
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci. , 11 January 2024
Sec. Zoological Medicine
Volume 10 - 2023 | https://doi.org/10.3389/fvets.2023.1283487
This article is part of the Research Topic Insights in Zoological Medicine: 2023 View all 9 articles
Despite the importance of antimicrobial resistance, only a few studies on the antimicrobial susceptibility on wild animals have been conducted owing to their population, accessibility, and characteristics. The objective of this study was to investigate the prevalence and characteristics of antimicrobial resistance pattern in Escherichia coli and Enterococcus faecalis isolated from the feces of captive wild animals in a zoo. A total of 61 captive wild animals were included in this study. E. coli was isolated from 58 of the 61 animals and E. faecalis was isolated from 29 animals. Among the isolated E. coli strains, ampicillin exhibited the highest resistance rate (27/29, 93.1%). Of these, 18 strains (18/29, 62%) showed multidrug resistance. The multilocus sequence typing (MLST) test showed that only ST155 was detected twice, while the other 16 strains showed different ST types. Among the E. faecalis strains, two were susceptible to all tested antimicrobials, whereas the remaining 27 strains showed resistance to one or more antimicrobials. Nine strains (9/27, 31%) showed multidrug resistance. Among the E. faecalis strains, resistance to quinupristin/dalfopristin was the highest at 96.3% (26/27), while the MLST of the nine MDR strains showed no predominant ST. Genetic association with human isolates or livestock products was observed in the isolated ST types. This indicates that antibiotic resistance in the zoo is responsible for the use of antibiotics and the partial horizontal transmission between humans and animals through feeding or contact.
Antimicrobial resistance is recognized as a major global health problem (1). The emergence of super bacteria (e.g., methicillin-resistant Staphylococcus aureus [MRSA], vancomycin-resistant S. aureus [VRSA], vancomycin-resistant Enterococcus [VRE], and Salmonella typhimurium DT104) that are resistant to antimicrobials and the emergence of multidrug-resistant bacteria that are resistant to various antimicrobials are major global health challenges (2–4). Antimicrobial-resistant bacteria pose the 21st century’s greatest public health threat, which the world is actively combating. The emergence of antimicrobial-resistant bacteria has been systematically researched by the U.S. Food and Drug Administration (FDA) since 1996. In Korea, the National Antimicrobial Resistance Safety Management Project began in 2003. Subsequently, research to analyze the distribution status and pattern of resistant bacteria by isolating various pathogenic bacteria from humans, livestock, fish, and the environment has been earnestly promoted.
Compared with antimicrobial resistance studies on livestock or companion animals in Korea, there are few studies that address the risk of antimicrobial resistance on wild animals. This is presumed to be due to the population of wild animals, conditions of conservation facilities, and the characteristics of individual wild species. While some studies have analyzed the antimicrobial resistance rates of pathogenic Escherichia coli isolated from wild animals in Korea (5, 6), there have been few studies on the antimicrobial resistance of indicator bacteria isolated from wild captive animals in Korean zoos.
Escherichia coli, which resides as a normal bacterium in the intestines of mammals such as humans and animals, is an opportunistic bacterium that is always exposed to antimicrobials and can cause disease when immunity is weakened. E. coli can easily acquire and transfer antimicrobial resistance and is considered a good bioindicator for observational studies on antimicrobial resistance (7, 8). Therefore, the antimicrobial resistance of target bacteria, such as those found in the environment, meat, and companion animals, are being actively studied. E. faecalis, like E. coli, is a normal bacterium in the mammalian intestine; however, nosocomial infections in hospitals have recently emerged in humans (9). Additionally, for some antimicrobials, there are intrinsic resistances that induce resistance in bacteria regardless of the use of antimicrobials. Acquired resistance due to the misuse of antimicrobials is also possible, serving as an indicator of antimicrobial resistance (10).
Recently, zoos have tended to focus on animal welfare and species conservation (11); however, some still use methods such as petting for zoo management and increasing public interest. It is possible to become infected with zoonotic pathogens through the ingestion of animal waste through the mouth, direct contact with animals, or contaminated surfaces (12). Another concern is animal-to-human transmission of antimicrobial-resistant bacteria (13). A strong positive correlation may exist between antibiotic use and antibiotic resistance in E. coli (14), and several studies have reported the horizontal transmission of zoonotic diseases from zoos or petting farms (15, 16).
The objectives of this study were: (1) to investigate antimicrobial resistance and (2) to analyze the relationship between commensal E. coli and E. faecalis, which are indicator bacteria, in captive wild animals at Seoul Zoo.
Samples were collected from animals that needed medical care, e.g., health checkups and anesthesia for movement, but did not show clinical symptoms. A total of 61 healthy animals belonging to 32 species were included in this study. There were 55 mammals of 27 species—including Barbary sheep (Ammotragus lervia) and Olive baboons (Papio Anubis)—5 birds of 4 species, and 1 reptile of 1 species (dwarf crocodile [Osteolaemus tetraspis]). Among these sampled individuals, only black-faced spoonbills (sample no. 11) and Siberian tigers (sample no. 46, 47, 48, with the same parents) were less than 1-year-old. All others were reproductive adults.
Samples were collected through rectal swabs using a sterile transport medium (Asan Pharm, Seoul, Korea) between March 2022 and September 2022. To differentiate between E. coli and E. faecalis remaining in the soil, only anal swabs were used in this study, and samples were not collected from feces. The patient information is presented in Table 1.
From the perspective of animal ethics, animals were not intentionally captured in this study. Captures and anal swabs were obtained only when medical care was required. Ethical clearance for this study was approved by 2019–008, 2022–004 at the Seoul Zoo IACUC. All sampling was conducted according to the committee criteria.
The swab samples were inoculated into thioglycollate medium (BD Difco™, Franklin, NJ, United States) and incubated at 37°C for 24 h, and then the cultured broth was inoculated into CHROMagar™ E. coli (CHROMagar™, Paris, France) and CHROMagar™ streptococcus (CHROMagar™) using a sterile loop needle. Bacterial colonies selected according to the criteria for each selective medium were enriched in trypticase soy agar containing 5% sheep blood (Asan Pharm, Seoul, Korea). Species identification was performed based on the sequences of the DNA gyraseB gene (E. coli) or the 16S ribosomal RNA gene (E. faecalis) according to the CLSI guidelines (17). The remaining DNA extract was stored at −20°C for multilocus sequence type (MLST) analysis.
The Kirby-Bauer disk diffusion method was performed according to CLSI guidelines (18). After the turbidity was adjusted as 0.5 McFaland standard, the bacterial suspension was smeared on Muller-Hilton agar (BD BBL™, Franklin, NJ, United States) antimicrobial disks (Oxoid, Hampshire, United Kingdom) were placed at equal intervals, followed by incubation at 37°C for 18 h (vancomycin for 24 h). Subsequently, the size of the inhibition zone was measured, and the presence or absence of antimicrobial resistance was determined according to CLSI guidelines (18). VRE measurements might have an error in the disk diffusion method (19); therefore, they were cross-validated using the E-TEST® strip (bioMerieux SA, Marcy-l’Étoile, France). For the-lactamase test, the double-disk diffusion method was used, and an amoxicillin-clavulanate disk was placed between cefepime and cefotaxime to observe diffusion. Quality control was performed using E. coli ATCC 25922, S. aureus ATCC 25923, and E. faecalis ATCC 29212.
