
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci. , 23 November 2023
Sec. Parasitology
Volume 10 - 2023 | https://doi.org/10.3389/fvets.2023.1296335
This article is part of the Research Topic Current Knowledge on Camelids Infectious and Parasitic Diseases View all 11 articles
 Igori Khatanbaatar1*
Igori Khatanbaatar1* Uranbileg Nyamdolgor2
Uranbileg Nyamdolgor2 Boldbaatar Chinchuluun2
Boldbaatar Chinchuluun2 Khandsuren Naranbaatar2
Khandsuren Naranbaatar2 Anja Taubert3
Anja Taubert3 Carlos R. Hermosilla3
Carlos R. Hermosilla3 Franz Suchentrunk4
Franz Suchentrunk4 Felix Knauer4
Felix Knauer4 Pamela A. Burger4*
Pamela A. Burger4* Gonchigoo Battsetseg2
Gonchigoo Battsetseg2Introduction: The two-humped Bactrian camel (Camelus bactrianus) is a large, even-toed ungulate native to the steppes of Central Asia. Domestic Bactrian camels are economically important in Mongolia and other Central Asian countries. These animals are used for transport, milk and meat production, and camel racing which is a great culture of nomads. Eimeriosis, also known as coccidiosis, is considered as an economically important parasitic diseases in Bactrian camels. There is still considerable lack of data concerning the spectrum of monoxenous Eimeria species, their epizootiology as well as their precise life cycles in Bactrian camels. This study was performed to determine the prevalence of Eimeria species in camelids from southern part of Mongolia.
Methods: A total of 536 fresh camel fecal samples (n = 536) collected from herds located in five different Aimags (provinces) of Mongolia were examined. Eimeria spp. oocysts were isolated using the sugar flotation technique, and after sporulation, oocysts were identified by morphometric evaluation.
Results: We identified the most common Eimeria species infecting Mongolian Bactrian camels: Eimeria cameli (22.3%), Eimeria rajasthani (37.3%) and Eimeria dromedarii (27.7%). Interestingly, mixed infections were detected in 24.8% (n = 133) of the samples, while 39.0% (n = 209) were negative for coccidian stages. To investigate the immunogenetic response of the Mongolian Bactrian camels to Eimeria spp. infection, we screened the genetic diversity in a functional important immune response gene of the major histocompatibility complex (MHC). We detected two polymorphic sites in the MHC class II DRA exon 2, which translated into one non-synonymous and one synonymous amino acid (aa) change.
Discussion: The resulting aa alleles were not significantly associated with any of the three detected Eimeria species infections, nor could we show heterozygote advantage in non-infected Mongolian Bactrian camels. Further investigations on molecular epidemiology, in vitro culture, pathogenicity and host–parasite interactions will be necessary to better understand the impact of eimeriosis in Bactrian camels.
Extant two-humped camel species are represented by the domestic (Camelus bactrianus) and wild (Camelus ferus) species. At present, wild camels are the only wild survivors of the Camelini tribe and inhabit northwestern China and southwestern Mongolia, especially within the Outer Altai Gobi Desert (1, 2). Domestic Bactrian camels are mainly distributed in the arid desert of Asian countries, such as Mongolia, China, Russia, Kazakhstan and Iran (3). These large animals are economically important in Mongolia where they are used for transport, entertainment (camel race, camel polo), and production of derived products such as fermented milk, meat, wool and skin (4), justifying the great socioeconomic importance of camels in the country.
Eimeriosis, also known as coccidiosis, is considered an important parasitic enteric disease of camels (5), but the occurrence of monoxenous (that lives within a single host during its whole life cycle) (6) Eimeria species and prevalence of eimeriosis is unknown in Bactrian camels in Mongolia. Among the five species known to infect Bactrian camels, i. e. Eimeria cameli, E. rajasthani, E. dromedarii, E. bactriani and E. pellerdyi. Nonetheless, E. cameli, E. rajasthani and E. dromedarii are considered as the most pathogenic species forming first generation macromeronts as reported for other highly pathogenic Eimeria species of domestic ruminants and New World camelids (7–10). Several studies showed the prevalence of different Eimeria species in camels. As such, Chineme (11) reported a case of dromedary (Camelus dromedarius) coccidiosis caused by E. cameli in Nigeria (11). In other studies, Kawasmeh and Elbihari (12), Yagoub (13), and Kasim et al. (14) found one or more species (E. rajasthani, E. dromedarii and E. cameli) with an overall prevalence of 14% in Saudi camels (C. dromedarius), 17.4% in Sudanese camels (C. dromedarius) and 41.6% in Saudi Arabian camels (C. dromedarius), respectively (12–14).
The report by Tafti et al. (15) indicated that the most important and frequent pathologic lesion in the digestive tract of camels is resulting from Eimeria spp. infections (63% of 100 slaughtered camels) (15). These pathological findings were in close agreement with reports from Hussein et al. (16), Kasim et al. (14) and Borji et al. (17) (14, 16, 17). Several cases of eimeriosis causing enteritis and mortality rates of up to 10% in young camels have been reported in few cases (11, 18, 19). Eimeria cameli, E. rajasthani, E. dromedarii are pathogenic to young camel calves causing enteritis (16). Infected young animals showed wasting, debility and diarrhea without mucus or blood. Older animals shedding oocysts in their faeces did not show any serious symptoms of eimeriosis (20). Considering the fact that all Eimeria infections are highly host and host cell-specific and that stage-specific innate as well as adaptive immune reactions are a common feature (21, 22), it appears essential that basic research is performed on different developmental stages in the respective hosts (8, 23).
Pathogen-mediated selection has been described as a driver for genetic diversity in host immune response genes, especially in the major histocompatibility complex (MHC). The MHC class I and class II genes are responsible for encoding molecules on the cell surface that recognize and present antigens (24). Therefore, these molecules are under strong selective pressure and have an important role for the adaptive immune response and for host-pathogen co-evolution (25). Studies in Inner Mongolian Brandt’s voles showed that MHC class II diversity is maintained by rare allele advantages and fluctuating selection, and that the association between intestinal parasite load and specific MHC class II DRB alleles varied between geographical regions (26).
