
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci., 08 September 2023
Sec. Animal Nutrition and Metabolism
Volume 10 - 2023 | https://doi.org/10.3389/fvets.2023.1255122
This article is part of the Research TopicGut Microbiota: Allied with Livestock Nutrition, Health, And WelfareView all 19 articles
 Jie Wang†
Jie Wang† Yang Chen†
Yang Chen† Mianying Li
Mianying Li Siqi Xia
Siqi Xia Kaisen Zhao
Kaisen Zhao Huimei Fan
Huimei Fan Jiale Ni
Jiale Ni Wenqiang Sun
Wenqiang Sun Xianbo Jia
Xianbo Jia Songjia Lai*
Songjia Lai*Pre-weaning is the most important period for the growth and development of calves. Intestinal morphology, microbial community and immunity are initially constructed at this stage, and even have a lifelong impact on calves. Early feeding patterns have a significant impact on gastrointestinal development and microbial communities. This study mainly analyzed the effects of three feeding methods on the gastrointestinal development of calves, and provided a theoretical basis for further improving the feeding mode of calves. it is very important to develop a suitable feeding mode. In this study, we selected nine newborn healthy Holstein bull calves were randomly selected and divided into three groups (n = 3), which were fed with starter + hay + milk (SH group), starter + milk (SF group), total mixed ration + milk (TMR group). After 80 days of feeding Feeding to 80 days of age after, the ileum contents and blood samples were collected, and the differences were compared and analyzed by metagenomic analysis and serum metabolomics analysis. Results show that compared with the other two groups, the intestinal epithelium of the SH group was more complete and the goblet cells developed better. The feeding method of SH group was more conducive to the development of calves, with higher daily gain and no pathological inflammatory reaction. The intestinal microbial community was more conducive to digestion and absorption, and the immunity was stronger. These findings are helpful for us to explore better calf feeding patterns. In the next step, we will set up more biological replicates to study the deep-seated reasons for the differences in the development of pre-weaning calves. At the same time, the new discoveries of neuro microbiology broaden our horizons and are the focus of our future attention.
Pre-weaning is the most important period for the growth and development of calves. Intestinal morphology, microbial community and immunity are initially constructed at this stage, and even have a lifelong impact on calves (1, 2). The living environment of the calf changed from a sterile uterus to the outside world, and the nutritional conditions changed from maternal provision to the calf’s own feeding of feed and milk. Due to the incomplete development of the immune ability and digestive system of calves, newborn calves are very sensitive to external stimuli, and any external interference will seriously affect the growth of calves (3, 4). If the appropriate feeding mode is not adopted at this time, it will lead to low daily weight gain, diarrhea (5). It is reported that 30% of the deaths of pre-weaning cows are caused by diarrhea, which highlights the importance of intestinal health (6). The development of the digestive tract of calves is a unique process. With the development of the gastrointestinal tract and the settlement of the microbial community, calves gradually change from pseudo-ruminant animals to functional ruminants in physiology. The morphology of intestinal epithelial villi, the development of intestinal smooth muscle and rumen volume are basically completed at this stage which was ac-companied by the development of salivary organs and the development of rumination behavior. These factors directly affect the feed intake, nutrient absorption rate and digestive ability of calves after weaning (7).
Studies have shown that early feeding patterns have a significant impact on gastrointestinal development and microbial communities. Differences in dietary composition and feed physical morphology will stimulate intestinal structure development and microbial community composition (8–10). For example, alfalfa hay can promote the development of intestinal epithelial villi; the feeding of high carbohydrate and low fiber feeding will be detrimental to the rumen development of calves; additives can directly or indirectly increase the proportion of probiotics (11–13). Research shows that intestinal microbial community can be directed to change, adjusting feeding patterns can help animals form beneficial microbial communities before weaning (14). This can not only promote the development of calf digestive ability, but also is indispensable for the development of intestinal immune function. Deng’s research on giant pandas has proved that Streptococcus alactolyticus can promote the dietary adaptation of giant pandas by participating in protein metabolism (15). Recent studies have also found that microbial communities also communicate biological information with the nervous system, it points to a new field: neuro microbiology, this further illustrates the huge impact of gut microbes on the host (16).
There is no fixed feeding mode for newborn calves. Appropriate feeding mode should be comprehensively formulated according to their nutritional needs, breed characteristics, feeding plan and other factors (17). For calves at this stage, growth is important, but more importantly, the healthy development of the digestive system based on the gastrointestinal tract. This will have a long-term impact on the subsequent growth and production of calves (18, 19). Therefore, it is very important to develop a suitable feeding mode. In this study, three different feeding modes were used to explore the effects of different feeding modes on the growth and development of ileum and blood metabolism of calves from the perspective of metagenomics and metabolomics. The relationship between intestinal development, digestive function and microbial community composition was explored to provide a theoretical basis for further improving the feeding pattern of calves. At the same time, it inspired us to explore the relationship between intestinal microorganisms and the nervous system from a new perspective of neurobiology.
This study was approved and conducted in accordance with the ethical standards of the Institutional Animal Care and Use Committee of the College of Animal Science and Technology, Sichuan Agricultural University, Sichuan, 611130, China.
Nine 7-day-old healthy male Holstein calves with a body weight of about 41.6 kg (standard deviation = 0.563) and similar physical condition were selected. Raised in Sichuan Xuebao Dairy Group HonSFeng cattle farm. Nine calves were randomly divided into three groups, SH group: starter feed + hay + milk, SF group: starter feed + milk, TMR group: total mixed ration + milk. The feed composition and nutrient composition of the starter feed are shown in Table 1. The hay in the SH group was composed of alfalfa and oat grass in a ratio of 3:2, cut into 1.5 cm long, mixed with starter and fed. TMR feed (starter: alfalfa: oat grass: water = 0.30: 0.12: 0.08: 0.50), the starter was crushed into powder, the hay was cut to about 1 cm, and mixed with water to paste. Other feeding and management methods were carried out according to the existing methods of cattle farms, and calves were free to feed and drink water during the experimental period. Continuous feeding to 80 days of age, using electric shock bloodletting method to slaughter and record the slaughter weight.
After the calves were fasted for 24 h, the external jugular vein blood collection method was adopted (June 10, 2022), and 5 mL blood collection vessels (EDTAK2, Jiangsu Kangjian Medical Device Co., Ltd., Nanjing, China) were used to collect some calf blood, and heparin sodium was used for anticoagulation. Some blood samples were taken to determine blood routine indexes such as red blood cell count and white blood cell count (Chengdu Li Lai Biotechnology Co., Ltd., Chengdu). The blood samples were centrifuged at 4°C, 3000 r / min for 5 min to obtain the upper serum. Some serum samples were sent to Novogene Bioinformatics Technology Co., Ltd. (Beijing, China) for metabolomics analysis, and then some serum samples were selected to determine digestive and immune indexes such as α-amylase (C016-1-1), lysozyme (A050-1-1) and trypsin (A080-2; the kits were provided by Nanjing Jiancheng Bioengineering Institute).
Slaughter of calves by electric bloodletting. The calves get up in the morning, use electric shock anesthesia after fasting blood collection, and then slaughter by carotid artery bloodletting. After slaughter, the contents of the ileum of the calf were quickly placed in a 2 mL frozen tube and stored in liquid nitrogen at −80°C. The unsealed ileum tissue was collected and fixed with 10% neutral formaldehyde solution. After dehydration, pruning, embedding, slicing, staining with hematoxylin and eosin (HE), sealing and other steps, the Pannoramic 250 digital slice scanner produced by Hungary 3DHISTECH company was used to collect images of the slices.
According to the Tiangen Magnetic Bead Kit (Tiangen Biotech, Beijing, China) instructions, microbial DNA was extracted from ileum content samples, and the purity and integrity of DNA were detected by agarose gel electrophoresis. The qualified DNA samples were broken into fragments of about 350 bp by ultrasonic crushing instrument. After repair, purification, PCR amplification and other steps, the preliminary database was completed. Qubit2.0 was used for quantification, and then NEBNext ® Ultra DNA Library Prep Kit for Illumina (NEB, USA) was used to construct a metagenomic library and sequenced on the Illumina HiSeq Xten platform. The library construction and sequencing work was completed by Novogene Bioinformatics Technology Co., Ltd. (Beijing, China). The clean date was obtained by filtering the raw date obtained by sequencing, and the clean date of each sample was assembled and analyzed by MEGAHIT software (20, 21) (v1.0.4-beta1). Then MetaGeneMark software (22) (V2.102) was used for gene prediction, and the genes predicted by each sample were put together to construct gene catalogue. Starting from the gene catalogue, the clean data of each sample was synthesized to obtain unigenes for subsequent analysis (23).
The obtained unigenes were compared with the NCBI’s NR database (24) (Version 2018-01-023) to determine the species annotation information of each unigenes. Combined with the gene abundance table, the abundance information of each sample in phylum, genus and species was obtained. PCA (25) (RADE4 package, version 2.15.3) and LEfSe (26) (LDA score default to 3) analysis were used to compare the different species between groups. Finally, unigenes were compared with KEGG database (27, 28) (version 2018-01-014), CAZy database (29) (version 201,8015) and eggNOG database (30) (version 4.56) to obtain the relative abundance and functional annotation differences between groups (31).
The metabolites in the ileum were studied based on LC–MS technology. After preliminary treatment, the supernatant was injected into the ultra-performance liquid chromatography–tandem mass spectrometry (UHPLC–MS/MS) system for analysis (32). Firstly, the raw data of mass spectrometry were imported into Compound discoverer 3.1 software for spectral processing and database retrieval, and the qualitative and quantitative results of metabolites were obtained. Then, the quality of data was controlled to ensure the accuracy and reliability of the data. Using high-resolution mass spectrometry (HRMS) technology, we can make the non-target metabolic group as much as possible to detect the molecular characteristic peaks in the sample. The raw data after offline is preprocessed by CD3.1 data processing software. In order to make the identification accurate, we extract the peaks according to the set of ppm, signal-to-noise ratio (S/N), additive ions, and other information and quantify the peak area. Then mzCloud, mzVault, and MassList databases were compared to identify metabolites. Finally, metabolites with a coefficient of variation of less than 30% in QC samples were retained as the final result. The metabolites were compared with KEGG, HMDB, and other databases to obtain the annotation results. Then, a multivariate statistical analysis of metabolites was performed, including principal component analysis (PCA), partial least squares discriminant analysis (PLS-DA) to establish the relationship between the expression of metabolites and samples (33). According to the results of Q2 and R2, the model was judged to reveal the differences in metabolic patterns between different groups. KEGG enrichment pathway analysis was performed on the differential metabolites to obtain clearer and more detailed differential analysis results.
The results of HE staining are shown in Figure 1. Compared with the other two groups, the epithelial villi in the SH group were relatively complete, the intestinal epithelial villi were high and well developed, the goblet cells were closely arranged, and some inflammatory cells were seen in the lamina propria. There were a large number of epithelial mucosal shedding in the SF group, and the inflammatory cells in the lamina propria were also more than those in the SH group, accompanied by congestion; the mucosal integrity of the TMR group was the worst, accompanied by inflammatory cells and congestion, and even obvious parasitic infection was observed in some sections.