While the resistance of E. coli was tested by disks of ampicillin (10 mcg), amoxicillin-clavulanate (30 mcg), cefepime (30 mcg), cefotaxime (30 mcg), meropenem (10 mcg), gentamicin (10 mcg), amikacin (30 mcg), sulfamethoxazole-trimethoprim (25 mcg), doxycycline (30 mcg), tetracycline (30 mcg), nitrofurantoin (300 mcg), chloramphenicol (30 mcg), and ciprofloxacin (5 mcg), the resistance of E. faecalis was tested by disks of penicillin G (10 U), ampicillin (10 mcg), erythromycin (15 mcg), vancomycin (30 mcg), doxycycline (30 mcg), tetracycline (30 mcg), nitrofurantoin (300 mcg), linezolid (30 mcg). chloramphenicol (30 mcg), rifampin (5 mcg), quinupristin/dalfopristin (15 mcg), and ciprofloxacin (5 mcg).
The proportion of antimicrobial-resistant strains was expressed as a percentage by dividing the number of resistant strains by the total number of positive strains. Multidrug-resistant bacteria were defined as strains showing resistance to three or more different classes of antimicrobials (20). The ratio of multi-drug resistant strains is expressed as a percentage of the total number of positive strains.
To evaluate the genetic relatedness of the isolated multidrug-resistant (MDR) bacterial clones, 7-gene MLST was conducted (Table 2). These 7 genes were amplified and sequenced to secure the nucleotide sequence of each gene, and the sequence type (ST) was determined by comparison with the PubMLST database.1 Based on the obtained ST type number, burst analysis was performed to analyze the genetic relationships between clones.2
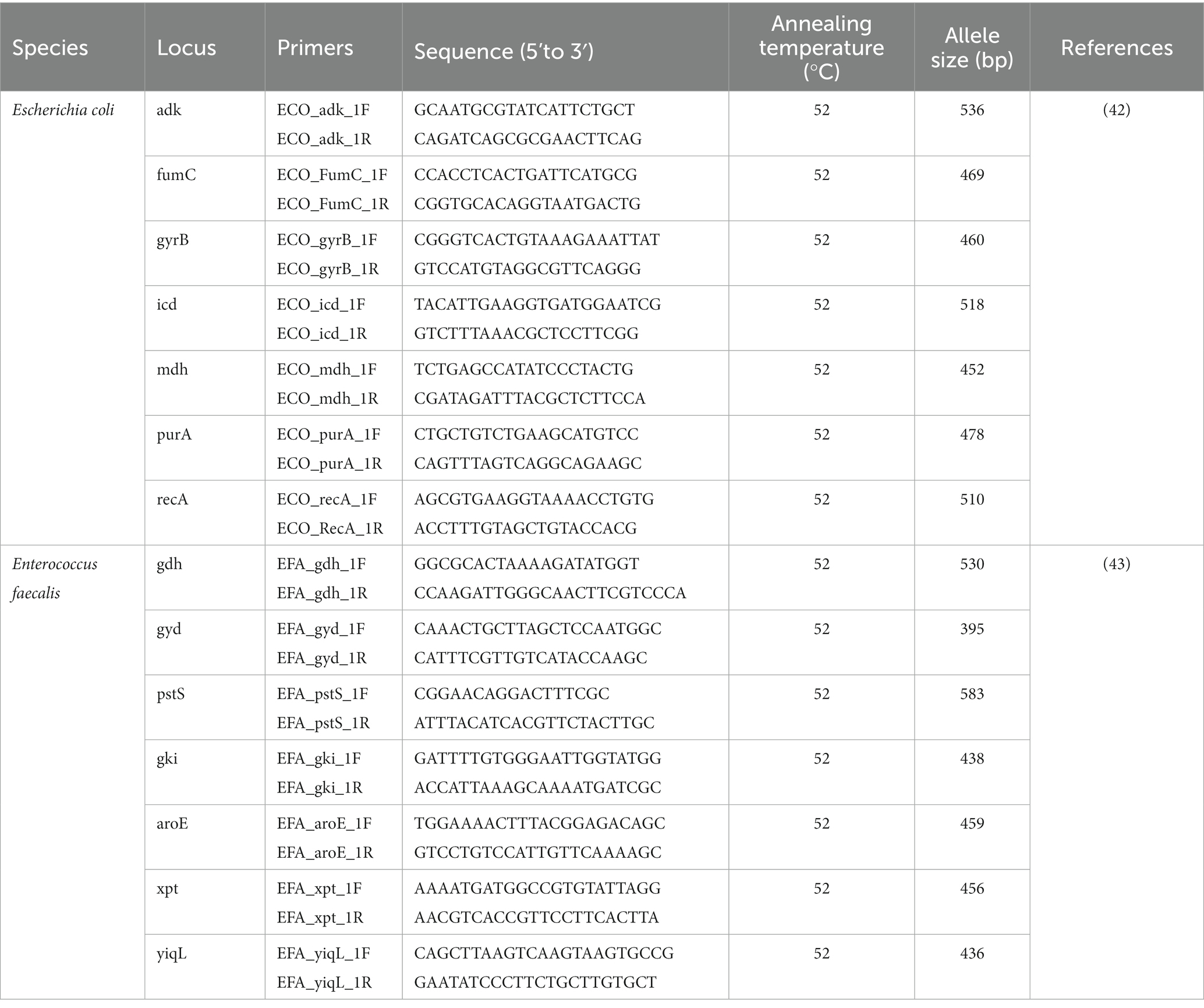
Table 2. Oligonucleotide and their reaction conditions of 7-gene multilocus sequence type analysis used in this study.
To evaluate the relatedness of the isolated E. coli strains, neighbor-joining phylogenetic analysis for gyrB gene sequence was performed using MEGA X (version 10.1).
The antimicrobial resistance of the isolated E. coli strains is shown in Table 1. E. coli was isolated from 58 of the 61 animals. Although 29 strains were susceptible to all tested antimicrobials (29/58, 50%), 29 strains were resistant to more than one. The 29 strains that showed resistance to more than one antimicrobial agent were ampicillin (27/29, 93.1%), tetracycline (16/29, 55.2%), sulfamethoxazole-trimethoprim (11/29, 37.9%), doxycycline (10/29, 34.5%), ciprofloxacin (8/29, 27.6%), and amoxicillin-clavulanate (8/29, 27.6%). Conversely, no resistance to meropenem and amikacin was observed. All strains were negative in the double-disk synergy test. Of the 61 samples, 18 strains (18/58, 31%) were MDR. Among the MDR E. coli strains, ampicillin resistance was the highest (17/18, 94%), followed by resistance to tetracycline and sulfamethoxazole-trimethoprim. Table 3 summarizes the results for MDR E. coli.