In the three extant Camelini species (C. bactrianus, C. ferus, C. dromedarius), the MHC is located on chromosome 20, with the class II region located closer to the centromere and the class I more distant (27). In camels, unexpectedly low diversity has been described in the MHC class II loci (27) as well as in all functional different groups (adaptive and innate) of immune response genes (28). So far, no study about the immunogenetic response in camels to intestinal parasite infection has been conducted.
To fill this knowledge gap, the aim of this study was to determine the prevalence of Eimeria species in domestic Bactrian camels in southern Mongolia as well as their adaptive immunogenetic response to Eimeria spp. infections. Due to its functional importance, specifically in connection with parasite infection, we focused on the exon 2 coding sequence of the MHCII DRA locus and investigated pathogen-mediated immunogenetic diversity in Mongolian Bactrian camels infected or non-infected with Eimeria spp.
Fecal samples were collected from 536 Mongolian Bactrian camels in the Umnugobi-, Bayankhongor-, Uvurkhangai-, Dundgobi– and Khovd Aimags, chosen at random (Figure 1). Collected samples were put separately into closed plastic containers and identified with numbers. In addition, 100 EDTA peripheral blood samples were collected in parallel to the fecal sample collection. All experimental protocols were approved by the Animal Care and Use Committee, Institute of Veterinary Medicine, Mongolian University of Life Sciences (MULS) (Agreement Number № MEBUS-16/01/05). Samples from local provinces were taken from live animals with official permission and under the supervision of the Provincial Veterinary Organization in accordance with the regulation of the Animal ethic committee, MULS.
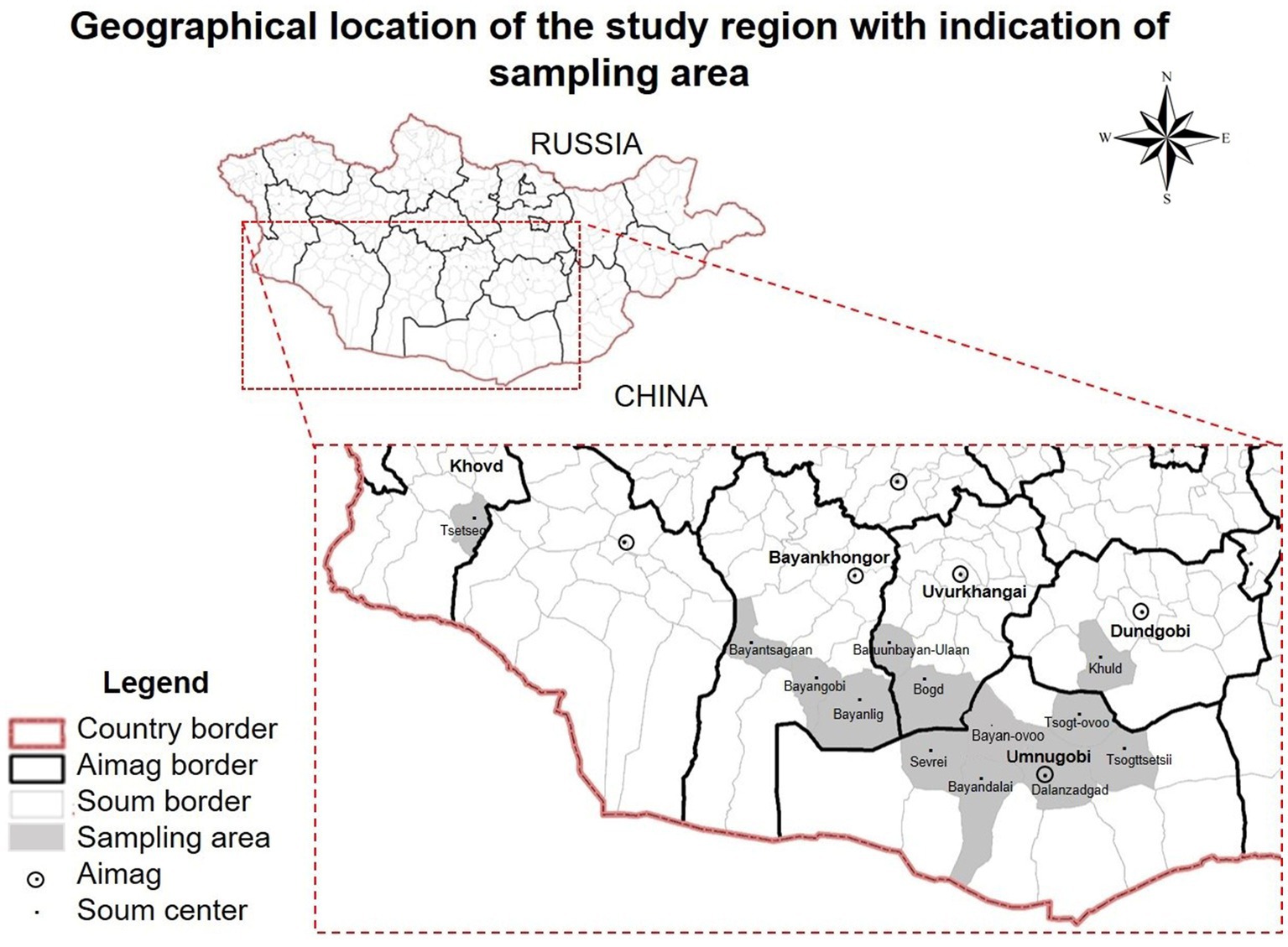
Figure 1. Geographical location of the study region with indication of Bactrian camel (Camelus bactrianus) sampling areas. Khovd, Bayankhongor, Uvurkhangai, Dundgobi, Umnugobi Aimags of Mongolia. This map has been provided by the ArcGIS 10.2 program.
