- 1Department of Comparative Pathobiology, Purdue University, West Lafayette, IN, United States
- 2Department of Veterinary Clinical Sciences, Washington State University, Pullman, WA, United States
MicroRNAs (miRNAs) are small non-coding RNAs that function by post-transcriptional regulation of gene expression. Their stability and abundance in tissue and body fluids makes them promising potential tools for both the diagnosis and prognosis of diseases and attractive therapeutic targets in humans and dogs. Studies of miRNA expression in normal and disease processes in dogs are scarce compared to studies published on miRNA expression in human disease. In this literature review, we identified 461 peer-reviewed papers from database searches using the terms “canine,” “dog,” “miRNA,” and “microRNA”; we screened 244 for inclusion criteria and then included a total of 148 original research peer-reviewed publications relating to specific miRNA expression in canine samples. We found an overlap of miRNA expression changes between the four groups evaluated (normal processes, non-infectious and non-inflammatory conditions, infectious and/or inflammatory conditions, and neoplasia) in 39 miRNAs, 83 miRNAs in three of the four groups, 110 miRNAs in two of the three groups, where 158 miRNAs have only been reported in one of the groups. Additionally, the mechanism of action of these overlapping miRNAs varies depending on the disease process, elucidating a need for characterization of the mechanism of action of each miRNA in each disease process being evaluated. Herein we also draw attention to the lack of standardization of miRNA evaluation, consistency within a single evaluation method, and the need for standardized methods for a direct comparison.
Introduction
MicroRNAs (miRNAs), first discovered in 1993 (1), are short (18–24 nucleotide) non-coding RNAs that perform regulatory functions through post-transcriptional regulation of gene expression (2–4). While individual miRNAs are not required for individual tissue development, they are often required to maintain homeostasis (2). MiRNAs are essential modulators of cell differentiation, proliferation, and function, and their expression is often altered in disease states such as cancer, metabolic disease, and with response to infectious agents (2). Gene expression studies have demonstrated such alterations, and functional studies have also linked miRNA dysregulation as a factor in disease progression (2).
The numerous alterations of miRNA in disease provide great potential for using miRNAs as diagnostic biomarkers (2). MiRNAs are stable and can be recovered from formalin-fixed, paraffin-embedded (FFPE) sections and other sources where overall RNA quality may be low (2). MiRNAs are often released from cells in exosomes and microvesicles or circulate bonded to lipoproteins and RNA-protein complexes and thus are available in body fluids, including plasma, saliva, and urine (2, 5–21). MiRNAs have also been detected in feces, tears, breast milk, bronchial lavage fluid, colostrum, seminal, amniotic, pleural, peritoneal, and cerebrospinal fluids (22). These characteristics make miRNAs suitable for diagnostic and prognostic testing. Additionally, as miRNA signatures in disease processes become known, efforts are being made to directly target dysregulated miRNAs to treat disease with either miRNA mimics or inhibitors, depending on the dysregulated miRNA and therapeutic goal (2, 23).
Although there are more studies published on miRNA expression in human disease than the veterinary counterpart, animal models are often used to elucidate the roles of miRNAs in oncogenesis and progression (24), as such, many similarities are found in miRNA expression between human and dog diseases. MiRNA expression studies have been performed on normal and abnormal canine tissues to evaluate physiological processes occurring during normal development and disease processes (24).
In dogs, aberrant miRNA expression has been identified in many cancer types, including but not limited to, lymphoma, mammary cancers, mast cell tumor, urothelial carcinoma, osteosarcoma, melanoma, and leukemia (24). While individual studies report miRNA expression in a specific disease process, miRNAs are often involved in numerous regulatory processes and altered expression may result in upregulation in one disease and downregulation in another (2–4). To date, most studies of miRNAs in dogs have generally evaluated one miRNA or a small group of miRNAs associated with a particular disease state or developmental process. The goal of this review was to describe what is reported concerning miRNA in dogs across normal physiologic processes (NP), non-infectious and non-inflammatory disease processes (NDP), infectious and/or inflammatory disease processes (IDP), and neoplasia. Here we concisely describe miRNA and their expression in both physiologic processes and disease states.
Materials and methods
Data acquisition
A search was performed on PubMed and Google Scholar for the terms “canine” OR “dog” AND “miRNA” OR “microRNA.” Publications available in the English language were considered. Review articles were not included in this study; however, original research publications cited within review articles were evaluated for inclusion in this review. Publications were screened for potential applicability.
Initial screening
A total of 461 reports were identified and further evaluated. Of the 461 reports identified, 224 were found to be not applicable (i.e., not evaluating dogs or not evaluating miRNAs) and 13 were unavailable for review. The remaining 224 were evaluated for inclusion eligibility. Criteria for inclusion in the review were 1. manuscript must be peer-reviewed, 2. manuscript must report evaluation of miRNA in canine samples, 3. manuscript must present original research (other literature review papers are not included in this literature review), and 4. manuscript must report expression evaluation relating to specific miRNA (i.e., blanket statements of miRNA being increased or decreased without discussion of individual miRNAs were not included in this review).
Results
A total of 148 peer-reviewed publications were included in this study (Figure 1). MiRNA expression in neoplasia is the most frequently published, with a total of 63 publications, while miRNA expression in NDP is the second most published, with a total of 55 publications. MiRNA expression in IDP represents 27 publications (Table 1). MiRNA expression in NP is the least represented in this review, with 10 publications (Table 1).
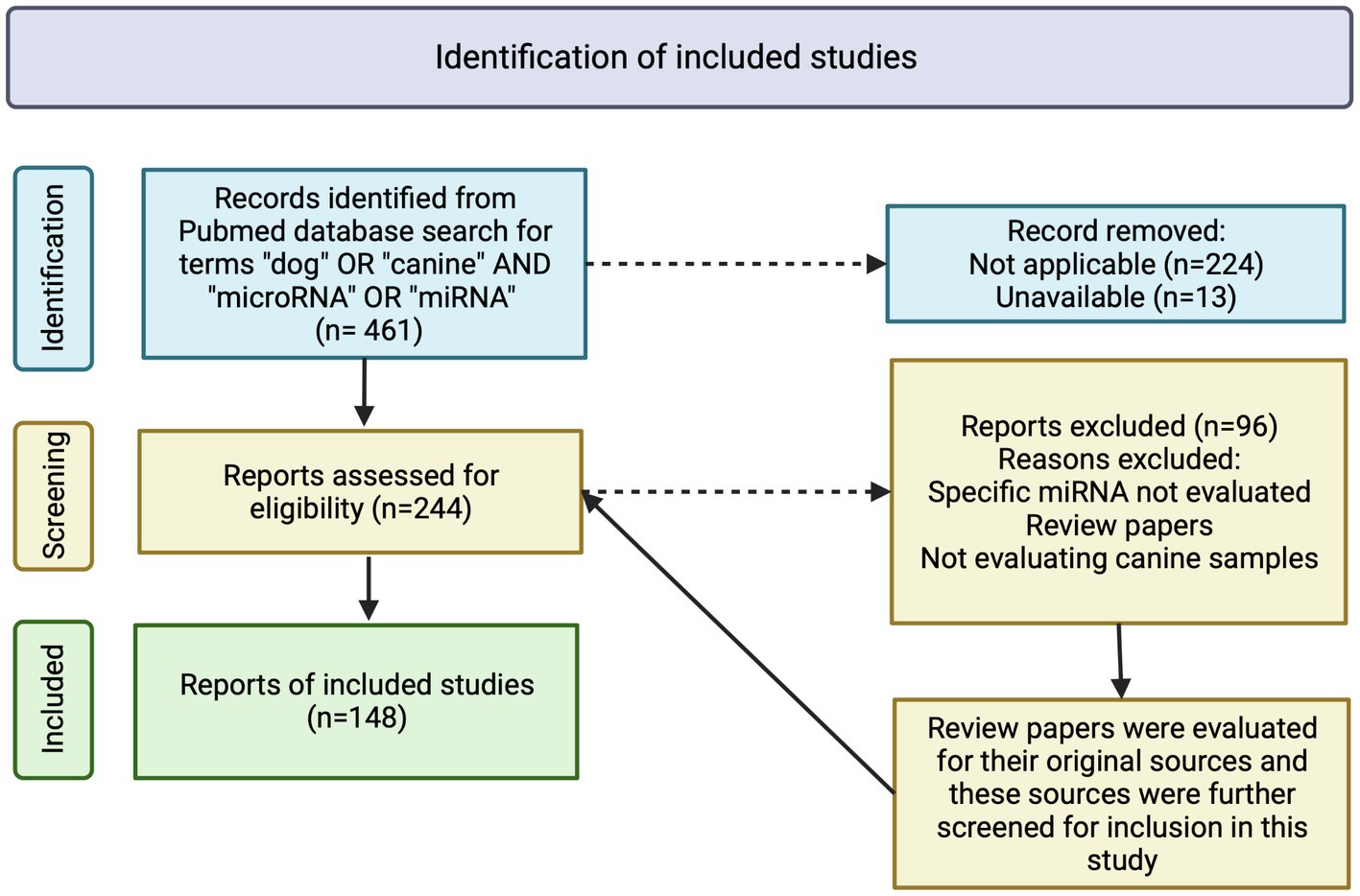
Figure 1. Flowchart of inclusion criteria and evaluation of peer-reviewed publications on detection and expression of microRNA in dogs. A total of 461 manuscripts were initially identified, with 237 excluded due to inapplicability or unavailability. Two-hundred and forty-four manuscripts were further assessed for eligibility with 96 being excluded due to inapplicability, not evaluating specific miRNA or the manuscripts being review papers. References of review papers were evaluated for applicability in this study. A total of 148 manuscripts were included in this review. Image created with biorender.com.