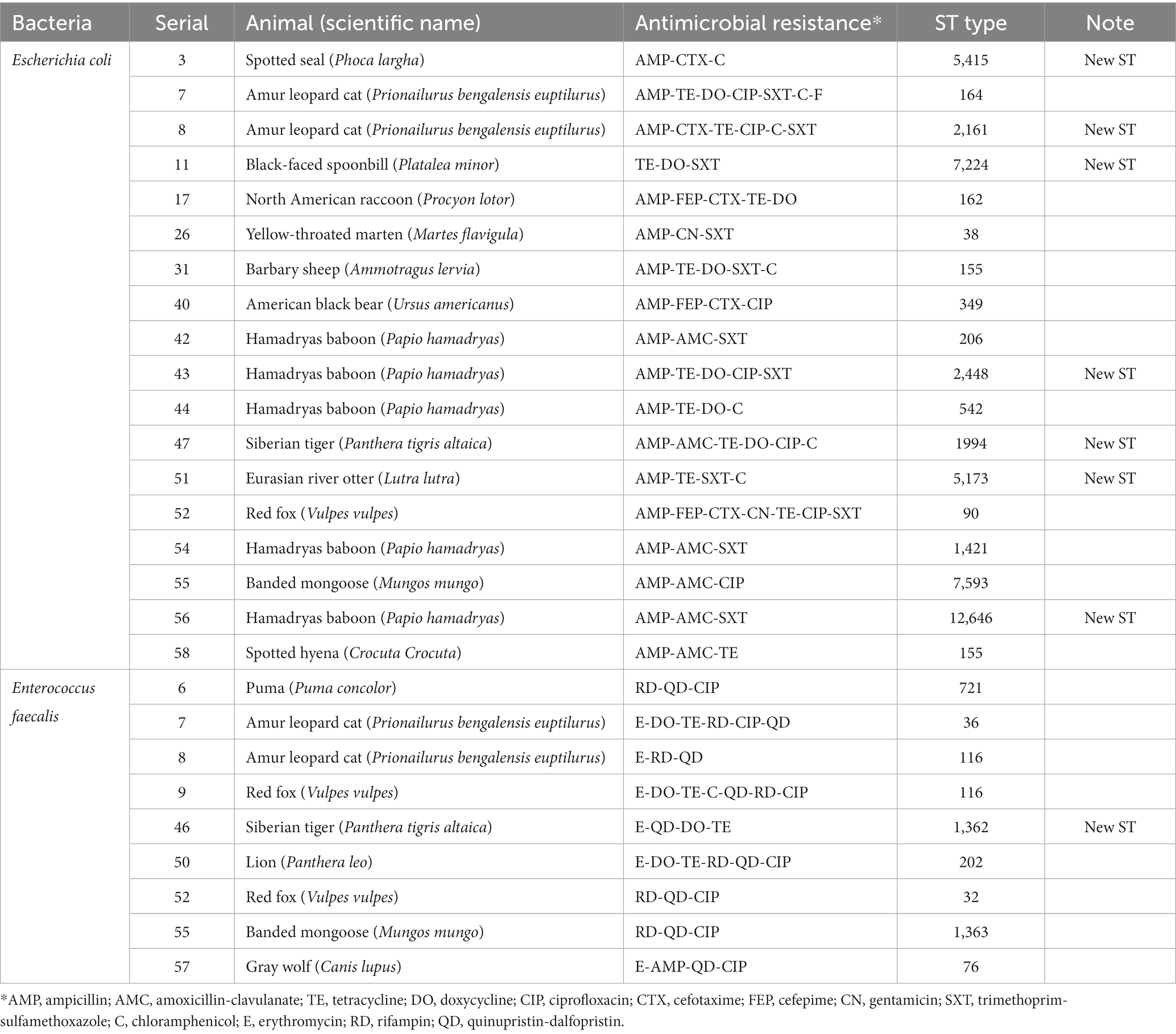
Table 3. Antimicrobial resistance and MLST type of multidrug-resistant E.coli and E. faecalis isolated in this study.
Among the species tested, Hamadryas baboons (Papio hamadryas; 5/6, 80.3%) and Amur leopard cats (Prionailurus bengalensis euptilurus; 2/2, 100%) showed the highest MDR strain retention. Contrastingly, MDR E. coli was isolated from only one of the 17 barbary sheep, despite the fact that barbary sheep were the largest population tested (17/61, 27.9%). Among the 18 MDR strains, 17 species were carnivores or omnivores and only one (Barbary sheep) was an herbivore.
The antimicrobial resistance of the isolated E. faecalis strains is shown in Table 1. E. faecalis was isolated from 29 of the 61 animals. While 2 strains were susceptible to all tested antimicrobials, 27 were resistant to one or more antimicrobials. Resistance to quinipristin/dalfopristin was the highest at 96.3% (26/27), followed by rifampin at 66.7% (18/27), and ciprofloxacin at 25.9% (7/27). No resistance to penicillin, nitrofurantoin, or linezolid was observed. Of the 29 strains, 9 (31.0%) showed MDR. Among the MDR E. faecalis, quinupristin/dalfopristin resistance was the highest at 100% (9/9), followed by resistance to rifampin and ciprofloxacin. Compared to the non-MDR E. faecalis strain, an increase in ciprofloxacin resistance was observed compared to that of the non-MDR E. faecalis strain. All the MDR E. faecalis were isolated from carnivores. No vancomycin resistance E. faecalis strains were isolated. Table 3 summarizes the results of the MDR E. faecalis.
Multilocus sequence typing (MLST) was performed on 18 MDR strains of E. coli and 9 MDR strains of E. faecalis. Seventeen STs were identified among MDR E. coli (Table 3). Except for ST155, which was simultaneously detected in Barbary sheep and spotted hyena, all ST types were detected only once. Of the 17 ST types, 7 ST types (ST1994, ST2448, ST5173, ST2161, ST5415, ST7224, and ST12646) were newly discovered. The 10 previously reported ST types have been reported in various sources such as humans, livestock (dogs, cows, pigs, chickens, and turkeys), environments (river, seawater, sewage, and wastewater), wild animals (vultures, elephants, guinea fowls, and hummingbirds), and plants (spinach), suggesting wide horizontal transmission of each clone worldwide (Table 4). Compared with the ST types reported in Korea, 7 ST types (ST90, ST155, ST162, ST206, ST2161, ST2448, and ST5415) showed single-locus variants with existing Korean isolates, and all strains showing SLV relationships were isolated from humans (Figure 1). Among the 5 MDR strains isolated from hamadryas baboons, 2 showed SLV relationships with each other (ST542 and ST12646), 1 (ST206) with a human isolate, and 1 (ST2161) MDR strain of the amur leopard cat showed SLV relationships with human isolates.
