- 1State Key Laboratory of Swine and Poultry Breeding Industry, Guangdong Key Laboratory of Animal Breeding and Nutrition, Institute of Animal Science, Guangdong Academy of Agricultural Sciences, Guangzhou, China
- 2Maoming Branch, Guangdong Laboratory for Lingnan Modern Agriculture, Maoming, China
- 3Guangzhou Customs Technical Center, Guangzhou, China
The current estrus detection method is generally time-consuming and has low accuracy. As such, a deeper understanding of the physiological processes during the estrous cycle accelerates the development of estrus detection efficiency and accuracy. In this study, the label-free acquisition mass spectrometry was used to explore salivary proteome profiles during the estrous cycle (day −3, day 0, day 3, and day 8) in pigs, and the parallel reaction monitoring (PRM) was applied to verify the relative profiles of protein expression. A total of 1,155 proteins were identified in the label-free analysis, of which 115 were identified as differentially expressed proteins (DEPs) among different groups (p ≤ 0.05). Functional annotation revealed that the DEPs were clustered in calcium ion binding, actin cytoskeleton, and lyase activity. PRM verified the relative profiles of protein expression, in which PHB domain-containing protein, growth factor receptor-bound protein 2, elongation factor Tu, carboxypeptidase D, carbonic anhydrase, and trefoil factor 3 were confirmed to be consistent in both label-free and PRM approaches. Comparative proteomic assays on saliva would increase our knowledge of the estrous cycle in sows and provide potential methods for estrus detection.
Introduction
As an indispensable part of sow reproductive management, prompt and accurate estrus detection directly determines the overall productivity of sow utilization by affecting the optimal insemination moment, reducing ineffective feeding, and timely eliminating sows with low reproductive performance (1). Several methods have been applied to detect estrus in sows, for instance, behavioral observation, teaser boar test, and steroids estimation (2). The most common method is manual recognition based on swelling of the vulva and behavioral signs to a back-pressure test or boar. The observation method is time-and labor-intensive, while accuracy depends heavily on the experience of the technician (2). Few steroids are present in boar saliva and are considered to contribute greatly to promoting estrus behaviors in sows (3). In this context, a boar saliva analog mixture of androstenone, androstanol, and quinoline was synthesized and shown effective for estrus detection (4, 5). As a steroid pheromone, the analog protects sows from boar exposure and probable disease infection by teaser boar (6, 7).
With the development of artificial intelligence, many researchers have focused on the automatic identification of sow estrus by sensing regular temperature changes (8, 9), activity (10) or the frequency and duration of the sow’s visit to a boar (11) after gathering massive images or videos (12, 13). These automatic identifications showed relatively lower or equivalent accuracy compared with manual observation (9, 11). A study conducted by Lei (11) first combined machine vision with a bionic boar model to identify sows in heat. Despite the high cost of machines, recognition has a high accuracy of 98.25% and other advantages, such as greater intelligence and biosafety, but less stress than manual detection methods. Estimations based on physical or biochemical parameters of vaginal mucus (14), hormone concentration or crystal forms in body fluids (15, 16) have also been investigated, but they were not widely used because of the high cost of assay kits or their unstable characteristics. It remains a vital concern for the breeding industry to explore timely, efficient, and sensitive estrus diagnostic methods. Meanwhile, knowledge gaps regarding estrous cycles physiology must be addressed.
As an important body secretion, saliva is rich in steroid hormones, nucleic acids, proteins, and polypeptides, portions of which can be passively diffused or actively transported from the blood, revealing high similarity with blood (17–19). Recently, saliva has become an effective, noninvasive, easily available, and convenient material for detecting the physiological status of animals. Saliva estimation has great application prospects for estrus detection (2). Herein, we compared salivary proteomics in sows during the estrous cycle and verified the relative expression of several proteins to supplement our understanding of estrus physiology and provide a reference for estrus recognition development.
Materials and methods
Animals and saliva collection
Eight healthy primiparous hybrid sows (Duroc × Tibetan crossbred pig) showing normal estrus were selected for this study. Animal welfare in this study was safeguarded and approved by the Animal Care and Use Committee of Guangdong Academy of Agricultural Sciences (Protocol number:2018001). The experimental sows were observed daily in the morning and evening to assess their estrus status. Estrus identification was conducted by external observation and the back pressure reaction. A sow standing still and accepting a boar crawling with a swelling vulva was considered estrus, and the day of estrus was defined as day 0. Saliva, representing four stages of the estrous cycle, was collected every morning before feeding, including ED-3 (proestrus, n = 3): 3 days before estrus; ED0 (estrus, n = 3): the day of estrus; ED3 (metestrus, n = 3): 3 days after estrus; and ED8 (diestrus, n = 3): 8 days after estrus. The number of saliva collections for each timepoint were three. Pigs were curious and chewed the hanging cotton bags wrapping up with absorbent cotton balls. Saliva infiltrated the absorbent cotton ball was squeezed out, released into the sample bag, and then transferred into clean tubes. The samples were subsequently centrifugated at 4000 × g for 5 min at 4°C and the supernatant was mixed with Protease Inhibitor Cocktail (10 μL/mL, Sigma) before storage at −80°C until quantitative proteomics assay.
Protein extraction, quality test, digestion and desalination
Protein extraction and LS-MS analysis were conducted by Novogene Co., Ltd. The sample was lysed with DB lysis buffer (8 M Urea, 100 mM TEAB, pH 8.5, Sigma) in a 1.5 mL centrifuge tube, and the lysate was centrifuged at 12000 × g for 15 min at 4°C. The supernatant was reduced with 10 mM DTT (Sigma) at 56°C for 1 h and alkylated with sufficient IAM (Sigma) for 1 h at room temperature in the dark. The total protein concentration was determined using the Bradford protein quantitative kit (Beyotime). Then, 20 μg of protein was loaded into each well for gel electrophoresis to assess protein quality. Next, 120 μg of protein samples were mixed with DB lysis buffer (8 M Urea, 100 mM TEAB, pH 8.5, Sigma), trypsin (Promega) and 100 mM TEAB buffer to make the final volume up to 100 μL. The mixture was incubated for 4 h at 37°C. Subsequently, trypsin and CaCl2 (Sinopharm chemical reagent Co., Ltd) were added, and the samples were incubated overnight. Formic acid (Thermo Fisher Scientific) was added to the digested samples, adjusted to pH < 3, and centrifuged at 12000 × g for 5 min at room temperature. The supernatant was loaded onto a C18 desalting column (Thermo Fisher Scientific), washed three times with washing buffer (0.1% formic acid, 3% acetonitrile, Thermo Fisher), eluted with elution buffer (0.1% formic acid, 70% acetonitrile), and the eluents of each sample were collected and lyophilized.
LC–MS analysis
The lyophilized proteins were dissolved in 10 μL of mobile phase A (0.1% formic acid in distilled deionized water), centrifuged at 14,000 × g for 20 min at 4°C, and 1 μL of supernatant was loaded into a homemade C18 Nano-Trap column (4.5 cm × 75 μm, 3 μm). Proteins were then linearly gradient fractionated using a homemade analytical column (15 cm × 150 μm, 1.9 μm) with mobile phases A and B (80% acetonitrile, 0.1% formic acid) and analyzed using a Q Exactive™ HF-X mass spectrometer (Thermo Fisher Scientific). Ions source of Nanospray Flex™ (ESI) were fully scanned, ranging from 350 to 1,500 m/z, in which the top 40 precursors of the highest abundance were fragmented by higher energy collisional dissociation (HCD) and analyzed by MS/MS using the following parameters: the resolution was 15,000 (at m/z 200), the automatic gain control (20) target value was 1 × 105, the maximum ion injection time was 45 ms, the normalized collision energy was set at 27%, the intensity threshold was 2.2 × 104, and the dynamic exclusion parameter was 20 s.
Identification and quantitation of proteins
The offline spectra were searched using Proteome Discoverer 2.2 (PD 2.2, Thermo Fisher Scientific), with a maximum of two missed cleavage sites allowed. Peptides with credibility greater than 99% were considered peptide spectrum matches (PSMs). A protein containing at least one unique peptide was defined as trusted. These preserved PSMs and trusted proteins were subjected to false discovery rate (FDR) analysis to remove data larger than 0.01. The general distribution character of all the identified proteins were analyzed with Jvenn online program.1 Relative quantitative differences were analyzed using the t-test. Proteins with a fold change ≥1.5 or ≤ 0.67 and p ≤ 0.05 were defined as DEPs.