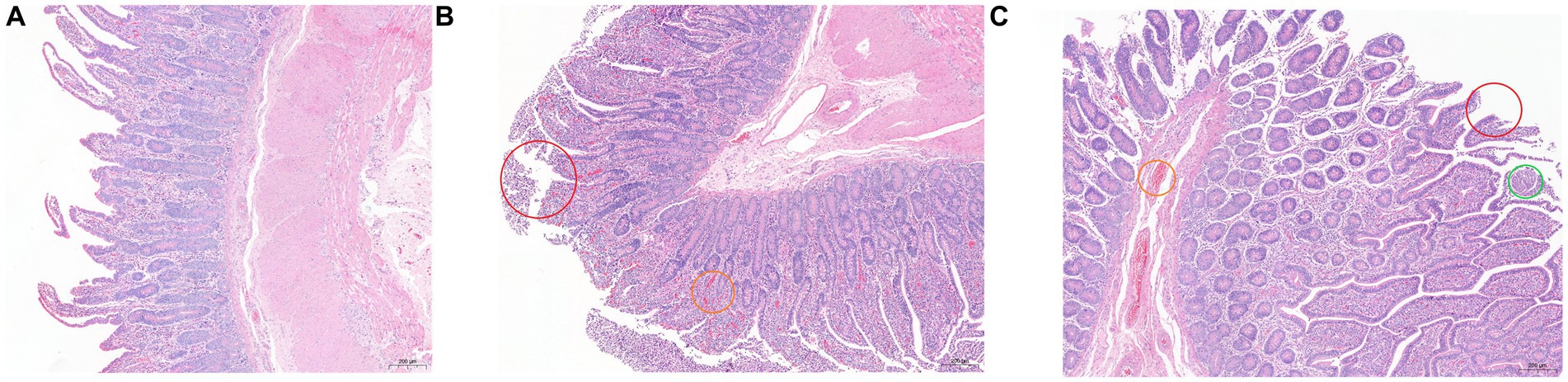
Figure 1. Hematoxylin and eosin (100×) -stained ileal tissue samples of Holstein calves fed different diets. (A) SH group; (B) SF group; (C) TMR group. Red circle marked mucosal epithelial shedding; the orange circle marked congestion; green circle marked as parasitic lesions.
In this experiment, 9 newborn, healthy and healthy male calves with no significant difference in birth weight were selected. The birth weight was about 41 kg. When the calves were slaughtered at 80 days of age, there were significant differences in weight among the three groups (p = 0.002). The average weight of SH group and SF group TMR group was 100.62 ± 1.94, 100.07 ± 1.92 and 90.77 ± 2.42. And the daily weight gain was 0.745 kg / day, 0.740 kg / day and 0.604 kg / day, respectively.
In Figure 2, some blood routine data. The results showed that the number of white blood cells SH was significantly less than that of SF group and TMR group, which may be due to the more severe inflammatory response in SF group and TMR group. The number of red blood cells in the TMR group was the highest. The hemoglobin content in the TMR group was also the highest in the three groups. The results of digestive enzymes and immunity showed that the contents of amylase, trypsin and lysozyme in SH were the highest in the three groups. The IgG content of SH group was also the highest in the three groups. These indicators show that the digestive ability and immune ability of SH group are higher than the other two groups.
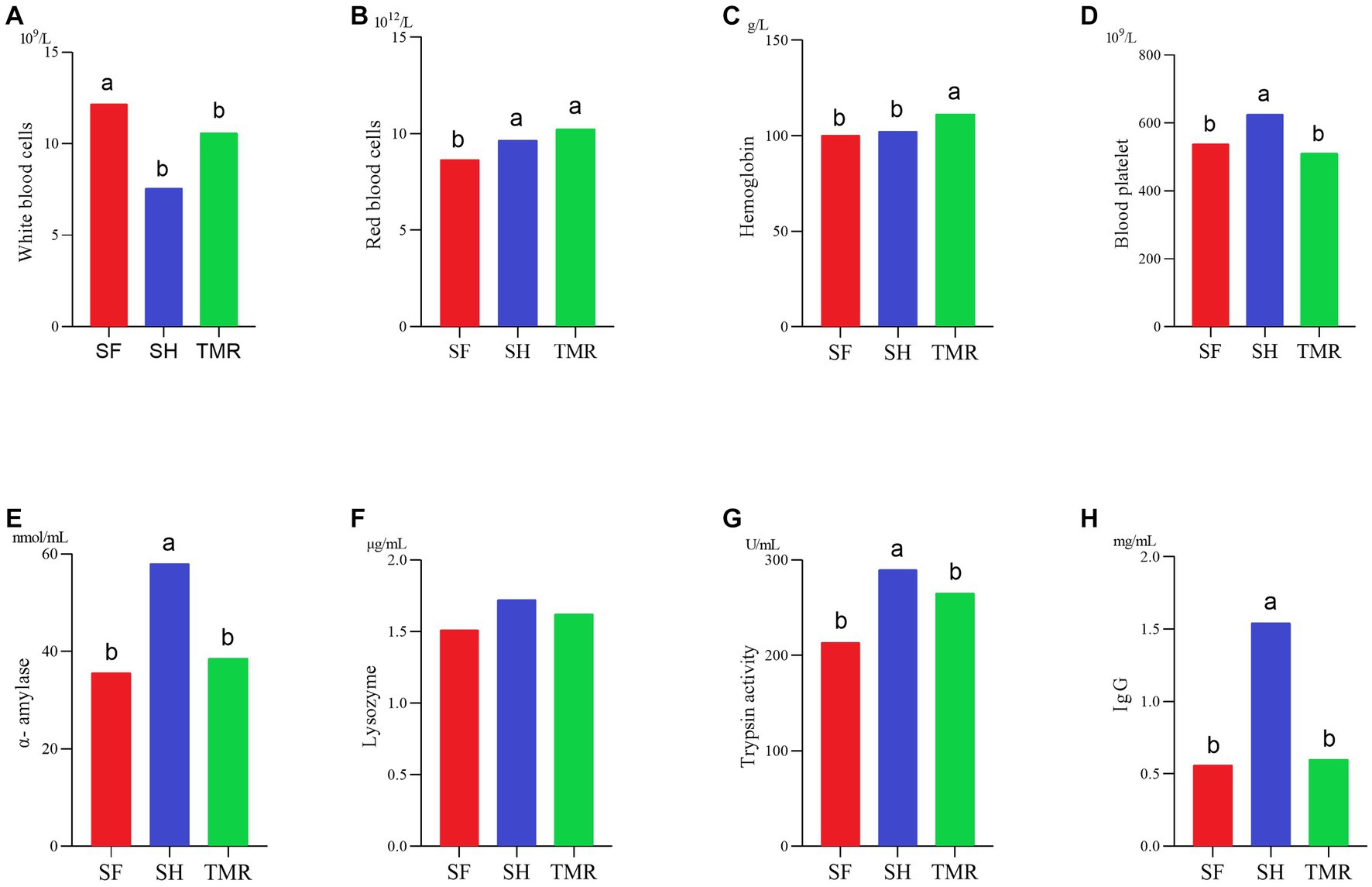
Figure 2. Some physiological indexes test results. (A–D) It’s blood routine determination results. (E–H) It’s digestive enzyme and immunoassay results.
As shown in Table 2, a total of 112,964.83 raw data were measured in 9 samples, and 112,791.07 clean data were obtained after Illumina pretreatment and filtering, with an average CG content of 45.87% and an effective value of more than 99.85%. The average length of scaftigs of 9 samples was 1434.85.
Based on the number of genes, correlation analysis was performed on 8 samples. The results are shown in Figure 3. The correlation between SF group and SH group was high, but the correlation between TMR group and SF group and SH group was low.
The processed genes were compared with the database to obtain the species annotation information of each sequence and compared after classification at each level. The results are shown in Figure 4. The dominant bacteria in the three groups were Firmicutes under the phylum level classification. The subdominant bacteria in group SF were Proteobacteria and Chlamydiae. The subdominant flora in SH group was Actinobacteria, more than the other two groups. The subdominant flora in the TMR group was Proteobacteria. Under the genus level classification, the dominant flora of group SF was Clostridium and Chlamydia; the dominant flora in SH group was Olsenella, which was more than that in TMR group and SF group, and the secondary dominant flora was Clostridium. The dominant flora in the TMR group was Clostridium secondary dominant flora Sarcina and more than that in the SF and SH groups.
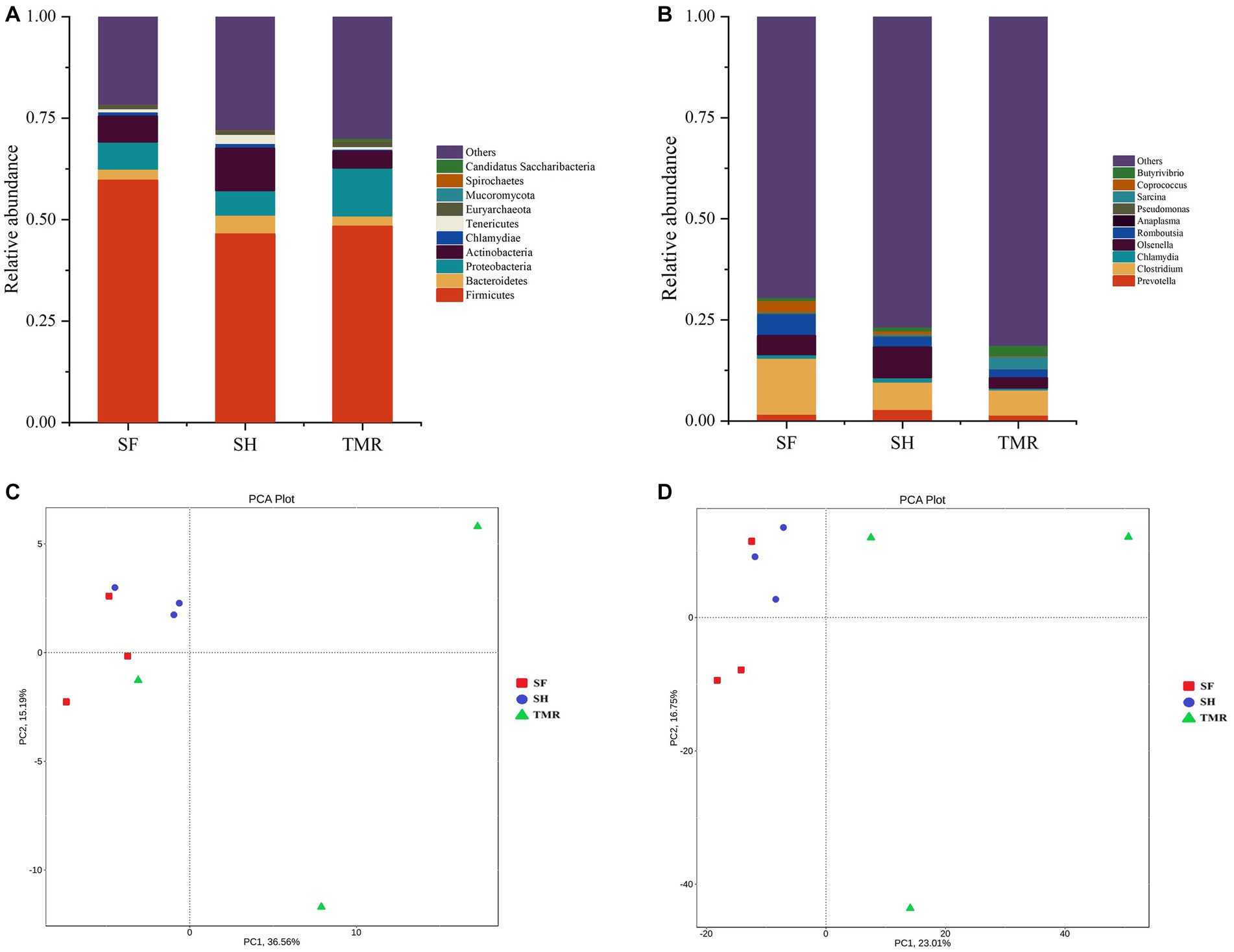
Figure 4. Analysis of microbial community differences between groups. (A) Relative abundance of species at phylum level TOP10. (B) Relative abundance of genus level species TOP10. (C) Phylum level species PCA analysis. (D) PCA analysis of genus-level species.