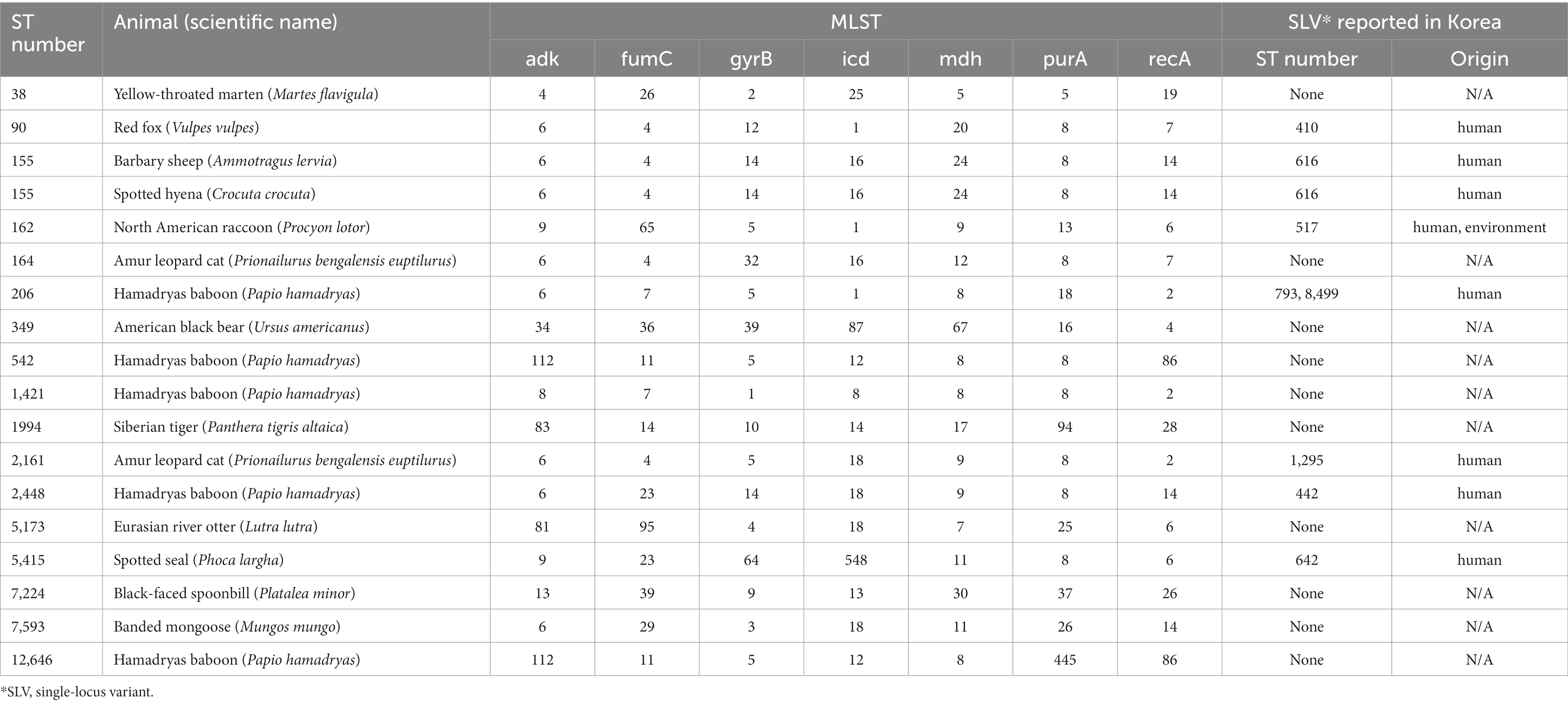
Table 4. Information of multilocus sequence type and the clonal complex relationship of multidrug-resistant E. coli isolated in this study.
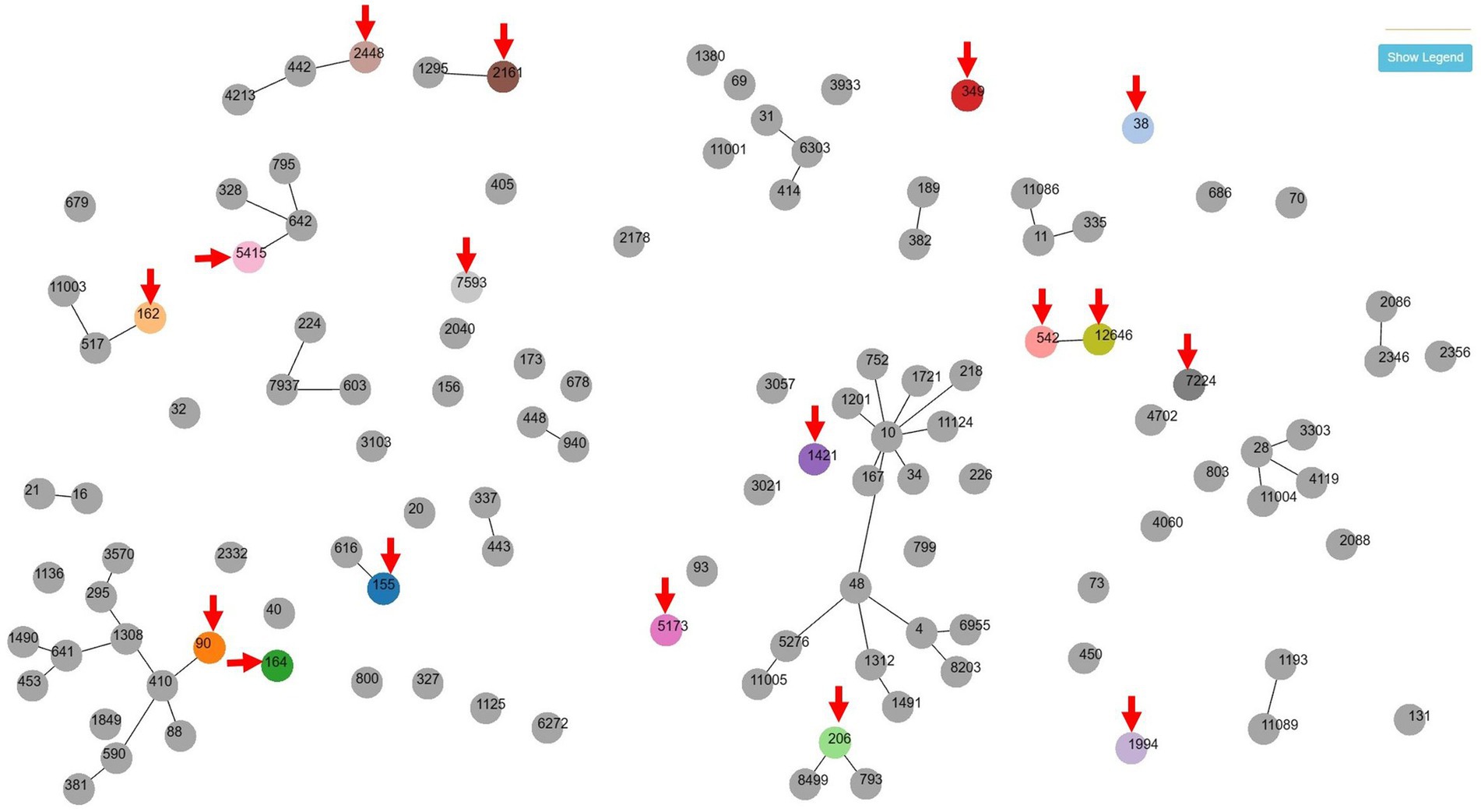
Figure 1. goeBURST (PHTLOViZ) analysis between multidrug-resistant Escherichia coli isolates of this study and strains reported in human in Korea. Seven zoo animal-origin ST types (ST90, ST155, ST162, ST206, ST2161, ST2448 and ST5415) showed single-locus variant relationships with human-origin existing Korean isolates while one ST type (ST38) was isolated in both of zoo animal (yellow-throated marten) and human. Arrows indicates the isolates of zoo animals in this study.
Among the MDR E. faecalis, 8 ST were identified, of which 1 (ST1362) was new (Table 3). Interestingly, all but 1 of the previously reported 7 ST types (ST721, ST36, ST116, ST202, ST32, and ST 76) have been reported in humans, except for 1 (ST1363), for which the source was unknown, and 1 (ST32) out of 6 was reported from hospitalized patient specimens in China, Spain, Cuba, Germany, and Portugal. Compared to the ST type reported in Korea, three major clonal complexes containing the ST isolated in this study were identified (Figure 2). Particularly, ST32, ST36, and ST202, isolated from a red fox, amur leopard cat, and lion, respectively, showed SLV relationships with various domestic ST types isolated from pig farms (Table 5). The Puma isolate (ST721) and the spotted hyena isolate (ST984) showed close relationships at the SLV stage.