The examined Bactrian camels followed traditional husbandry practices, with animals grazing during daytime. Camels were mainly crossbreeding and indigenous. For representative reasons, geographical locations of camels sampled are shown in Figure 1.
All fecal samples were examined by sugar flotation technique for isolation of camelid Eimeria spp. oocysts. Briefly, 3 g of fecal material were weighted and placed into a beaker and 15 mL of saturated sucrose solution (Sheather’s solution, specific gravity = 1.28) were added and homogenized. Thereafter, the fecal suspension was transferred into a 15 mL centrifuge tube and centrifuged at 2,000 rpm for 10 min at room temperature (RT) (29–37).
Samples were investigated by means of light microscopy and all oocysts within the microscope slides were considered in this study.
For species identification, oocysts from each individual sample were allowed to sporulate in 2.5% potassium dichromate under constant oxygenation (38). Eimeria species identification was based on the morphological and morphometric features of sporulated oocysts such as the size, shape, colour and texture of oocyst wall, presence or absence of micropyle, polar cap, among others, with the aid of taxonomic keys according to Levine and Ivens (1970), (6, 29, 39).
DNA was extracted using the QIAmp® blood mini kit (Qiagen, Vienna, Austria) from 100 EDTA peripheral blood samples following the manufacture’s instruction. The 246 bp long MHCII DRA exon 2 was amplified with the camel specific DNA primer pairs (DRA-ex2-F-TGAGAATTTTGGGTTTGCTTATGGCA/ DRA-ex2-R-CCTCTGAGCAACACG AACGTC CTTCA) with an annealing temperature of 57°C (27). The PCR reactions were performed in a reaction volume of 15 μL including 0.2 mM dNTPs, 25 mM MgCl2, 0.5 μM of forward and reverse primer, 1x Amplitaq Gold buffer, and 0.5 U of Amplitaq Gold Hotstart polymerase (ThermoFisher Scientific, Vienna, Austria). Please note that we tried to sequence also MHCII DRB exon 2 following Plasil et al. (27), however, only few samples yielded PCR products, which might be due to the longer amplicon of 852 bp (27). Successfully amplified PCR products were purified with FastAP™ and Exonuclease I (ThermoFischer Scientific) following the manufacture’s guide for PCR product clean-up prior to sequencing. The purified PCR products were Sanger sequenced in both directions with the BigDye™ Terminator v3.1 Cycle Sequencing Kit (ThermoFischer Scientific) on an ABI sequencer at the Research Institute of Wildlife Ecology, Vetmeduni Vienna, Austria. Sequences were visualised, aligned and translated into amino acids (aa) using CodonCode Aligner 11.0.1 (CodonCode Cooperation, Centerville, USA). We applied DNAsp 5.10.1 (40) to phase ambiguous (heterozygous) sequences and to determine haplotype (Hd) and nucleotide diversity (Pi).
To test for potential associations between Eimeria infection and the MHCII DRA exon2 aa alleles as well as for heterozygote advantage, we applied the generalized linear model with a logit link function in IBM® SPSS® Statistics version 29.0.1. (IBM Corp., Armonk, NY, USA). We tested six models with each of the three detected Eimeria species as binary dependent variable and the aa alleles, heterozygosity, gender and the respective other Eimeria spp. (Table 1) as binary, location (soum) as categorical, and age as continuous predictor variables, respectively. We applied strict Bonferroni correction for multiple model (k = 6) testing for a nominal alpha error of 0.05. In case of a significant effects of co-infection with two Eimeria species (p < 0.0083, Bonferroni corrected) we used Crosstabs statistics in SPSS to calculate the association coefficient Phi, which is a chi-square-based measure of association between nominal data (41).
In total, 327 fecal samples (n = 327) had Eimeria oocysts with an overall prevalence of 61% in Bactrian camels from Bayankhongor-, Uvurkhangai-, Umnugobi-, Dundgobi– and Khovd Aimags. Eimeria parasites were found in all five investigated Aimags (Figure 1). Three different camelid-specific Eimeria species were identified, being E. rajasthani 200 (37.3%) and E. dromedarii 149 (27.7%) the most prevalent ones, followed by E. cameli 120 (22.3%). In 209 samples (39%) no Eimeria oocysts were observed (Table 2; 3). Mixed Eimeria spp. infections with two or three Eimeria species, were detected in 133 samples (24.8%) (Table 3). Out of 327 positive samples, 194 (36.1%) samples presented single infection, 125 (23.3%) samples had two species, and only 9 (1.6%) samples had mixed infections with all three species (see Figure 2).