The results of PCA analysis showed that under the classification of phylum level and genus level, the intra-group difference of SH group was the smallest, the intra-group difference of SF group was slightly larger than that of SH group, and the intra-group difference of TMR group was the largest. There were significant differences among the three groups, and the intra-group difference of TMR was greater than the inter-group difference, which indicate that after feeding the total mixture, the ileum microbes of calves were greatly different. In order to screen species with significant differences between groups, the results of LDA analysis (Figure 5) showed that the main differences between SH group and TMR group were p _ Firmicutes and o _ Mycoplasmatales in SH group and p _ Sarcina in TMR group. The main differential flora between SF group and TMR group were o _ Veillonellaceae, g _ Prevotella in SF group and p _ Sarcina in TMR group.
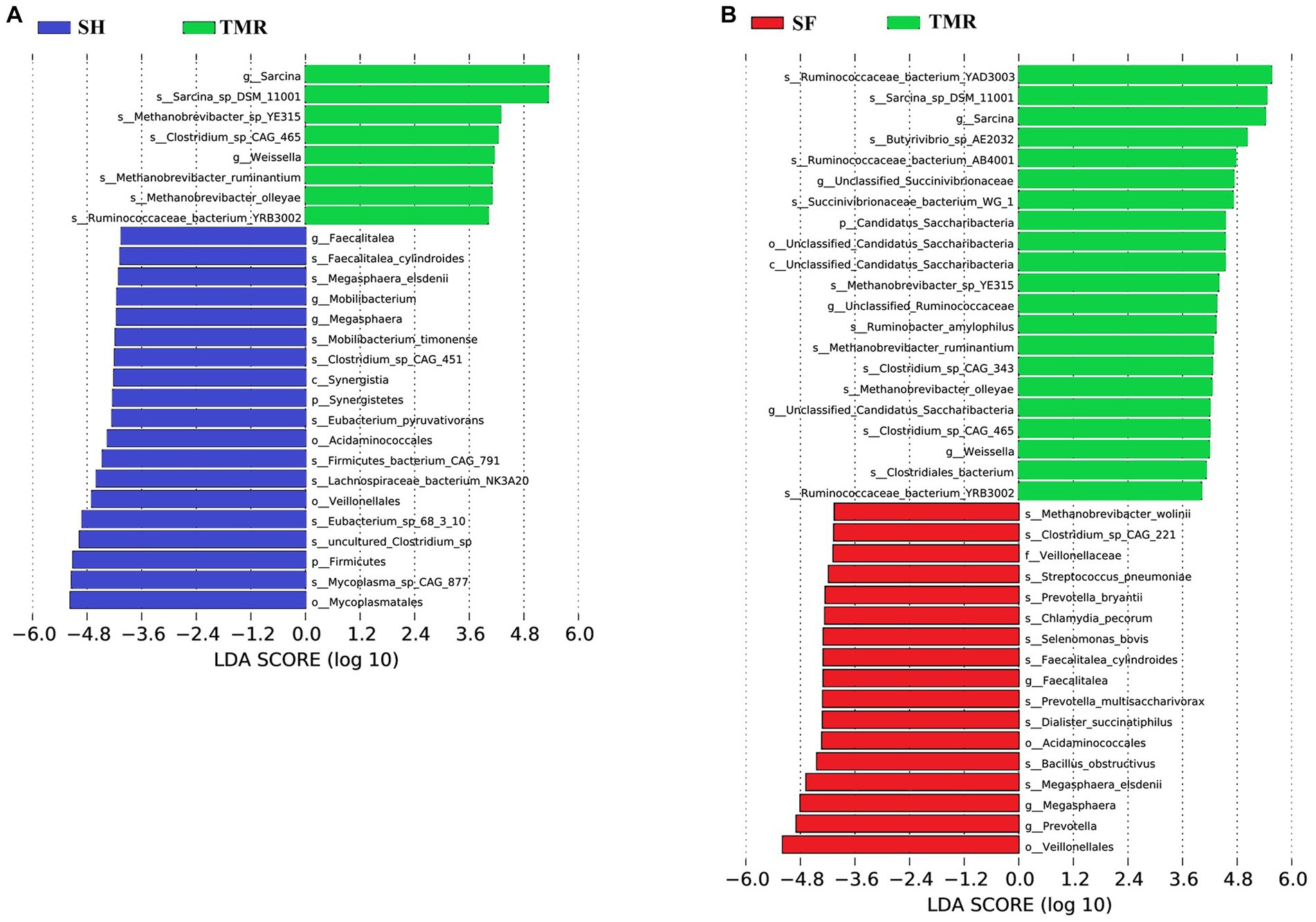
Figure 5. LDA score of LEfSe-PICRUSt. (A) LDA score analysis diagram of different species between SH group and TMR group. (B) LDA score analysis diagram of different species between SF group and TMR group.
The relative abundance of KEGG pathway enrichment in the three groups was analyzed (Figure 6). It was found that among the 45 pathways under the second level classification, the SH group was mainly enriched in carbohydrate metabolism, followed by amino acid metabolism, translation and membrane transport; the dominant pathways in the TMR group were carbohydrate metabolism and amino acid metabolism. It is worth noting that the TMR group was enriched in cell growth and death and drug resistance: antimicrobial pathway higher than SH group and SF group. These genes were further compared in the metabolic pathways of the third level, and were enriched in 302 pathways. The three groups all enriched more genes in ABC transporters (pathway ID: ko 02010), ribosome (pathway ID: ko03010), purine metabolism (pathway ID: ko 00230) and pyrimidine metabolism (pathway ID: ko00240), but the SF group was more and the TMR group was the least. Interestingly, the number of genes enriched in Cell cycle-Caulobacter (pathway ID: ko04112), necroptosis (pathway ID: ko04217) and other pathways under the classification of cell growth and death pathway was the least in SH group, and more in TMR group and SF group. The Cationic antimicrobial peptide (CAMP) resistance (pathway ID: ko01503) under the Drug resistance: Antimicrobial classification was similar.
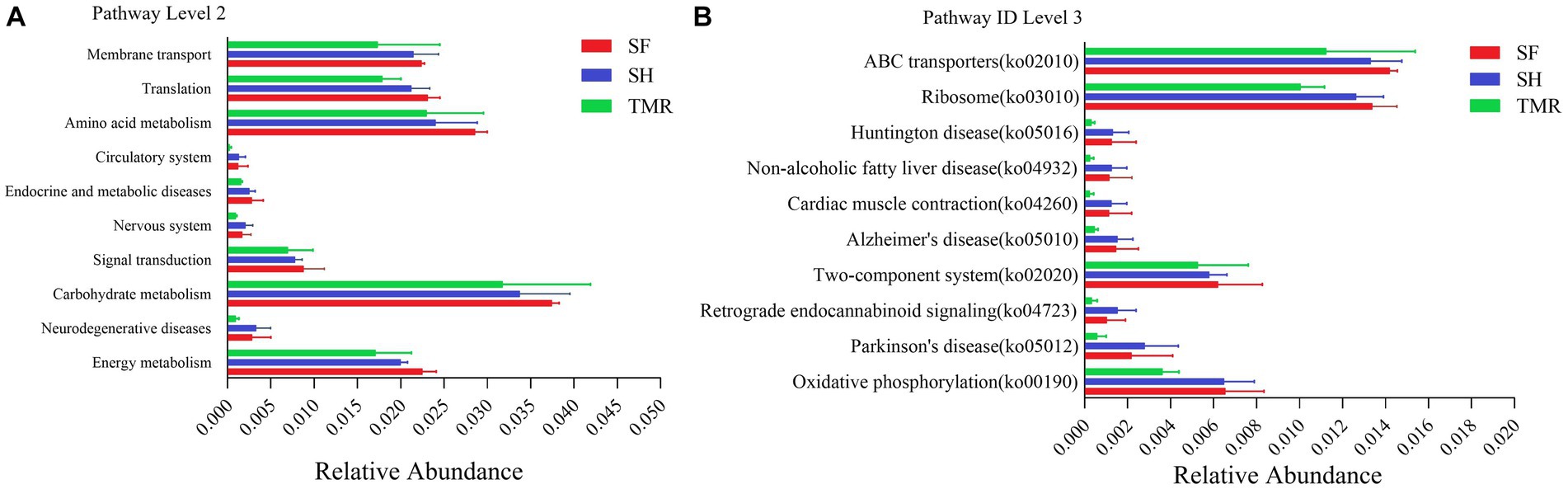
Figure 6. KEGG pathway enrichment analysis. (A) KEGG level 2 enrichment analysis top 10 between groups. (B) KEGG level 3 enrichment analysis top 10 between groups.
The results of EggNOG analysis (Figure 7) showed that the most abundant genes were enriched in Replication, recombination and repair, followed by Amino acid transport and metabolism and Translation, ribosomal structure and biogenesis. After comparing the relative abundance of genes annotated in eggNOG in the three groups, it was found that the main enrichment pathways of the three groups were similar, with the most in SF group and the least in TMR group. The relative abundance of secondary metabolites biosynthesis, transport and catabolism, Cell motility and RNA processing and modification in TMR group was higher than that in the other two groups.
After comparing the relative abundance of CAZy genes in the three groups (Figure 8), it was found that Glycoside Hydrolases (GH) were the main ones, and there were differences in CAZy family under the second level classification. The CAZy family of SH group was more evenly distributed, and the most enriched families were GT2 family and GH1 family, among which GH1, GH3 and GH25 family were higher than the other two groups. GH19, GT2 and GH13 family were more enriched in group SF than in the other two groups. There were more GH19, GH24 and GT2 family in TMR group, and GH24 was more than the other two groups. The CAZy families that attract our attention are GH1 and GH19. The differences between the three groups are also mainly reflected in these two families. The enrichment of GH1 in SH group was more than that in SF group and TMR group, mainly due to the β-glucosidase (EC 3.2.1.21) of GH1 family under level 3 classification. GH19 was more enriched in SF group and TMR group than in SH group, mainly chitinase (EC 3.2.1.14) of GH19 family under level 3 classification.