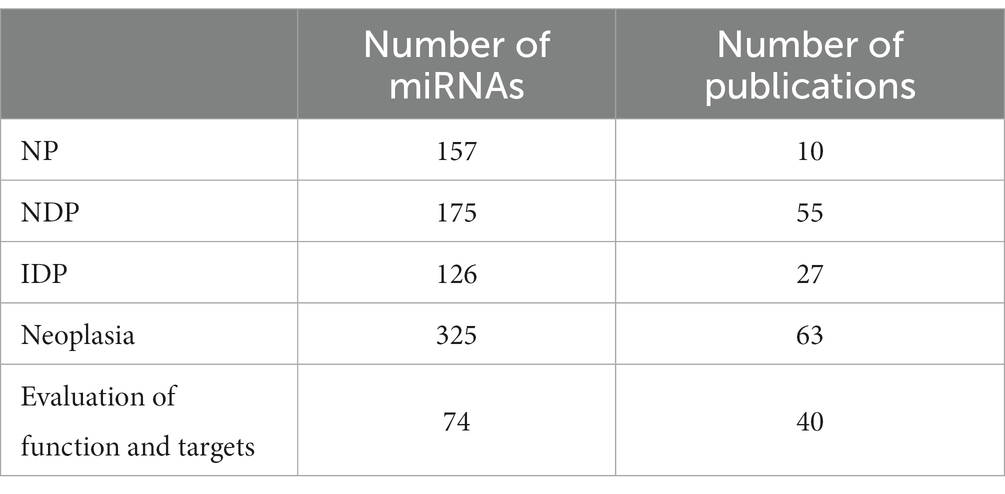
Table 1. Stratification of miRNAs and publications within the four groups: normal processes, noninfectious/non-inflammatory disease processes, infectious/inflammatory disease processes (NP, NDP, IDP, respectively) and neoplasia and experimental evaluation of functions and miRNA targets.
The publications included in this review reported the expression of a total of 391 miRNAs. Of those, 157 miRNAs are categorized in NP (Table 1; Supplementary Table S1), 175 miRNAs in NDP (Table 1; Supplementary Table S2), 126 miRNAs in IDP (Table 1; Supplementary Table S3), and 325 miRNAs in neoplasia (Table 1; Supplementary Table S4). Regarding overlapping, 158 miRNAs were studied in only one of the categories (NP, NDP, IDP, or neoplasia), 110 miRNAs were reported in two of the categories, 83 miRNAs reported in three, and 39 miRNAs were reported in all four groups (Table 1; Figure 2).
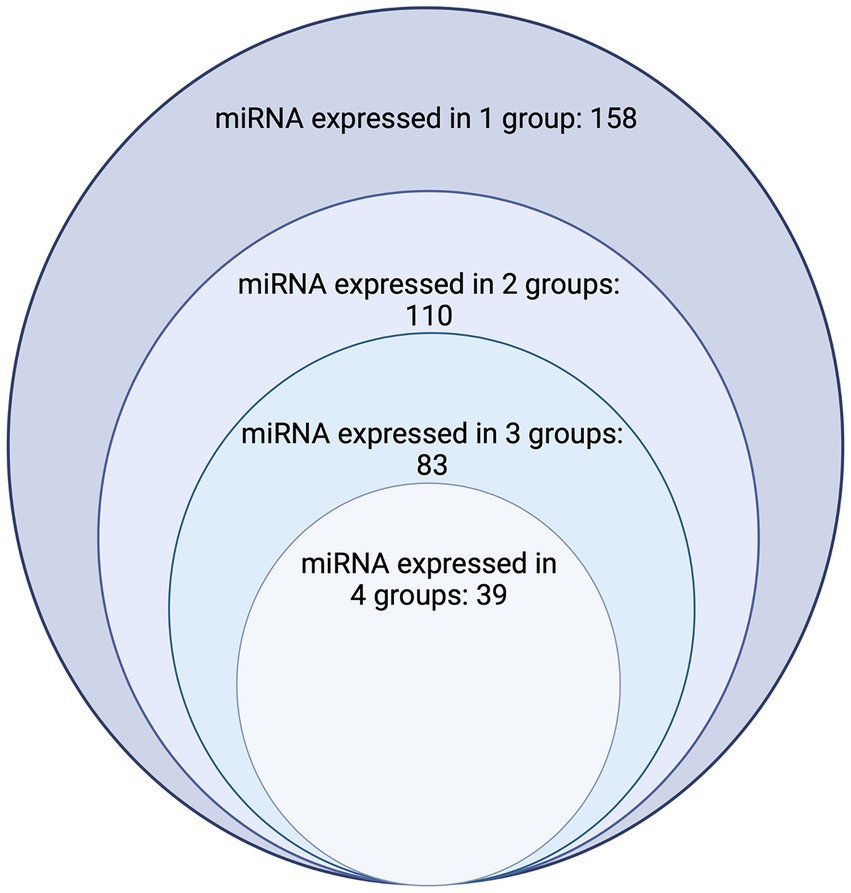
Figure 2. Chart depicting the number of dog miRNAs with expression documented in one, two, three, and four groups. The largest circle represents the number of miRNAs where expression has been found in only one group (n = 158), the second largest circle represents the number of miRNAs which have had expression characterized in two of the four groups (Normal processes, noninfectious/noninflammatory disease processes, infectious/inflammatory disease processes, and neoplasia) considered (n = 110). The third largest/s smallest circle represents the number of miRNAs which have had expression characterized in three of the four groups (n = 83). The smallest circle represents the number of miRNAs who have had expression characterized in all four groups considered (n = 39) Image created with biorender.com.
Forty articles included in this review also characterized the miRNA functions, targets, and/or affected pathways (Supplementary Table S5) of various miRNAs, including 14/39 of the miRNAs reported in all four of the groups.
MiR-1, expressed with both upregulation and downregulation in NDP (25–28) and neoplasia (29–36), expression in NP (37–39), and downregulation in IDP (40), targets both MET in hepatocellular carcinoma (33) and EDFR in mammary carcinoma (35, 41), and was shown to inhibit cell proliferation in a MET dependent manner (33). MiR-20a, reported to be expressed in NP (37, 42), upregulated in various neoplasms (7, 41, 43, 44), and downregulated in both NDP (45) and IDP (46), targets TGF-β affecting epithelial to mesenchymal transition and fibrosis in myxomatous mitral valve disease (45). MiR-214 was found to be both upregulated and downregulated in NP (37, 47) and neoplasia (7, 8, 41, 44, 48–52), downregulated in NDP (28), upregulated in IDP (40, 53), targets COP1, affecting the P53 pathways for apoptosis in hemangiosarcoma (50). Both miR-148a (enriched in NP (38), upregulated in NDP (54–56) and IDP (57), and both upregulated and downregulated in neoplasia (35, 41, 58–60)) and miR-205 [enriched in NP (38), downregulated in IDP (40), and both upregulated and downregulated in NDP (61–63) and neoplasia (24, 35, 41, 52, 60, 64–67)] target ERBB3 in mammary carcinoma and oral melanoma, respectively (41, 52, 58, 64, 65) while miR-205 has also been shown to target EGFR in mammary carcinoma (35, 41) and ECFC affecting NOTCH2 and promoting angiogenesis in distraction osteogenesis (61).
The majority of studies used only a single method of evaluation including reverse transcription-quantitative polymerase chain reaction (RT-qPCR) (8, 10–13, 16, 17, 19, 27, 31, 33, 36, 37, 40–43, 47, 49, 50, 52, 54, 55, 59, 60, 62, 68–121, 25–), microarray (44, 53, 122–125), or next generation sequencing (NGS) (32, 38, 126–131), Some studies evaluated the miRNA through two methods including NGS with validation via RT-qPCR (29, 30, 35, 39, 40, 45, 46, 51, 56, 61, 63, 65, 67, 132–144),microarray with validation via RT-qPCR (5, 9, 14, 18, 28, 57, 58, 64, 66, 145–149), one study utilized NGS with validation using digital drop PCR (ddPCR) (7), one study utilized nanostring with qRT-PCR (20), and two studies utilized in situ hybridization (ISH) for validation of their findings (39, 109).
