- 1Department of Zoology, Abdul Wali Khan University Mardan, Mardan, Pakistan
- 2Department of Pharmacology and Toxicology, College of Pharmacy, King Saud University, Riyadh, Saudi Arabia
- 3King Abdulaziz City for Science and Technology, Riyadh, Saudi Arabia
- 4Laboratory of Infectious Diseases, Joint Faculty of Veterinary Medicine, Kagoshima University, Kagoshima, Japan
Public health is a major concern for several developing countries due to infectious agents transmitted by hematophagous arthropods such as ticks. Health risks due to infectious agents transmitted by ticks infesting butcher-associated stray dogs (BASDs) in urban and peri-urban regions have been neglected in several developing countries. To the best of the authors’ knowledge, this is the first study assessing public health risks due to ticks infesting BASDs in Pakistan’s urban and peri-urban areas. A total of 575 ticks (390 from symptomatic and 183 from asymptomatic BASDs) were collected from 117 BASDs (63 symptomatic and 54 asymptomatic); the ticks belonged to 4 hard tick species. A subset of each tick species’ extracted DNA was subjected to polymerase chain reaction (PCR) to amplify the 16S rDNA and cox1 sequences of the reported tick species, as well as bacterial and protozoal agents. The ticks’ 16S rDNA and cox1 sequences showed 99–100% identities, and they were clustered with the sequence of corresponding species from Pakistan and other countries in phylogenetic trees. Among the screened 271 ticks’ DNA samples, Anaplasma spp. was detected in 54/271 (19.92%) samples, followed by Ehrlichia spp. (n = 40/271, 14.76%), Rickettsia spp. (n = 33/271, 12.17%), Coxiella spp. (n = 23/271, 4.48%), and Hepatozoon canis (n = 9/271, 3.32%). The obtained sequences and phylogenetic analyzes revealed that the pathogens detected in ticks were Ehrlichia minasensis, Ehrlichia sp., Hepatozoon canis, Coxiella burnetii, Coxiella sp., Anaplasma capra, Anaplasma platys, Anaplasma sp., Rickettsia massiliae, “Candidatus Rickettsia shennongii” and Rickettsia aeschlimannii. Tick-borne pathogens such as E. minasensis, H. canis, A. capra, A. platys, and R. aeschlimannii, were detected based on the DNA for the first time in Pakistan. This is the first report on public health risks due to ticks infesting BASDs. These results not only provided insights into the occurrence of novel tick-borne pathogens in the region but also revealed initial evidence of zoonotic threats to both public health and domestic life.
Introduction
The availability of survival resources (i.e., shelter, food, and water) affects the dog population in a region and directly correlates with the human population (1). Stray dogs are highly vulnerable to vector-borne diseases (VBDs) of zoonotic concern and, consequently, pose threats to human and animal health (2). Although the exact number globally is unknown, 75% out of 700 million dogs are considered stray dogs (3). They are the first farmed animals and are closely associated with humans on cultural, social, and economic levels (4). Pakistan is a developing country, and contemporary social issues have imposed an epidemic of stray dogs that may affect both the welfare of the animals and the public’s health, especially in urban and peri-urban areas (5). The exact number of these free-roaming stray dogs in the country is unknown, and the estimated stray dog population may have reached up to 3 million (6).
Tick species infesting stray dogs in the urban and peri-urban areas include Rhipicephalus sanguineus senso lato (s.l.), Haemaphysalis elliptica, Rhipicephalus microplus, Haemaphysalis erinacei, and Haemaphysalis parva (7, 8). Knowledge of tick identification is vital for surveillance to minimize the zoonotic consequences associated with tick-borne pathogens (9). Despite the morphological identification, genetic characterization using various genetic markers such as 16S rDNA and cox1 is a suitable approach to determine the taxonomic status of a tick species (10–16).