After analyzing the blood metabolites of the three groups, a total of 403 cathode metabolites were found. PLS-DA analysis (Figure 9) of these metabolites between groups showed that the separation between groups was obvious, the degree of polymerization in the group was high, and the model parameters of each group also met the standard, indicating that the results were stable and reliable. Further analysis and screening of differential metabolites (Table 3) showed that there were 87 differential metabolites in the cathode of SF group and SH group, 17 were significantly up-regulated and 70 were significantly down-regulated. There were 152 differential metabolites in the cathode of SF group and TMR group, 27 were significantly up-regulated and 125 were significantly down-regulated. There were 108 differential metabolites in the cathode of SH group and TMR group, 26 were significantly up-regulated and 82 were significantly down-regulated.
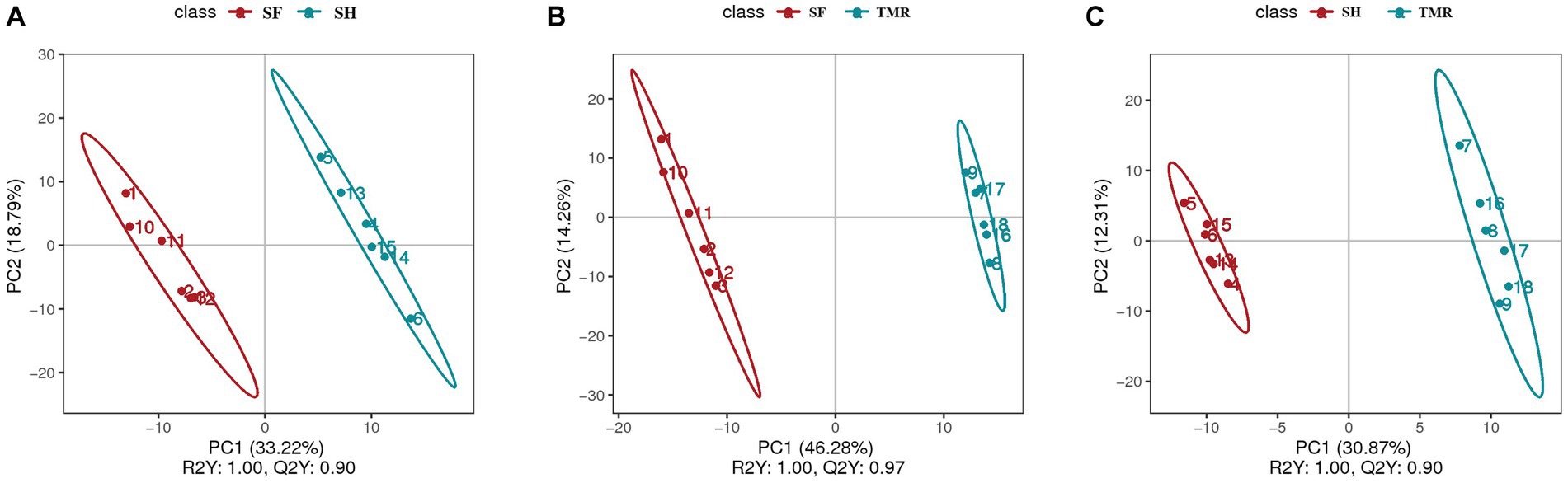
Figure 9. PLS-DA analysis of serum metabolites between groups. (A) PLS-DA analysis of SF group and SH group; (B) PLS-DA analysis of SF group and TMR group; (C) PLS-DA analysis of SH group and TMR group.
The selected differential metabolites were subjected to KEGG enrichment analysis to determine the effects of differential metabolites on key blood metabolic pathways. The results showed that the differential metabolites between SF group and SH group were mainly enriched in cortisol synthesis and secretion (value of p = 0.032). The differential metabolites between SF group and TMR group were mainly enriched in cAMP signaling pathway (value of p = 0.043), dopaminergic synapse (value of p = 0.043), Taste transduction (value of p = 0.043). The differential metabolites between SH group and TMR group were mainly enriched in cAMP signaling pathway (value of p = 0.024), dopaminergic synapse (value of p = 0.024), taste transduction (value of p = 0.024), tyrosine metabolism (value of p = 0.046). The specific pathway information and differential metabolites enriched in the pathway are shown in the Table 4.
The ileum is located in the posterior part of the small intestine and is the main place for digestion and absorption of nutrients. Therefore, the development of ileal function is essential for the health and growth of calves. We observed from the sections after HE staining that the intestinal epithelial villi in the SF and SH groups were better developed, the villus height was higher, and the goblet cells in the villi were more closely arranged. The goblet cells can secrete mucus and form a physical barrier on the surface of the intestinal mucosa, which is the main component of the intestinal barrier (34). These characteristics mean that the ileum of SF group and SH group has a larger absorption and digestion area, and the intestinal morphology is more conducive to the absorption of nutrients, and has a stronger barrier function, which is the characteristic of healthy development of calf intestine, which is conducive to the development of digestive ability and gastrointestinal health of calves (35, 36). The research of Ma showed that the intestinal tract of calves must undergo development, physical and physiological changes, before they can better adapt to the transition from milk to solid feed (37). Studies have shown that the healthy development of intestinal epithelial environment is conducive to the development of intestinal probiotics and the stability of microbial communities (38). We observed a more obvious inflammatory response in the TMR group, and the intestinal wall in the SH group was relatively complete. After subsequent analysis, we believe that it may be due to the digestive ability of calves and maturity, which cannot adapt to the inflammatory response caused by TMR feed. The parasite phenomenon may be caused by the invasion of pathogens. Whether it is related to the total mixed feed needs further study.
Metagenomic analysis compared the microbial community differences among the three groups. The results showed that SH group was enriched in Actinobacteria and more than the other two groups. In previous studies, except for a small number of pathogenic bacteria, Actinobacteria is usually a probiotic that is conducive to the digestion and absorption of organic matter. In some farms, probiotics are used to selectively stimulate the growth of Actinobacteria (39). Moreover, Actinobacteria can also produce secondary metabolites such as natural drugs, enzymes, and active factors, and is considered to be a potential probiotic (40). Bifidobacterium under the classification of Actinobacteria plays an important role in regulating the immune system and preventing intestinal diseases. It can reduce the inflammatory response by producing short-chain fatty acids and regulating inflammatory factors. The proliferation of Actinobacteria may be the key to regulating immunity and reducing inflammatory response in SH group. Some studies have shown that Actinobacteria also plays a role in reducing pathogens such as Escherichia-Shigella, indicating that it can also have a positive effect on preventing diarrhea (41). Olsenella has obvious advantages in SH enrichment. Olsenella belongs to probiotics under the classification of lactic acid bacteria. Studies have shown that Olsenella is positively correlated with butyrate. Butyrate is one of the main volatile fatty acids produced by rumen fermentation, and ileum is also the main part of absorbing butyrate. Butyrate can provide energy for Olsenella and can strengthen the intestinal barrier function, which is conducive to intestinal health (42). The study of Zhang also showed that individuals with higher relative abundance of Olsenella have a stronger ability to produce short-chain fatty acids, which can effectively affect the structure of the intestinal epithelial mucus layer, stimulate the development of intestinal epithelial tissue, nourish the development of intestinal epithelial cells such as goblet cells, and reduce the risk of inflammatory bowel disease in individuals (43).
The dominant bacteria in the SF group were Chlamydiae and Proteobacteria. Chlamydiae is a pathogen that may cause enteritis (44, 45), but no diarrhea symptoms were found in this individual, and there may be a case of invisible infection of Chlamydiae (46). At the same time, other pathogens with high correlation with Chlamydiae, such as Campylobacter, were not found. Chlamydiae, as a potential pathogen, may be related to the local inflammation observed in the SF group, which may lead to impaired intestinal absorption and induce other diseases (47). Timely treatment is needed. The study of Xia in weaned piglets showed that the addition of plant essential oils containing substances such as tributyrin to the feed can resist Chlamydia, regulate microbial morphology, and improve intestinal villus morphology (48).
Proteobacteria in both TMR group and SF group have high abundance. Previous studies have shown that many intestinal pathogenic bacteria belong to Proteobacteria, and the relative abundance of Proteobacteria is too high, which may cause inflammation and cause calf diarrhea (49, 50). Shin used the enrichment of Proteobacteria as one of the important markers of intestinal microflora imbalance in healthy individuals. In general, Proteobacteria will not be significantly enriched in healthy individuals (51). The study of Han on weaned piglets showed that Proteobacteria could activate inflammation-related cytokines and cause intestinal inflammation damage. At the same time, the daily weight gain of piglets with enrichment of Proteobacteria was significantly reduced, which indicated that the intestinal nutrition absorption function was also affected (52). The study of Varada on newborn buffaloes found that feeding probiotics can regulate the composition of intestinal microbial communities by reducing the abundance of Proteobacteria (53). Numerous studies have shown that the enrichment of Proteobacteria can lead to inflammatory response and cause intestinal epithelial cell dysfunction (54, 55). This may be one of the reasons why the TMR group had a large number of inflammatory cell infiltration, intestinal epithelial cell shedding, and even diarrhea symptoms. Sarcina enriched in TMR is a pathogen that has been reported many times. Katharine found that Sarcina was associated with gastric ulcer and fungal infection in calves and goats, which may be the cause of gastritis and enteritis (56). The study of Simpson also found that Sarcina may cause necrosis of the intestinal epithelial surface and may be related to parasitic infection (57). Many studies have pointed out that Sarcina is associated with diarrhea, inflammation and other symptoms. Sarcina is significantly enriched in the intestinal tract of horses parasitized by ascaris, and Sarcina is often found in goats, rabbits and other animals with diarrhea symptoms or fungal infection (58, 59). The study of Zhuo on piglets directly pointed out that the enrichment of Sarcina is not conducive to the healthy development of the intestinal tract, affects the production performance and feed absorption rate, and increases the probability of intestinal pathogen infection. They also pointed out that Sarcina has a great relationship with the reduction of microorganisms that produce short-chain fatty acids and feeding patterns (60). This is similar to our findings. Therefore, we believe that the inflammatory response, parasitic infection and diarrhea symptoms in the TMR group are closely related to Sarcina.
