectively. Zhao et al. (22) detected five significant high PRV gE seroprevalence groups in China for the first time in their data from 2017 to 2021. However, research on PRV spatiotemporal clustering in China is still relatively limited. As Zhao et al. (40) stated in his study, conducting spatiotemporal analysis of PRRSV infections can assist in identifying clusters with high PRRSV prevalence and examining the evolving patterns of PRRSV infections. This analysis can aid policy makers in developing more accurate and cost-effective interventions for future PRRSV control in China. Similarly, conducting spatiotemporal analysis of pseudorabies infections is also significant in this regard.
From a quarterly perspective, the seroprevalence rate of PRV-gE antibodies fluctuated throughout the year, reaching its peak in the second quarter (Q2), and Q2 has the highest risk of infection. This finding is consistent with a previous report (33). This could be attributed to the significant temperature variation between day and night during this period.
We conducted a seroepidemiologic survey for wild type PRV gE in China from January 2022 to December 2022. During this period, we collected 161,880 blood samples from 556 swine farms belonging to 106 companies across 16 provinces. These samples were then tested for PRV gE antibodies using ELISA. At the animal level and the farm level, the overall seroprevalence of PRV gE was 12.36% (95% CI, 12.20–12.52%) and 46.22% (95% CI, 42.08–50.37%), respectively. In addition, the seroprevalence of PRV varied significantly among provinces and herds. We conducted a comprehensive study utilizing logistic regression analysis to identify the related factors associated with Pseudorabies virus (PRV) serologic status on farms. These factors include the geographic location of the farm, herd type, and season. We identified five distinct PRV gE seroprevalence clusters in China from April 1, 2022, to December 31, 2022. Additionally, we found that the months with higher PRV gE seropositivity in China in 2022 were from April to June. Our findings serve as a valuable addition to existing research on the seroprevalence, related factors, and temporal clustering of PRV gE in China. Furthermore, our study provides a reference point for the development of effective strategies for the prevention and control of pseudorabies and wild virus outbreaks.
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
The animal studies were approved by the Animal Welfare Guidelines of the World Organization for Animal Health. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent was obtained from the owners for the participation of their animals in this study.
WG: Conceptualization, Data curation, Investigation, Software, Visualization, Writing—original draft. XJ: Data curation, Investigation, Software, Visualization, Writing—original draft. ZH: Validation, Writing—review & editing. QW: Data curation, Writing—original draft. YS: Data curation, Writing—original draft. XT: Investigation, Writing—original draft. MQ: Investigation, Writing—original draft. JZ: Writing—review & editing. YL: Validation, Writing—review & editing. XL: Conceptualization, Formal analysis, Funding acquisition, Methodology, Project administration, Resources, Supervision, Validation, Writing—review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was funded by the Taishan Industry Leadership Talent Project of Shandong Province in China, and the earmarked fund for CARS (CARS-35).
We would like to thank New Hope Liuhe Co., Ltd. for providing us with the testing information.
WG, XJ, ZH, QW, YS, XT, YL, and XL were employed by Shandong New Hope Liuhe Co., Ltd. WG, XJ, ZH, QW, YS, XT, MQ, JZ, YL, and XL were employed by New Hope Liuhe Co., Ltd. XL was employed by Shandong New Hope Liuhe Agriculture and Animal Husbandry Technology Co., Ltd.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Mettenleiter TC. Aujeszky's disease (pseudorabies) virus: the virus and molecular pathogenesis—state of the art, June 1999. Vet Sci. (2000) 31:99–115. doi: 10.1051/vetres:2000110
2. Muller T, Hahn EC, Tottewitz F, Kramer M, Klupp BG, Mettenleiter TC, et al. Pseudorabies virus in wild swine: a global perspective. Arch Virol. (2011) 156:1691–705. doi: 10.1007/s00705-011-1080-2
3. Liu A, Xue T, Zhao X, Zou J, Pu H, Hu X, et al. Pseudorabies virus associations in wild animals: review of potential reservoirs for cross-host transmission. Viruses. (2022) 14:2254. doi: 10.3390/v14102254
4. Moreno A, Sozzi E, Grilli G, Gibelli LR, Gelmetti D, Lelli D, et al. Detection and molecular analysis of pseudorabies virus strains isolated from dogs and a wild boar in Italy. Vet Microbiol. (2015) 177:359–65. doi: 10.1016/j.vetmic.2015.04.001