Discussion
Several miRNAs have been evaluated in canine tissues from cytologic samples, histologic, and fecal samples, and body fluids, including plasma and serum, as well as canine cell lines. As reported in this literature review, an overlap of miRNA expression has been found in various disease processes.
The overlap in expression profiles of miRNA in various disease processes and normal tissues elucidates the need for further characterization of the mechanisms the miRNA used to regulate each disease process and their function in normal tissues to better utilize them as diagnostic, therapeutic, and prognostic tools. A single miRNA may negatively regulate multiple target proteins through interaction with different target mRNAs (23). Defining the targets of miRNAs becomes vital for understanding the biological role of the miRNAs and for identifying potential uses for therapeutic and diagnostic agents (2).
In this review we did not consider the effects differences of geographic location on miRNA expression and how differening treatment protocols and disease prevalences within different global locations may affect miRNA expression, as this is beyond the scope of this review. However, it should be considered that the treatment and management method of individual disease processes as well as environmental factors, which often vary by geographical location, may alter that pathophysiologic behavior of any one process which then could be reflected by a different expression panel of miRNAs.
Another limitation of this review is the variety of methods for evaluating miRNAs in dogs. In addition to a lack of standardization within the methods for evaluating miRNA, there is also a lack of standardization in how the normalization of RT-qPCR is performed. The differing normalization methods may reflect differences in the outcome of miRNA expression findings (150). Additionally, several studies utilized RNU6 as a reference gene when normalizing their RT-qPCR data (5, 10, 16, 18, 25, 28, 33, 39, 46, 47, 60–63, 65, 66, 79, 80, 82, 84, 85, 87, 92, 97, 101, 107–109, 113, 114, 116, 120, 135, 139–141, 143, 147, 149), which, since RNU6 is not a miRNA and therefore may not behave as a miRNA, has been suggested to lead to inaccurate or skewed results (151, 152). Additionally, some studies chose to normalize their data to their exogenous reference gene or spike in control (19, 55, 142). Recently, it has been recommended to evaluate each tissue individually to find the best miRNA to use as a reference gene using programs like NormFinder or GeNORM (153). Additional normalization has also been reported using the mean of miRNAs expressed (154). The lack of standardization in evaluating of miRNA raises concerns for studies with results that are not repeatable by independent sources.
Conclusion
This literature review characterizes the peer-reviewed literature on miRNA expression in dogs across four categories (NP, NDP, IDP, and neoplasia). Herein we have highlighted the overlap of miRNA expression in various disease processes tissues and that miRNA expression is dependent on disease process.
Author contributions
MV: Conceptualization, Formal analysis, Investigation, Methodology, Project administration, Visualization, Writing – original draft, Writing – review & editing. AS: Conceptualization, Project administration, Supervision, Visualization, Writing – review & editing.
Funding
The authors declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2023.1261085/full#supplementary-material
References
1. Feinbaum, R, Ambros, V, and Lee, R. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cells. (2004) 116:843–54. doi: 10.1016/0092-8674(93)90529-y
2. Hammond, SM. An overview of microRNAs. Adv Drug Deliv Rev. (2015) 87:3–14. doi: 10.1016/j.addr.2015.05.001
3. Reddy, KB. MicroRNA (miRNA) in cancer. Cancer Cell Int. (2015) 15:4–9. doi: 10.1186/s12935-015-0185-1
4. Rupaimoole, R, and Slack, FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. (2017) 16:203–21. doi: 10.1038/nrd.2016.246
5. Bragato, JP, Melo, LM, Venturin, GL, Rebech, GT, Garcia, LE, Lopes, FL, et al. Relationship of peripheral blood mononuclear cells miRNA expression and parasitic load in canine visceral Leishmaniasis. PLoS One. (2018) 13:1–16. doi: 10.1371/journal.pone.0206876
6. Dirksen, K, Verzijl, T, Grinwis, GC, Favier, RP, Penning, LC, Burgener, IA, et al. Use of serum MicroRNAs as biomarker for hepatobiliary diseases in dogs. J Vet Intern Med. (2016) 30:1816–23. doi: 10.1111/jvim.14602
7. Fish, EJ, Martinez-Romero, EG, DeInnocentes, P, Koehler, JW, Prasad, N, Smith, AN, et al. Circulating microRNA as biomarkers of canine mammary carcinoma in dogs. J Vet Intern Med. (2020) 34:1282–90. doi: 10.1111/jvim.15764
8. Heishima, K, Ichikawa, Y, Yoshida, K, Iwasaki, R, Sakai, H, Nakagawa, T, et al. Circulating microRNA-214 and -126 as potential biomarkers for canine neoplastic disease. Sci Rep. (2017) 7:1–14. doi: 10.1038/s41598-017-02607-1
9. Jeanson-Leh, L, Lameth, J, Krimi, S, Buisset, J, Amor, F, le Guiner, C, et al. Serum profiling identifies novel muscle miRNA and cardiomyopathy-related miRNA biomarkers in golden retriever muscular dystrophy dogs and duchenne muscular dystrophy patients. Am J Pathol. (2014) 184:2885–98. doi: 10.1016/j.ajpath.2014.07.021
10. Kent, MS, Zwingenberger, A, Westropp, JL, Barrett, LE, Durbin-Johnson, BP, Ghosh, P, et al. MicroRNA profiling of dogs with transitional cell carcinoma of the bladder using blood and urine samples. BMC Vet Res. (2017) 13:1–13. doi: 10.1186/s12917-017-1259-1
11. Li, Q, Freeman, LM, Rush, JE, and Laflamme, DP. Expression profiling of circulating microRNAs in canine myxomatous mitral valve disease. Int J Mol Sci. (2015) 16:14098–108. doi: 10.3390/ijms160614098
12. Di Loria, A, Dattilo, V, Santoro, D, Guccione, J, De Luca, A, Ciaramella, P, et al. Expression of serum exosomal miRNA 122 and lipoprotein levels in dogs naturally infected by Leishmania infantum: a preliminary study (2020) 10:1–10. doi: 10.3390/ani10010100
13. Marioni-Henry, K, Zaho, D, Amengual-Batle, P, Rzechorzek, NM, and Clinton, M. Expression of microRNAs in cerebrospinal fluid of dogs with central nervous system disease. Acta Vet Scand. (2018) 60:80. doi: 10.1186/s13028-018-0434-0
14. Nakata, K, Heishima, K, Sakai, H, Yamato, O, Furusawa, Y, Nishida, H, et al. Plasma microRNA miR-26b as a potential diagnostic biomarker of degenerative myelopathy in Pembroke welsh corgis. BMC Vet Res. (2019) 15:1–9. doi: 10.1186/s12917-019-1944-3
15. Narita, M, Nishida, H, Asahina, R, Nakata, K, Yano, H, Dickinson, PJ, et al. Expression of micrornas in plasma and in extracellular vesicles derived from plasma for dogs with glioma and dogs with other brain diseases. Am J Vet Res. (2020) 81:355–60. doi: 10.2460/ajvr.81.4.355
16. Ramadan, ES, Salem, NY, Emam, IA, AbdElKader, NA, Farghali, HA, and Khattab, MS. MicroRNA-21 expression, serum tumor markers, and immunohistochemistry in canine mammary tumors. Vet Res Commun. (2022) 46:377–88. doi: 10.1007/s11259-021-09861-9