Stray dogs being important reservoirs for tick-borne zoonotic agents may pose a threat to public health by causing anaplasmosis, rickettsiosis, babesiosis, query fever (Q fever), and ehrlichiosis (17–19). The genus Ehrlichia has seven determined species: Ehrlichia canis, Ehrlichia chaffeensis, Ehrlichia ruminantium, Ehrlichia muris, Ehrlichia ewingii, Ehrlichia ovina, and Ehrlichia minasensis (20, 21). Ticks responsible for the transmission of Ehrlichia spp. include species of the genera Rhipicephalus, Hyalomma, Dermacentor, Amblyomma, and Ixodes (20, 22). Canine monocytic ehrlichiosis (CME) is the most common tick-borne zoonotic disease that affects dogs globally (23). The brown dog tick, Rh. sanguineus, primarily vectors the etiological agent of CME (23). Hepatozoon canis causes canine hepatozoonosis, which is widely distributed in South America, Africa, Asia, and South Europe (24). Q fever is a tick-borne infection caused by Coxiella burnetii (25). Tick species of the genera Rhipicephalus, Hyalomma, Amblyomma, Dermacentor, and Ixodes transmit this pathogen (26). Anaplasmataceae, consisting of different genera, such as Anaplasma, Ehrlichia, Neorickettsia, and Wolbachia, contains vector-borne bacteria that are mainly transmitted by ticks (27). Tick species belonging to the genera Amblyomma, Dermacentor, Haemaphysalis, Ixodes, and Rhipicephalus transmit Anaplasma spp. to various hosts, including dogs (8, 28). Anaplasma platys and Anaplasma phagocytophilum are the most commonly detected species in dogs responsible for zoonotic diseases (2). Dogs are reservoir hosts for various spotted fever group agents (SFG) transmitted by Rh. sanguineus ticks (20). Rickettsial infections transmitted by Rh. sanguineus include the spotted fever group rickettsiosis caused by R. massiliae (20, 29), Mediterranean spotted fever (MSF) caused by Rickettsia conorii (30), and Rocky Mountain spotted fever (RMSF) caused by Rickettsia rickettsii (23). Dogs and their associated ticks play a major role in the ecology and epidemiology of several Rickettsia species, including Rickettsia aeschlimannii (23, 30).
The percentage of stray or semi-domesticated dogs is inversely correlated with the gross domestic product (GDP) per capita. Additionally, the burden of canine zoonotic parasites has been correlated with poverty, but in some cases, accurate and updated estimates regarding this data are lacking (31). Raising community knowledge, raising awareness of unhealthy practices, and ensuring preventive