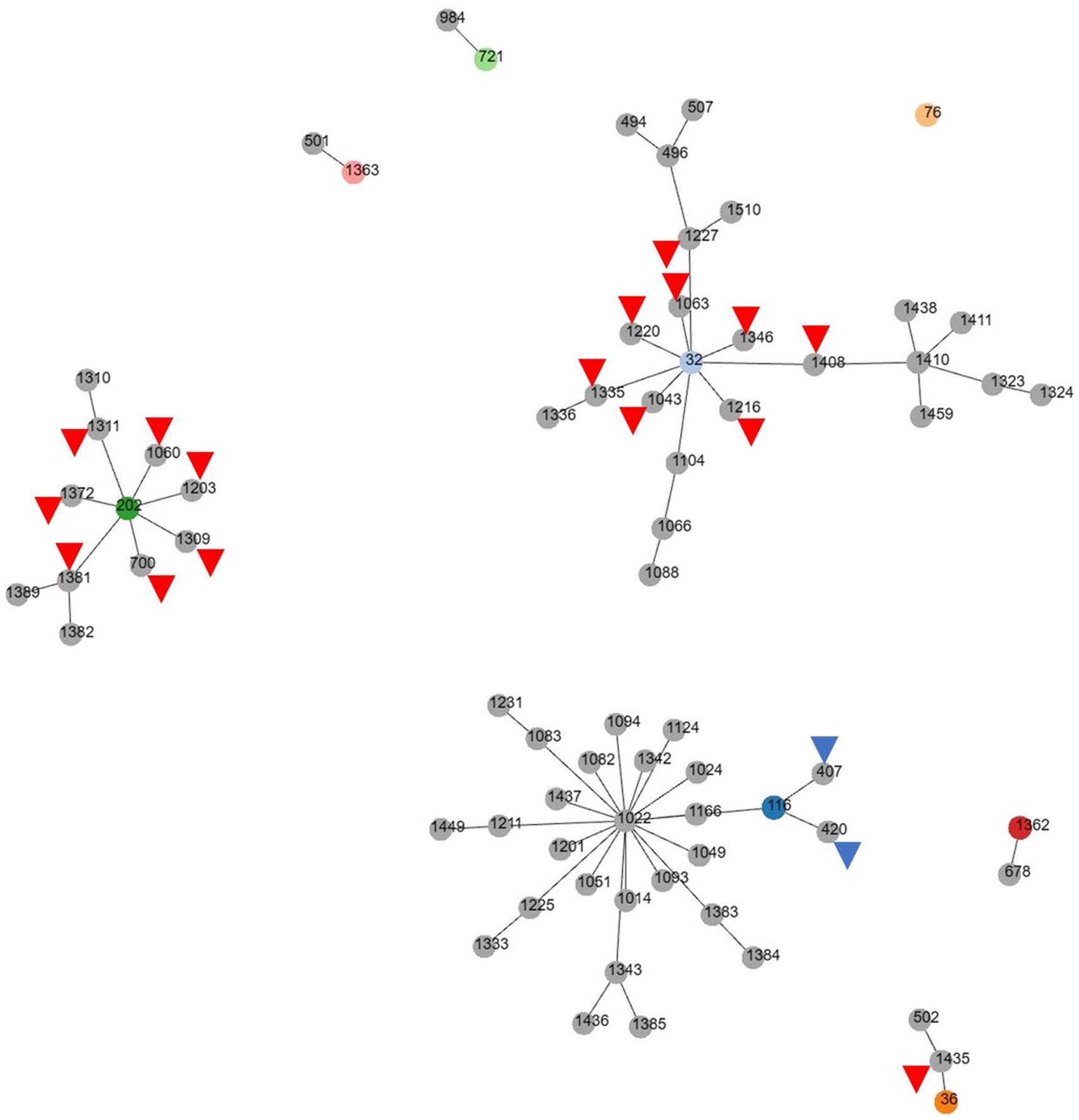
Figure 2. goeBURST (PHTLOViZ) analysis of multidrug-resistant Enterococcus faecalis reported in Korea. While three major clonal complexes containing the ST type isolated in this study are identified, ST32, ST116 and ST202 show SLV relationships with various domestic ST types isolated from pig farms (red arrowheads) or chicken (blue arrowheads).
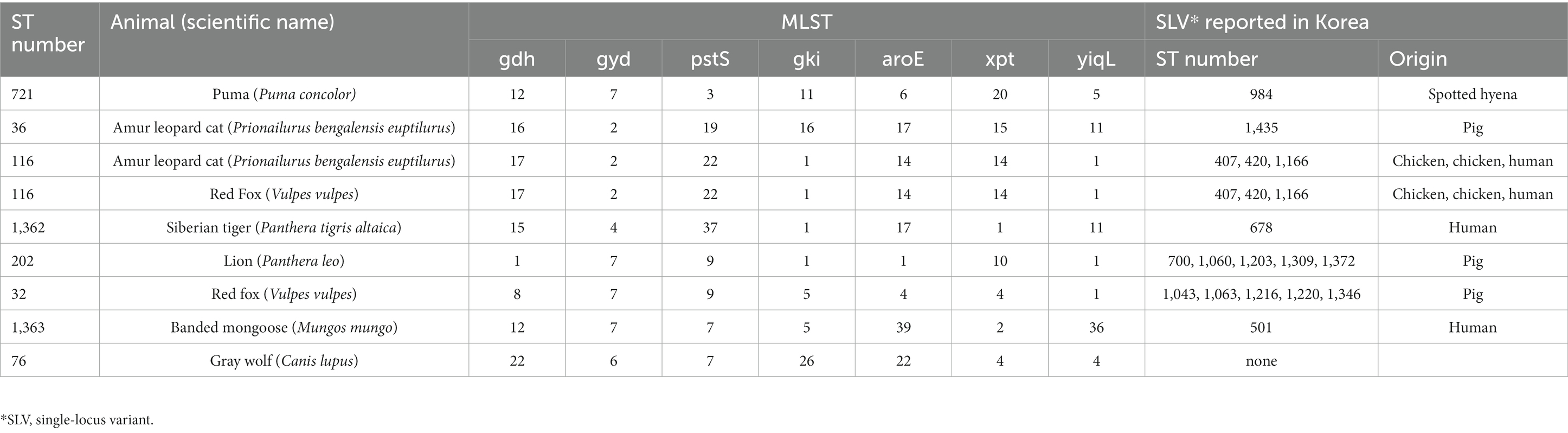
Table 5. Information of multilocus sequence type and the clonal complex relationship of multidrug-resistant E. faecalis isolated in this study.
Among 58 E. coli strains, a total of 52 E. coli strains were analyzed, including 18 MDR E coli strains. As a result of the analysis, the MDR strains were found to belong to the same clade except for one (ST5415; Figure 3). The 17 E. coli strains belonging to the same clade were composed of various species and animals with various feeding habits, showing contradictory results to MLST.