17. Sanders, K, Veldhuizen, A, Kooistra, HS, Slob, A, Timmermans-Sprang, EPM, Riemers, FM, et al. Circulating MicroRNAs as non-invasive biomarkers for canine Cushing’s syndrome. Front Vet Sci. (2021) 8:1–12. doi: 10.3389/fvets.2021.760487
18. Steudemann, C, Bauersachs, S, Weber, K, and Wess, G. Detection and comparison of microRNA expression in the serum of Doberman pinschers with dilated cardiomyopathy and healthy controls. BMC Vet Res. (2013) 9:12. doi: 10.1186/1746-6148-9-12
19. Sun, B, Qu, Z, Cheng, GL, Yang, YW, Miao, YF, Chen, XG, et al. Urinary microRNAs miR-15b and miR-30a as novel noninvasive biomarkers for gentamicin-induced acute kidney injury. Toxicol Lett. (2021) 338:105–13. doi: 10.1016/j.toxlet.2020.12.006
20. Vansteenkiste, DP, Fenger, JM, Fadda, P, Martin-Vaquero, P, and da Costa, RC. MicroRNA expression in the cerebrospinal fluid of dogs with and without cervical spondylomyelopathy. J Vet Intern Med. (2019) 33:2685–92. doi: 10.1111/jvim.15636
21. Yang, VK, Loughran, KA, Meola, DM, Juhr, CM, Thane, KE, Davis, AM, et al. Circulating exosome microRNA associated with heart failure secondary to myxomatous mitral valve disease in a naturally occurring canine model. J Extracell Vesicles. (2017) 6:1350088. doi: 10.1080/20013078.2017.1350088
22. Cortez, MA, Bueso-ramos, C, Ferdin, J, Lopez-berestein, G, Sood, AK, and Calin, GA. MicroRNAs in body fluids--the mix of hormones and biomarkers. Nat Rev Clin Oncol. (2012) 8:467–77. doi: 10.1038/nrclinonc.2011.76
23. Wang, V, and Wu, W. MicroRNA-BAsed therapeutics for cancer. BioDrugs. (2009) 23:15–23. doi: 10.2165/00063030-200923010-00002
24. Sahabi, K, Selvarajah, GT, Abdullah, R, Cheah, YK, and Tan, GC. Comparative aspects of microRNA expression in canine and human cancers. J Vet Sci. (2018) 19:162–71. doi: 10.4142/jvs.2018.19.2.162
25. Chen, Y, Wakili, R, Xiao, J, Wu, CT, Luo, X, Clauss, S, et al. Detailed characterization of microRNA changes in a canine heart failure model: relationship to arrhythmogenic structural remodeling. J Mol Cell Cardiol. (2014) 77:113–24. doi: 10.1016/j.yjmcc.2014.10.001
26. Mizuno, H, Nakamura, A, Aoki, Y, Ito, N, Kishi, S, Yamamoto, K, et al. Identification of muscle-specific MicroRNAs in serum of muscular dystrophy animal models: promising novel blood-based markers for muscular dystrophy. PLoS One. (2011) 6:14–9. doi: 10.1371/journal.pone.0018388
27. Nakata, K, Namiki, M, Kobatake, Y, Nishida, H, Sakai, H, Yamato, O, et al. Up-regulated spinal microRNAs induce aggregation of superoxide dismutase 1 protein in canine degenerative myelopathy. Res Vet Sci. (2021) 135:479–85. doi: 10.1016/j.rvsc.2020.11.018
28. Qin, H, Chen, G, Liang, MY, Rong, J, Yao, J-p, Liu, H, et al. The altered expression profile of microRNAs in cardiopulmonary bypass canine models and the effects of mir-499 on myocardial ischemic reperfusion injury. J Transl Med. (2013) 11:154. doi: 10.1186/1479-5876-11-154
29. Chen, H-W, Lai, Y-C, Rahman, MM, Husna, AA, Hasan, MN, Hatai, H, et al. NGS-identified miRNAs in canine mammary gland tumors show unexpected expression alterations in qPCR analysis. In Vivo. (2022) 36:1628–36. doi: 10.21873/invivo.12873
30. Chen, H-W, Lai, Y-C, Rahman, MM, Husna, AA, Hasan, MN, and Miura, N. Micro RNA differential expression profile in canine mammary gland tumor by next generation sequencing. Gene. (2022) 818:146237. doi: 10.1016/j.gene.2022.146237
31. Fenger, JM, Roberts, RD, Iwenofu, OH, Bear, MD, Zhang, X, Couto, JI, et al. MiR-9 is overexpressed in spontaneous canine osteosarcoma and promotes a metastatic phenotype including invasion and migration in osteoblasts and osteosarcoma cell lines. BMC Cancer. (2016) 16:1–19. doi: 10.1186/s12885-016-2837-5
32. Koehler, J, Sandey, M, Prasad, N, Levy, SA, Wang, X, and Wang, X. Differential expression of miRNAs in hypoxia (“HypoxamiRs”) in three canine high-grade glioma cell lines. Front Vet Sci. (2020) 7:1–12. doi: 10.3389/fvets.2020.00104
33. Lai, YC, Ushio, N, Rahman, MM, Katanoda, Y, Ogihara, K, Naya, Y, et al. Aberrant expression of microRNAs and the miR-1/MET pathway in canine hepatocellular carcinoma. Vet Comp Oncol. (2018) 16:288–96. doi: 10.1111/vco.12379
34. Leonardo, L, Laura, P, and Serena, BM. miR-1 and miR-133b expression in canine osteosarcoma. Res Vet Sci. (2018) 117:133–7. doi: 10.1016/j.rvsc.2017.12.002
35. Lutful Kabir, FM, DeInnocentes, P, and Bird, RC. Altered microRNA expression profiles and regulation of INK4A/CDKN2A tumor suppressor genes in canine breast Cancer models. J Cell Biochem. (2015) 116:2956–69. doi: 10.1002/jcb.25243
36. Shukla, P, Vogl, C, Wallner, B, Rigler, D, Müller, M, and Macho-Maschler, S. High-throughput mRNA and miRNA profiling of epithelial-mesenchymal transition in MDCK cells. BMC Genomics. (2015) 16:1–19. doi: 10.1186/s12864-015-2036-9
37. Kasimanickam, VR, and Kasimanickam, RK. Differential expression of microRNAs in sexually immature and mature canine testes. Theriogenology. (2015) 83:394–398.e1. doi: 10.1016/j.theriogenology.2014.10.003
38. Koenig, EM, Fisher, C, Bernard, H, Wolenski, FS, Gerrein, J, Carsillo, M, et al. The beagle dog MicroRNA tissue atlas: identifying translatable biomarkers of organ toxicity. BMC Genomics. (2016) 17:1–13. doi: 10.1186/s12864-016-2958-x
39. Vacchi-Suzzi, C, Hahne, F, Scheubel, P, Marcellin, M, Dubost, V, Westphal, M, et al. Heart structure-specific transcriptomic atlas reveals conserved microRNA-mRNA interactions. PLoS One. (2013) 8:1–13. doi: 10.1371/journal.pone.0052442
40. Zou, Y, Bin, ZW, He, JJ, Elsheikha, HM, Zhu, XQ, and Lu, YX. Toxocara canis differentially affects hepatic MicroRNA expression in beagle dogs at different stages of infection. Front Vet Sci. (2020) 7:587273. doi: 10.3389/fvets.2020.587273
41. Fish, EJ, Irizarry, KJ, DeInnocentes, P, Ellis, CJ, Prasad, N, Moss, AG, et al. Malignant canine mammary epithelial cells shed exosomes containing differentially expressed microRNA that regulate oncogenic networks. BMC Cancer. (2018) 18:1–20. doi: 10.1186/s12885-018-4750-6
42. Cirera, S, Willumsen, LM, Johansen, TT, and Nielsen, LN. Evaluation of microRNA stability in feces from healthy dogs. Vet Clin Pathol. (2018) 47:115–21. doi: 10.1111/vcp.12566
43. Garnica, TK, Lesbon, JCC, Ávila, ACFCM, Rochetti, AL, Matiz, ORS, Ribeiro, RCS, et al. Liquid biopsy based on small extracellular vesicles predicts chemotherapy response of canine multicentric lymphomas. Sci Rep. (2020) 10:1–11. doi: 10.1038/s41598-020-77366-7
44. Uhl, E, Schliekelman, P, Thompkins, SM, and Suter, S. Identification of altered MicroRNA expression in canine lymphoid cell lines and cases of B- and T-cell lymphomas. Genes Chromosom Cancer. (2011) 50:950–67. doi: 10.1002/gcc.20917
45. Yang, VK, Tai, AK, Huh, TP, Meola, DM, Juhr, CM, Robinson, NA, et al. Dysregulation of valvular interstitial cell let-7c, MIR-17, MIR-20a, and MIR-30d in naturally occurring canine myxomatous mitral valve disease. PLoS One. (2018) 13:1–17. doi: 10.1371/journal.pone.0188617
46. Wu, X, Chen, X, Mi, W, Wu, T, Gu, Q, and Huang, H. MicroRNA sequence analysis identifies microRNAs associated with peri-implantitis in dogs. Biosci Rep. (2017) 37:1–12. doi: 10.1042/BSR20170768