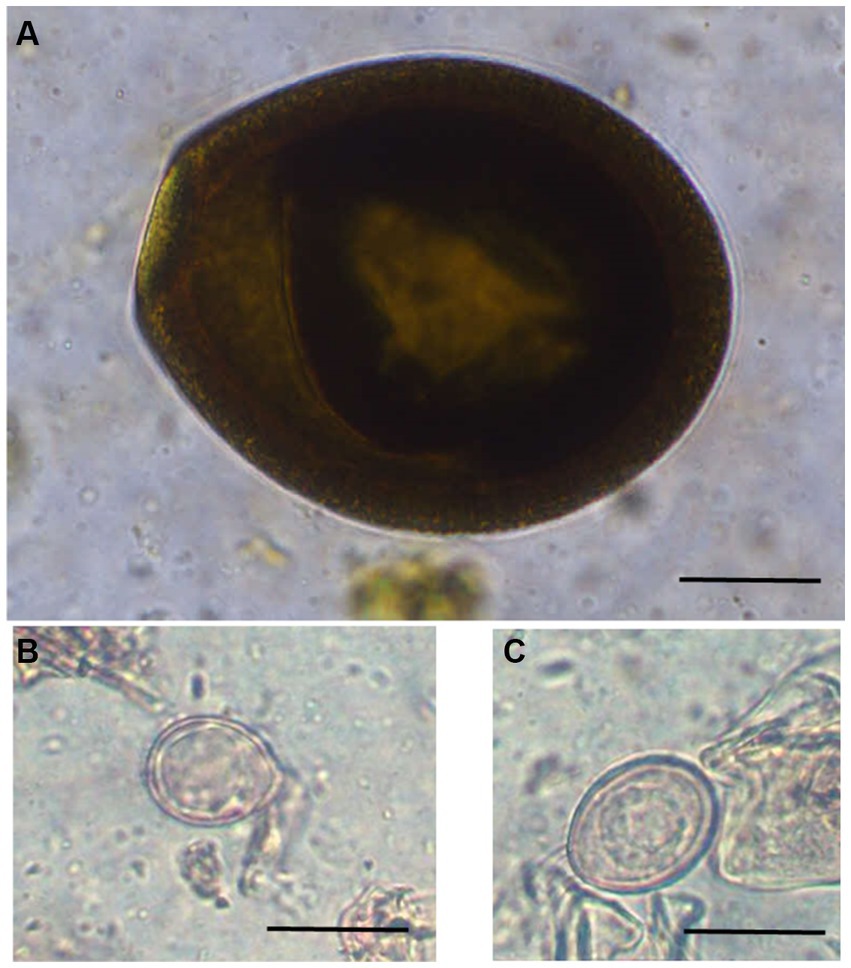
Figure 2. Photomicrographs of different unsporulated Eimeria oocysts of Bactrian camels (Camelus bactrianus) in Mongolia. (A) Eimeria cameli. The oocysts were truncated ovoid, dark brown to black in colour. The oocyst wall was composed of 3 layers: outer, dark brown in colour with tiny projections. The middle layer was thin smooth and yellowish in colour. The inner layer was dark brown. Length x width 96.5 × 82 μm, (B) Eimeria dromedarii. The oocyst shape ranged from subspherical to ovoid, with rough walls composed of two distinct layers: outer, pale yellow and inner, dark green. Length x width = 23 × 17 μm and, (C) Eimeria rajasthani. The oocysts were ellipsoidal in shape with smooth walls that were composed of two layers: outer, pale yellow and inner, yellowish green in colour. The size length x width 29 × 21.6 μm. Scale bar = 25 μm.
We successfully amplified and sequenced the 246 bp long MHC class II DRA exon 2 in 70 (out of 100) samples (Table 1). We screened the DRA exon 2 for genetic diversity and detected two polymorphic nucleotides (nt) at the positions nt = 58 and nt =143 in the 246 bp long fragment. Phasing the 70 individual sequences resulted in three haplotypes (h = 3) with a haplotype (gene) diversity Hd = 0.620 (± 0.018), nucleotide diversity Pi = 0.004 (± 0.00006) and an average number of nucleotide differences k = 0.983. At position nt = 58, the change from T to A (T58A) led to a non-synonymous amino acid (aa) change from phenylalanine (F) to tyrosine (Y), both hydrophobic, while the nucleotide change G143T was synonymous (no aa change). This resulted in two different aa alleles (haplotypes; H1 and H2) identified in Mongolian Bactrian camels as shown in Figure 3. While 19 camels were homozygous for H1 and 11 individuals for H2, respectively, the majority of 40 Bactrian camels was heterozygous and harboured both aa alleles of the MHCII DRA exon 2. The complete sample, genotype and aa allele information is presented in Table 3. The sequence alignment of MHCII DRA exon 2 for all Bactrian camel samples is provided in Supplementary file S1.

Figure 3. MHCII DRA exon 2 amino acid haplotypes identified in Mongolian Bactrian camels. The polymorphic amino acid change is highlighted in bold.
We investigated a potential statistical effect between the MHC DRA exon 2 aa alleles H1, H2 or H1/H2 on the three different Eimeria spp. infections in the Bactrian camels, using a generalised linear model approach. However, we could not identify any significant (p < 0.0083 after Bonferroni correction) association between the MHCII DRA aa alleles and any of the Eimeria spp. infections (Table 4). Similarly, we did not detect a significant effect of the homozygote or heterozygote genotypes on Eimeria spp. infections, respectively. However, we identified a significant (p = 0.003) positive effect with a moderate association coefficient Phi = 0.532 (p < 0.001) between two Eimeria species, E. cameli and E. rajasthani, independent from all other tested predictor variables and factors, i. e., location, age or gender did not show any significant effect on the prevalence of E. cameli and E. rajasthani in the respective models under strict Bonferroni correction (Table 4).