5. Liu Q, Wang X, Xie C, Ding S, Yang H, Guo S, et al. A novel human acute encephalitis caused by pseudorabies virus variant strain. Clin Infect Dis. (2021) 73:e3690–e700. doi: 10.1093/cid/ciaa987
6. Verpoest S, Cay AB, De Regge N. Molecular characterization of belgian pseudorabies virus isolates from domestic swine and wild boar. Vet Microbiol. (2014) 172:72–7. doi: 10.1016/j.vetmic.2014.05.001
7. Cheng Z, Kong Z, Liu P, Fu Z, Zhang J, Liu M, et al. Natural infection of a variant pseudorabies virus leads to bovine death in China. Transbound Emerg Dis. (2020) 67:518–22. doi: 10.1111/tbed.13427
8. Pomeranz LE, Reynolds AE, Hengartner CJ. Molecular biology of pseudorabies virus: impact on neurovirology and veterinary medicine. Microbiol Mol Biol Rev. (2005) 69:462–500. doi: 10.1128/MMBR.69.3.462-500.2005
9. Ming F, Yang J, Chu P, Ma M, Shi J, Cai H, et al. Immunization of aged pigs with attenuated pseudorabies virus vaccine combined with CpG oligodeoxynucleotide restores defective Th1 immune responses. PLoS ONE. (2013) 8:e65536. doi: 10.1371/journal.pone.0065536
10. Yu X, Sun Q, Ku X, He D, Li Z, Ghonaim AH, et al. The epidemiological investigation of co-infection of major respiratory bacteria with pseudorabies virus in intensive pig farms in China. Vet Med Sci. (2021) 7:175–83. doi: 10.1002/vms3.289
11. Ye C, Zhang QZ, Tian ZJ, Zheng H, Zhao K, Liu F, et al. Genomic characterization of emergent pseudorabies virus in China reveals marked sequence divergence: evidence for the existence of two major genotypes. Virology. (2015) 483:32–43. doi: 10.1016/j.virol.2015.04.013
12. Zhou M, Wu X, Jiang D, Sui C, Chen L, Cong X, et al. Characterization of a moderately pathogenic pseudorabies virus variant isolated in China, 2014. Infect Genet Evol. (2019) 68:161–71. doi: 10.1016/j.meegid.2018.12.016
13. Luo Y, Li N, Cong X, Wang CH, Du M, Li L, et al. Pathogenicity and genomic characterization of a pseudorabies virus variant isolated from Bartha-K61-vaccinated swine population in China. Vet Microbiol. (2014) 174:107–15. doi: 10.1016/j.vetmic.2014.09.003
14. Xiang S, Zhou Z, Hu X, Li Y, Zhang C, Wang J, et al. Complete genome sequence of a variant pseudorabies virus strain isolated in central China. Genome Announc. (2016) 4:e00149–16. doi: 10.1128/genomeA.00149-16
15. Tong W, Liu F, Zheng H, Liang C, Zhou YJ, Jiang YF, et al. Emergence of a pseudorabies virus variant with increased virulence to piglets. Vet Microbiol. (2015) 181:236–40. doi: 10.1016/j.vetmic.2015.09.021
16. Stegeman A. Pseudorabies virus eradication by area-wide vaccination is feasible. Vet Q. (1995) 17:150–6. doi: 10.1080/01652176.1995.9694556
17. An TQ, Peng JM, Tian ZJ, Zhao HY, Li N, Liu YM, et al. Pseudorabies virus variant in Bartha-K61-vaccinated pigs, China, 2012. Emerg Infect Dis. (2013) 19:1749–55. doi: 10.3201/eid1911.130177
18. Yan W, Li C, Yu X, He Q, Yang H. The Immunoprotection efficacy of Bartha K61 strain attenuated vaccine against pseudorabies variani strain and traditional strains. Chin J Vet Med. (2020) 56:15-18+23+2 [in Chinese].
19. Liu Q, Kuang Y, Li Y, Guo H, Zhou C, Guo S, et al. The epidemiology and variation in pseudorabies virus: a continuing challenge to pigs and humans. Viruses. (2022) 14:1463. doi: 10.3390/v14071463
20. Wang CH, Yuan J, Qin HY, Luo Y, Cong X, Li Y, et al. A novel gE-deleted pseudorabies virus (Prv) provides rapid and complete protection from lethal challenge with the PRV variant emerging in Bartha-K61-vaccinated swine population in China. Vaccine. (2014) 32:3379–85. doi: 10.1016/j.vaccine.2014.04.035
21. Wu R, Bai C, Sun J, Chang S, Zhang X. Emergence of virulent pseudorabies virus infection in Northern China. J Vet Sci. (2013) 14:363–5. doi: 10.4142/jvs.2013.14.3.363
22. Zhao P, Wang Y, Zhang P, Du F, Li J, Wang C, et al. Epidemiological investigation, risk factors, spatial-temporal cluster, and epidemic trend analysis of pseudorabies virus seroprevalence in China (2017 to 2021). Microbiol Spectr. (2023) 11:e0529722. doi: 10.1128/spectrum.05297-22
23. Zhou X, Li N, Luo Y, Liu Y, Miao F, Chen T, et al. Emergence of African swine fever in China, 2018. Transbound Emerg Dis. (2018) 65:1482–4. doi: 10.1111/tbed.12989
24. Gong L, Xu R, Wang Z, Deng Q, Wang H, Zhang G. African swine fever recovery in China. Vet Med Sci. (2020) 6:890–3. doi: 10.1002/vms3.299
25. Zhu G. Study and application of key technologies for pseudorabies control and eradication in large scale pig farms in Xinyang City (Master's degree). Henan Agricultural University, Zhengzhou, China (2020).