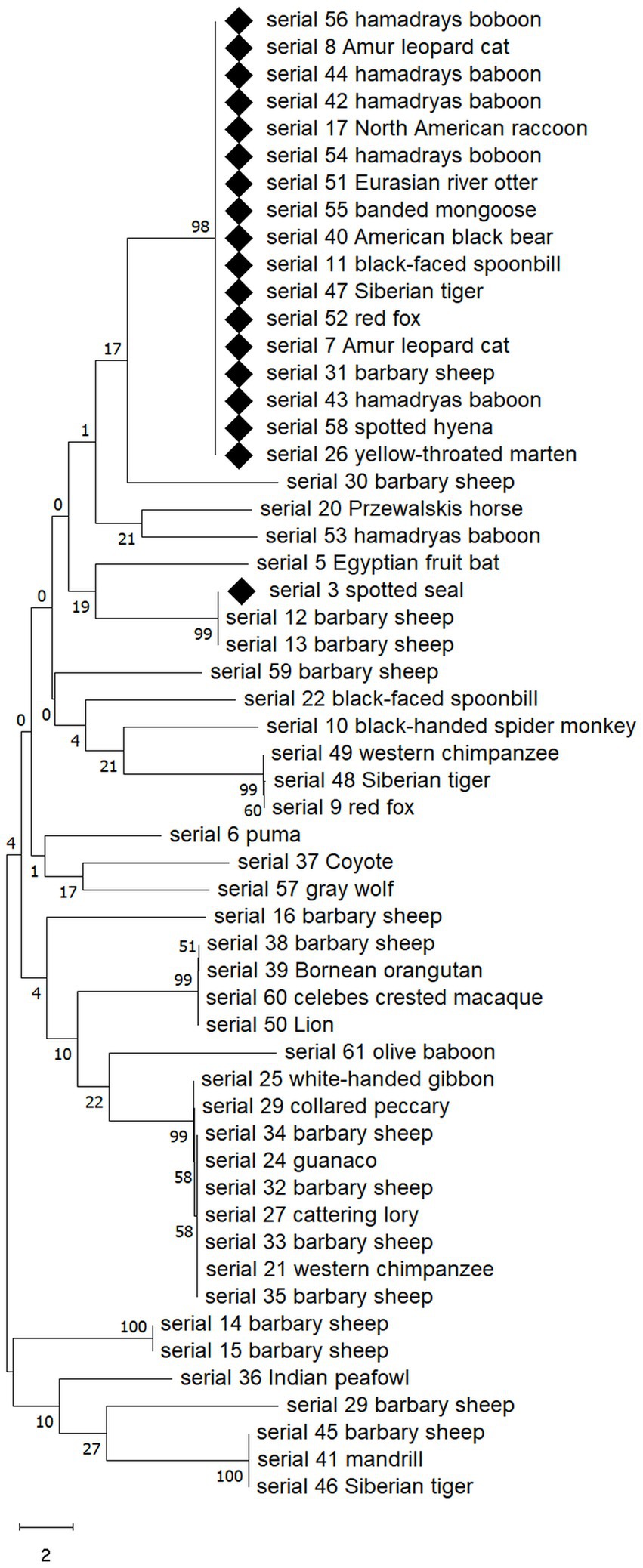
Figure 3. Neighbor-joining phylogenetic analysis of isolated 52 E. coli strains showing MDR E. coli appears to belong to the same clade except for one. Sequences of MDR strains are marked with diamonds. The analysis was performed by MEGA v10.0. Numbers on branches indicate bootstrap values based on 1,000 replicates.
This study was conducted to evaluate the degree and characteristics of antibiotic resistance in E. coli and E. faecalis in the intestines of clinically healthy zoo-fed wild animals. The results showed that both the isolated E. coli and E. faecalis were highly resistant to specific antibiotics (ampicillin, tetracycline, trimethoprim/sulfomethoxazole, and ciprofloxacin), besides the intrinsic resistance. Particularly, the incidence of MDR appears to be approximately 30% for both bacteria, suggesting that zoos cannot be an exception to the public health management of antibiotic resistance.
Three reasons have been suggested to explain why bacteria isolated from zoo animals acquire antibiotic resistance. First, owing to the characteristics of zoo animals, it is difficult to properly select antibiotics and administer them at an appropriate dose and duration. In zoo animals, it is often difficult to conduct appropriate tests in a timely manner when there is a need for antibiotics, and it is difficult to evaluate treatment progress. As a result, if the appropriate antibiotic is not used or if it is not used for a sufficient period and dose, it is easy for residual bacteria to acquire antibiotic resistance (21, 22). The E. coli isolated in this study showed high frequency of resistance to ampicillin, tetracycline, and trimethoprim/sulfamethoxazole. Owing to their wide application range and easy accessibility, these antibiotics have been widely used as empirical antibiotics in zoos, and the results of this study seem to reflect this. Among similar overseas zoo studies, a study at Chinese zoos showed that ampicillin, tetracycline, sulfamethoxazole-trimethoprim, and doxycycline have the highest E. coli resistance, which is similar to our results (23). However, a significant difference was that the ampicillin resistance rate in this study was 93% (27/29), which was higher than the Chinese study result (54.3%, 540/995). In the Petting Zoo of Canada, a study found that tetracycline and ampicillin resistance were the highest in captive wild animals (llamas and birds) targeting the non-O157 STEC serogroup (24). According to the results of a 2012 antimicrobial study conducted in a Japanese zoo, tetracycline, streptomycin, and ampicillin were the most common antibiotic resistance (25), which is largely consistent with the results of this study. Similar to the results of antimicrobial susceptibility studies in livestocks (26), resistance to tetracycline and ampicillin was high. The multidrug resistance rate was also higher than that of cattle (16%) but lower than that of pigs (69.7%) and chickens (82.6%) (26).
Second, food supplied to zoo animals is introduced in a state contaminated with resistant bacteria or resistant genes. The fact that antibiotic-resistant strains are more common in carnivorous and omnivorous mammals than in herbivorous mammals suggests that accumulation in the body of animals higher up in the food chain is also involved in antibiotic resistance. Among the MDR E. coli isolates in this study, previously reported ST types were all isolated from livestock products (chicken and cattle) and, for MDR E. faecalis, 4 of 7 cases were also reported from livestock products (chicken). Additionally, 17 of the 18 MDR E. coli strains were isolated from either carnivores or omnivores, and the fact that all E. faecalis isolates were isolated only from carnivores seems to reflect this. In zoos, there are cases where cheap imported food is supplied to breeding animals due to economic factors, and antibiotic resistance can be transferred through this; therefore, management measures for the current status of antibiotic resistance in the importing country or contamination of imported meat are needed.
The third factor is the potential for horizontal transmission between contact groups of animals, including zoo workers. In the case of methicillin-resistant Staphylococcus pseudintermedius isolated from dogs (Canis lupus familiaris), widespread horizontal transmission of a specific clone has been reported in the United States and Europe but not in Korea (27). A zoo is a closed and distinct ecosystem, a space in which direct contact between working veterinarians, zookeepers, and animals or indirect contact with objects, tools, and food occurs continuously, thus mutual horizontal transmission is possible. Based on the MLST and goeBurst analyses conducted in this study, 7 MDR E. coli isolates and three MDR E. faecalis isolates were found to be single-locus variants of strains reported from humans in Korea, suggesting the possibility that it is a change that occurs during the process of horizontal propagation in humans and animals. Therefore, continuous monitoring and investigation for the transmission are required to manage antibiotic resistance.