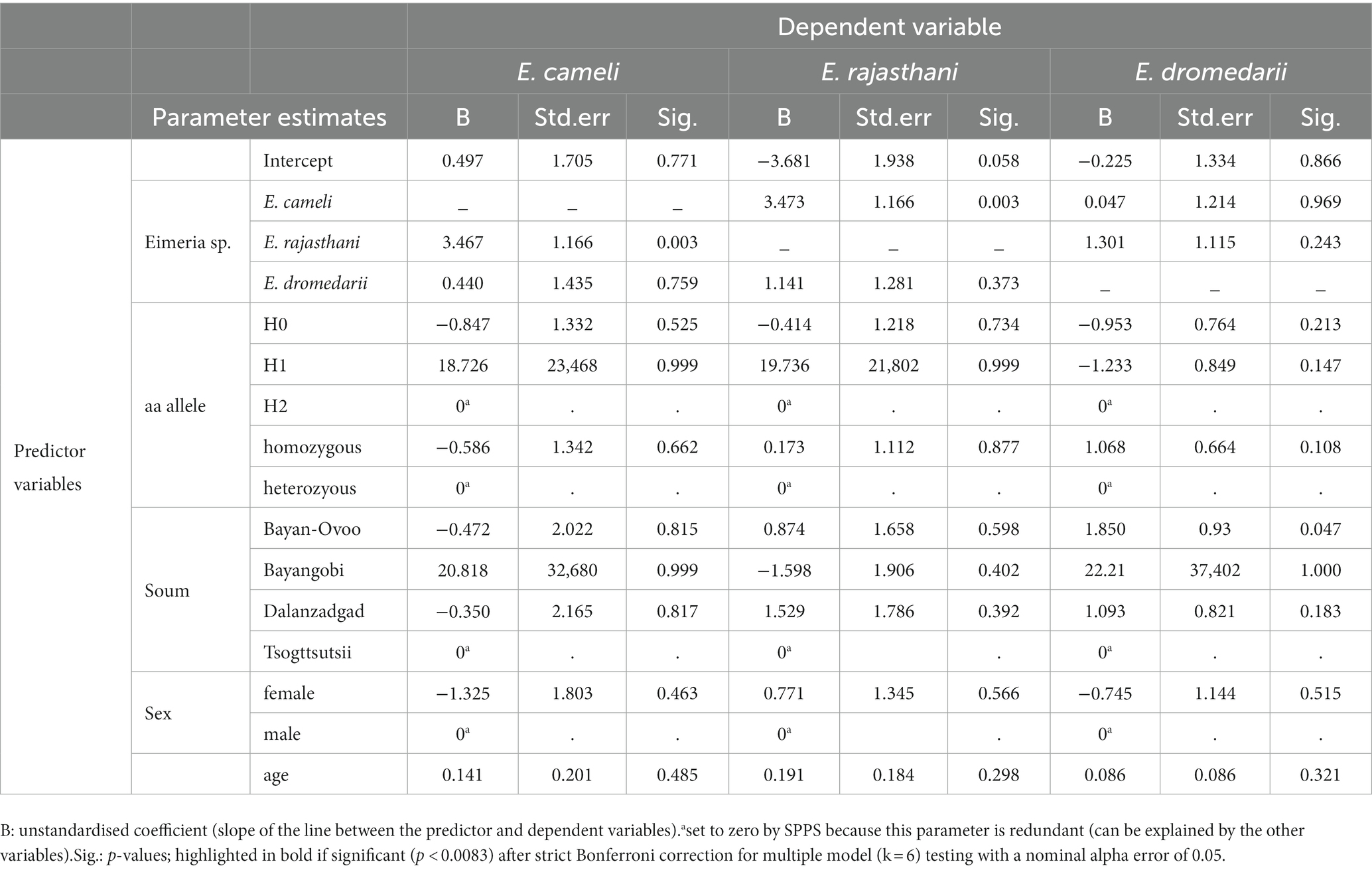
Table 4. Parameter estimates of the generalized linear model testing between MHCII DRA exon 2 amino acid (aa) alleles and Eimeria spp. infections.
Eimeriosis, in Camelini worldwide has been recently summarized (5). However, no reports from Mongolia were included since there were no available studies at the time. Here, we report the prevalence of Eimeria spp. infections in southern Mongolia. Due to the rather high prevalence of camel coccidiosis, it could be assumed that Eimeria infections are widely spread in the country, and it may play an important role as underestimated subclinical or clinical disease affecting the growth rate performance of mainly young Mongolian Bactrian camels as reported for other hosts. Eimeria spp. prevalence up to 50% was reported in Bactrian camels from Inner Mongolia in China (42). In total, three monoxenous Eimeria species were found: E. dromerdarii, E. rajasthani and E. cameli, which are considered pathogenic for camels (20). Mixed infections with two or three species were here observed, presenting a higher frequency then previously reported by Yakhchali and Athari (2010) (43). The authors described the identification of four Eimeria species including E. bactriani (52.4%), E. cameli (19.3%), E. pellerdyi (15.6%) and E. dromedarii (12.5%) and mixed infections (up to four Eimeria species) in 10.54% of investigated camels.
Regarding pathogeny of camel coccidiosis several studies confirm its importance. Iyer et al. (44) reported an outbreak of gastroenteritis in camels in Punjab, India, affecting hundreds of camels with 1–40% mortality during summer months (44). Two dead camels were examined at necropsy. Gastroenteritis was the predominant finding and affected abomasum, duodenum, and cecum; jejunum and ileum were not examined. Endogenous stages (schizonts, gamonts, and oocysts) were detected in duodenum, and cecum (15, 45, 46). Same conclusion applies to a similar case of haemonchosis and E. cameli-associated gastroenteritis in one year old camel from India (45). Rangarao and Sharma (47) noted diarrhea-associated with the presence of E. rajasthani oocysts in all eight calves in India (47). An eimeriosis-like illness was diagnosed histologically in 27 of 38 camels submitted in 1996 for postmortem examination to the Central Veterinary Research Laboratory (CVRL), Dubai, UAE (48). Of these 27, illness was severe in 21 and mild in 6 animals, respectively. Severe hemorrhagic enteritis with eosinophilia of small intestine (mostly jejunum and ileum and rarely duodenum) was associated with numerous stages of E. cameli whereas large intestines were not affected.
Camel eimeriosis has mostly been associated with younger camels (20, 49). It is an important disease in pre-weaned and recently weaned camels (20). While nearly animals of all ages are exposed to infectious sporulated Eimeria oocysts in the environment, they may not show obvious signs of the disease. In the majority of the hosts, the parasite coexists causing minimal damage to the infected host (20). Clinical eimeriosis usually occurs if the host is subject to a heavy infection, with high number of infectious oocysts ingested, or if its resistance is lowered (20), and its immune status is not adequate to cope with a coccidian infection.
Another critical time for the infection of camels could be the time immediately preceding the period of dryness (peak from July to October), when the short winter rains cause camels to crowd from the surrounding desert to limited water holes and springs in oases (12), like in Saudi Arabia, potentially promoting the spread of the disease. Further studies with camel Eimeria spp., including molecular characterization and establishment of suitable in vitro culture systems will allow detail investigations on sporozoite-host cell interactions and early host innate immune reactions as reported for ruminant eimeriosis (8, 38).