After comparing the enrichment of KEGG signaling pathway, we found that in Cell cycle-Caulobacter (pathway ID: ko04112) and Necroptosis (pathway ID: ko04217) signaling pathways, SH group was less, SF group and TMR group were more. Cell cycle-Caulobacter is a key pathway that can regulate DNA replication, cell cycle and cell topology. Cell cycle response regulator is the core of his network. Cell cycle-Caulobacter regulates the asymmetric division of bacterial cells, resulting in stalked cells and swarmer cells with different fates (61). The stalked type of progeny cell type will enter the S phase, while the swarmer type will remain in the G1 phase (62). We compared the metabolites annotated in the three groups, and found that the unique metabolites in the SF group and the TMR group eventually pointed to polar morphogenesis and pili biogenesis, while the unique metabolites in the SH group pointed to flagellar ejection holdfast biogenesis, stalk formation, that is, the cells in the SF group, TMR group and SH group went to different divisions, and different daughter cells had different cell membrane structures and cell morphology. This leads to differences in subsequent functions (63, 64). Studies have shown that phages with pili and flagella structures are more susceptible to the host through CtrA regulation (65). This molecular network regulation has a direct relationship with bacterial infection. McAdams ‘s study pointed out that the function within the Cell cycle-Caulobacter network is conserved, but after asymmetric division, due to differences in cell function and protein, downstream specific coupling species will be very different (66). Therefore, we boldly speculate that the Cell cycle-Caulobacter pathway in the SH group is not as active as the other two groups, and the final cell division results are different, which may be one of the important reasons for the differences in bacterial infection and inflammatory response between the SH group and the TMR group and the SF group. The difference in Necroptosis is clearer. SH has no unique metabolites, while the unique metabolites of SH group and TMR group are the same, which are H2AX, VDAC, Drp1, ESCRT-III. These metabolites directly or indirectly point to inflammatory response, mitochondrial damage, etc., and their activity may lead to cell damage, and ultimately necroptosis (67). Yi’s study found that plasma pathogens such as Staphylococcus aureus can lead to cell necroptosis by affecting mitochondrial membrane potential and membrane permeability. This necroptosis is regulated by calmodulin-dependent protein kinase II (CaMKII) and mixed lineage kinase domain-like (MLKL) (68). The unique metabolites we found in TMR group and SF group were involved in the above reaction process. Huang’s study also pointed out that activation of MLKL and CaMKII can lead to mitochondrial dysfunction and pro-inflammatory response (69). We also found that the unique metabolites of TMR group and SF group were concentrated in cytochrome c oxidase \ reductase after comparing the Oxidative phosphorylation (pathway ID: ko00190) of the three groups. We believe that this may be due to bacterial infection that activates pathways such as MLKL, which leads to necroptosis caused by mitochondrial dysfunction and promotes inflammatory response, and ultimately leads to damage to intestinal epithelial structure (70). In addition, intestinal flora may also interact with metabolic intermediates in Oxidative phosphorylation. Existing studies have found that NAD (nicotinamide adenine dinucleotide) can promote the secretion of intestinal mucin 2 (MUC2), enhance the ability of antibacterial infection, and alleviate intestinal inflammation by inhibiting NF-kB. Oxidative phosphorylation abnormalities and mitochondrial damage are likely to affect the levels of metabolites such as NAD, increasing the risk of intestinal inflammation and bacterial infection (71).
In the two signaling pathways of Purine metabolism (pathway ID: ko 00230) and Pyrimidine metabolism (pathway ID: ko00240), the three groups have more enrichment, but their metabolic process is different. As shown in Figure 10, some of the reactions in the TMR and SF groups were more active than SH in these two pathways, including several key enzymes that catalyze one-way reactions, such as apyrase (EC: 3.6.1.5) and AMP deaminase (EC: 3.5.4.6), which are unique to the SF and TMR groups. Studies have shown that when the body is in an inflammatory or stress state, the demand for nucleotides will increase to promote intestinal cell proliferation and tissue repair (72, 73). The activity of apyrase (EC: 3.6.1.5) will lead to excessive metabolic intermediates such as GMP and AMP. Their final products all point to uric acid, but the enzyme for the next metabolism of uric acid is not active. Although nucleotide metabolism in the SF and TMR groups may be an autonomous repair of inflammation, it also increases the risk of accumulation of substances such as uric acid. In general, purine catabolism produces metabolites with potential toxicity (74, 75). Excessive accumulation of uric acid may lead to the formation of sodium salt deposition in shutdown, skin and other parts, causing pain. It may also cause kidney damage and lead to the occurrence of uric acidemia, which is common in long-term high protein intake of livestock and poultry (76, 77); in serious cases, it may also lead to atherosclerosis (78). Apyrase is involved in the dephosphorylation of GTP, ATP, UTP, CTP and other substances, and is a one-way reaction. ATP, GTP and ITP were dephosphorylated to produce Uric acid. UTP and CTP were also dephosphorylated to produce Uracil, which was converted into β-Alanine to participate in the next reaction. After dephosphorylation, dTTP is dephosphorylated to generate dTMP, which will further generate Thymine, and then generate (R)-3-Amino-2-methylpropanoate to participate in Valine, leucine and isoleucine degradation pathway. Therefore, we inferred that the Nucleotide metabolism of the three groups was more enriched because the intestine of the calves was in the developmental stage, and the metabolism was vigorous because it responded to stimuli such as feed differences and microbial flora changes. However, the SF group and the SH group showed uniqueness in some metabolic processes due to the need for anti-inflammatory and epithelial cell repair. The activity of enzymes such as apyrase (EC: 3.6.1.5), and the enzymes not annotated to uric acid catabolism may lead to the risk of uric acid accumulation in the SF group and the TMR group.
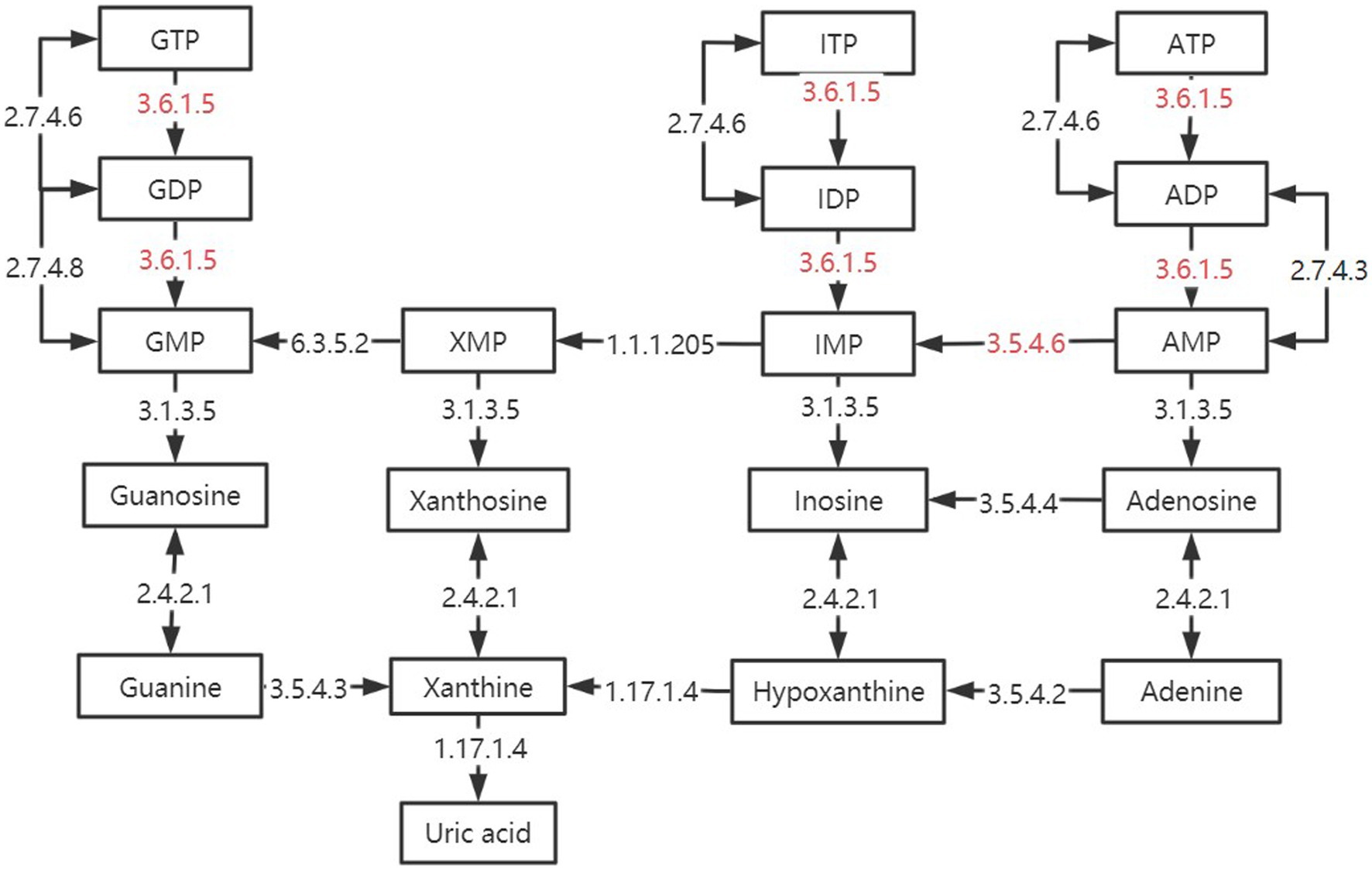
Figure 10. Differences in purine metabolic pathways between groups. The black box is marked as the intermediate product in the purine metabolic pathway. The black arrow is marked with enzymes annotated in the kegg enzyme database. The red-labeled enzyme is unique in SF and TMR.
By comparing the analysis results of the three groups of CAZy databases. The differences are mainly reflected in the GH1 family and the GH19 family. Under level 3 classification, the enrichment of β-glucosidase (EC 3.2.1.21) belonging to GH1 family in SH group was more than that in the other two groups. β-glucosidase mainly plays a role in glycolipid metabolism and is a key enzyme that catalyzes cellulose to produce glucose. We believe that the higher β-glucosidase activity in the SH group is closely related to its microbial community. Studies have shown that adding β-glucosidase to feed or activating β-glucosidase through diet can improve the digestive capacity of the gastrointestinal tract and benefit animal growth (79). β-glucosidase can also increase the activity of intestinal amylase (80). This is also consistent with our found, the amylase content of SH group was the highest. Zhang ‘s study found that castration changed the cecal digestion environment of dairy cows, significantly reduced the activity of β-glucosidase, reduced the relative abundance of Ruminococcaceae and other bacteria related to β-glucosidase, and reduced the daily gain of dairy cows (81). Therefore, the enrichment of β-glucosidase has a positive effect on calves. The chitinase (EC 3.2.1.14) in GH19 family showed more obvious differences. The enrichment of chitinase in SH group was much less than that in SF group and TMR group. Chitinase has strong specificity and can cleave chitin glycosidic bonds. Chitin is a component of invertebrates, arthropods, fungi, etc., but mammals can hardly synthesize chitin themselves. Therefore, attention should be paid when excessive chitinase is enriched in mammals, and it may be food containing chitin or microbial pathogens invade the body (82). Recent studies have shown that chitin protein is related to gastrointestinal diseases. Chitinase can play an important role in gastrointestinal digestion and diseases due to its specificity, and it is likely to become a new idea for the treatment of gastrointestinal diseases (83, 84). The study of Beier found that microorganisms can also produce chitinase, including Firmicutes, Actinobacteria, etc. Chitinase can specifically hydrolyze chitin and make it lose pathogenicity, which is particularly critical for mammals (85). We observed local inflammatory reactions in both the SF group and the TMR group. Parasites also appeared in the TMR group. It is most likely that foreign pathogens produce chitin, which stimulates some of the bacteria in the intestine that can release chitinase to respond, causing differences in intestinal flora.