Bioinformatics analysis
Both identified proteins and DEPs were subjected to bioinformatics analysis. Functional prediction of Gene Ontology (GO) and InterPro (21) was conducted using an interproscan program (version 5.22–61.0) against the non-redundant protein databases, including Pfam, PRINTS, ProDom, SMART, ProSite, and PANTHER (22). Protein family and pathway predictions were analyzed using the Clusters of Orthologous Groups (23) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases. Potential protein–protein interactions were assayed using the STRING-db server (24).2
Validation of relative expression level by PRM
To verify the relative expression levels of DEPs identified in label-free quantitative proteomic analysis, saliva samples collected on days −3, 0,3, 8 for each day (n = 3) during the estrous cycle were confirmed by PRM with an acquired MS/MS spectrum. Proteins extracted from saliva were quantity and quality tested, enzymatically hydrolyzed, desalted, and lyophilized, as described above. For LC–MS/MS analysis pre-experiment, 1 μg of the mixture was eluted as a “label-free” method using the EASY-nLCTM1200 UHPLC system (Thermo Fisher Scientific) coupled with a Q Exactive series mass spectrometer (Thermo Fisher Scientific). The raw data were searched by PD2.2 software with full scan mode, and PRM pattern sequentially, and the selected peptides were determined using Skyline software based on reproducibility and stability. In the LC–MS/MS formal experiments, equivalent peptides pretreated with trypsin were spiked with the same amount of isotope-labeled peptide as an internal standard. Samples were analyzed by full scan and PRM patterns as described above. For offline data analysis, the peak area of each target protein was corrected according to the internal standard peptide to make it available for subsequent evaluation of relative abundance.
Data analysis
The relative expression levels of proteins were represented by fold changes by comparing the abundance of proteins during the estrous cycle. Statistical analysis was carried out by GraphPad Prism, version 8. Data comparisons between two groups were analyzed using the t-test. Data comparisons among three or more groups were carried out using one-way ANOVA, followed by Tukey multiple range test. Statistical significance was set at p < 0.05.
Results
Identification of salivary proteins during the estrous cycle
A total of 8,091 peptides and 1,155 proteins were identified in salivary samples, including 885, 923, 942, and 908 proteins in the ED-3, ED0, ED3, and ED8 groups, respectively. The general distribution character of all the identified proteins were analyzed with Jvenn online program, as shown in Figure 1. The number of shared proteins in four groups was 791, and the number of specific proteins for ED-3, ED0, ED3, and ED8 were 6, 17, 9, and 4, respectively (Figure 1).
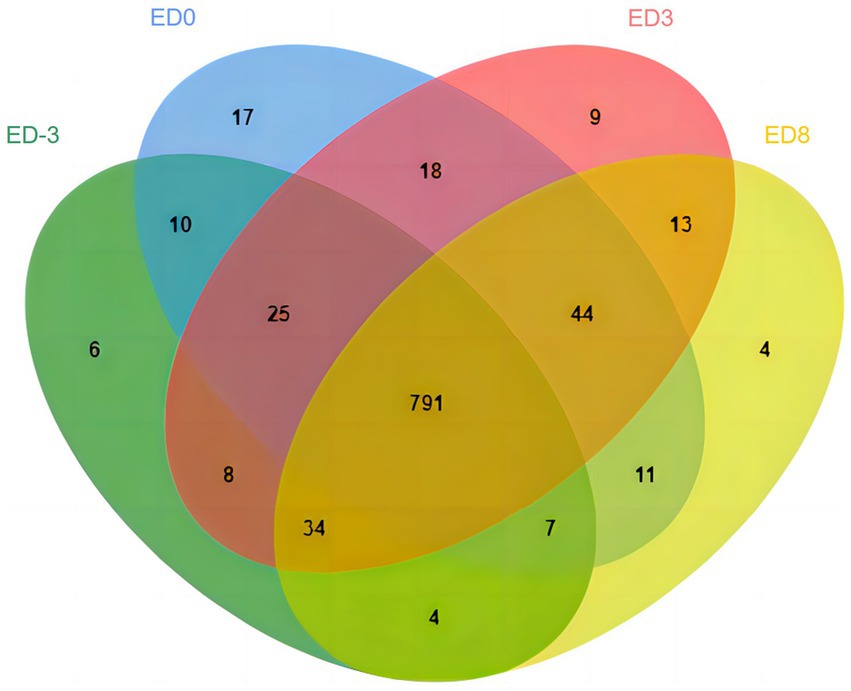
Figure 1. Venn diagram of identified proteins at proestrus (ED-3), estrus (ED0), metestrus (ED3), and diestrus (ED8) stages in pig saliva.
Functional prediction of total proteins
Functional prediction and classification of total proteins were conducted using the interproscan program, as shown in Figure 2. The results of GO enrichment indicated that all proteins were classified into three domains:666 proteins in molecular function, 399 proteins in biological processes, and 202 proteins in cellular component (Figure 2A). Molecular function had the largest number of items, and the top three items were protein binding (n = 96), calcium ion binding (n = 44), and serine-type endopeptidase activity (n = 36). The biological process results indicated that proteins were involved in proteolysis (n = 57), oxidation–reduction process (n = 47) and carbohydrate metabolic process (n = 23). According to cellular component results, most proteins are involved in the extracellular region (n = 43), membrane (n = 23), intermediate filament (n = 21) and extracellular space (n = 20). For COG analysis (Figure 2B), the top three functional classes were classified as O (posttranslational modification, protein turnover, chaperones), R (General function prediction only), and G (carbohydrate transport and metabolism) with 159, 75, and 42, respectively. KEGG analysis (Figure 2C) revealed that most proteins were involved in metabolism (n = 362), organismal (n = 300), and cellular processes (n = 208), while environmental information processing (n = 117) and signal transduction (n = 94) enriched fewer proteins. In the IPR prediction (Figure 2D), the immunoglobulin V-set domain (n = 50) contained the most proteins.
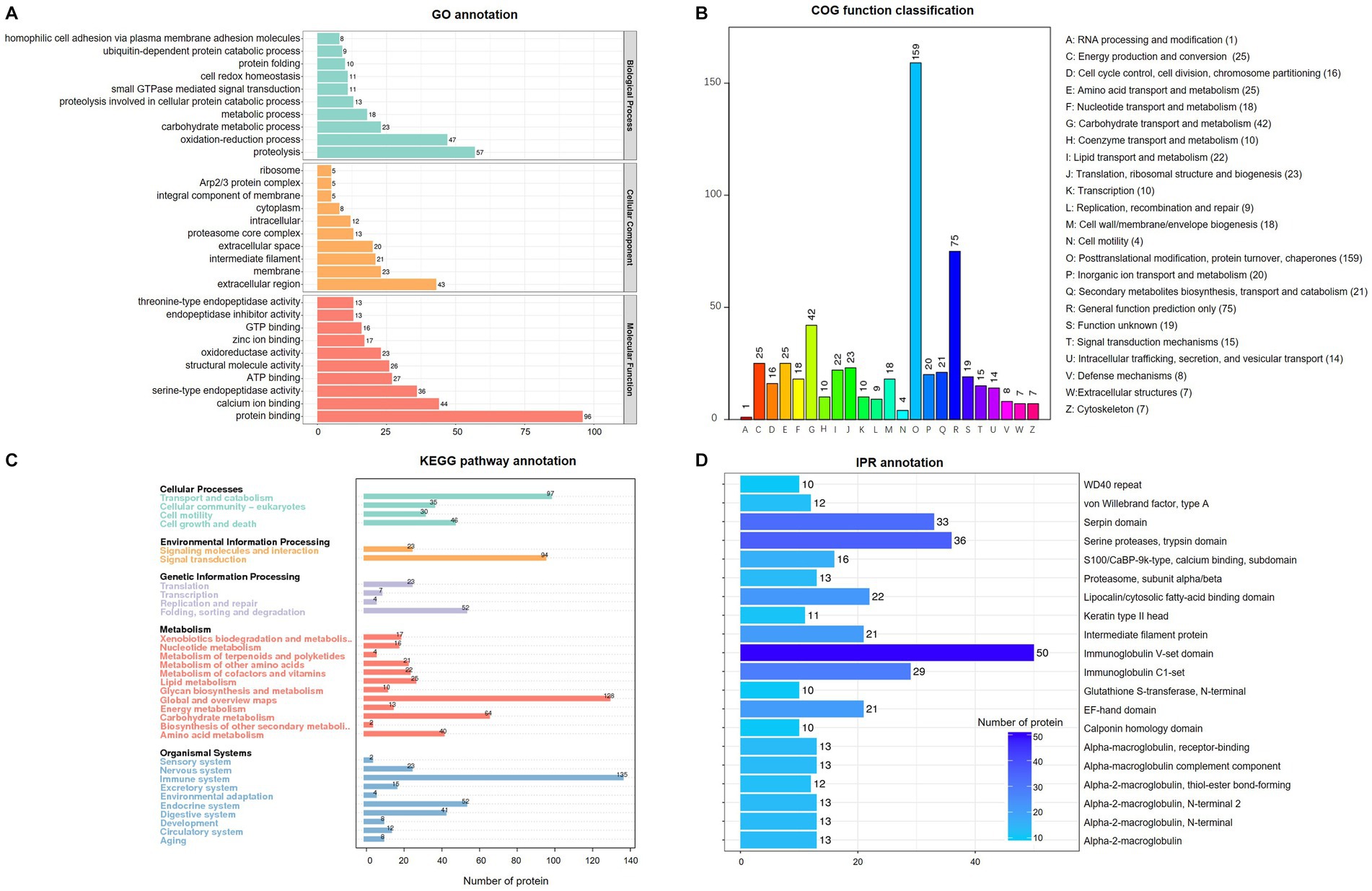
Figure 2. Enrichment analysis of identified total proteins in pig saliva during the estrous cycle. (A) Gene ontology (GO) analysis; (B) Clusters of Orthologous Groups of proteins enrichment; (C) Kyoto Encyclopedia of Genes and genomes (KEGG) pathway annotation. (D) InterPro (IPR) functional analysis.