26. Gu J, Hu D, Peng T, Wang Y, Ma Z, Liu Z, et al. Epidemiological investigation of pseudorabies in Shandong Province from 2013 to 2016. Transbound Emerg Dis. (2018) 65:890–8. doi: 10.1111/tbed.12827
27. Zhou H, Pan Y, Liu M, Han Z. Prevalence of porcine pseudorabies virus and its coinfection rate in Heilongjiang Province in China from 2013 to 2018. Viral Immunol. (2020) 33:550–4. doi: 10.1089/vim.2020.0025
28. Lin Y, Tan L, Wang C, He S, Fang L, Wang Z, et al. Serological investigation and genetic characteristics of pseudorabies virus in Hunan Province of China from 2016 to 2020. Front Vet Sci. (2021) 8:762326. doi: 10.3389/fvets.2021.762326
29. Zhang C, Cui H, Zhang W, Meng L, Chen L, Wang Z, et al. Epidemiological investigation of porcine pseudorabies virus in Hebei Province, China, 2017-2018. Front Vet Sci. (2022) 9:930871. doi: 10.3389/fvets.2022.930871
30. Zhai X, Zhao W, Li K, Zhang C, Wang C, Su S, et al. Genome characteristics and evolution of pseudorabies virus strains in Eastern China from 2017 to 2019. Virol Sin. (2019) 34:601–9. doi: 10.1007/s12250-019-00140-1
31. Yao J, Li J, Gao L, He Y, Xie J, Zhu P, et al. Epidemiological investigation and genetic analysis of pseudorabies virus in Yunnan Province of China from 2017 to 2021. Viruses. (2022) 14:50895. doi: 10.3390/v14050895
32. Zheng HH, Bai YL, Xu T, Zheng LL, Li XS, Chen HY, et al. Isolation and phylogenetic analysis of reemerging pseudorabies virus within pig populations in central China during 2012 to 2019. Front Vet Sci. (2021) 8:764982. doi: 10.3389/fvets.2021.764982
33. Chen X, Li H, Zhu Q, Chen H, Wang Z, Zheng L, et al. Serological investigation and genetic characteristics of pseudorabies virus between 2019 and 2021 in Henan Province of China. Viruses. (2022) 14:1685. doi: 10.3390/v14081685
34. Zhang L, Ren W, Chi J, Lu C, Li X, Li C, et al. Epidemiology of porcine pseudorabies from 2010 to 2018 in Tianjin, China. Viral Immunol. (2021) 34:714–21. doi: 10.1089/vim.2021.0069
35. Tan L, Zhou Y, Qiu Y, Lei L, Wang C, Zhu P, et al. Pseudorabies in pig industry of China: epidemiology in pigs and practitioner awareness. Front Vet Sci. (2022) 9:973450. doi: 10.3389/fvets.2022.973450
36. Zimmerman JJ, Karriker LA, Ramirez A, Schwaetz KJ, Stevenson GW, Zhang J. Diseases of Swine. 11th, ed. Yang HC trans. Shenyang: Liaoning Science and Technology Press (2021). p. 566–77.
37. Liu Y, Zhang S, Xu Q, Wu J, Zhai X, Li S, et al. Investigation on pseudorabies prevalence in Chinese swine breeding farms in 2013-2016. Trop Anim Health Prod. (2018) 50:1279–85. doi: 10.1007/s11250-018-1555-1
38. Allepuz A, Saez M, Alba A, Napp S, Casal J. Exploratory spatial analysis of Aujeszky's disease during four phases of the eradication programme in Catalonia, Spain (2003-2007). Prev Vet Med. (2008) 86:164–75. doi: 10.1016/j.prevetmed.2008.04.005
39. Berke O, Grosse Beilage E. Spatial relative risk mapping of pseudorabies-seropositive pig herds in an animal-dense region. J Vet Med B Infect Dis Vet Public Health. (2003) 50:322–5. doi: 10.1046/j.1439-0450.2003.00689.x
Keywords: pseudorabies virus, seroprevalence, epidemiological investigation, related factors, spatial-temporal clustering, China
Citation: Gao W, Jiang X, Hu Z, Wang Q, Shi Y, Tian X, Qiao M, Zhang J, Li Y and Li X (2023) Epidemiological investigation, determination of related factors, and spatial-temporal cluster analysis of wild type pseudorabies virus seroprevalence in China during 2022. Front. Vet. Sci. 10:1298434. doi: 10.3389/fvets.2023.1298434
Received: 21 September 2023; Accepted: 13 November 2023;
Published: 04 December 2023.
Edited by:
Moh A. Alkhamis, Kuwait University, KuwaitReviewed by:
Xiangdong Li, Yangzhou University, ChinaCopyright © 2023 Gao, Jiang, Hu, Wang, Shi, Tian, Qiao, Zhang, Li and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaowen Li, bHh3ODI3MkAxNjMuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.