47. Kasimanickam, VR, Kasimanickam, RK, and Dernell, WS. Dysregulated microRNA clusters in response to retinoic acid and CYP26B1 inhibitor induced testicular function in dogs. PLoS One. (2014) 9:e99433. doi: 10.1371/journal.pone.0099433
48. Heishima, K, Meuten, T, Yoshida, K, Mori, T, and Thamm, DH. Prognostic significance of circulating microRNA-214 and -126 in dogs with appendicular osteosarcoma receiving amputation and chemotherapy. BMC Vet Res. (2019) 15:1–13. doi: 10.1186/s12917-019-1776-1
49. Heishima, K, Mori, T, Ichikawa, Y, Sakai, H, Kuranaga, Y, Nakagawa, T, et al. MicroRNA-214 and microRNA-126 are potential biomarkers for malignant endothelial proliferative diseases. Int J Mol Sci. (2015) 16:25377–91. doi: 10.3390/ijms161025377
50. Heishima, K, Mori, T, Sakai, H, Sugito, N, Murakami, M, Yamada, N, et al. MicroRNA-214 promotes apoptosis in canine hemangiosarcoma by targeting the COP1-p53 axis. PLoS One. (2015) 10:1–19. doi: 10.1371/journal.pone.0137361
51. Varvil, MS, Bailey, TW, Dhawan, D, Deborah, W, and Ramos-vara, JA. The miRNome of canine invasive urothelial carcinoma. Front Vet Sci. (2022) 9:945638. doi: 10.3389/fvets.2022.945638
52. Zamarian, V, Catozzi, C, Ressel, L, Finotello, R, Ceciliani, F, Vilafranca, M, et al. MicroRNA expression in formalin-fixed, paraffin-embedded samples of canine cutaneous and Oral melanoma by RT-qPCR. Vet Pathol. (2019) 56:848–55. doi: 10.1177/0300985819868646
53. Soares, MF, Melo, LM, Bragato, JP, Furlan, AO, Scaramele, NF, Lopes, FL, et al. Differential expression of miRNAs in canine peripheral blood mononuclear cells (PBMC) exposed to Leishmania infantum in vitro. Res Vet Sci. (2021) 134:58–63. doi: 10.1016/j.rvsc.2020.11.021
54. Alawneh, KZ, Raffee, LA, Alshehabat, MAM, Haddad, H, and Jaradat, SA. Characterizing and profiling micrornas in dogs undergoing induced ischemic brain stroke after middle cerebral artery occlusion under fluoroscopic guidance. Vasc Health Risk Manag. (2021) 17:543–50. doi: 10.2147/VHRM.S317861
55. Dirksen, K, Verzijl, T, van den Ingh, TSGAM, Vernooij, JCM, van der Laan, LJW, Burgener, IA, et al. Hepatocyte-derived microRNAs as sensitive serum biomarkers of hepatocellular injury in Labrador retrievers. Vet J. (2016) 211:75–81. doi: 10.1016/j.tvjl.2016.01.010
56. Rouse, R, Rosenzweig, B, Shea, K, Knapton, A, Stewart, S, Xu, L, et al. MicroRNA biomarkers of pancreatic injury in a canine model. Exp Toxicol Pathol. (2017) 69:33–43. doi: 10.1016/j.etp.2016.11.001
57. Melo, LM, Bragato, JP, Venturin, GL, Rebech, GT, Costa, SF, Garcia, LE, et al. Induction of miR 21 impairs the anti-Leishmania response through inhibition of IL-12 in canine splenic leukocytes. PLoS One. (2019) 14:1–19. doi: 10.1371/journal.pone.0226192
58. Bulkowska, M, Rybicka, A, Senses, KM, Ulewicz, K, Witt, K, Szymanska, J, et al. MicroRNA expression patterns in canine mammary cancer show significant differences between metastatic and non-metastatic tumours. BMC Cancer. (2017) 17:1–17. doi: 10.1186/s12885-017-3751-1
59. Craig, KKL, Wood, GA, Keller, SM, Mutsaers, AJ, and Wood, RD. MicroRNA profiling in canine multicentric lymphoma. PLoS One. (2019) 14:1–24. doi: 10.1371/journal.pone.0226357
60. Kobayashi, M, Saito, A, Tanaka, Y, Michishita, M, Kobayashi, M, Irimajiri, M, et al. Microrna expression profiling in canine prostate cancer. J Vet Med Sci. (2017) 79:719–25. doi: 10.1292/jvms.16-0279
61. Jiang, W, Zhu, P, Zhang, T, Liao, F, Yu, Y, Liu, Y, et al. MicroRNA-205 mediates endothelial progenitor functions in distraction osteogenesis by targeting the transcription regulator NOTCH2. Stem Cell Res Ther. (2021) 12:1–14. doi: 10.1186/s13287-021-02150-x
62. Wei, J, Zhang, Y, Li, Z, Wang, X, Chen, L, du, J, et al. GCH1 attenuates cardiac autonomic nervous remodeling in canines with atrial-tachypacing via tetrahydrobiopterin pathway regulated by microRNA-206. Pacing Clin Electrophysiol. (2018) 41:459–71. doi: 10.1111/pace.13289
63. Zhang, Y, Zheng, S, Geng, Y, Xue, J, Wang, Z, Xie, X, et al. MicroRNA profiling of atrial fibrillation in canines: MiR-206 modulates intrinsic cardiac autonomic nerve remodeling by regulating SOD1. PLoS One. (2015) 10:1–16. doi: 10.1371/journal.pone.0122674
64. Noguchi, S, Mori, T, Hoshino, Y, Yamada, N, Maruo, K, and Akao, Y. MicroRNAs as tumour suppressors in canine and human melanoma cells and as a prognostic factor in canine melanomas. Vet Comp Oncol. (2013) 11:113–23. doi: 10.1111/j.1476-5829.2011.00306.x
65. Rahman, MM, Lai, YC, Husna, AA, Chen, HW, Tanaka, Y, Kawaguchi, H, et al. Micro RNA transcriptome profile in canine oral melanoma. Int J Mol Sci. (2019) 20:4832. doi: 10.3390/ijms20194832
66. Ushio, N, Rahman, MM, Maemura, T, Lai, Y‑C, Iwanaga, T, Kawaguchi, H, et al. Identification of dysregulated microRNAs in canine malignant melanoma. Oncol Lett. (2019) 17:1080–8. doi: 10.3892/ol.2018.9692
67. Zamarian, V, Ferrari, R, Stefanello, D, Ceciliani, F, Grieco, V, Minozzi, G, et al. miRNA profiles of canine cutaneous mast cell tumours with early nodal metastasis and evaluation as potential biomarkers. Sci Rep. (2020) 10:1–13. doi: 10.1038/s41598-020-75877-x
68. Albonico, F, Mortarino, M, Avallone, G, Gioia, G, Comazzi, S, and Roccabianca, P. The expression ratio of miR-17-5p and miR-155 correlates with grading in canine splenic lymphoma. Vet Immunol Immunopathol. (2013) 155:117–23. doi: 10.1016/j.vetimm.2013.06.018
69. Bagardi, M, Ghilardi, S, Zamarian, V, Ceciliani, F, and Brambilla, PG. Circulating miR-30b-5p is Upregulatedregulated in cavalier king Charles spaniels affected by early myxomatous mitral valve disease. PLoS One. (2022) 17:e0266208. doi: 10.1371/journal.pone.0266208
70. Boggs, RM, Wright, ZM, Stickney, MJ, Porter, WW, and Murphy, KE. MicroRNA expression in canine mammary cancer. Mamm Genome. (2008) 19:561–9. doi: 10.1007/s00335-008-9128-7
71. Børresen, B, Nielsen, LN, Jessen, LR, Kristensen, AT, Fredholm, M, and Cirera, S. Circulating let-7g is Down-regulated in Bernese Mountain dogs with disseminated histiocytic sarcoma and carcinomas – a prospective study. Vet Comp Oncol. (2017) 15:525–33. doi: 10.1111/vco.12196
72. Bragato, JP, Rebech, GT, de Freitas, JH, Santos, MO, Costa, SF, Eugênio, FR, et al. miRNA-21 regulates CD69 and IL-10 expression in canine leishmaniasis. PLoS One. (2022) 17:1–15. doi: 10.1371/journal.pone.0265192
73. Braman, A, Weber, PS, Tritten, L, Geary, T, Long, M, Beachboard, S, et al. Further characterization of molecular markers in canine dirofilaria immitis infection. J Parasitol. (2018) 104:697–701. doi: 10.1645/18-12
74. Buffi, G, Diotallevi, A, Ceccarelli, M, Bruno, F, Castelli, G, Vitale, F, et al. The host micro-RNA cfa-miR-346 is induced in canine leishmaniasis. BMC Vet Res. (2022) 18:1–11. doi: 10.1186/s12917-022-03359-5
75. Clark, SD, Song, W, Cianciolo, R, Lees, G, Nabity, M, and Liu, S. Abnormal expression of miR-21 in kidney tissue of dogs with X-linked hereditary nephropathy: a canine model of chronic kidney disease. Vet Pathol. (2019) 56:93–105. doi: 10.1177/0300985818806050