Functional annotation and prediction of DEPs
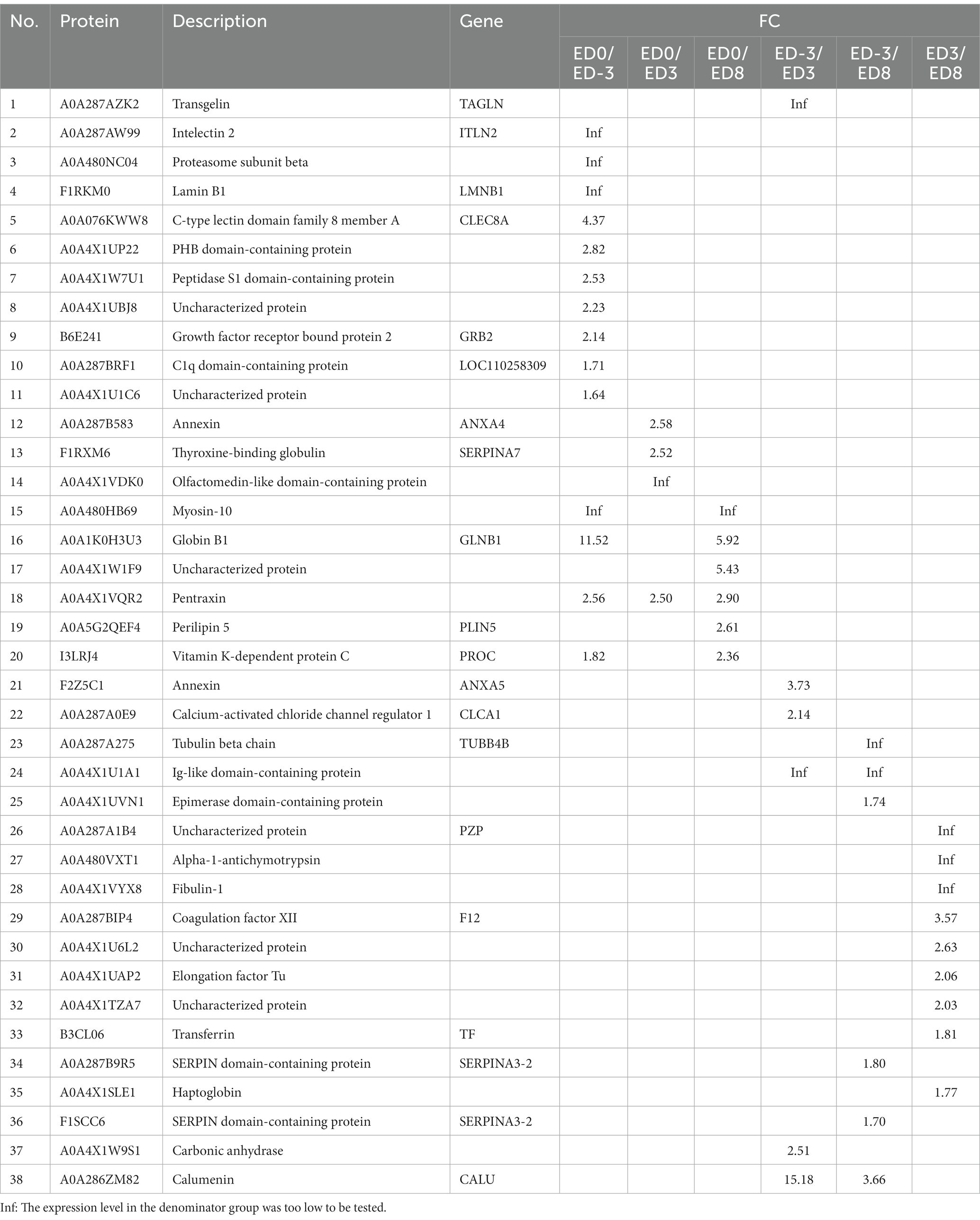
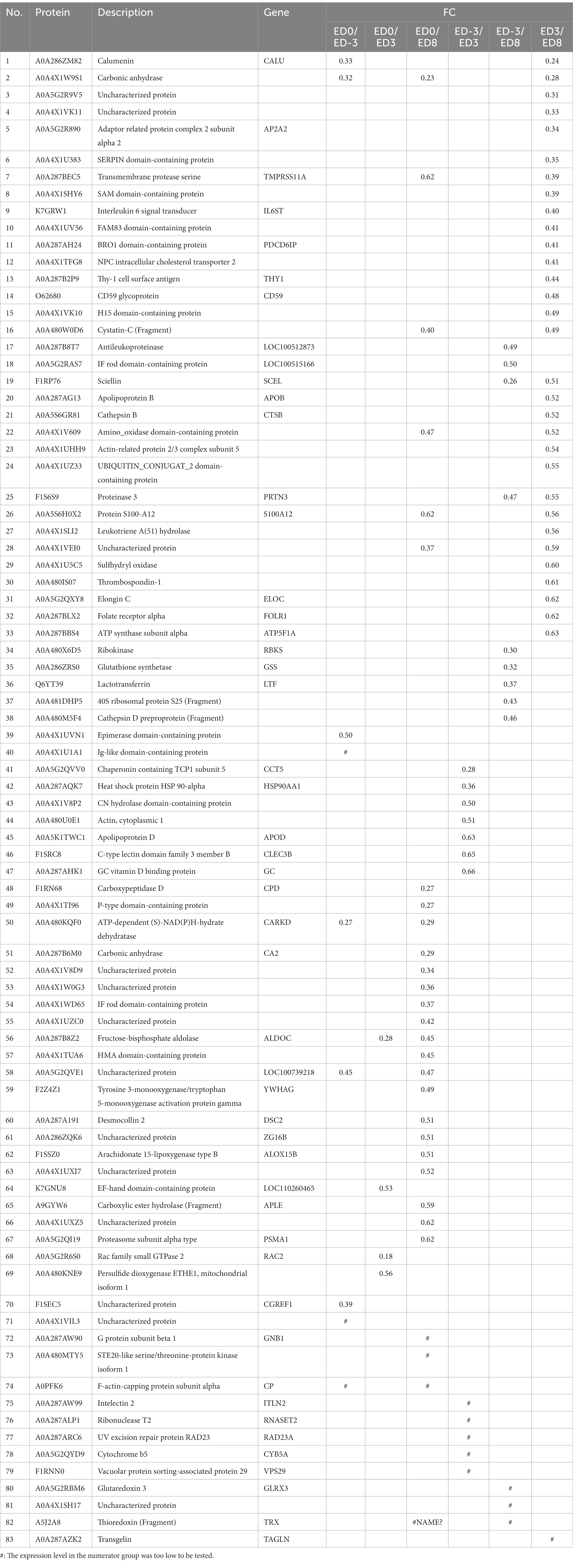
Protein quantity was compared among the different groups to isolate DEPs (FC ≥ 1.5, p ≤ 0.05). In total, 93 proteins were identified as DEPs, of which the expression of 38 proteins were upregulated and 83 were downregulated. The relative fold changes are shown in Tables 1, 2. The comparison ED3 vs. ED8 and ED0 vs. ED8 showed the largest number of proteins with downregulated expressions, with 30 and 34, respectively. Most proteins with upregulated expressions came from groups ED0 vs. ED-3 and ED3 vs. ED8 (15 and 11, respectively). In ED0 vs. ED-3 and ED0 vs. ED8, the expressions of 20 proteins were upregulated, and those of 12 were downregulated with FC > 2. All DEPs were divided into six sets according to time-series expression patterns, in which 21, 14, 10, 36, 17, and 17 proteins were clustered (Figure 3).