Concerning immunogenic response, in the 246 bp exon 2 of the MHCII DRA locus sequenced in 70 Mongolian Bactrian camels infected or non-infected with Eimeria spp. we detected the same two nucleotide polymorphisms as described before in a global set of Bactrian camels (27). These synonymous and non-synonymous polymorphisms, which produce three different aa alleles are also shared between dromedaries (C. dromedarius) and wild camels (C. ferus) (27). The frequency (0.57) of the heterozygous allele in the here investigated Mongolian Bactrian camels was similar to the frequency described in global Bactrian camels (0.53), lower than in wild camels (0.63) and higher than in dromedaries (0.32) (27). MHC diversity is often maintained by pathogen-mediated balancing selection. It is generally assumed that heterozygous individuals have an advantage, e.g., higher fitness than individuals that are homozygous at the same locus (50, 51). Although we observed twice as many heterozygous individuals (n = 40) for the aa alleles at position nt 58 than homozygotes for either the reference (n = 19) or alternative (n = 11) allele, we could not identify a statistically significant heterozygosity effect on the Eimeria spp. infection, in terms of prevalence. No evidence for MHC class II DRB heterozygote advantage in relation to intestinal parasite infection has been described in Brandt’s voles from Inner Mongolia (26). The lack of such a heterozygosity effect in our study might also be explained by the relatively low number (n = 70) of successfully phenotyped (Eimeria spp. infection) and genotyped (MHCII DRA exon 2) samples. In addition, the Eimeria infection status was evaluated in a qualitative way, i.e., presence or absence, while a quantitative assessment (i.e., intensities of infection) would have provided more refined infection data to be included into our statistical models. Contrary to many other species, Old World camels and specifically Bactrian camels have a low number of MHC II DRB alleles. In fact, the DRB exon 2 showed only four alleles in 43 previously investigated Bactrian camels, which translated into two haplotypes at the amino acid level with one haplotype present in 74% of the samples (27). As our attempt to amplify MHCII DRB exon 2 unfortunately failed, we cannot exclude that we might have identified new DRB alleles in the investigated Mongolian Bactrian camels. However, considering our findings of the DRA exon 2 identifying exactly the previously described alleles (27), we probably might neither expect novel DRB alleles in the population. We did not find any age-associated effect on Eimeria spp. prevalence, even though older individuals may be expected to have higher prevalence than younger ones, simply due to the longer lifetime that accumulates their change of infection. This might suggest that adult may cope with the infection successfully and clear it, possibly also in connection with their overall immunogenetic status.
Interestingly, we detected a moderate positive association between the two Eimeria species E. cameli and E. rajasthani. Co-evolution between two parasites (E. cameli and E. rajasthani) has been observed, and these two species were dominant in Mongolian camels. While at a low dose inoculate a linear reproduction is observed, at higher doses the reproduction of the parasite becomes impaired (the so-called ‘crowding effect’), mainly due to the damage of cells or lower availability of nutrients (52). Resistance of the host to a pathogen is a very important factor in the evolutionary arms race between host and parasite. Although the host evolves at a much slower rate than the parasite is capable of, the host has developed ways to reduce susceptibility to infection. We argue that studying questions of host–parasite interaction in camelids can be well approached with an Eimeria parasite infection system in Bactrian camels.
In conclusion, this study revealed that E. cameli, E. rajasthani, and E. dromedarii infections frequently occur in Mongolian Bactrian camel. Given that clinical and subclinical Eimeria spp. infections are well known to dampen camel production, regular monitoring including diagnosis of species, quantitative description of the parasite infection load, and MHC and other immunogenetic loci could help to prevent future Eimeria-induced economic losses in Mongolian camel rearing.
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
The animal studies were approved by Animal Care and Use Committee, Institute of Veterinary Medicine, Mongolian University of Life Sciences (Agreement Number № MEBUS-16/01/05). The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent was obtained from the owners for the participation of their animals in this study.
IK: Conceptualization, Formal analysis, Funding acquisition, Writing – original draft, Data curation, Investigation. UN: Data curation, Investigation, Visualization, Writing – review & editing. BC: Data curation, Investigation, Writing – review & editing. KN: Data curation, Formal analysis, Investigation, Writing – review & editing. AT: Conceptualization, Supervision, Validation, Writing – review & editing. CH: Conceptualization, Supervision, Validation, Writing – review & editing. FS: Formal analysis, Methodology, Writing – review & editing. FK: Methodology, Software, Validation, Writing – review & editing. PB: Conceptualization, Formal analysis, Funding acquisition, Project administration, Resources, Supervision, Writing – original draft. GB: Conceptualization, Funding acquisition, Supervision, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. We would like to thank the Yak and Camel Foundation for funding the genetic lab work. This work was partially funded by Mongolian Foundation of Science and Technology (Project №ShUSS 2020/23). IK acknowledges funding from the Ernst-Mach grant by the Austrian’s Agency for Education and Internationalisation (OeAD). Open Access costs were funded by the Open Access Fonds of the University of Veterinary Medicine Vienna.
We would like to thank the Yak and Camel Foundation for funding the genetic lab work.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2023.1296335/full#supplementary-material
2. The Bactrian Camels Genome Sequencing and Analysis ConsortiumJirimutu,, Wang, Z, Ding, G, Chen, G, Sun, Y, et al. Genome sequences of wild and domestic bactrian camels. Nat Commun. (2012) 3:1202. doi: 10.1038/ncomms2192
3. Yagil, R. Camels and camel Milk animal production and health report. FAO (Food and Agricultural Organization of the UN). (1982) 26
4. Bayasgalan, C, Chultemdorj, T, Roth, F, Zinsstag, J, Hattendorf, J, Badmaa, B, et al. Risk factors of brucellosis seropositivity in Bactrian camels of Mongolia. BMC Vet Res. (2018) 14:342. doi: 10.1186/s12917-018-1664-0
5. Dubey, JP, and Schuster, RK. A review of coccidiosis in Old World camels. Vet Parasitol. (2018) 262:75–83. doi: 10.1016/j.vetpar.2018.08.008
7. Levine, ND, and Ivens, V. The coccidian parasites (protozoa, sporozoa) of ruminants. Illinois: Illinois Biological Monographs No 44 University of Illinois Press. (1970):1–278.