76. Daldaban, F, Karaca Bekdik, İ, Aslan, Ö, Akyüz, B, Akçay, A, and Arslan, K. Investigation of TLR1-9 genes and miR-155 expression in dogs infected with canine distemper. Comp Immunol Microbiol Infect Dis. (2021) 79:101711. doi: 10.1016/j.cimid.2021.101711
77. Dawson, K, Wakili, R, Ördög, B, Clauss, S, Chen, Y, Iwasaki, Y, et al. MicroRNA29: a mechanistic contributor and potential biomarker in atrial fibrillation. Circulation. (2013) 127:1466–75. doi: 10.1161/CIRCULATIONAHA.112.001207
78. Von Deetzen, M-C, Schmeck, BT, Gruber, AD, and Klopfleisch, R. Malignancy associated MicroRNA expression changes in canine mammary Cancer of different malignancies. ISRN Vet Sci. (2014) 2014:1–5. doi: 10.1155/2014/148597
79. Deng, J, Zhang, Y, He, G, Lu, H, Zhao, Y, Li, Y, et al. Arterial wall injury and miRNA expression induced by stent retriever thrombectomy under stenotic conditions in a dog model. J Neurointerv Surg. (2021) 13:563–7. doi: 10.1136/neurintsurg-2020-016347
80. El-Sebaey, AM, and Abramov, PN. Hepatocyte-derived canine familiaris-microRNAs as serum biomarkers of hepatic steatosis or fibrosis as implicated in the pathogenesis of canine cholecystolithiasis. Vet Clin Pathol. (2022) 50:37–46. doi: 10.1111/vcp.12942
81. El-Sebaey, AM, Abramov, PN, and Abdelhamid, FM. Clinical characteristics, serum biochemical changes, and expression profile of serum CFA-mirnas in dogs confirmed to have congenital portosystemic shunts accompanied by liver pathologies. Vet Sci. (2020) 7:35. doi: 10.3390/vetsci7020035
82. Elshafie, NO, Nascimento, NCD, Lichti, NI, Kasinski, AL, Childress, MO, and Do, SAP. MicroRNA biomarkers in canine diffuse large B-cell lymphoma. Vet Pathol. (2021) 58:34–41. doi: 10.1177/0300985820967902
83. Fenger, JM, Bear, MD, Volinia, S, Lin, TY, Harrington, BK, London, CA, et al. Overexpression of miR-9 in mast cells is associated with invasive behavior and spontaneous metastasis. BMC Cancer. (2014) 14:1–16. doi: 10.1186/1471-2407-14-84
84. Fujiwara-Igarashi, A, Igarashi, H, Mizutani, N, Goto-Koshino, Y, Takahashi, M, Ohno, K, et al. Expression profile of circulating serum microRNAs in dogs with lymphoma. Vet J. (2015) 205:317–21. doi: 10.1016/j.tvjl.2015.04.029
85. Gaitero, L, Russell, SJ, Monteith, G, and LaMarre, J. Expression of microRNAs miR-21 and miR-181c in cerebrospinal fluid and serum in canine meningoencephalomyelitis of unknown origin. Vet J. (2016) 216:122–4. doi: 10.1016/j.tvjl.2016.07.014
86. Gioia, G, Mortarino, M, Gelain, ME, Albonico, F, Ciusani, E, Forno, I, et al. Immunophenotype-related microRNA expression in canine chronic lymphocytic leukemia. Vet Immunol Immunopathol. (2011) 142:228–35. doi: 10.1016/j.vetimm.2011.05.020
87. Gogulski, M, Cieślak, A, Grabska, J, Ardois, M, Pomorska-Mól, M, Kołodziejski, PA, et al. Effects of silybin supplementation on nutrient digestibility, hematological parameters, liver function indices, and liver-specific mi-RNA concentration in dogs. BMC Vet Res. (2021) 17:228. doi: 10.1186/s12917-021-02929-3
88. Gourbault, O, and Llobat, L. Micrornas as biomarkers in canine osteosarcoma: a new future? Vet Sci. (2020) 7:146. doi: 10.3390/vetsci7040146
89. Herrera Uribe, J, Vitger, AD, Ritz, C, Fredholm, M, Bjørnvad, CR, and Cirera, S. Physical training and weight loss in dogs lead to transcriptional changes in genes involved in the glucose-transport pathway in muscle and adipose tissues. Vet J. (2016) 208:22–7. doi: 10.1016/j.tvjl.2015.11.002
90. Hulanicka, M, Garncarz, M, Parzeniecka-Jaworska, M, and Jank, M. Plasma miRNAs as potential biomarkers of chronic degenerative valvular disease in dachshunds. BMC Vet Res. (2014) 10:205. doi: 10.1186/s12917-014-0205-8
91. Jeong, SJ, Lee, KH, Nam, AR, and Cho, JY. Genome-wide methylation profiling in canine mammary tumor reveals miRNA candidates associated with human breast cancer. Cancers (Basel). (2019) 11:1466. doi: 10.3390/cancers11101466
92. Konstantinidis, AO, Pardali, D, Adamama-Moraitou, KK, Gazouli, M, Dovas, CI, Legaki, E, et al. Colonic mucosal and serum expression of microRNAs in canine large intestinal inflammatory bowel disease. BMC Vet Res. (2020) 16:1–14. doi: 10.1186/s12917-020-02287-6
93. Koury, J, Ramirez, A, Xie, C, Harb, J, Dong, C, Maki, C, et al. Phosphodiesterase 4D, miR-203 and selected cytokines in the peripheral blood are associated with canine atopic dermatitis. PLoS One. (2019) 14:e0218670. doi: 10.1371/journal.pone.0218670
94. Lee, HB, Park, HK, Choi, HJ, Lee, S, Lee, SJ, Lee, JY, et al. Evaluation of circulating microrna biomarkers in the acute pancreatic injury dog model. Int J Mol Sci. (2018) 19:3048. doi: 10.3390/ijms19103048
95. Li, H, Li, S, Yu, B, and Liu, S. Expression of miR-133 and miR-30 in chronic atrial fibrillation in canines. Mol Med Rep. (2012) 5:1457–60. doi: 10.3892/mmr.2012.831
96. Van Middendorp, LB, Kuiper, M, Munts, C, Wouters, P, Maessen, JG, van Nieuwenhoven, FA, et al. Local microRNA-133a downregulation is associated with hypertrophy in the dyssynchronous heart. ESC Hear Fail. (2017) 4:241–51. doi: 10.1002/ehf2.12154
97. Morlang, MI, Weber, K, von Bomhard, W, and Mueller, RS. Cutaneous microRNA expression in healthy Labrador and Golden retrievers and retrievers with allergic and inflammatory skin diseases. Vet Dermatol. (2021) 32:331–e92. doi: 10.1111/vde.12971
98. Mortarino, M, Gioia, G, Gelain, ME, Albonico, F, Roccabianca, P, Ferri, E, et al. Identification of suitable endogenous controls and differentially expressed microRNAs in canine fresh-frozen and FFPE lymphoma samples. Leuk Res. (2010) 34:1070–7. doi: 10.1016/j.leukres.2009.10.023
99. Noguchi, S, Kumazaki, M, Mori, T, Baba, K, Okuda, M, Mizuno, T, et al. Analysis of microRNA-203 function in CREB/MITF/RAB27a pathway: comparison between canine and human melanoma cells. Vet Comp Oncol. (2016) 14:384–94. doi: 10.1111/vco.12118
100. Noguchi, S, Inoue, M, Ichikawa, T, Kurozumi, K, Matsumoto, Y, Nakamoto, Y, et al. The NRG3/ERBB4 signaling cascade as a novel therapeutic target for canine glioma. Exp Cell Res. (2021) 400:112504. doi: 10.1016/j.yexcr.2021.112504
101. Noguchi, S, Mori, T, Hoshino, Y, Yamada, N, Nakagawa, T, Sasaki, N, et al. Comparative study of anti-oncogenic MicroRNA-145 in canine and human malignant melanoma. J Vet Med Sci. (2012) 74:1–8. doi: 10.1292/jvms.11-0264
102. Noguchi, S, Tanimoto, N, Nishida, R, and Matsui, A. Functional analysis of the miR-145/Fascin1 cascade in canine oral squamous cell carcinoma. Oral Dis. (2022) 29:1495–504. doi: 10.1111/odi.14143
103. Oosthuyzen, W, ten Berg, PWL, Francis, B, Campbell, S, Macklin, V, Milne, E, et al. Sensitivity and specificity of microRNA-122 for liver disease in dogs. J Vet Intern Med. (2018) 32:1637–44. doi: 10.1111/jvim.15250
104. Pazzaglia, L, Leonardi, L, Conti, A, Novello, C, Quattrini, I, Montanini, L, et al. MiR-196a expression in human and canine osteosarcomas: a comparative study. Res Vet Sci. (2015) 99:112–9. doi: 10.1016/j.rvsc.2014.12.017