In addition to this, there is a more critical phenomenon, we found that in the TMR group, a unique enrichment of many neurotransmitters and their precursors: Dopamine, Noradrenaline, Levodopa, etc., which may point to a new research field: neuro microbiology. In recent years, more and more studies have shown that microorganisms not only have the functions of assisting digestion and maintaining barriers, but also participate in the regulation of the enteric nervous system and the central nervous system by producing neurotransmitters or neurotransmitter precursors (86). It is reported that intestinal microorganisms can synthesize neurotransmitters such as dopamine, norepinephrine, 5-hydroxytryptamine and compounds involved in the metabolism of neurotransmitters in the body, which can act on the local intestinal nervous system and regulate the intestinal environment (87, 88). Taking the synthesis mechanism of dopamine (phenylalanine-tyrosine-L-dopa-dopamine) as an example, phenylalanine hydroxylase converts L-phenylalanine into L-tyrosine, which can cross the blood–brain barrier and enter the brain. In the brain, it is converted to (s)-3,4-dihydroxyphenylalanine (L-DOPA) by tyrosine hydroxylase, and then L-DOPA is converted to dopamine by dopa decarboxylase (89, 90). Although the pathway by which these neurotransmitters are synthesized by the gut microbiome is unclear, it is likely that the gut microbiome also has the ability to convert local dopamine to norepinephrine, such as dopamine-beta oxidase (91). Intestinal microorganisms can also synthesize neurotransmitter precursors such as L-tyrosine. Due to the lack of relevant transporters, some neurotransmitters circulating in the blood, such as dopamine and norepinephrine, cannot cross the blood–brain barrier. However, the precursors of these neurotransmitters can cross the blood–brain barrier, synthesize related substances in the brain, and then regulate, which may be the way that microorganisms connect the central nervous system.
Intestinal flora communicates with the central nervous system, sympathetic nerve and vagus nerve through neurotransmitters, regulates the level of neurotransmitters such as auxin, and affects the growth and development, gastrointestinal digestion and immune function of animals (92). For example, slight changes in the vagus nerve will lead to dramatic changes in the release of downstream neurotransmitters, which will affect digestive level, intestinal permeability, intestinal motility and immune response (93, 94). The central nervous system receives signals from these neurotransmitters and regulates the digestive environment and microbial community composition in the gastrointestinal tract by producing neurochemicals such as 5-hydroxytryptamine, cholecystokinin, and dopamine. Microorganisms also stimulate intestinal endocrine cells to produce hormones and complete feedback to the central nervous system through neural afferent fibers. For example, in the presence of dopamine and norepinephrine, Escherichia coli will proliferate faster and may also enhance its motility and toxicity (95). Liu ‘s study found that abnormal casein synthesis aggravated enteritis, disrupted microbial flora and its metabolic function, and led to abnormal immune response (96). The intermediate compound of neurotransmitters: short-chain fatty acids, also participate in the regulation of microorganisms and neurotransmitters. Studies have shown that short-chain fatty acids can regulate the metabolism of many neurotransmitters by acting on the response element binding proteins in the cAMP signaling pathway (97, 98). Taking dopamine as an example, short-chain fatty acids can regulate the production of dopamine and the conversion of dopamine and norepinephrine by regulating the expression of tyrosine carboxylase and dopamine-β-carboxylase (99). These metabolic pathways are reflected in the differential metabolic pathways in the TMR group. Especially for ruminant mammals such as calves, the short-chain fatty acids in the body will be more abundant, and the regulation mechanism of short-chain fatty acids will be more complicated. In some animal model experiments, it has also been found that metabolic abnormalities caused by intestinal microbial community disorders and the release of monoamine substances may lead to depression (100). This proves the deep relationship between intestinal microbial changes and host mental disorders. In animal husbandry, whether it will affect animal feed intake and estrus cycle needs further study.
In recent years, many studies have shown that intestinal microorganisms can communicate and regulate with the gastrointestinal tract and brain through neurotransmitters, and interact to maintain the dynamic balance of the intestinal environment. One of the difficulties in the field of neurobiology is to identify whether the source of various neuroactive compounds is host or microorganism. Secondly, the influence between intestinal microorganisms and neurotransmitters, and the complex communication between intestinal microorganisms and the brain are difficult. Combining the analysis results of metabolomics and metagenomics will help us understand the operation relationship between the microbial-intestinal-brain axis from the perspective of neuro microbiology. In the future, specific microbial targeted interventions can be performed on the host from the perspective of neuro microbiology, which can be used to adjust the intestinal flora and improve digestion. It is even possible to regulate specific microbial communities by understanding the relationship between gut microbes and brain health, and to treat neurological diseases through microbial regulation of the central nervous system.
In summary, our study found that the feeding mode of ‘starter + hay + milk ‘was more conducive to the development of calves. The calves in the SH group had better intestinal development, intestinal microbial community was more conducive to digestion and absorption, and immunity was stronger. The feeding mode of the SH group was conducive to the healthy development of the calves. Therefore, the calves in the SH group had higher daily weight gain, stronger digestive ability, and no pathological inflammatory reaction was found. For calves, daily weight gain is important, and more importantly, there can be a stronger digestive system and immune system to ensure that calves can have better growth and development after weaning, which is the advantage of SH group compared with SF group. The enrichment of neurotransmitters found in the TMR group also inspired us to study intestinal microorganisms from a new perspective. The research ideas of neuro microbiology will be one of our future focuses. Therefore, the results of this study provide a basis for improving the feeding mode of calves, and inspire us to explore the relationship between microorganisms and hosts from the perspective of neural metabolites.
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
The animal study was approved by the Institutional Animal Care and Use Committee of the College of Animal Science and Technology. The study was conducted in accordance with the local legislation and institutional requirements.
JW: Data curation, Writing – review & editing, Writing – original draft. YC: Data curation, Writing – original draft, Writing – review & editing. ML: Data curation, Investigation, Project administration, Writing – review & editing. SX: Investigation, Software, Writing – review & editing. KZ: Investigation, Software, Writing – review & editing. HF: Investigation, Software, Writing – review & editing. JN: Investigation, Software, Writing – review & editing. WS: Conceptualization, Investigation, Software, Writing – review & editing. XJ: Conceptualization, Investigation, Software, Writing – review & editing. SL: Methodology, Project administration, Supervision, Writing – review & editing.
This study was supported by the key research and development project of Sichuan beef cattle genome selection technology system (2022YFYZ0006).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. ^https://github.com/voutcn/megahit
2. ^http://topaz.gatech.edu/GeneMark/
1. Qi-yu, D, Rong, Z, and Yan, T. Current research progresses on calf rearing and nutrition in China. J Integr Agric. (2017) 16:2805–14. doi: 10.1016/S2095-3119(17)61767-2
2. Soberon, F, Raffrenato, E, Everett, RW, and Amburgh, M. Preweaning milk replacer intake and effects on long-term productivity of dairy calves. J Dairy Sci. (2012) 95:783–93. doi: 10.3168/jds.2011-4391
3. Alawneh, JI, Barreto, M, Moore, R, Soust, M, Al-Harbi, H, James, A, et al. Systematic review of an intervention: the use of probiotics to improve health and productivity of calves. Prev Vet Med. (2020) 183:105147. doi: 10.1016/j.prevetmed.2020.105147
4. Osorio, J. Gut health, stress, and immunity in neonatal dairy calves: the host side of host-pathogen interactions. J Anim Sci Biotechnol. (2020) 11:105. doi: 10.1186/s40104-020-00509-3
5. Malmuthuge, N, Griebel, PJ, and Guan, LL. The gut microbiome and its potential role in the development and function of newborn calf gastrointestinal tract. Front Vet Sci. (2015) 2:36. doi: 10.3389/fvets.2015.00036
6. Urie, NJ, Lombard, JE, Shivley, CB, Kopral, CA, Adams, AE, Earleywine, TJ, et al. Preweaned heifer management on US dairy operations: part V. factors associated with morbidity and mortality in preweaned dairy heifer calves. J Dairy Sci. (2018) 101:9229–44. doi: 10.3168/jds.2017-14019
7. Soberon, F, and Amburgh, MV. Lactation biology symposium: the effect of nutrient intake from milk or milk replacer of preweaned dairy calves on lactation milk yield as adults: a meta-analysis of current data. J Anim Sci. (2013) 91:706–12. doi: 10.2527/jas.2012-5834
8. Garcia-Mazcorro, JF, Ishaq, SL, Rodriguez-Herrera, MV, Garcia-Hernandez, CA, Kawas, JR, and Nagaraja, TG. Review: are there indigenous Saccharomyces in the digestive tract of livestock animal species? Implications for health, nutrition and productivity traits. Animal. (2020) 14:22–30. doi: 10.1017/s1751731119001599
9. Guzman, CE, Bereza-Malcolm, LT, De Groef, B, and Franks, AE. Uptake of milk with and without solid feed during the monogastric phase: effect on fibrolytic and methanogenic microorganisms in the gastrointestinal tract of calves. Anim Sci J. (2016) 87:378–88. doi: 10.1111/asj.12429