However, our results suggest that antibiotic resistance in the zoos investigated was not caused by a single factor. For example, in the case of food, carnivores receive meat provided from the same source at the same time daily; therefore, if food is the main entry route for resistance, carnivores should have the same or similar ST type, but this has not been the case. Rather, Barbary sheep and spotted hyenas with the same ST (ST155) have different diets and completely separate breeding areas, suggesting that the possibility of transmission by antibiotic use or contact with people or tools is higher than that by feeding. As a result of MLST for the MDR strain, a wide variety of ST types was detected, and only some shared the same ST, making it difficult to view horizontal transmission through direct or indirect contact between animals and humans or between animals and animals as the main route of antibiotic resistance acquisition. Resistance due to the use of antibiotics had a clear influence on the occurrence of resistance, given that resistance to antibiotics used by zoos was generally high in both strains investigated.
Of the 61 samples, Barbary sheep accounted for the majority (17 cases). The zoo investigated had about 55 Barbary sheep and regularly performed hoof care on about 20 Barbary sheep annually; therefore, the largest number of samples could be tested. All Barbary sheep ate the same feed in the same enclosure; there was little E. coli and E. faecalis resistance (5.9%, 1/17). However, as described in the results, Hamadryas baboons were the species that have highest frequency of MDR E. coli (83.3% 5/6). Similarly, MDR E. faecalis has been isolated from all carnivores, including pumas, Amur leopard cats, red foxes, Siberian tigers, lions, banded mongooses, and gray wolves. Various studies have shown that carnivores have higher multidrug resistance than omnivores or herbivores (28). Another factor that makes carnivores more resistant than other species in zoos is that, unlike herbivores, carnivores received more antimicrobials because antibiotics can be hidden in their food and can be easily administered. However, given that the biggest difference in feeding management between herbivores and carnivores is meat, the possibility that resistance factors (bacteria or gene) originate from meat cannot be ruled out (29, 30) and additional research is needed.
In this study, 7 MDR strains of E. coli showed SLV with human isolates in Korea while 6 MDR strains of E. faecalis showed SLV with human isolates in Korea. Unlike E. coli, where it was difficult to determine the origin of SLV because of the lack of data, it was confirmed that three ST types (ST32, ST202 and ST116) of E. faecalis linked several SLVs previously identified in pig (ST 32 and ST202) or chicken/human (ST116) in Korea. Interestingly, ST32 was reported to be isolated from chicken meat in Korea (31), and in this study, it was confirmed to be the branched to several ST types isolated from pigs along with ST202. On the other hand, in the case of ST116, it was confirmed that it branched from a human isolate (ST1166) and into two isolates reported in chickens (ST407 and ST420), suggesting the possibility that humans were involved in the introduction into the zoo. However, the fact that different results are obtained from the same species points out that there are various factors involved in the introduction and spread of the bacteria.
What was interesting in this study was that among the 52 isolates of E. coli included in the phylogenetic tree analysis, the strains that showed MDR belonged to the same clade, except for one. In particular, they belonged to the same clade regardless of animal taxon, species, or feeding habits, which was contrary to the diversity of MDR E. coli ST types shown in MLST. This result suggests that among E. coli isolates from the zoo, strains belonging to a specific clade evaluated based on the gyrB gene acquired MDR more easily, or that a specific strain, although the origin is unclear, gradually spread and differentiated after acquiring MDR.
Although Enterococcus species is a commonly found bacterium in the intestine, E. faecalis was isolated from only 48% (29/61) of the animals in this study. While E. faecalis and E. faecium are known to be the most abundant species in humans (32), the distribution of Enterococcus species in animals has been reported to vary (33). In this study, we attempted to isolate and culture E. faecalis from various animals raised in zoos, but no consistent pattern was observed in animal taxa, species, or type of food consumed. However, considering that Enterococcus species may have a relationship of exchanging resistance with each other, future studies also need to confirm the resistance patterns of major Enterococcus species in each animal.
This study investigated the frequency and characteristics of antibiotic-resistant bacteria in zoo wild animals kept in limited spaces. As a result of the investigation, high MDR bacterial isolation was observed in carnivores, and clones isolated from human infection sites were detected, so continuous investigation into the introduction route appears to be necessary.
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
The animal study was approved by Institutional Animal Care and Use Committee in Seoul zoo. The study was conducted in accordance with the local legislation and institutional requirements.
MiK: Data curation, Formal analysis, Investigation, Methodology, Resources, Writing – original draft. MyK: Data curation, Investigation, Writing – original draft, Formal analysis, Software, Visualization. Y-GY: Data curation, Investigation, Writing – original draft, Resources. Y-TL: Writing – original draft, Data curation, Investigation. J-IH: Conceptualization, Formal analysis, Funding acquisition, Investigation, Methodology, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by the National Institute of Wildlife Disease Control and Prevention as a “Specialized Graduate School Support Project for Wildlife Diseases Specialists.”
All authors thank the staff of the Seoul zoo for their helps on collecting samples.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Robinson, TP , Bu, DP , Carrique-Mas, J , Fèvre, EM , Gilbert, M , Grace, D, et al. Antibiotic resistance is the quintessential one health issue. Trans R Soc Trop Med Hyg. (2016) 110:377–80. doi: 10.1093/trstmh/trw048
2. Ozawa, Y , Tanimoto, K , Nomura, T , Yoshinaga, M , Arakawa, Y , and Ike, Y . Vancomycin-resistant enterococci in humans and imported chickens in Japan. Appl Environ Microbiol. (2002) 68:6457–61. doi: 10.1128/AEM.68.12.6457-6461.2002
3. Albertini, MT , Benoit, C , Berardi, L , Berrouane, Y , Boisivon, A , Cahen, P, et al. Surveillance of methicillin-resistant Staphylococcus aureus (MRSA) and Enterobacteriaceae producing extended-spectrum beta-lactamase (ESBLE) in northern France: a five-year multicentre incidence study. J Hosp Infect. (2002) 52:107–13. doi: 10.1053/jhin.2002.1286
4. Murthy, R . Implementation of strategies to control antimicrobial resistance. Chest. (2001) 119:405S–11S. doi: 10.1378/chest.119.2_suppl.405s
5. Kwak, HJ , Lee, WW , Kim, JH , Chung, KT , Woo, BG , Lee, BG, et al. The antimicrobial susceptibility and plasmid profile of E coli isolates from wild bird. Korean J Vet Ser. (2006) 29:37–46.
6. Kim, KH , Lim, HS , Lee, JW , Park, DH , Yang, CR , and Cho, JK . Antimicrobial resistance of Escherichia coli isolated from wild birds in Daegu. Korean J Vet Ser. (2021) 44:209–16. doi: 10.7853/kjvs.2021.44.4.209
7. Elena, SF , Whittam, TS , Winkworth, CL , Riley, MA , and Lenski, RE . Genomic divergence of Escherichia coli strains: evidence for horizontal transfer and variation in mutation rates. Int Microbiol. (2005) 8:271–8.