105. Qi, XY, Huang, H, Ordog, B, Luo, X, Naud, P, Sun, Y, et al. Fibroblast inward-rectifier potassium current upregulation in profibrillatory atrial remodeling. Circ Res. (2015) 116:836–45. doi: 10.1161/CIRCRESAHA.116.305326
106. Qiao, G, Xia, D, Cheng, Z, and Zhang, G. miR-132 in atrial fibrillation directly targets connective tissue growth factor. Mol Med Rep. (2017) 16:4143–50. doi: 10.3892/mmr.2017.7045
107. Ren, X, Fan, Y, Shi, D, Xu, E, and Liu, Y. MicroRNA-124 inhibits canine mammary carcinoma cell proliferation, migration and invasion by targeting CDH2. Res Vet Sci. (2022) 146:5–14. doi: 10.1016/j.rvsc.2022.03.004
108. Ro, WB, Kang, MH, Song, DW, Kim, HS, Lee, GW, and Park, HM. identification and characterization of circulating MicroRNAs as novel biomarkers in dogs with heart diseases. Front Vet Sci. (2021) 8:729929. doi: 10.3389/fvets.2021.729929
109. Robriquet, F, Babarit, C, Larcher, T, Dubreil, L, Ledevin, M, Goubin, H, et al. Identification in GRMD dog muscle of critical miRNAs involved in pathophysiology and effects associated with MuStem cell transplantation. BMC Musculoskelet Disord. (2016) 17:209. doi: 10.1186/s12891-016-1060-5
110. Sakai, M, Spee, B, Grinwis, GCM, Penning, LC, Wolferen, ME, Laan, LJW, et al. Association of circulating microRNA-122 and microRNA-29a with stage of fibrosis and progression of chronic hepatitis in Labrador retrievers. J Vet Intern Med. (2019) 33:151–7. doi: 10.1111/jvim.15366
111. Shan, H, Zhang, Y, Lu, Y, Zhang, Y, Pan, Z, Cai, B, et al. Downregulation of miR-133 and miR-590 contributes to nicotine-induced atrial remodelling in canines. Cardiovasc Res. (2009) 83:465–72. doi: 10.1093/cvr/cvp130
112. Shibasaki, H, Imamura, M, Arima, S, Tanihata, J, Kuraoka, M, Matsuzaka, Y, et al. Characterization of a novel microRNA, miR-188, elevated in serum of muscular dystrophy dog model. PLoS One. (2019) 14:e0211597. doi: 10.1371/journal.pone.0211597
113. Sterenczak, KA, Eckardt, A, Kampmann, A, Willenbrock, S, Eberle, N, Länger, F, et al. HMGA1 and HMGA2 expression and comparative analyses of HMGA2, Lin28 and let-7 miRNAs in oral squamous cell carcinoma. BMC Cancer. (2014) 14:1–11. doi: 10.1186/1471-2407-14-694
114. Vinall, RL, Kent, MS, and Devere White, RW. Expression of microRNAs in urinary bladder samples obtained from dogs with grossly normal bladders, inflammatory bladder disease, or transitional cell carcinoma. Am J Vet Res. (2012) 73:1626–33. doi: 10.2460/ajvr.73.10.1626
115. Wieszczeczyński, M, Krakowski, L, Opielak, G, Krakowska, I, Furmaga, J, Brodzki, P, et al. MicroRNA and vascular endothelial growth factor (VEGF) as new useful markers in the diagnosis of benign prostatic hyperplasia in dogs. Theriogenology. (2021) 171:113–8. doi: 10.1016/j.theriogenology.2021.05.017
116. Willenbrock, S, Wagner, S, Reimann-Berg, N, Moulay, M, Hewicker-Trautwein, M, Nolte, I, et al. Generation and characterisation of a canine EGFP-HMGA2 prostate cancer in vitro model. PLoS One. (2014) 9:1–12. doi: 10.1371/journal.pone.0098788
117. Wu, X, Gu, Q, Chen, X, Mi, W, Wu, T, and Huang, H. MiR-27a targets DKK2 and SFRP1 to promote reosseointegration in the regenerative treatment of peri-implantitis. J Bone Miner Res. (2019) 34:123–34. doi: 10.1002/jbmr.3575
118. Yang, C, Liu, X, Zhao, K, Zhu, Y, Hu, B, Zhou, Y, et al. MiRNA-21 promotes osteogenesis via the PTEN/PI3K/Akt/HIF-1α pathway and enhances bone regeneration in critical size defects. Stem Cell Res Ther. (2019) 10:1–11. doi: 10.1186/s13287-019-1168-2
119. Zhang, T, Feng, X, Zhou, T, Zhou, N, Shi, X, Zhu, X, et al. miR-497 induces apoptosis by the IRAK2/NF-κB axis in the canine mammary tumour. Vet Comp Oncol. (2021) 19:69–78. doi: 10.1111/vco.12626
120. Zhou, P, Tu, L, Lin, X, Hao, X, Zheng, Q, Zeng, W, et al. Cfa-miR-143 promotes apoptosis via the p53 pathway in canine influenza virus H3N2-infected cells. Viruses. (2017) 9:360. doi: 10.3390/v9120360
121. Zhou, X, Wen, W, Shan, X, Qian, J, Li, H, Jiang, T, et al. MiR-28-3p as a potential plasma marker in diagnosis of pulmonary embolism. Thromb Res. (2015) 138:91–5. doi: 10.1016/j.thromres.2015.12.006
122. Joos, D, Leipig-Rudolph, M, and Weber, K. Tumour-specific microRNA expression pattern in canine intestinal T-cell-lymphomas. Vet Comp Oncol. (2020) 18:1–7. doi: 10.1111/vco.12570
123. Osaki, T, Sunden, Y, Sugiyama, A, Azuma, K, Murahata, Y, Tsuka, T, et al. Establishment of a canine mammary gland tumor cell line and characterization of its miRNA expression. J Vet Sci. (2016) 17:385–90. doi: 10.4142/jvs.2016.17.3.385
124. Powers, JC, Sabri, A, al-Bataineh, D, Chotalia, D, Guo, X, Tsipenyuk, F, et al. Differential microRNA-21 and microRNA-221 upregulation in the biventricular failing heart reveals distinct stress responses of right versus left ventricular fibroblasts. Circ Hear Fail. (2020) 13:1–6. doi: 10.1161/CIRCHEARTFAILURE.119.006426
125. Ro, W, Bin, KMH, Song, DW, Lee, SH, and Park, HM. Expression profile of circulating MicroRNAs in dogs with cardiac hypertrophy: a pilot study. Front Vet Sci. (2021) 8:1–10. doi: 10.3389/fvets.2021.652224
126. Beaumier, A, Robinson, SR, Robinson, N, Lopez, KE, Meola, DM, Barber, LG, et al. Extracellular vesicular microRNAs as potential biomarker for early detection of doxorubicin-induced cardiotoxicity. J Vet Intern Med. (2020) 34:1260–71. doi: 10.1111/jvim.15762
127. Grimes, JA, Prasad, N, Levy, S, Cattley, R, Lindley, S, Boothe, HW, et al. A comparison of microRNA expression profiles from splenic hemangiosarcoma, splenic nodular hyperplasia, and normal spleens of dogs. BMC Vet Res. (2016) 12:1–12. doi: 10.1186/s12917-016-0903-5
128. Ichii, O, Ohta, H, Horino, T, Nakamura, T, Hosotani, M, Mizoguchi, T, et al. Urinary exosome-derived microRNAs reflecting the changes of renal function and histopathology in dogs. Sci Rep. (2017) 7:1–11. doi: 10.1038/srep40340
129. Ichii, O, Otsuka, S, Ohta, H, Yabuki, A, Horino, T, and Kon, Y. MicroRNA expression profiling of cat and dog kidneys. Res Vet Sci. (2014) 96:299–303. doi: 10.1016/j.rvsc.2014.01.003
130. Luo, W, Fang, M, Xu, H, Xing, H, Fu, J, and Nie, Q. Comparison of miRNA expression profiles in pituitary-adrenal axis between beagle and Chinese field dogs after chronic stress exposure. PeerJ. (2016) 2016:1–20. doi: 10.7717/peerj.1682
131. Penso-Dolfin, L, Swofford, R, Johnson, J, Alföldi, J, Lindblad-Toh, K, Swarbreck, D, et al. An improved microRNA annotation of the canine genome. PLoS One. (2016) 11:1–18. doi: 10.1371/journal.pone.0153453
132. Asada, H, Tomiyasu, H, Uchikai, T, Ishihara, G, Goto-Koshino, Y, Ohno, K, et al. Comprehensive analysis of miRNA and protein profiles within exosomes derived from canine lymphoid tumour cell lines. PLoS One. (2019) 14:1–15. doi: 10.1371/journal.pone.0208567
133. Chu, CP, Liu, S, Song, W, Xu, EY, and Nabity, MB. Small RNA sequencing evaluation of renal microRNA biomarkers in dogs with X-linked hereditary nephropathy. Sci Rep. (2021) 11:1–12. doi: 10.1038/s41598-021-96870-y
134. Guelfi, G, Iaboni, M, Sansone, A, Capaccia, C, Santoro, MM, and Diverio, S. Extracellular circulating miRNAs as stress-related signature to search and rescue dogs. Sci Rep. (2022) 12:1–12. doi: 10.1038/s41598-022-07131-5