10. Suarez-Mena, FX, Heinrichs, AJ, Jones, CM, Hill, TM, and Quigley, JD. Straw particle size in calf starters: effects on digestive system development and rumen fermentation. J Dairy Sci. (2016) 99:341–53. doi: 10.3168/jds.2015-9884
11. Khan, MA, Bach, A, Weary, DM, and Keyserlingk, MAGV. Invited review: transitioning from milk to solid feed in dairy heifers. J Dairy Sci. (2016) 99:885–902. doi: 10.3168/jds.2015-9975
12. O'Hara, E, Kelly, A, McCabe, MS, Kenny, DA, Guan, LL, and Waters, SM. Effect of a butyrate-fortified milk replacer on gastrointestinal microbiota and products of fermentation in artificially reared dairy calves at weaning. Sci Rep. (2018) 8:14901. doi: 10.1038/s41598-018-33122-6
13. Wang, H, Yu, Z, Gao, Z, Li, Q, Qiu, X, Wu, F, et al. Effects of compound probiotics on growth performance, rumen fermentation, blood parameters, and health status of neonatal Holstein calves. J Dairy Sci. (2022) 105:2190–200. doi: 10.3168/jds.2021-20721
14. O'Hara, E, Neves, ALA, Song, Y, and Guan, LL. The role of the gut microbiome in cattle production and health: driver or passenger? Annu Rev Anim Biosci. (2020) 8:199–220. doi: 10.1146/annurev-animal-021419-083952
15. Deng, F, Wang, C, Li, D, Peng, Y, Deng, L, Zhao, Y, et al. The unique gut microbiome of giant pandas involved in protein metabolism contributes to the host’s dietary adaption to bamboo. Microbiome. (2023) 11:180. doi: 10.1186/s40168-023-01603-0
16. Agirman, G, and Hsiao, EY. SnapShot: the microbiota-gut-brain axis. Cells. (2021) 184:2524–2524.e1. doi: 10.1016/j.cell.2021.03.022
17. Diao, Q, Zhang, R, and Fu, T. Review of strategies to promote rumen development in calves. Animals. (2019) 9:490. doi: 10.3390/ani9080490
18. Dill-McFarland, KA, Breaker, JD, and Suen, G. Microbial succession in the gastrointestinal tract of dairy cows from 2 weeks to first lactation. Sci Rep. (2017) 7:40864. doi: 10.1038/srep40864
19. Wu, ZH, Azarfar, A, Simayi, A, Li, SL, Jonker, A, and Cao, ZJ. Effects of forage type and age at which forage provision is started on growth performance, rumen fermentation, blood metabolites and intestinal enzymes in Holstein calves. Anim Prod Sci. (2018) 58:2288–99. doi: 10.1071/AN16576
20. Nielsen, H, Almeida, M, Juncker, A, Rasmussen, S, Li, J, Sunagawa, S, et al. Identification and assembly of genomes and genetic elements in complex metagenomic samples without using reference genomes. Nat Biotechnol. (2014) 32:822–8. doi: 10.1038/nbt.2939
21. Scher, JU, Andrew, S, Longman, RS, Nicola, S, Carles, U, Craig, B, et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. elife. (2013) 2:e01202. doi: 10.7554/eLife.01202
22. Karlsson, FH, Fåk, F, Nookaew, I, Tremaroli, V, Fagerberg, B, Petranovic, D, et al. Symptomatic atherosclerosis is associated with an altered gut metagenome. Nat Commun. (2012) 3:1245. doi: 10.1038/ncomms2266
23. Villar, E, Farrant, GK, Follows, M, Garczarek, L, Speich, S, Audic, S, et al. Ocean plankton. Environmental characteristics of Agulhas rings affect interocean plankton transport. Science. (2015) 348:1261447. doi: 10.1126/science.1261447
24. Karlsson, FH, Tremaroli, V, Nookaew, I, Bergström, G, Behre, CJ, Fagerberg, B, et al. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature. (2013) 498:99–103. doi: 10.1038/nature12198
25. Avershina, E, Frisli, T, and Rudi, K. De novo semi-alignment of 16S rRNA gene sequences for deep phylogenetic characterization of next generation sequencing data. Microbes Environ. (2013) 28:211–6. doi: 10.1264/jsme2.me12157
26. Segata, N, Izard, J, Waldron, L, Gevers, D, Miropolsky, L, Garrett, WS, et al. Metagenomic biomarker discovery and explanation. Genome Biol. (2011) 12:R60. doi: 10.1186/gb-2011-12-6-r60
27. Kanehisa, M, Furumichi, M, Tanabe, M, Sato, Y, and Morishima, K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. (2017) 45:D353–d361. doi: 10.1093/nar/gkw1092
28. Kanehisa, M, Goto, S, Hattori, M, Aoki-Kinoshita, KF, Itoh, M, Kawashima, S, et al. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. (2006) 34:D354–7. doi: 10.1093/nar/gkj102
29. Cantarel, BL, Coutinho, PM, Rancurel, C, Bernard, T, Lombard, V, and Henrissat, B. The carbohydrate-active enzymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. (2009) 37:D233–8. doi: 10.1093/nar/gkn663
30. Huerta-Cepas, J, Szklarczyk, D, Forslund, K, Cook, H, Heller, D, Walter, MC, et al. eggNOG 4.5: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. (2016) 44:D286–93. doi: 10.1093/nar/gkv1248
31. Kanehisa, M, Goto, S, Sato, Y, Kawashima, M, Furumichi, M, and Tanabe, M. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. (2014) 42:D199–205. doi: 10.1093/nar/gkt1076
32. Wilson, ID, Gika, H, Theodoridis, G, Plumb, RS, Shockcor, J, Holmes, E, et al. Global metabolic profiling procedures for urine using UPLC-MS. Nat Protoc. (2010) 5:1005–18. doi: 10.1038/nprot.2010.50
33. Boulesteix, AL, and Strimmer, K. Partial least squares: a versatile tool for the analysis of high-dimensional genomic data. Brief Bioinform. (2007) 8:32–44. doi: 10.1093/bib/bbl016
34. Van Soest, B, Weber Nielsen, M, Moeser, A, Abuelo, A, and VandeHaar, M. Transition milk stimulates intestinal development of neonatal Holstein calves. J Dairy Sci. (2022) 105:7011–22. doi: 10.3168/jds.2021-21723
35. van Keulen, P, Khan, M, Dijkstra, J, Knol, F, and McCoard, S. Effect of arginine or glutamine supplementation and milk feeding allowance on small intestine development in calves. J Dairy Sci. (2020) 103:4754–64. doi: 10.3168/jds.2019-17529
36. Welboren, A, Hatew, B, Renaud, J, Leal, L, Martín-Tereso, J, and Steele, M. Intestinal adaptations to energy source of milk replacer in neonatal dairy calves. J Dairy Sci. (2021) 104:12079–93. doi: 10.3168/jds.2021-20516
37. Ma, J, Shah, A, Shao, Y, Wang, Z, Zou, H, and Kang, K. Dietary supplementation of yeast cell wall improves the gastrointestinal development of weaned calves. Anim Nutr. (2020) 6:507–12. doi: 10.1016/j.aninu.2020.06.003
38. Ma, T, O'Hara, E, Song, Y, Fischer, AJ, He, Z, Steele, MA, et al. Altered mucosa-associated microbiota in the ileum and colon of neonatal calves in response to delayed first colostrum feeding. J Dairy Sci. (2019) 102:7073–86. doi: 10.3168/jds.2018-16130
39. Bai, Y, Zhou, Y, Zhang, R, Chen, Y, Wang, F, and Zhang, M. Gut microbial fermentation promotes the intestinal anti-inflammatory activity of Chinese yam polysaccharides. Food Chem. (2023) 402:134003. doi: 10.1016/j.foodchem.2022.134003
40. Barczynska, R, Slizewska, K, Litwin, M, Szalecki, M, and Kapusniak, J. Effects of dietary fiber preparations made from maize starch on the growth and activity of selected bacteria from the Firmicutes, Bacteroidetes, and Actinobacteria phyla in fecal samples from obese children. Acta Biochim Pol. (2016) 63:261–6. doi: 10.18388/abp.2015_1068
41. Barczynska, R, Jurgoński, A, Slizewska, K, Juśkiewicz, J, and Kapusniak, J. Effects of potato dextrin on the composition and metabolism of the gut microbiota in rats fed standard and high-fat diets. J Funct Foods. (2017) 34:398–407. doi: 10.1016/j.jff.2017.05.023
42. Yi, S, Lee, H, So, K, Kim, E, Jung, Y, Kim, M, et al. Effect of feeding raw potato starch on the composition dynamics of the piglet intestinal microbiome. Anim Biosci. (2022) 35:1698–710. doi: 10.5713/ab.22.0045
43. Zhu, Y, Mu, C, Liu, S, and Zhu, W. Dietary citrus pectin drives more ileal microbial protein metabolism and stronger fecal carbohydrate fermentation over fructooligosaccharide in growing pigs. Anim Nutr. (2022) 11:252–64. doi: 10.1016/j.aninu.2022.08.005
44. Heijne, M, van der Goot, J, Buys, H, Dinkla, A, Roest, H, van Keulen, L, et al. Pathogenicity of Chlamydia gallinacea in chickens after oral inoculation. Vet Microbiol. (2021) 259:109166. doi: 10.1016/j.vetmic.2021.109166
45. Reinhold, P, Sachse, K, and Kaltenboeck, B. Chlamydiaceae in cattle: commensals, trigger organisms, or pathogens? Vet J. (2011) 189:257–67. doi: 10.1016/j.tvjl.2010.09.003
46. Müller, U, Sachse, K, Kemmerling, K, Rietz, C, and Sauerwein, H. Identification of certain management practices and health data associated with Chlamydia infection status in German dairy herds. Vet J. (2013) 197:905–7. doi: 10.1016/j.tvjl.2013.06.003
47. Anstey, SI, Quigley, BL, Polkinghorne, A, and Jelocnik, M. Chlamydial infection and on-farm risk factors in dairy cattle herds in south East Queensland. Aust Vet J. (2019) 97:505–8. doi: 10.1111/avj.12879
48. Xia, B, Wu, W, Fang, W, Wen, X, Xie, J, and Zhang, H. Heat stress-induced mucosal barrier dysfunction is potentially associated with gut microbiota dysbiosis in pigs. Anim Nutr. (2022) 8:289–99. doi: 10.1016/j.aninu.2021.05.012
49. Dorbek-Kolin, E, Husso, A, Niku, M, Loch, M, Pessa-Morikawa, T, Niine, T, et al. Faecal microbiota in two-week-old female dairy calves during acute cryptosporidiosis outbreak – association with systemic inflammatory response. Res Vet Sci. (2022) 151:116–27. doi: 10.1016/j.rvsc.2022.07.008
50. Zaman, S, Gohar, M, Kanwal, H, Chaudhary, A, and Imran, M. Impact of probiotic Geotrichum candidum QAUGC01 on health, productivity, and gut microbial diversity of dairy cattle. Curr Microbiol. (2022) 79:376. doi: 10.1007/s00284-022-03074-2
51. Shin, N, Whon, T, and Bae, J. Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. (2015) 33:496–503. doi: 10.1016/j.tibtech.2015.06.011
52. Han, Y, Tang, C, Zhao, Q, Fan, S, Yang, P, and Zhang, J. Butyrate mitigates lipopolysaccharide-induced intestinal morphological changes in weanling piglets by regulating the microbiota and energy metabolism, and alleviating inflammation and apoptosis. Microorganisms. (2022) 10:2001. doi: 10.3390/microorganisms10102001
53. Varada, V, Kumar, S, Chhotaray, S, and Tyagi, A. Host-specific probiotics feeding influence growth, gut microbiota, and fecal biomarkers in buffalo calves. AMB Express. (2022) 12:118. doi: 10.1186/s13568-022-01460-4
54. Lu, C, Liu, Y, Ma, Y, Wang, S, Cai, C, Yang, Y, et al. Comparative evaluation of the ileum microbiota composition in piglets at different growth stages. Front Microbiol. (2021) 12:765691. doi: 10.3389/fmicb.2021.765691
55. Piccolo, B, Mercer, K, Bhattacharyya, S, Bowlin, A, Saraf, M, Pack, L, et al. Early postnatal diets affect the bioregional small intestine microbiome and Ileal metabolome in neonatal pigs. J Nutr. (2017) 147:1499–509. doi: 10.3945/jn.117.252767