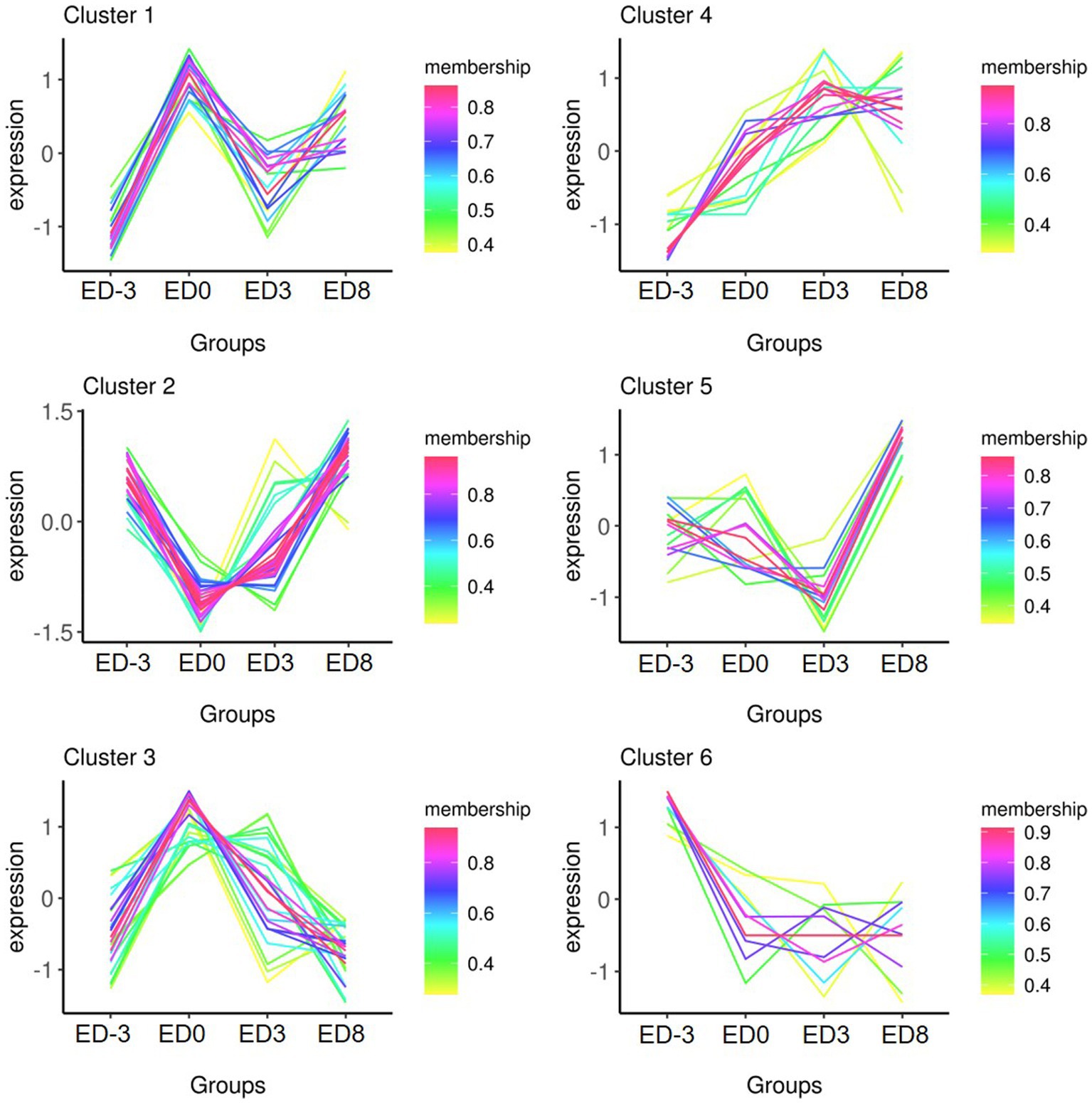
Figure 3. Time- series expression trends of six clusters of DEPs in pig saliva during the estrous cycle.
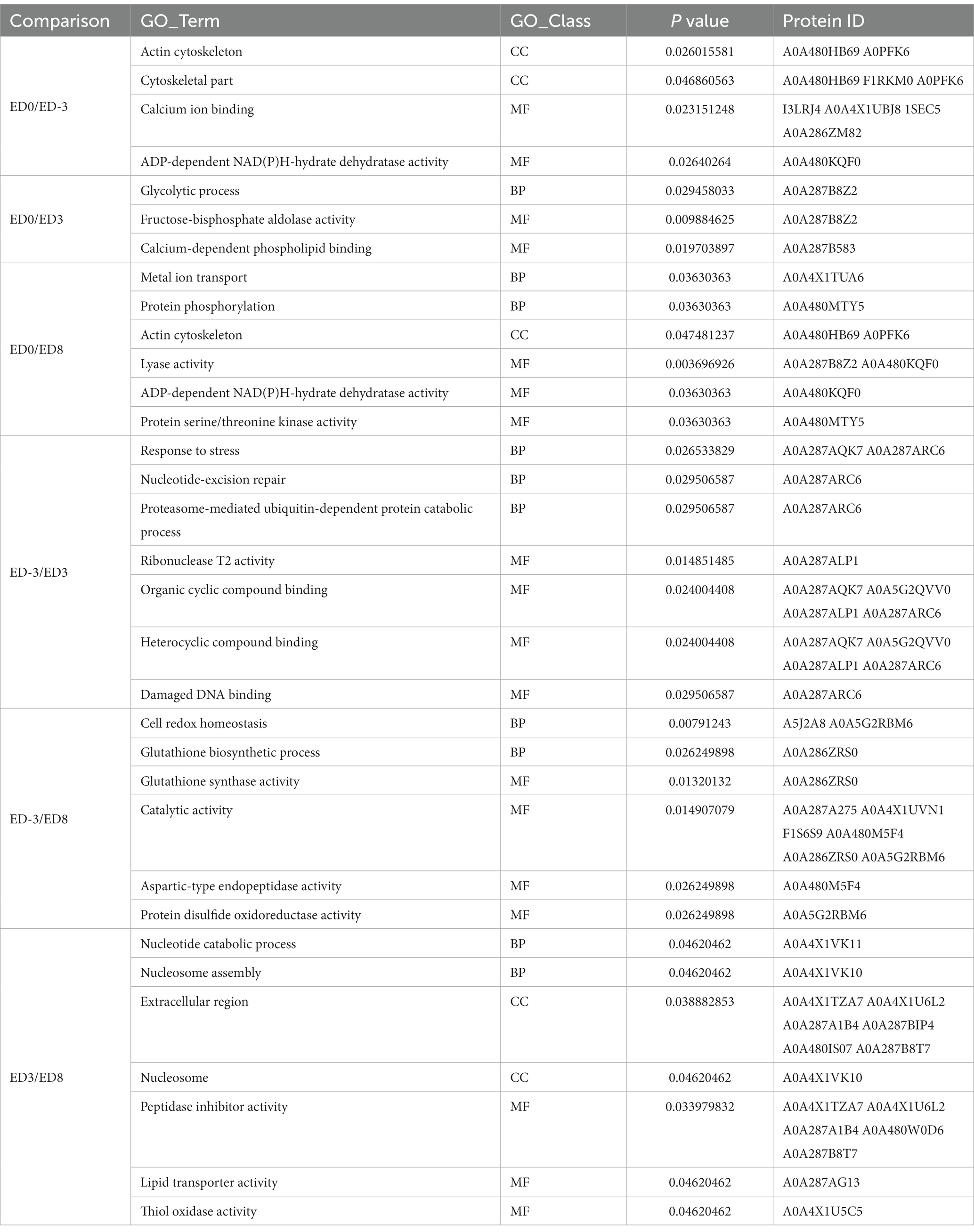
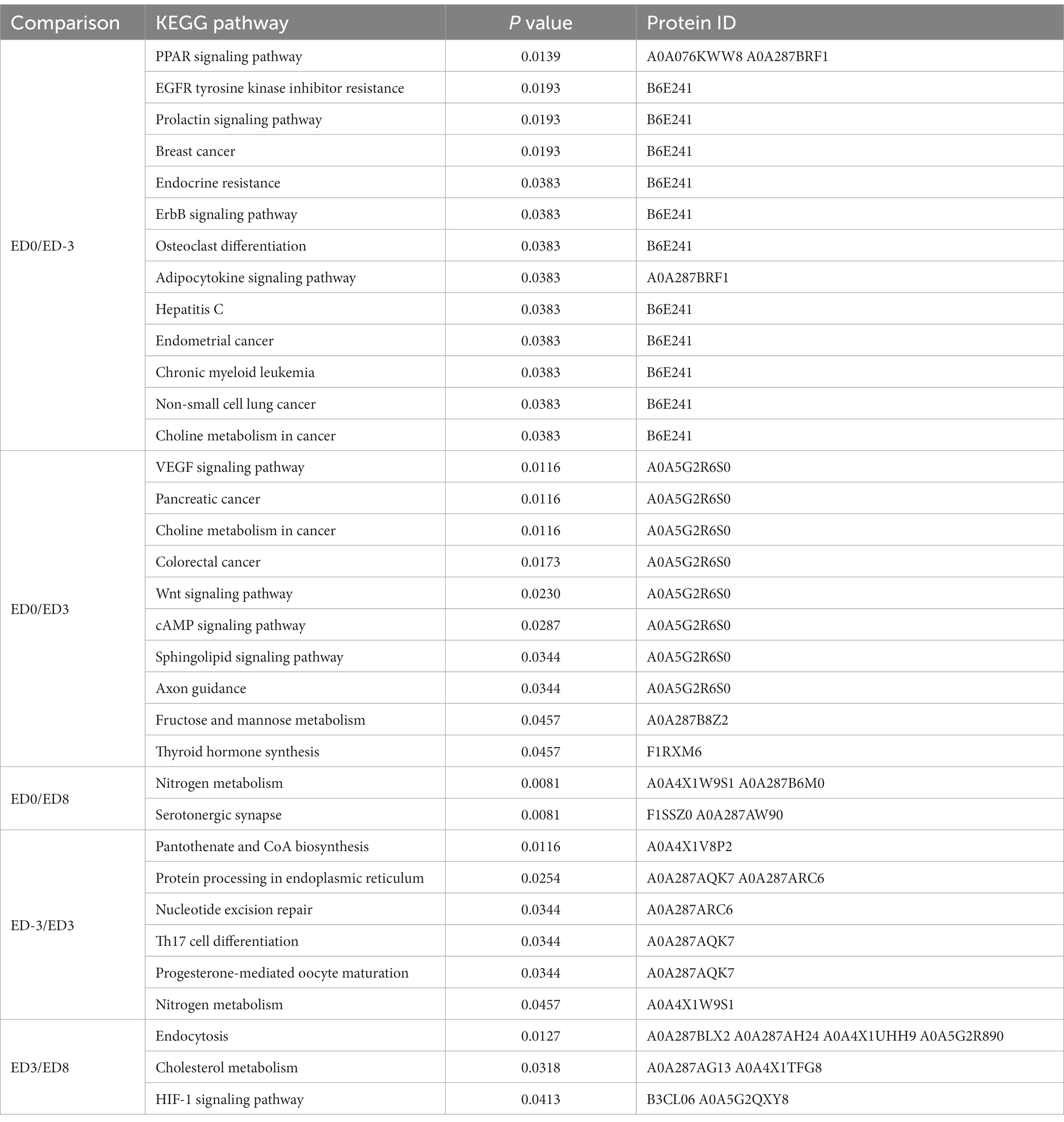
Functional annotation was performed using GO and KEGG analyzes. According to the enrichment analysis (Table 3), the significant GO items generated among different comparisons with ED0 were mainly classified by molecular function, including actin cytoskeleton, calcium ion binding, ADP-dependent NAD(P)H-hydrate dehydratase activity, glycolytic process, fructose-bisphosphate aldolase activity, calcium-dependent phospholipid binding, and lyase activity. In the KEGG enrichment analysis (Table 4), the significant pathways were mainly related to the peroxisome proliferator-activated receptors (PPAR) signaling pathway, epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor resistance, prolactin signaling pathway, vascular endothelial growth factor (VEGF) signaling pathway, Wnt signaling pathway, cAMP signaling pathway, nitrogen metabolism, and serotonergic synapse.
Relative expression verification by PRM
The 12-sample (3 replicates) cohort derived from four representative time-points of the estrous cycle was sent for PRM assay to verify the relative expression. PRM quantitative analysis of DEPs exhibited high agreement with the label-free results (Figure 4). The relative expression trends of six proteins, including stomatin-domain-containing protein (STOM, A0A4X1UP22), growth factor receptor bound protein 2 (Grb2, B6E241), elongation factor Tu (EFTU, A0A4X1UAP2), carboxypeptidase D (CPD, F1RN68), carbonic anhydrase (CA, A0A4X1W9S1), and trefoil factor 3 (TFF3, A0A4X1TI96), were validated using the PRM method and exhibited similar expression trends as label-free. The first three proteins exhibited an upgrade, followed by a descending trend, with a maximum expression level of 1.34 × 107, 1.48 × 106, 1.30 × 107, respectively, for label-free analysis, and 0.0017, 0.00044, 0.00063, respectively, for PRM analysis, at estrus. Instead, the other three proteins showed a significant ‘V’ trend, with the lowest level of 6.6 × 105, 1.91 × 109, 1.10 × 107, respectively, for label-free analysis, and 0.00043, 0.15, 0.0027, respectively, for PRM analysis, appearing at estrus.
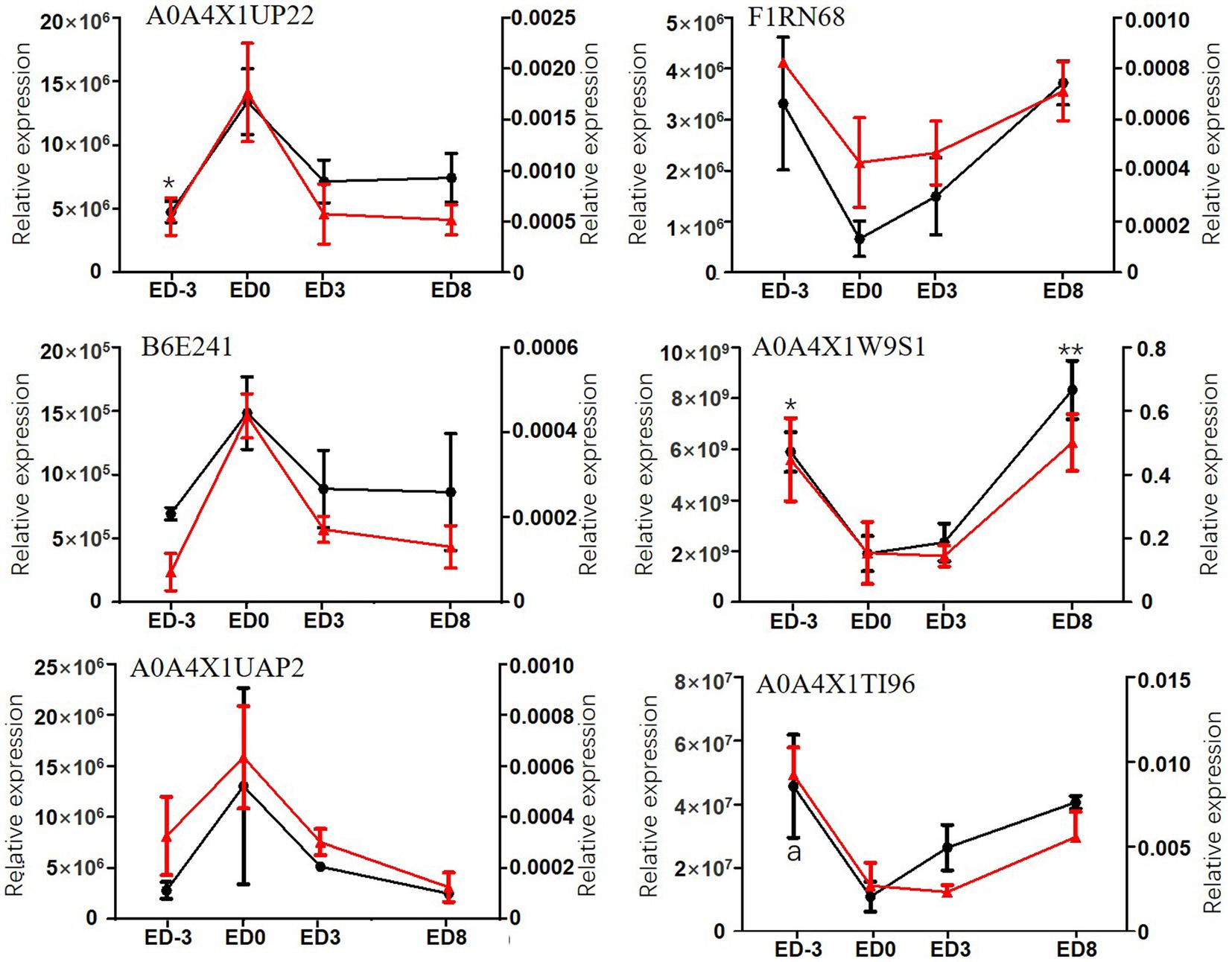
Figure 4. Relative expression levels of six DEPs assayed by label-free method and PRM in pig saliva during the estrous cycle. The primary coordinate represents label-free assay, left, red lines; * and ** indicate significant differences compared with ED0, p < 0.05, p < 0.01, respectively. The secondary coordinate represents PRM assay, right, black lines; a represents the significant difference compared with ED0.