8. Miller, K , O’Neill, AJ , and Chopra, I . Escherichia coli mutators present an enhanced risk for emergence of antibiotic resistance during urinary tract infections. Antimicrob Agents Chemother. (2004) 48:23–9. doi: 10.1128/AAC.48.1.23-29.2004
9. Murray, BE . Diversity among multidrug-resistant enterococci. Emerg Infect Dis. (1998) 4:37–47. doi: 10.3201/eid0401.980106
10. Sava, IG , Heikens, E , and Huebner, J . Pathogenesis and immunity in enterococcal infections. Clin Microbiol Infect. (2010) 16:533–40. doi: 10.1111/j.1469-0691.2010.03213.x
11. Ward, SJ , Sherwen, S , and Clark, FE . Advances in applied zoo animal welfare science. J Appl Anim Welf Sci. (2018) 21:23–33. doi: 10.1080/10888705.2018.1513842
12. Conrad, CC , Stanford, K , Narvaez-Bravo, C , Callaway, T , and McAllister, T . Farm fairs and petting zoos: a review of animal contact as a source of zoonotic enteric disease. Foodborne Pathog Dis. (2017) 14:59–73. doi: 10.1089/fpd.2016.2185
13. Collignon, PJ , Conly, JM , Andremont, A , McEwen, SA , and Aidara-Kane, AWHO-AGISAR, et al. World Health Organization ranking of antimicrobials according to their importance in human medicine: a critical step for developing risk management strategies for the use of antimicrobials in food production animals. Clin Infect Dis. (2016) 63:1087–93. doi: 10.1093/cid/ciw475
14. Chantziaras, I , Boyen, F , Callens, B , and Dewulf, J . Correlation between veterinary antimicrobial use and antimicrobial resistance in food-producing animals: a report on seven countries. J Antimicrob Chemother. (2014) 69:827–34. doi: 10.1093/jac/dkt443
15. McMillian, M , Dunn, JR , Keen, JE , Brady, KL , and Jones, TF . Risk behaviors for disease transmission among petting zoo attendees. J Am Vet Med Assoc. (2007) 231:1036–8. doi: 10.2460/javma.231.7.1036
16. Stirling, J , Griffith, M , Dooley, JG , Goldsmith, CE , Loughrey, A , Lowery, CJ, et al. Zoonoses associated with petting farms and open zoos. Vector Borne Zoonotic Dis. (2008) 8:85–92. doi: 10.1089/vbz.2006.0639
17. Petti, CA , Brandt, ME , Church, DL , Emler, S , Simmon, K , and Zelazny, AM . Interpretive criteria for identification of Bacteria and Fungi by targeted DNA sequencing. Wayne: Clinical and Laboratory Standards Institute (2018).
18. Lewis, JS , Weinstein, MP , Bobenchik, AM , Campeau, S , Cullen, SK , Dingle, T, et al. Performance standards for antimicrobial susceptibility testing. Wayne: Clinical and Laboratory Standards Institute (2022).
19. Lee, SY , Park, JH , Park, HS , Lee, MA , Kang, ES , and Hong, KS . Comparison of antimicrobial susceptibility testing methods to detect glycopeptide resistance in enterococci-E-test, Vitek, disk diffusion and agar dilution method. Korean J Clin Pathol. (2000) 20:301–7.
20. Magiorakos, AP , Srinivasan, A , Carey, RB , Carmeli, Y , Falagas, ME , Giske, CG, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. (2012) 18:268–81. doi: 10.1111/j.1469-0691.2011.03570.x
21. Blue, JL , and Wooley, RE . Antibacterial sensitivity patterns of bacteria isolated from dogs with otitis externa. J Am Vet Med Assoc. (1977) 171:362–3.
22. Rodrigues da Costa, M , and Diana, AA . A systematic review on the link between animal welfare and antimicrobial use in captive animals. Animals (Basel). (2022) 12:1025. doi: 10.3390/ani12081025
23. Zhu, Z , Jiang, S , Qi, M , Liu, H , Zhang, S , Liu, H, et al. Prevalence and characterization of antibiotic resistance genes and integrons in Escherichia coli isolates from captive non-human primates of 13 zoos in China. Sci Total Environ. (2021) 798:149268. doi: 10.1016/j.scitotenv.2021.149268
24. Conrad, CC , Stanford, K , Narvaez-Bravo, C , Neumann, NF , Munns, K , Tymensen, L, et al. Zoonotic fecal pathogens and antimicrobial resistance in Canadian petting zoos. Microorganisms. (2018) 6:70. doi: 10.3390/microorganisms6030070
25. Ishihara, K , Hosokawa, Y , Makita, K , Noda, J , Ueno, H , Muramatsu, Y, et al. Factors associated with antimicrobial-resistant Escherichia coli in zoo animals. Res Vet Sci. (2012) 93:574–80. doi: 10.1016/j.rvsc.2011.09.006
26. Lim, SK , Nam, HM , Moon, DC , Jang, GC , Jung, SC , and Korean, V . Antimicrobial resistance of Escherichia coli isolated from healthy animals during 2010-2012. Korean J Vet Res. (2014) 54:131–7. doi: 10.14405/kjvr.2014.54.3.131
27. Han, JI , Rhim, H , Yang, CH , and Park, HM . Molecular characteristics of new clonal complexes of Staphylococcus pseudintermedius from clinically normal dog. Vet Q. (2017) 38:14–20. doi: 10.1080/01652176.2017.1400710
28. Bamunusinghage, NPD , Neelawala, RG , Magedara, HP , Ekanayaka, NW , Kalupahana, RS , Silva-Fletcher, A, et al. Antimicrobial resistance patterns of fecal Escherichia coli in wildlife, urban wildlife, and livestock in the eastern region of Sri Lanka, and differences between carnivores, omnivores, and herbivores. J Wildl Dis. (2022) 58:380–3. doi: 10.7589/JWD-D-21-00048
29. Rahman, MA , Rahman, AKMA , Islam, MA , and Alam, MM . Antimicrobial resistance of Escherichia coli isolated from milk, beef and chicken meat in Bangladesh. Bangl J Vet Med. (2017) 15:141–6. doi: 10.3329/bjvm.v15i2.35525
30. White, DG , Zhao, S , Simjee, S , Wagner, DD , and McDermott, PF . Antimicrobial resistance of foodborne pathogens. Microbes Infect. (2002) 4:405–12. doi: 10.1016/s1286-4579(02)01554-x
31. Kim, YB , Seo, HJ , Seo, KW , Jeon, HY , Kim, DK , Kim, SW, et al. Characteristics of high-level ciprofloxacin-resistant Enterococcus faecalis and Enterococcus faecium from retail chicken meat in Korea. J Food Prot. (2018) 81:1357–63. doi: 10.4315/0362-028X.JFP-18-046
32. Zaheer, R , Cook, SR , Barbieri, R , Goji, N , Cameron, A , Petkau, A, et al. Surveillance of Enterococcus spp. reveals distinct species and antimicrobial resistance diversity across a one-health continuum. Sci Rep. (2020) 10:3937. doi: 10.1038/s41598-020-61002-5
Keywords: antimicrobial resistance, zoo animals, genotype, Escherichia coli, Enterococcus faecalis
Citation: Kim M, Kim M, Yeo Y-G, Lee Y-T and Han J-I (2024) Antimicrobial resistance of commensal Escherichia coli and Enterococcus faecalis isolated from clinically healthy captive wild animals in Seoul zoo. Front. Vet. Sci. 10:1283487. doi: 10.3389/fvets.2023.1283487
Received: 26 August 2023; Accepted: 20 December 2023;
Published: 11 January 2024.
Edited by:
Marta Martinez Aviles, Instituto Nacional de Investigación y Tecnología Agroalimentaria (INIA), SpainReviewed by:
Marta Dec, University of Life Sciences of Lublin, PolandCopyright © 2024 Kim, Kim, Yeo, Lee and Han. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jae-Ik Han, amloYW5AamJudS5hYy5rcg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.