135. Hino, Y, Rahman, MM, Lai, YC, Husna, AA, Chen, HW, Hasan, MN, et al. Hypoxic miRNAs expression are different between primary and metastatic melanoma cells. Gene. (2021) 782:145552. doi: 10.1016/j.gene.2021.145552
136. Husna, AA, Rahman, MM, Lai, YC, Chen, HW, Hasan, MN, Nakagawa, T, et al. Identification of melanoma-specific exosomal miRNAs as the potential biomarker for canine oral melanoma. Pigment Cell Melanoma Res. (2021) 34:1062–73. doi: 10.1111/pcmr.13000
137. Jung, SW, and Bohan, A. Genome-wide sequencing and quantification of circulating microRNAs for dogs with congestive heart failure secondary to myxomatous mitral valve degeneration. Am J Vet Res. (2018) 79:163–9. doi: 10.2460/ajvr.79.2.163
138. Lecchi, C, Zamarian, V, Borriello, G, Galiero, G, Grilli, G, Caniatti, M, et al. Identification of altered miRNAs in cerumen of dogs affected by otitis externa. Front Immunol. (2020) 11:1–13. doi: 10.3389/fimmu.2020.00914
139. Lopez, CM, Yu, PY, Zhang, X, Yilmaz, AS, London, CA, and Fenger, JM. MiR-34a regulates the invasive capacity of canine osteosarcoma cell lines. PLoS One. (2018) 13:1–23. doi: 10.1371/journal.pone.0190086
140. Saengchoowong, S, Khongnomnan, K, Poomipak, W, Praianantathavorn, K, Poovorawan, Y, Zhang, Q, et al. High-throughput microRNA profiles of permissive madin-Darby canine kidney cell line infected with influenza B viruses. Viruses. (2019) 11:1–16. doi: 10.3390/v11110986
141. Santoro, D, di Loria, A, Mirante, T, Oliveira, DM, Laudanna, C, Malanga, D, et al. Identification of differentially expressed microRNAs in the skin of experimentally sensitized naturally affected atopic beagles by next-generation sequencing. Immunogenetics. (2020) 72:241–50. doi: 10.1007/s00251-020-01162-w
142. Shing, JC, Schaefer, K, Grosskurth, SE, Vo, AH, Sharapova, T, Bodié, K, et al. Small RNA sequencing to discover circulating MicroRNA biomarkers of testicular toxicity in dogs. Int J Toxicol. (2021) 40:26–39. doi: 10.1177/1091581820961515
143. Zhao, FR, Su, S, Zhou, DH, Zhou, P, Xu, TC, Zhang, LQ, et al. Comparative analysis of microRNAs from the lungs and trachea of dogs (Canis familiaris) infected with canine influenza virus. Infect Genet Evol. (2014) 21:367–74. doi: 10.1016/j.meegid.2013.11.019
144. Zheng, Y, Fu, X, Wang, L, Zhang, W, Zhou, P, Zhang, X, et al. Comparative analysis of MicroRNA expression in dog lungs infected with the H3N2 and H5N1 canine influenza viruses. Microb Pathog. (2018) 121:252–61. doi: 10.1016/j.micpath.2018.05.015
145. Genini, S, Guziewicz, KE, Beltran, WA, and Aguirre, GD. Altered miRNA expression in canine retinas during normal development and in models of retinal degeneration. BMC Genomics. (2014) 15:1–17. doi: 10.1186/1471-2164-15-172
146. Jin, K, Su, KK, Li, T, Zhu, XQ, Wang, Q, Ge, RS, et al. Hepatic premalignant alterations triggered by human nephrotoxin aristolochic acid i in canines. Cancer Prev Res. (2016) 9:324–34. doi: 10.1158/1940-6207.CAPR-15-0339
147. Liu, D, Yang, M, Yao, Y, He, S, Wang, Y, Cao, Z, et al. Cardiac fibroblasts promote Ferroptosis in atrial fibrillation by secreting Exo-miR-23a-3p targeting SLC7A11. Oxidative Med Cell Longev. (2022) 2022:3961495. doi: 10.1155/2022/3961495
148. Starkey, MP, Compston-Garnett, L, Malho, P, Dunn, K, and Dubielzig, R. Metastasis-associated microRNA expression in canine uveal melanoma. Vet Comp Oncol. (2018) 16:81–9. doi: 10.1111/vco.12315
149. Xie, X, Pang, M, Liang, S, Lin, Y, Zhao, Y, Qiu, D, et al. Cellular microRNAs influence replication of H3N2 canine influenza virus in infected cells. Vet Microbiol. (2021) 257:109083. doi: 10.1016/j.vetmic.2021.109083
150. Faraldi, M, Gomarasca, M, Sansoni, V, Perego, S, Banfi, G, and Lombardi, G. Normalization strategies differently affect circulating miRNA profile associated with the training status. Sci Rep. (2019) 9:1–13. doi: 10.1038/s41598-019-38505-x
151. Roberts, TC, Coenen-Stass, AML, and Wood, MJA. Assessment of RT-qPCR normalization strategies for accurate quantification of extracellular microRNAs in murine serum. PLoS One. (2014) 9:237. doi: 10.1371/journal.pone.0089237
152. Schwarzenbach, H, Da Silva, AM, Calin, G, and Pantel, K. Data normalization strategies for microRNA quantification. Clin Chem. (2015) 61:1333–42. doi: 10.1373/clinchem.2015.239459
153. Becker, C, Hammerle-Fickinger, A, Riedmaier, I, and Pfaffl, MW. mRNA and microRNA quality control for RT-qPCR analysis. Methods. (2010) 50:237–43. doi: 10.1016/j.ymeth.2010.01.010
154. Mestdagh, P, van Vlierberghe, P, de Weer, A, Muth, D, Westermann, F, Speleman, F, et al. A novel and universal method for microRNA RT-qPCR data normalization. Genome Biol. (2009) 10:R64. doi: 10.1186/gb-2009-10-6-r64
155. Song, G, and Wang, L. A conserved gene structure and expression regulation of miR-433 and miR-127 in mammals. PLoS One. (2009) 4:e7829. doi: 10.1371/journal.pone.0007829
156. Ding, X, Yu, C, Liu, Y, Yan, S, Li, W, Wang, D, et al. Chronic obstructive sleep apnea accelerates pulmonary remodeling via TGF-β/miR-185/CoLA1 signaling in a canine model. Oncotarget. (2016) 7:57545–55. doi: 10.18632/oncotarget.11296
157. Eman, SR, Kubesy, AA, Baraka, TA, Torad, FA, Shaymaa, IS, and Mohammed, FF. Evaluation of hepatocyte-derived microRNA-122 for diagnosis of acute and chronic hepatitis of dogs. Vet World. (2018) 11:667–7. doi: 10.14202/vetworld.2018.667-673
158. Ramadan, ES, Kubesy, AA, Baraka, TA, Torad, FA, Salem, SI, and Salem, NY. Expression of blood hepatocyte-derived microRNA-122 in canine multicentric lymphoma with hepatic involvement. Vet Res Commun. (2019) 43:231–8. doi: 10.1007/s11259-019-09764-w
159. Leonardi, L, Benassi, MS, Pollino, S, Locaputo, C, and Pazzaglia, L. MiR-106B-25 cluster expression: a comparative human and canine osteosarcoma study. Vet Rec Open. (2020) 7:1–8. doi: 10.1136/vetreco-2019-000379
160. Wang, K, Huang, D, Zhou, P, Su, X, Yang, R, Shao, C, et al. Bisphenol a exposure triggers the malignant transformation of prostatic hyperplasia in beagle dogs via cfa-miR-204/KRAS axis. Ecotoxicol Environ Saf. (2022) 235:113430. doi: 10.1016/j.ecoenv.2022.113430
161. Noguchi, S, Ogusu, R, Wada, Y, Matsuyama, S, and Mori, T. PTEN, a target of microrna-374b, contributes to the radiosensitivity of canine oral melanoma cells. Int J Mol Sci. (2019) 20:4631. doi: 10.3390/ijms20184631
Keywords: microRNA, miRNA, canine, neoplasia, cancer, infectious diseases, inflammatory disease, developmental disease
Citation: Varvil MS and dos Santos AP (2023) A review on microRNA detection and expression studies in dogs. Front. Vet. Sci. 10:1261085. doi: 10.3389/fvets.2023.1261085
Edited by:
Francisco Javier Salguero, UK Health Security Agency, United KingdomReviewed by:
Michal Aureliusz Jank, Warsaw University of Life Sciences, PolandPaola Modesto, Experimental Zooprophylactic Institute for Piedmont, Italy
Copyright © 2023 Varvil and dos Santos. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Andrea Pires dos Santos, U2FudG9zMUBwdXJkdWUuZWR1
 Mara S. Varvil
Mara S. Varvil Andrea Pires dos Santos
Andrea Pires dos Santos