Discussion
The present study focused on characterizing the overall proteomics during the estrous cycle in pig saliva and searched for estrus relevant or specific proteins. Given the evident advantages of saliva in protein species diversity, high abundance, and immense popularity in biological diagnosis, it is reasonable to hypothesize that exploring protein profiles and estrus-specific proteins in saliva may assist in developing novel non-invasive estrus diagnostic methods. Recent data showed that 1,155 total proteins were found in four representative groups, and up to 115 proteins were differentially expressed among the different groups. Both the total number and DEPs were larger than those obtained by Li (2), which may be caused by different breeds and comparisons. PRM quantity validated the expression patterns of the six proteins including stomatin-domain-containing protein, growth factor receptor bound protein, elongation factor Tu, carboxypeptidase, carbonic anhydras, and trefoil factor 3, displaying consistent expression trends with the label-free method.
Protein levels changed over time during the estrous cycle. In this study, many DEPs were upregulated at estrus, such as C-type lectin domain family 8 member A, STOM, Grb2, annexin A4, thyroxine-binding globulin, globin B1, pentraxin, perilipin 5 (Plin5), Vitamin K-dependent protein C. The expression of pentraxin in cumulus cells is positively correlated with in vivo fertilization by affecting the expansion of the cumulus oophorus, the quality of the corresponding oocytes, and ovulation (25). Plin5 bidirectionally regulates the dynamic balance of lipid metabolism in oxidative tissues by controlling the activity of adipose triglyceride lipase. Plin5 inhibits the lipolysis in the basic state, while the suppressed lipolysis process would be accelerated once the stress occurred (26). However, there are few studies regarding the function of the other upregulated proteins at estrus.
Notably, the expressions of many proteins were downregulated during diestrus. Carbonic anhydrase (CA), as a zinc-containing enzyme, is widely distributed in mammal tissues, including the reproductive tract (27). CA plays key roles in body fluids balance by influencing metabolic carbon dioxide transport (28).LTF is a major component of mucosal fluids defense because of the antimicrobial and immunomodulating properties. It is evident that the bimodal distribution of LTF in cervical immune components is determined by the menstrual cycle (29). Besides, LTF is expressed correlated with the circulating level of 17β-estradiol (E2), and selectively synthesized in the uterine epithelium dependent on estrogen receptor α (ERα) (30). For several other downregulated proteins during diestrus, e.g., calumenin, SERPIN domain-containing protein, sciellin, ribokinase, glutathione synthetase, few reports are available for their roles in reproduction.
GO function classification of DEPs established their close association with biological process, cellular components, and molecular function. Calcium ion binding was enriched in the greatest number of proteins in the ED0 vs. ED-3 group. As a second intracellular messenger, calcium ions are crucial in cellular physiological processes, and are essential for mammalian oocyte growth, meiosis, hormone production, growth factor secretion, and meiotic maturation (31). In this category, the four enriched proteins, I3LRJ4, A0A4X1UBJ8, 1SEC5, and A0A286ZM82, suggested their involvement in oocyte maturation. The actin cytoskeleton contributes to oocyte meiotic maturation by mediating spindle assembly, length and chromosome segregation (32). Overlapping of the actin cytoskeleton in the ED0 vs. ED8 and ED0 vs. ED-3 groups emphasized their importance during the estrus stage.
KEGG enrichment analysis of DEPs was performed to identify the functional pathways during the reproductive cycle. Compared with ED0, the most enriched pathways included the PPAR signaling pathway, EGFR tyrosine kinase inhibitor resistance, prolactin signaling pathway, VEGF signaling pathway, Wnt signaling pathway, cAMP signaling pathway, nitrogen metabolism, and serotonergic synapse. The PPAR pathway was demonstrated to be pivotal in canine gametogenesis, ovulation, or CL regression during the estrous cycle (33). VEGF signaling is a key pathway for granulosa cell proliferation, steroid hormone synthesis, apoptosis inhibition, and ovarian angiogenesis during follicular development and ovulation (34). The Wnt pathway is required for embryonic and adult ovarian development. Wnt-4 stimulates granulosa cell differentiation and female sexual development during the embryonic stage. In contrast, adult ovarian function and fertility rely on the synchronized actions of Wnt family genes and hormones (35). The inhibitory effects of nitric oxide (NO), a representative product of nitrogen metabolism, on steroidogenesis have been reported in several animal models. The controversial effects of NO during folliculogenesis, which protects or induces follicular apoptosis, were concentration-dependent. Moreover, the NO pathway exerts a central influence on mammalian oocyte meiotic maturation among the numerous mediator molecules (36). The strong relationship between these pathways and reproductive physiology strengthens the reliability of our enrichment results.
Both the label-free and PRM groups consistently revealed the maximum levels of STOM, Grb2 and EFTU during estrus. There are five members in the stomatin-domain-containing protein family: STOM, STOML1, STOML2, STOML3, and podocin (37). The proliferation and invasion of human trophoblasts can be mediated by STOML2 through modulating mitochondrial function (38). A significant increase in STOML2 expression could promote endometrial stromal cell proliferation and decidualization in mice and human (39). Grb2 induces various cellular events as an adaptor protein in signal transduction pathways (40). The Grb2/Sos complex activates Ras in response to cellular growth factors. Grb2 is involved in vulvar development and sex myoblast migration in Caenorhabditis. elegans (40). In Xenopus oocytes, Grb2 induced the re-initiation of meiosis (40). A study identified the Grb2 as a key candidate gene in progesterone production during ovulation through bioinformatics analysis and verified its function in progestogenic pathways in rat COCs (41). In the current study, Grb2 showed maximum expression during estrus, which might be related to ovulation. EFTU increased from day 12 to 16 in corpus luteum of cyclic ewes (42), which may assist corpus luteum regression during estrus. In contrast, EFTU was most abundant on the day of estrus in our study, indicating its potential effects on follicular development, ovulation, and LH synthesis. This distinct tendency may be attributed to different organisms or tissues.
Simultaneously, minimal expression of CPD, CA and TFF3 was observed in label-free and PRM. CPD is a carboxypeptidases (CPs) family member characterized by a conserved metallocarboxypeptidase domain. The CPs family in mammals includes CPH, CPM, CPE, and nine other proteins (43). CPD signals are significantly high in bovine dominant follicles and are considered a reliable marker for dominant follicles (44). CPD has great functional and structural similarities with other CPs (45); however, limited studies are available on CPD function in reproduction (44, 46). In our study, CPD showed the lowest level of saliva on estrus day, suggesting that CPD may regulate estrus physiology by modulating the precursors processes of hormones and neuropeptides. A previous study explored the localization of CA in the genitalia of female pigs. CA was positive in the blood vessel endothelium and absent in the ovarian parenchyma. In the oviduct, conspicuous CA activity was observed in the surface epithelium of the tubal ampulla, where fertilization and early embryo development occurred. Moreover, CA was stained in the uterotubal junction-tubal isthmus, which acts as a sperm reservoir. Both the intensity and localization of CA remained constant during the estrous cycle. The localization of CA may indicate its crucial role in maintaining acid–base homeostasis in the luminal fluid (47). Likewise, the involvement of CA in bicarbonate secretion during the estrous cycle was also observed in mouse and rat uterine (48, 49) since carbonic anhydrase 2(CAR2, CA2) had the maximum expression at estrus during the estrous cycle when the resting uterine surface pH was significantly higher than that at diestrus; nevertheless, the high pH could be reduced significantly by the CA inhibitor (48). Remarkably, estrogen induced a parallel increase in CAR2 expression and endometrial surface pH. Our data showed the opposite result; CA had the lowest expression at estrus. Hence, the expression pattern of CA may change with species. In human, the expression of TFF3 is upregulated on day 4 vs. Day 2 of menstruation and is implicated in endometrial regeneration and repair during menstruation (50). The presence of TFF3 in saliva may be associated with the endometrium, whereas the underlying function and mechanisms of estrus are not yet known and need to be resolved.