8. Hermosilla, C, Ruiz, A, and Taubert, A. Eimeria bovis: an update on parasite-host cell interactions. Int J Med Microbiol. (2012) 302:210–5. doi: 10.1016/j.ijmm.2012.07.002
9. Silva, LMR, Chávez-Maya, F, Macdonald, S, Pegg, E, Blake, DP, Taubert, A, et al. A newly described strain of Eimeria arloingi (strain a) belongs to the phylogenetic group of ruminant-infecting pathogenic species, which replicate in host endothelial cells in vivo. Vet Parasitol. (2017) 248:28–32. doi: 10.1016/j.vetpar.2017.10.014
10. Dubey, JP. A review of coccidiosis in south American camelids. Parasitol Res. (2018) 117:1999–2013. doi: 10.1007/s00436-018-5890-y
11. Chineme, CN. A case report of coccidiosis caused by Eimeria cameli in a camel (Camelus dromedarius) in Nigeria. J Wildl Dis. (1980) 16:377–80. doi: 10.7589/0090-3558-16.3.377
12. Kawasmeh, ZA, and Eimeria Cameli, Elbihari S. (Henry and Masson, 1932) Reichenow, 1952: redescription and prevalence in the Eastern Province of Saudi Arabia. Cornell Vet 1983:73.
13. Yagoub, IA. Coccidiosis in Sudanese camels (Camelus dromedarius): 1 —first record and description of Eimeria spp. Harboured by camels in the eastern region of Sudan. J Protozool. (1989) 36:422–3. doi: 10.1111/j.1550-7408.1989.tb05539.x
14. Kasim, AA, Hussein, HS, and Shawa, YRA. Coccidia in Camels (Camelus dromedarius) in Saudi Arabia. J Protozool. (1985) 32:202–3. doi: 10.1111/j.1550-7408.1985.tb03039.x
15. Tafti, AK, Maleki, M, Oryan, A, and Mozafari, AA. Pathological study of digestive system lesions of camels (Camelus dromedarius) slaughtered in Iran. 19-22nd September In: Proceedings of 18th meeting of the European Society of Veterinary Pathology : Amsterdam, The Netherlands (2000) 245–5.
16. Hussein, HS, Kasim, AA, and Shawa, YR. The prevalence and pathology of Eimeria infections in camels in Saudi Arabia. J Comp Pathol. (1987) 97:293–7. doi: 10.1016/0021-9975(87)90093-4
17. Borji, H, RG, MAR, NAG, MM. Prevalence of Cryptosporidium and Eimeria infections in dromedary (Camelus dromedarius) in abattoir of Mashhad. Iran: J Camel Pract Res (2009) 2009 p.
18. Gruvel, J, and Graber, M. Coccidia and Coccidiosis. 2nd ed. Budapest: Akademia Kiado (1969). 698 p.
20. Yakhchalim, M, and Cheraghi, E. Eimeriosis in bactrian and dromedary camels in the Miandoab region. Iran Acta Vet Brno. (2007) 57:545. doi: 10.2298/AVB0706545Y
21. Muñoz-Caro, T, Mena Huertas, S, Conejeros, I, Alarcón, P, Hidalgo, MA, Burgos, RA, et al. Eimeria bovis-triggered neutrophil extracellular trap formation is cd11b-, ERK 1/2-, p38 MAP kinase- and soce-dependent. Vet Res. (2015) 46:23. doi: 10.1186/s13567-015-0155-6
22. Hamid, PH, Hirzmann, J, Kerner, K, Gimpl, G, Lochnit, G, Hermosilla, CR, et al. Eimeria bovis infection modulates endothelial host cell cholesterol metabolism for successful replication. Vet Res. (2015) 46:100. doi: 10.1186/s13567-015-0230-z
23. Conejeros, I, López-Osorio, S, Zhou, E, Velásquez, ZD, del Río, MC, Burgos, RA, et al. Glycolysis, monocarboxylate transport, and purinergic signaling are key events in Eimeria bovis-induced NETosis. Front Immunol. (2022) 13:13. doi: 10.3389/fimmu.2022.842482
24. Janeway, C, Travers, P, Walport, M, Shlomchik, MJ, et al. Immunobiology: The immune system in health and disease: The major histocompatibility complex and its functions. The Immune System in Health and Disease: Immunobiology (2001).
25. Meyer, D, and Thomson, G. How selection shapes variation of the human major histocompatibility complex: a review. Ann Hum Genet. (2001) 65:1–26. doi: 10.1046/j.1469-1809.2001.6510001.x
26. Zhang, M, and He, H. Parasite-mediated selection of major histocompatibility complex variability in wild brandt’s voles (Lasiopodomys brandtii) from Inner Mongolia. China BMC Evol Biol. (2013) 13:149–9. doi: 10.1186/1471-2148-13-149
27. Plasil, M, Mohandesan, E, Fitak, RR, Musilova, P, Kubickova, S, Burger, PA, et al. The major histocompatibility complex in Old World camelids and low polymorphism of its class II genes. BMC Genomics. (2016) 17:167. doi: 10.1186/s12864-016-2500-1
28. Lado, S, Elbers, JP, Rogers, MF, Melo-Ferreira, J, Yadamsuren, A, Corander, J, et al. Nucleotide diversity of functionally different groups of immune response genes in Old World camels based on newly annotated and reference-guided assemblies. BMC Genomics. (2020) 21:4. doi: 10.1186/s12864-020-06990-4
29. Anne MZGAConboy. Veterinary clinical parasitology, 8th edition. American Association of Veterinary Parasitologists. 8th ed (2012).