56. Marchesi, J, Adams, D, Fava, F, Hermes, G, Hirschfield, G, Hold, G, et al. The gut microbiota and host health: a new clinical frontier. Gut. (2016) 65:330–9. doi: 10.1136/gutjnl-2015-309990
57. Simpson, K, Callan, R, and Van Metre, D. Clostridial Abomasitis and enteritis in ruminants. Vet Clin North Am Food Anim Pract. (2018) 34:155–84. doi: 10.1016/j.cvfa.2017.10.010
58. Cain, J, Norris, J, Ripley, N, Suri, P, Finnerty, C, Gravatte, H, et al. The microbial community associated with Parascaris spp. infecting juvenile horses. Parasit Vectors. (2022) 15:408. doi: 10.1186/s13071-022-05533-y
59. Yuan, X, Liu, J, Hu, X, Yang, S, Zhong, S, Yang, T, et al. Alterations in the jejunal microbiota and fecal metabolite profiles of rabbits infected with Eimeria intestinalis. Parasit Vectors. (2022) 15:231. doi: 10.1186/s13071-022-05340-5
60. Zhuo, Y, Huang, Y, He, J, Hua, L, Xu, S, Li, J, et al. Effects of corn and broken Rice extrusion on the feed intake, nutrient digestibility, and gut microbiota of weaned piglets. Animals. (2022) 12:818. doi: 10.3390/ani12070818
61. Fatima, N, Fazili, K, and Bhat, N. Proteolysis dependent cell cycle regulation in Caulobacter crescentus. Cell Div. (2022) 17:3. doi: 10.1186/s13008-022-00078-z
62. Beroual, W, Prévost, K, Lalaouna, D, Ben Zaina, N, Valette, O, Denis, Y, et al. The noncoding RNA CcnA modulates the master cell cycle regulators CtrA and GcrA in Caulobacter crescentus. PLoS Biol. (2022) 20:e3001528. doi: 10.1371/journal.pbio.3001528
63. Aldridge, P, Paul, R, Goymer, P, Rainey, P, and Jenal, U. Role of the GGDEF regulator PleD in polar development of Caulobacter crescentus. Mol Microbiol. (2003) 47:1695–708. doi: 10.1046/j.1365-2958.2003.03401.x
64. Paul, R, Jaeger, T, Abel, S, Wiederkehr, I, Folcher, M, Biondi, E, et al. Allosteric regulation of histidine kinases by their cognate response regulator determines cell fate. Cells. (2008) 133:452–61. doi: 10.1016/j.cell.2008.02.045
65. Mascolo, E, Adhikari, S, Caruso, SM, deCarvalho, T, Folch Salvador, A, Serra-Sagristà, J, et al. The transcriptional regulator CtrA controls gene expression in Alphaproteobacteria phages: evidence for a lytic deferment pathway. Front Microbiol. (2022) 13:918015. doi: 10.3389/fmicb.2022.918015
66. McAdams, H, and Shapiro, L. The architecture and conservation pattern of whole-cell control circuitry. J Mol Biol. (2011) 409:28–35. doi: 10.1016/j.jmb.2011.02.041
67. Liu, L, Liu, Y, Cheng, X, and Qiao, X. The alleviative effects of quercetin on cadmium-induced necroptosis via inhibition ROS/iNOS/NF-κB pathway in the chicken brain. Biol Trace Elem Res. (2021) 199:1584–94. doi: 10.1007/s12011-020-02563-4
68. Yi, Y, Gao, K, Lin, P, Chen, H, Zhou, D, Tang, K, et al. Staphylococcus aureus-induced necroptosis promotes mitochondrial damage in goat endometrial epithelial cells. Animals. (2022) 12:2218–8. doi: 10.3390/ani12172218
69. Huang, Y, Liang, Q, Zhang, H, Chen, S, Xu, L, Zeng, B, et al. Baicalin inhibits necroptosis by decreasing oligomerization of phosphorylated MLKL and mitigates caerulein-induced acute pancreatitis in mice. Int Immunopharmacol. (2022) 108:108885. doi: 10.1016/j.intimp.2022.108885
70. Li, S, Liu, R, Xia, S, Wei, G, Ishfaq, M, Zhang, Y, et al. Protective role of curcumin on aflatoxin B1-induced TLR4/RIPK pathway mediated-necroptosis and inflammation in chicken liver. Ecotoxicol Environ Saf. (2022) 233:113319. doi: 10.1016/j.ecoenv.2022.113319
71. Zhou, Y, Jin, H, Wu, Y, Chen, L, Bao, X, and Lu, C. Gallic acid protects against ethanol-induced hepatocyte necroptosis via an NRF2-dependent mechanism. Toxicol In Vitro. (2019) 57:226–32. doi: 10.1016/j.tiv.2019.03.008
72. Fitzsimmons Liam, F, Liu, L, Kim, J-S, Jones-Carson, J, and Vázquez-Torres, A. Salmonella reprograms nucleotide metabolism in its adaptation to Nitrosative stress. MBio. (2018) 9:e00211–8. doi: 10.1128/mBio.00211-18
73. Lane, A, and Fan, T. Regulation of mammalian nucleotide metabolism and biosynthesis. Nucleic Acids Res. (2015) 43:2466–85. doi: 10.1093/nar/gkv047
74. Shi, C, Zhou, Z, Chi, X, Xiu, S, Yi, C, Jiang, Z, et al. Recent advances in gout drugs. Eur J Med Chem. (2023) 245:114890. doi: 10.1016/j.ejmech.2022.114890
75. Tong, S, Zhang, P, Cheng, Q, Chen, M, Chen, X, Wang, Z, et al. The role of gut microbiota in gout: is gut microbiota a potential target for gout treatment. Front Cell Infect Microbiol. (2022) 12:1051682. doi: 10.3389/fcimb.2022.1051682
76. Zhang, L, Wang, Y, Jia, H, Liu, X, Zhang, R, and Guan, J. Transcriptome and metabolome analyses reveal the regulatory effects of compound probiotics on cecal metabolism in heat-stressed broilers. Poult Sci. (2023) 102:102323. doi: 10.1016/j.psj.2022.102323
77. Zhao, R, Li, Z, Sun, Y, Ge, W, Wang, M, Liu, H, et al. Escherichia coliengineered Nissle 1917 with urate oxidase and an oxygen-recycling system for hyperuricemia treatment. Gut Microbes. (2022) 14:2070391. doi: 10.1080/19490976.2022.2070391
78. Wu, Z, Zhang, H, Li, Z, Li, H, Miao, X, Pan, H, et al. Mutual effect of homocysteine and uric acid on arterial stiffness and cardiovascular risk in the context of predictive, preventive, and personalized medicine. EPMA J. (2022) 13:581–95. doi: 10.1007/s13167-022-00298-x
79. Qian, LC, and Sun, JY. Effect of β-glucosidase as a feed supplementary on the growth performance, digestive enzymes and physiology of broilers. Asian-australas J Anim Sci. (2009) 22:260–6. doi: 10.5713/ajas.2009.80416
80. Long, C, Qi, X, and Venema, K. Chemical and nutritional characteristics, and microbial degradation of rapeseed meal recalcitrant carbohydrates: a review. Front Nutr. (2022) 9:948302. doi: 10.3389/fnut.2022.948302
81. Li, Z, Shi, J, Lei, Y, Wu, J, Zhang, R, Zhang, X, et al. Castration alters the cecal microbiota and inhibits growth in Holstein cattle. J Anim Sci. (2022) 100:skac367. doi: 10.1093/jas/skac367
82. Chang, D, Sharma, L, and Dela Cruz, CS. Chitotriosidase: a marker and modulator of lung disease. Eur Respir Rev. (2020) 29:190143. doi: 10.1183/16000617.0143-2019
83. Allonsius, C, Vandenheuvel, D, Oerlemans, E, Petrova, M, Donders, G, Cos, P, et al. Inhibition of Candida albicans morphogenesis by chitinase from Lactobacillus rhamnosus GG. Sci Rep. (2019) 9:2900. doi: 10.1038/s41598-019-39625-0
84. Marzena, M, Anna, Z, M, GM, Jacek, O, and Jakub, F. Chitinases and Chitinase-like proteins as therapeutic targets in inflammatory diseases, with a special focus on inflammatory bowel diseases. Int J Mol Sci. (2021) 22:6966. doi: 10.3390/ijms22136966
85. Beier, S, and Bertilsson, S. Bacterial chitin degradation-mechanisms and ecophysiological strategies. Front Microbiol. (2013) 4:149. doi: 10.3389/fmicb.2013.00149
86. Miri, S, Yeo, J, Abubaker, S, and Hammami, R. Neuromicrobiology, an emerging neurometabolic facet of the gut microbiome? Front Microbiol. (2023) 14:1098412. doi: 10.3389/fmicb.2023.1098412
87. Cox, LM, and Weiner, HL. Microbiota signaling pathways that influence neurologic disease. Neurotherapeutics. (2018) 15:135–45. doi: 10.1007/s13311-017-0598-8
88. Cryan, J, O'Riordan, K, Sandhu, K, Peterson, V, and Dinan, T. The gut microbiome in neurological disorders. Lancet Neurol. (2020) 19:179–94. doi: 10.1016/s1474-4422(19)30356-4
89. Nagatsu, T, Nakashima, A, Ichinose, H, and Kobayashi, K. Human tyrosine hydroxylase in Parkinson's disease and in related disorders. J Neural Transm. (2019) 126:397–409. doi: 10.1007/s00702-018-1903-3
90. Zhou, C, Nutt, DJ, and Davies, SJ. Visualizing classification of drugs used in psychotic disorders: A 'subway map' representing mechanisms, established classes and informal categories. J Psychopharmacol. (2022) 36:1007–15. doi: 10.1177/02698811221115758
91. Maini Rekdal, V, Bess, EN, Bisanz, JE, Turnbaugh, PJ, and Balskus, EP. Discovery and inhibition of an interspecies gut bacterial pathway for levodopa metabolism. Science. (2019) 364:eaau6323. doi: 10.1126/science.aau6323
92. T, DLM. Gut Bacteria and neurotransmitters. Microorganisms. (2022) 10:1838. doi: 10.3390/microorganisms10091838
93. Bonaz, B, Bazin, T, and Pellissier, S. The Vagus nerve at the Interface of the microbiota-gut-brain Axis. Front Neurosci. (2018) 12:49. doi: 10.3389/fnins.2018.00049
94. Breit, S, Kupferberg, A, Rogler, G, and Hasler, G. Vagus nerve as modulator of the brain-gut axis in psychiatric and inflammatory disorders. Front Psych. (2018) 9:44. doi: 10.3389/fpsyt.2018.00044
95. Hughes, DT, and Sperandio, V. Inter-kingdom signalling: communication between bacteria and their hosts. Nat Rev Microbiol. (2008) 6:111–20. doi: 10.1038/nrmicro1836
96. Liu, X, Liu, S, Tang, Y, Pu, Z, Xiao, H, Gao, J, et al. Intragastric Administration of Casein Leads to nigrostriatal disease progressed accompanied with persistent nigrostriatal-intestinal inflammation Activited and intestinal microbiota-metabolic disorders induced in MPTP mouse model of Parkinson's disease. Neurochem Res. (2021) 46:1514–39. doi: 10.1007/s11064-021-03293-2
97. Dalile, B, Van Oudenhove, L, Vervliet, B, and Verbeke, K. The role of short-chain fatty acids in microbiota-gut-brain communication. Nat Rev Gastroenterol Hepatol. (2019) 16:461–78. doi: 10.1038/s41575-019-0157-3
98. van de Wouw, M, Boehme, M, Lyte, JM, Wiley, N, Strain, C, O'Sullivan, O, et al. Short-chain fatty acids: microbial metabolites that alleviate stress-induced brain-gut axis alterations. J Physiol. (2018) 596:4923–44. doi: 10.1113/jp276431
99. Fung, T, Vuong, H, Luna, C, Pronovost, G, Aleksandrova, A, Riley, N, et al. Intestinal serotonin and fluoxetine exposure modulate bacterial colonization in the gut. Nat Microbiol. (2019) 4:2064–73. doi: 10.1038/s41564-019-0540-4
Keywords: calves, feeding patterns, metagenomics, metabolomics, neuro microbiology
Citation: Wang J, Chen Y, Li M, Xia S, Zhao K, Fan H, Ni J, Sun W, Jia X and Lai S (2023) The effects of differential feeding on ileum development, digestive ability and health status of newborn calves. Front. Vet. Sci. 10:1255122. doi: 10.3389/fvets.2023.1255122
Received: 08 July 2023; Accepted: 29 August 2023;
Published: 08 September 2023.
Edited by:
Balamuralikrishnan Balasubramanian, Sejong University, Republic of KoreaReviewed by:
Tianliu Zhang, Henan Agricultural University, ChinaCopyright © 2023 Wang, Chen, Li, Xia, Zhao, Fan, Ni, Sun, Jia and Lai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Songjia Lai, bGFpc2o1Nzk0QDE2My5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.