Conclusion
The present study compared global saliva proteome profiles during the estrous cycle based on the label-free quantitative proteomics in a crossbred pig. The identification and enrichment analysis of the differentially expressed proteins indicated the involvement of many proteins in pig estrus physiological functions. The expression trends of the six proteins were confirmed using PRM. This study provided new insights into the physiology of the reproductive cycle and served as a reference for developing estrus detection kits or strips. Nevertheless, further investigations are needed to uncover the confusion regarding DEPs generation, secretion, function mechanisms, and practical potential in estrus detection.
Data availability statement
The data presented in the study are deposited in the ProteomeXchange Consortium repository, accession number PXD043141.
Author contributions
HX conceived the study, designed and performed the experiments, conducted data analysis, prepared figures and tables, and wrote the manuscript. BL, BH, and YW prepared materials and collected samples. SW and FM modified the manuscript. JL supervised the study. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the following projects: Guangdong Provincial Key Research and Development Program (2022B0202090003); Pig Innovation team of Guangdong Modern Agricultural Industrial Technology system (2022KJ126); Agricultural Superior Industry Scientific Team-building Project of Guangdong Academy of Agricultural Sciences (202118TD); Guangdong provincial rural revitalization strategy special funds seeding industry revitalization project (2021-44000-24010202-8887); Guangdong Xiaoerhua Pig Seed Industry Innovation Park (2022-4408X1-43010402-0019).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
1. Endo, N. Possible causes and treatment strategies for the estrus and ovulation disorders in dairy cows. J Reprod Dev. (2022) 68:85–9. doi: 10.1262/jrd.2021-125
2. Li, C, Song, C, Qi, K, Liu, Y, Dou, Y, Li, X, et al. Identification of estrus in sows based on salivary proteomics. Animals (Basel). (2022) 12:1656. doi: 10.3390/ani12131656
3. Sankarganesh, D, Kirkwood, RN, Nagnan-Le Meillour, P, Angayarkanni, J, Achiraman, S, and Archunan, G. Pheromones, binding proteins, and olfactory systems in the pig (Sus scrofa): an updated review. Front Vet Sci. (2022) 9:989409. doi: 10.3389/fvets.2022.989409
4. McGlone, JJ, Garcia, A, and Rakhshandeh, A. Multi-farm analyses indicate a novel boar pheromone improves sow reproductive performance. Animals (Basel). (2019) 9:37. doi: 10.3390/ani9020037
5. Mcglone, JJ, Devaraj, S, and Garcia, A. A novel boar pheromone mixture induces sow estrus behaviors and reproductive success. Appl Anim Behav Sci. (2019) 219:104832. doi: 10.1016/j.applanim.2019.104832
6. Sankarganesh, D, Kirkwood, R, Angayarkanni, J, Achiraman, S, and Archunan, G. Pig pheromones and behaviors: a review. Theriogenology. (2021) 175:1–6. doi: 10.1016/j.theriogenology.2021.08.032
7. Van den Berghe, F, Paris, MCJ, Sarnyai, Z, Vlamings, B, Millar, RP, Ganswindt, A, et al. Dog appeasing pheromone prevents the androgen surge and may reduce contact dominance and active submission after stressful interventions in African wild dogs (Lycaon pictus). PLoS One. (2019) 14:e0212551. doi: 10.1371/journal.pone.0212551
8. Simões, VG, Lyazrhi, F, Picard-Hagen, N, Gayrard, V, Martineau, GP, and Waret-Szkuta, A. Variations in the vulvar temperature of sows during proestrus and estrus as determined by infrared thermography and its relation to ovulation. Theriogenology. (2014) 82:1080–5. doi: 10.1016/j.theriogenology.2014.07.017
9. Sykes, DJ, Couvillion, JS, Cromiak, A, Bowers, S, Schenck, E, Crenshaw, M, et al. The use of digital infrared thermal imaging to detect estrus in gilts. Theriogenology. (2012) 78:147–52. doi: 10.1016/j.theriogenology.2012.01.030
10. Freson, L, Godrie, S, Bos, N, Jourquin, J, and Geers, R. Validation of an infra-red sensor for oestrus detection of individually housed sows. Comput Electron Agric. (1998) 20:21–9. doi: 10.1016/S0168-1699(98)00005-2
11. Lei, K, Zong, C, Du, X, Teng, G, and Feng, F. Oestrus analysis of sows based on bionic boars and machine vision technology. Animals (Basel). (2021) 11:1485. doi: 10.3390/ani11061485
12. Chen, C, Zhu, W, Ma, C, Guo, Y, Huang, W, and Ruan, C. Image motion feature extraction for recognition of aggressive behaviors among group-housed pigs. Comput Electron Agric. (2017) 142:380–7. doi: 10.1016/j.compag.2017.09.013
13. Kim, J, Chung, Y, Choi, Y, Sa, J, Kim, H, Chung, Y, et al. Depth-based detection of standing-pigs in moving noise environments. Sensors (Basel). (2017) 17:2757. doi: 10.3390/s17122757
14. Agbugba, LC, Oyewunmi, AO, Ogundumade, TP, and Leigh, OO. Investigation of vaginal mucus parameters: development of models for staging the oestrous cycle of the Bunaji cow. Reprod Domest Anim. (2020) 55:1044–53. doi: 10.1111/rda.13694
15. Talukder, MRI, Hasan, M, Rosy, TA, Bari, FY, and Juyena, NS. Monitoring vaginal electrical resistance, follicular waves, and hormonal profile during Oestrous cycle in the transition period in Bangladeshi sheep. J Vet Res. (2018) 62:571–9. doi: 10.2478/jvetres-2018-0080
16. Chavan, NB, Kumaresan, A, Chhillar, S, Nayak, S, Prakash, MA, Lathika, S, et al. Salivary crystallization pattern: a possible unconventional tool for timing of insemination and early pregnancy diagnosis in zebu cows. J Dairy Res. (2023) 90:21–5. doi: 10.1017/s0022029922000887
17. Chojnowska, S, Baran, T, Wilińska, I, Sienicka, P, Cabaj-Wiater, I, and Knaś, M. Human saliva as a diagnostic material. Adv Med Sci. (2018) 63:185–91. doi: 10.1016/j.advms.2017.11.002
18. Lindsay, A, and Costello, JT. Realising the potential of urine and saliva as diagnostic tools in sport and exercise medicine. Sports Med. (2017) 47:11–31. doi: 10.1007/s40279-016-0558-1
19. Michalke, B, Rossbach, B, Göen, T, Schäferhenrich, A, and Scherer, G. Saliva as a matrix for human biomonitoring in occupational and environmental medicine. Int Arch Occup Environ Health. (2015) 88:1–44. doi: 10.1007/s00420-014-0938-5
20. Green, JA, Kim, JG, Whitworth, KM, Agca, C, and Prather, RS. The use of microarrays to define functionally-related genes that are differentially expressed in the cycling pig uterus. Soc Reprod Fertil Suppl. (2006) 62:163–76.