measures against disease-causing agents are crucial for effectively controlling ticks and tick-borne pathogens (32). Stray dogs survive through edible wastes from humans, especially from butchered surroundings in urban and peri-urban areas, mostly in developing countries. An overabundance of butcher trash appears to result from the lack of proper waste management and disposal, mostly in low-income countries (33). The abundance of discarded meat trash that is readily available for feed mostly results in the presence of large numbers of butcher-associated stray dogs (BASDs) close to butcher shops. Pakistan is a low and middle-income country (LMIC) that is heavily populated and socioeconomically underdeveloped (6, 11, 29). Due to the lack of veterinary supervision and management of butcher waste, it attracts hungry stray dogs who feed upon it. In Pakistan, butcher shops on road and street sides are major reasons for the spread of ticks that infest BASDs. No previous study has assessed public health risks and the epidemiological status of ticks and tick-borne pathogens on BASDs. This study aimed to identify the ticks infesting BASDs and to molecularly detect associated tick-borne pathogens that pose public health threats in urban and peri-urban areas.
Materials and methods
Study model
This study was designed to detect infectious agents harbored by ticks infesting BASDs. Stray dogs were observed in or near butcher shops. To test the hypothesis that meat buyers in the study area might be at risk due to tick-borne pathogens, ticks were observed on BASDs mostly in or near butcher shops. Ticks were collected from these BASDs and subjected to molecular detection for various bacterial and protozoal pathogens (Figure 1).
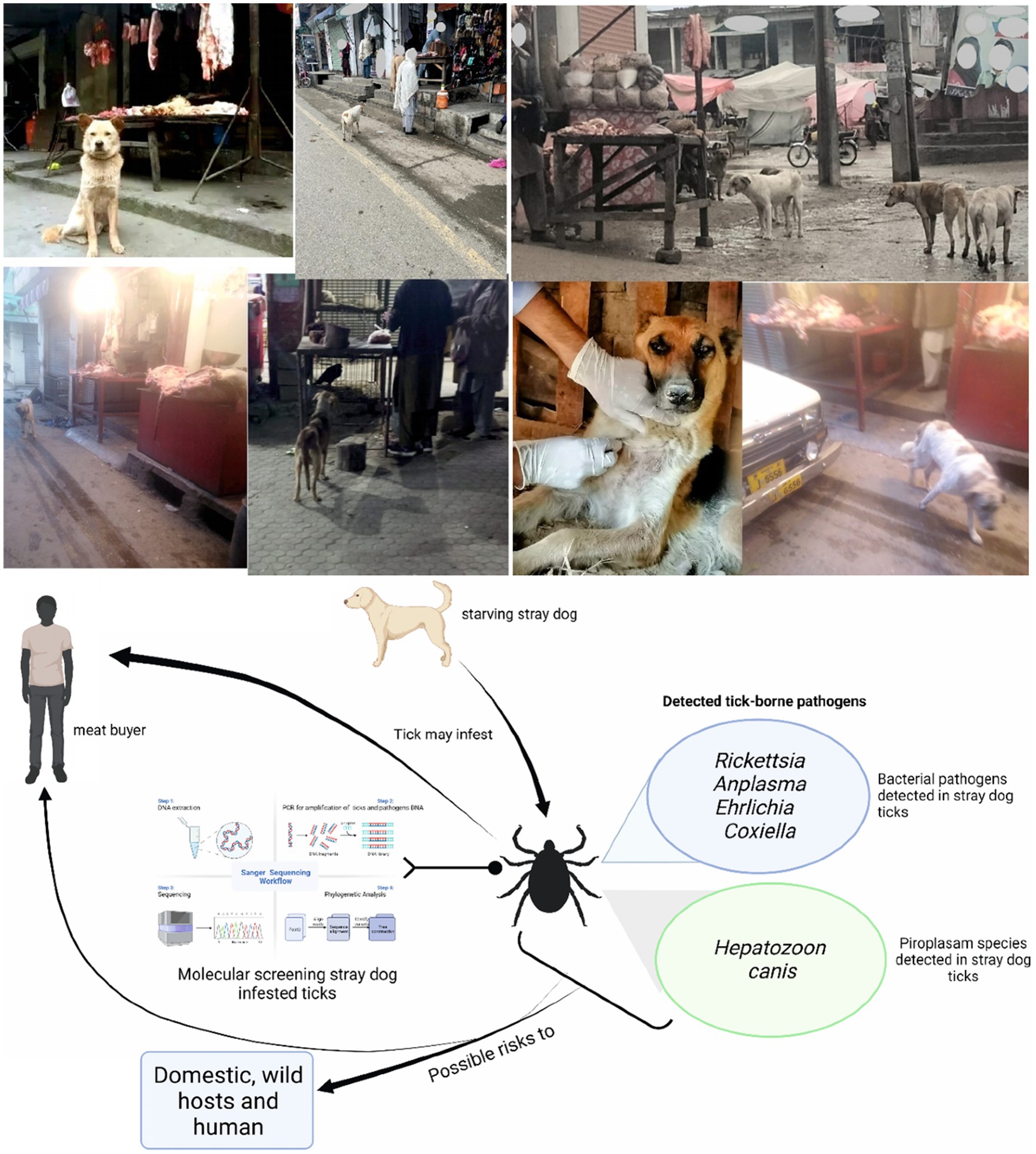
Figure 1. Study model describing the possible risks related to butcher-associated stray dogs. Parts of the figures were drawn by using pictures from Servier Medical Art (http://smart.servier.com/), licensed under a Creative Commons Attribution 3.0 Unported License (accessed 02 May 2023).
Study area
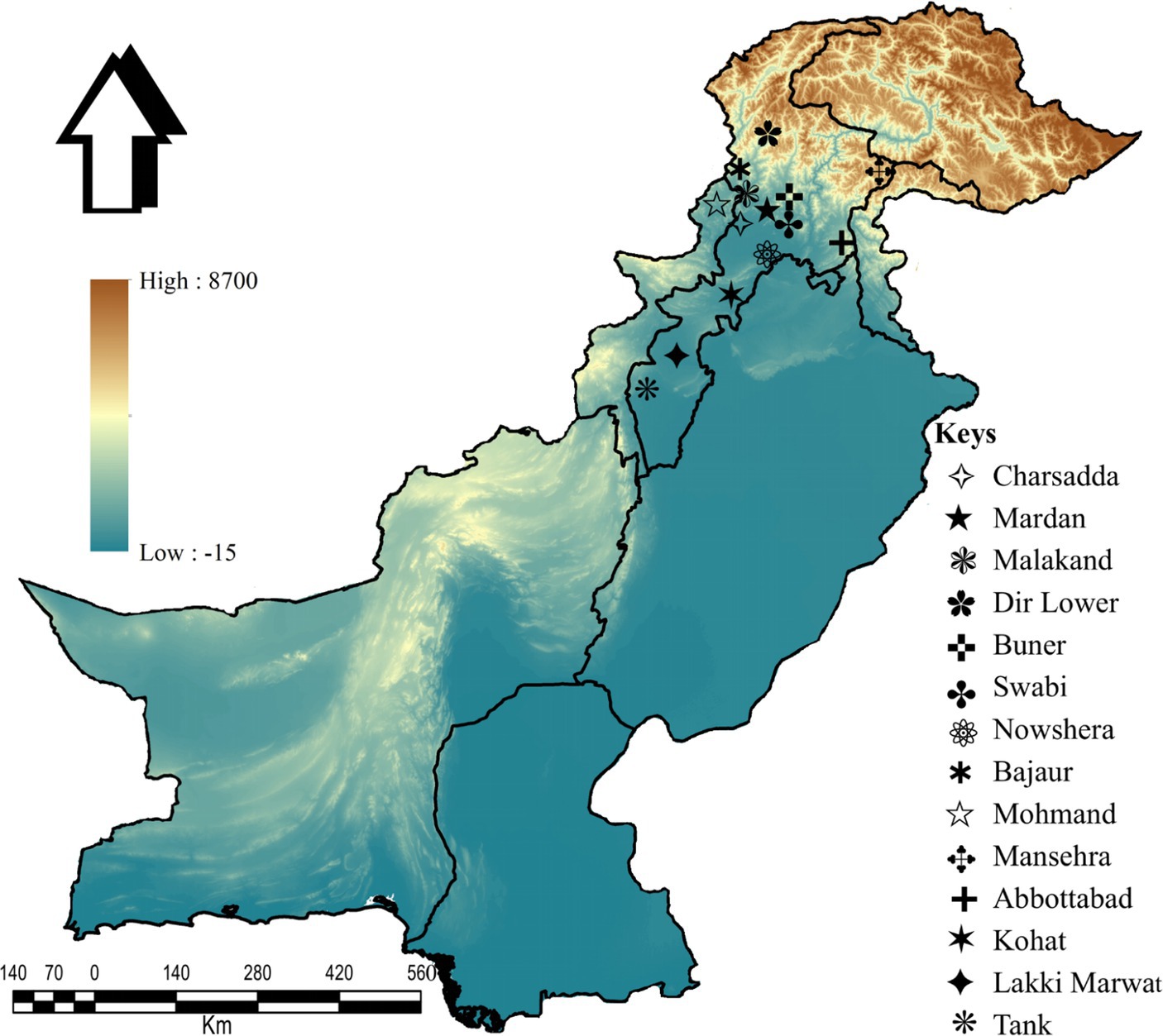
This study was conducted in 14 districts: Charsadda (34°09′46.2”N, 71°45′08.2″E), Mardan (34°11′41.8”N, 72°03′03.3″E), Malakand (34°33′31.4”N, 71°56′50.1″E), Lower Dir (34°53′20.1”N, 71°53′15.2″E), Buner (34°28′48.9”N, 72°31′13.5″E), Swabi (34°07′44.0”N, 72°27′23.9″E), Nowshera (34°00′12.4”N, 71°59′42.1″E), Bajaur (34°45′55.6”N, 71°30′54.0″E), Mohmand (34°29′05.3”N, 