30. Pauling, CD, Oller, AR, and Jackson, V. Fecal parasite identification by microscopy and PCR in scimitar-horned oryx, Oryx dammah, managed at two sites. Int J Parasitol Parasites Wildl. (2016) 5:312–20. doi: 10.1016/j.ijppaw.2016.11.001
31. Charles, M, and Hendrix, ER. Diagnostic parasitology for veterinary technicians. 4th ed printed in United States of America (2012).
33. Eckert, J, RBMWSPCoudert. Biotechnology guidelines on techniques in coccidiosis research. Brussels Luxembourg: ECSC-EC-EAEC (1995).
34. Levine, ND, and Iyens, V. The coccidian parasites (Protozoa, Sporozoa) of ruminants. Chicago, and London: University of illinois press Urbana (1970).
35. Radfar, MH, and Aminzadeh, GM. Common gastrointestinal parasites of indigenous camels (Camelus dromedarius) with traditional husbandry management (free-ranging system) in central deserts of Iran. J Parasit Dis. (2013) 37:225–30. doi: 10.1007/s12639-012-0170-8
36. Soulsby, EL. Helminths, arthropods and protozoa of domesticated animals, 7th edition. 7th ed. London: Bailliere Tindall (1982).
37. Obanda, V, Maingi, N, Muchemi, G, Ng’ang’a, CJ, Angelone, S, and Archie, EA. Infection dynamics of gastrointestinal helminths in sympatric non-human primates, livestock and wild ruminants in Kenya. PLoS One. (2019) 14:e0217929. doi: 10.1371/journal.pone.0217929
38. Hermosilla, C, Barbisch, B, Heise, A, Kowalik, S, and Zahner, H. Development of Eimeria boris in vitro: suitability of several bovine, human and porcine endothelial cell lines, bovine fetal gastrointestinal, Madin-Darby bovine kidney (MDBK) and African green monkey kidney (VERO) cells. Parasitol Res. (2002) 88:301–7. doi: 10.1007/s00436-001-0531-1
39. Soulsby, EJL. Helminths, arthropods and Protozoa of domesticated animals. 7th ed. London: Balliere, Tindall and Cassel, London (1982). 614 p.
40. Librado, P, and Rozas, J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. (2009) 25:1451–2. doi: 10.1093/bioinformatics/btp187
41. R Core Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing (2023).
42. Wei, JG, and Wang, Z. A survey of the Eimeria species in the double-humped camels of the inner Mongolian autonomous region. China Chin J Vet Med. (1990) 16:23–4.
43. Yakhchali, M, and Atari, A. A study on prevalence of Eimeria spp. infection in camels of Tabriz region. Arch Razi Inst. (2010) 65. doi: 10.22092/ARI.2010.103858
44. Iyer, PK, Ramachandran, S, and Joshi, TP. An outbreak of haemorrhagic gastro-enteritis in camels (Camelus dromedarius). Ann Parasitol Hum Comp. (1968) 43:5–14. doi: 10.1051/parasite/1968431005
45. Narnaware, SD, Kumar, S, Dahiya, SS, and Patil, NV. Concurrent infection of coccidiosis and haemonchosis in a dromedary camel calf from Rajasthan, India. In: Journal of Camel Practice and Research. (2017) 24:225. doi: 10.5958/2277-8934.2017.00038.8
46. Dubey, JP, Schuster, RK, and Kinne, J. Gametogony of Eimeria cameli in the small intestine of one-humped camel (Camelus dromedarius). Parasitol Res. (2018) 117:3633–8. doi: 10.1007/s00436-018-6064-7
47. Sharma, RL. Intestinal coccidiosis due to eimeria rajasthani in camel (camelus dromedarius). Indian. Vet J. (1997) 74.
48. Kinne, J, and Wernery, U. Severe outbreak of camel coccidiosis in the United Arab Emirates. Journal of Camel Practice and Research. (1997) 4.
49. Kauffman, J. Parasitic infections of domestic animals: A diagnostic manual. Birkhäuser Verlag, Schweiz; (1996) 262–263.
50. Hedrick, PW. Pathogen resistance and genetic variation at MHC loci. Evolution (NY). (2002). doi: 10.1111/j.0014-3820.2002.tb00116.x
51. Spurgin, LG, and Richardson, DS. How pathogens drive genetic diversity: MHC, mechanisms and misunderstandings, Proceedings of the Royal Society B: Biological Sciences. (2010) 979–988.
Keywords: Camelus bactrianus, coccidiosis, Eimeria cameli, Eimeria rajasthani, Eimeria dromedarii
Citation: Khatanbaatar I, Nyamdolgor U, Chinchuluun B, Naranbaatar K, Taubert A, Hermosilla CR, Suchentrunk F, Knauer F, Burger PA and Battsetseg G (2023) Prevalence of Eimeria spp. infections and major histocompatibility complex class II DRA diversity in Mongolian Bactrian camels (Camelus bactrianus). Front. Vet. Sci. 10:1296335. doi: 10.3389/fvets.2023.1296335
Received: 18 September 2023; Accepted: 02 November 2023;
Published: 23 November 2023.
Edited by:
Alireza Sazmand, Bu-Ali Sina University, IranReviewed by:
Giovanni Sgroi, Experimental Zooprophylactic Institute of Southern Italy (IZSM), ItalyCopyright © 2023 Khatanbaatar, Nyamdolgor, Chinchuluun, Naranbaatar, Taubert, Hermosilla, Suchentrunk, Knauer, Burger and Battsetseg. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Igori Khatanbaatar, a2hhdGFuYmFhdGFyQG11bHMuZWR1Lm1u; Pamela A. Burger, UGFtZWxhLkJ1cmdlckB2ZXRtZWR1bmkuYWMuYXQ=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.