21. Rodrigues, AD, Cooke, RF, Cipriano, RS, Silva, LGT, Cerri, RLA, Cruppe, LH, et al. Impacts of estrus expression and intensity during a timed-AI protocol on variables associated with fertility and pregnancy success in Bos indicus-influenced beef cows. J Anim Sci. (2018) 96:236–49. doi: 10.1093/jas/skx043
22. Jones, P, Binns, D, Chang, HY, Fraser, M, Li, W, McAnulla, C, et al. Inter ProScan 5: genome-scale protein function classification. Bioinformatics. (2014) 30:1236–40. doi: 10.1093/bioinformatics/btu031
23. Fedorka, CE, Scoggin, KE, Boakari, YL, Hoppe, NE, Squires, EL, Ball, BA, et al. The anti-inflammatory effect of exogenous lactoferrin on breeding-induced endometritis when administered post-breeding in susceptible mares. Theriogenology. (2018) 114:63–9. doi: 10.1016/j.theriogenology.2018.03.017
24. Hermawan, A, and Putri, H. Bioinformatics studies provide insight into possible target and mechanisms of action of Nobiletin against cancer stem cells. Asian Pac J Cancer Prev. (2020) 21:611–20. doi: 10.31557/apjcp.2020.21.3.611
25. Zheng, X, Zheng, Y, Qin, D, Yao, Y, Zhang, X, Zhao, Y, et al. Regulatory role and potential importance of GDF-8 in ovarian reproductive activity. Front Endocrinol (Lausanne). (2022) 13:878069. doi: 10.3389/fendo.2022.878069
26. Zhang, X, Xu, W, Xu, R, Wang, Z, Zhang, X, Wang, P, et al. Plin5 Bidirectionally regulates lipid metabolism in oxidative tissues. Oxidative Med Cell Longev. (2022) 2022:4594956. doi: 10.1155/2022/4594956
27. Karahan, SC, Guven, S, Mentese, A, Bacak, A, Kopuz, M, and Ozeren, M. Serum anti-carbonic anhydrase I and II antibodies and idiopathic recurrent pregnancy loss. Reprod Biomed Online. (2009) 19:859–63. doi: 10.1016/j.rbmo.2009.09.030
28. Göktuğ Kadıoğlu, B, Nalçakan, A, and Dilek, E. Relationship between zinc content and carbonic anhydrase activity in blood of anemic pregnant women in Turkey. J Obstet Gynaecol Res. (2020) 46:2612–7. doi: 10.1111/jog.14506
29. Santacroce, L, Palmirotta, R, Bottalico, L, Charitos, IA, Colella, M, Topi, S, et al. Crosstalk between the resident microbiota and the immune cells regulates female genital tract health. Life (Basel). (2023) 13:1531. doi: 10.3390/life13071531
30. Winuthayanon, W, Hewitt, SC, Orvis, GD, Behringer, RR, and Korach, KS. Uterine epithelial estrogen receptor α is dispensable for proliferation but essential for complete biological and biochemical responses. Proc Natl Acad Sci U S A. (2010) 107:19272–7. doi: 10.1073/pnas.1013226107
31. Dua, D, Nagoorvali, D, Chauhan, MS, Palta, P, Mathur, P, and Singh, MK. Calcium ionophore enhanced developmental competence and apoptotic dynamics of goat parthenogenetic embryos produced in vitro. In Vitro Cell Dev Biol Anim. (2019) 55:159–68. doi: 10.1007/s11626-019-00322-x
32. Li, Z, Guo, Y, Ndandala, CB, Chen, H, Huang, C, Zhao, G, et al. Analysis of circ RNA and miRNA expression profiles in IGF3-induced ovarian maturation in spotted scat (Scatophagus argus). Front Endocrinol (Lausanne). (2022) 13:998207. doi: 10.3389/fendo.2022.998207
33. Socha, BM, Socha, P, Szóstek-Mioduchowska, AZ, Nowak, T, and Skarżyński, DJ. Peroxisome proliferator-activated receptor expression in the canine endometrium with cystic endometrial hyperplasia-pyometra complex. Reprod Domest Anim. (2022) 57:771–83. doi: 10.1111/rda.14121
34. Xiao, L, Wang, Z, Lu, N, He, Y, Qiao, L, Sheng, X, et al. GPER mediates the IL6/JAK2/STAT3 pathway involved in VEGF expression in swine ovary GCs. J Mol Endocrinol. (2021) 68:23–33. doi: 10.1530/jme-21-0125
35. Zhang, G, Michener, CM, and Yang, B. Low-grade ovarian stromal tumors with genetic alterations of the Wnt/β-catenin pathway that is crucial in ovarian follicle development and regulation. Cancers (Basel). (2022) 14:5622. doi: 10.3390/cancers14225622
36. Budani, MC, and Tiboni, GM. Novel insights on the role of nitric oxide in the ovary: a review of the literature. Int J Environ Res Public Health. (2021) 18:18. doi: 10.3390/ijerph18030980
37. Gonzalez-Velandia, KY, Hernandez-Clavijo, A, Menini, A, Dibattista, M, and Pifferi, S. Expression pattern of Stomatin-domain proteins in the peripheral olfactory system. Sci Rep. (2022) 12:11447. doi: 10.1038/s41598-022-15572-1
38. Zhang, X, Li, BY, Fu, LJ, Adu-Gyamfi, EA, Xu, BR, Liu, TH, et al. Stomatin-like protein 2 (SLP2) regulates the proliferation and invasion of trophoblast cells by modulating mitochondrial functions. Placenta. (2020) 100:13–23. doi: 10.1016/j.placenta.2020.07.008
39. Feng, Q, Hu, ZY, Liu, XQ, Zhang, X, Lan, X, Geng, YQ, et al. Stomatin-like protein 2 is involved in endometrial stromal cell proliferation and differentiation during decidualization in mice and humans. Reprod Biomed Online. (2017) 34:191–202. doi: 10.1016/j.rbmo.2016.11.009
40. Bai, X, Lee, JY, Kim, TI, Dai, F, Lee, TJ, and Hong, SJ. Molecular cloning and characterization of growth factor receptor bound-protein in Clonorchis sinensis. PLoS One. (2014) 9:e85577. doi: 10.1371/journal.pone.0085577
41. Yang, F, Wang, M, Zhang, B, Xiang, W, Zhang, K, Chu, M, et al. Identification of new progestogen-associated networks in mammalian ovulation using bioinformatics. BMC Syst Biol. (2018) 12:36. doi: 10.1186/s12918-018-0577-7
42. Arianmanesh, M, McIntosh, RH, Lea, RG, Fowler, PA, and Al-Gubory, KH. Ovine corpus luteum proteins, with functions including oxidative stress and lipid metabolism, show complex alterations during implantation. J Endocrinol. (2011) 210:47–58. doi: 10.1530/joe-10-0336
43. Reznik, SE, and Fricker, LD. Carboxypeptidases from a to z: implications in embryonic development and Wnt binding. Cell Mol Life Sci. (2001) 58:1790–804. doi: 10.1007/pl00000819
44. Sisco, B, Hagemann, LJ, Shelling, AN, and Pfeffer, PL. Isolation of genes differentially expressed in dominant and subordinate bovine follicles. Endocrinology. (2003) 144:3904–13. doi: 10.1210/en.2003-0485
45. Cawley, NX, Wetsel, WC, Murthy, SR, Park, JJ, Pacak, K, and Loh, YP. New roles of carboxypeptidase E in endocrine and neural function and cancer. Endocr Rev. (2012) 33:216–53. doi: 10.1210/er.2011-1039
46. Singh, U, Yu, Y, Kalinina, E, Konno, T, Sun, T, Ohta, H, et al. Carboxypeptidase E in the mouse placenta. Differentiation. (2006) 74:648–60. doi: 10.1111/j.1432-0436.2006.00093.x
47. Rodriguez-Martinez, H, Ekstedt, E, and Ridderstråle, Y. Histochemical localization of carbonic anhydrase in the female genitalia of pigs during the oestrous cycle. Acta Anat (Basel). (1991) 140:41–7. doi: 10.1159/000147036
48. He, Q, Chen, H, Wong, CH, Tsang, LL, and Chan, HC. Regulatory mechanism underlying cyclic changes in mouse uterine bicarbonate secretion: role of estrogen. Reproduction. (2010) 140:903–10. doi: 10.1530/rep-10-0178
49. Gholami, K, Muniandy, S, and Salleh, N. In-vivo functional study on the involvement of CFTR, SLC26A6, NHE-1 and CA isoenzymes II and XII in uterine fluid pH, volume and electrolyte regulation in rats under different sex-steroid influence. Int J Med Sci. (2013) 10:1121–34. doi: 10.7150/ijms.5918
Keywords: sow, estrus detection, saliva, quantitative proteomics, differentially expressed proteins, reproduction efficiency
Citation: Xin H, Li B, Meng F, Hu B, Wang S, Wang Y and Li J (2023) Quantitative proteomic analysis and verification identify global protein profiling dynamics in pig during the estrous cycle. Front. Vet. Sci. 10:1247561. doi: 10.3389/fvets.2023.1247561
Edited by:
Regiane R. Santos, Schothorst Feed Research, NetherlandsReviewed by:
Danuza Leão, Instituto de Desenvolvimento Sustentável Mamirauá, BrazilDagan Mao, Nanjing Agricultural University, China
Copyright © 2023 Xin, Li, Meng, Hu, Wang, Wang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianhao Li, MzU2MjA0Nzc2M0BxcS5jb20=
 Haiyun Xin
Haiyun Xin Baohong Li1
Baohong Li1 Bin Hu
Bin Hu