71°21′41.3″E), Mansehra (34°19′44.4”N, 73°12′15.3″E), Abbottabad (34°10′39.4”N, 73°14′09.4″E), Kohat (33°33′46.2”N, 71°28′20.4″E), Lakki Marwat (32°36′12.4”N, 70°54′22.1″E), and Tank (32°12′52.5”N, 70°23′09.8″E) in KP, Pakistan. The Global Positioning System (GPS) was used for the geographical coordinates of the collection sites, and the study map was designed using ArcGIS v. 10.3.1 (ESRI, Redlands, CA, United States; Figure 2).
Tick collection and preservation
Ticks were collected from BASDs in or near butcher shops from June 2021 to May 2022 in 14 selected districts. They were carefully isolated from the host body using tweezers to avoid external damage to the specimens. The specimens were rinsed in distilled water, followed by 70% ethanol, and preserved in 100% ethanol.
The examined BASDs consisted of both symptomatic and asymptomatic dogs. Symptomatic dogs were characterized by accompanying symptoms such as diarrhea, vomiting, limb swelling, rashes, itching, seizures, fatigue, scabs or skin lesions, and hair loss.
Morphology of ticks
The collected tick specimens were identified morphologically at the species level under a stereomicroscope (SZ61, Olympus Corporation, Tokyo, Japan) using standard morphological identification keys (29, 34).
DNA extraction and PCR
A total of 271 ticks (193 and 78 from symptomatic and asymptomatic BASDs, respectively) were randomly selected for molecular analyzes. Before DNA extraction, the specimens were washed in distilled water, followed by absolute ethanol. Then, they were kept in an incubator (30–45 min) until dry (20–30 min). Tick specimens were cut with sterile scissors and homogenized in phosphate-buffered saline using a micro pestle, and DNA was extracted by employing the phenol-chloroform method (35). A NanoDrop (Nano-Q, Optizen, Daejeon, South Korea) was used to quantify the DNA, and the DNA samples were stored at −20°C for further examination.
The extracted DNA was subjected to a polymerase chain reaction (PCR; GE-96G, BIOER, Hangzhou, China) to amplify mitochondrial 16S ribosomal DNA (16S rDNA) and cytochrome C oxidase 1 (cox1) fragments for tick identification. Each PCR reaction mixture contained 25 μL volume, comprising 1 μL each primer (forward and reverse; 10 μM), 8.5 μL PCR water “nuclease-free,” 12.5 μL DreamTaq green MasterMix (2X; Thermo Scientific, Waltham, MA, United States), and 2 μL of template DNA (100 ng/μl).
The extracted DNA was utilized for screening various tick-borne pathogens based on different partial genes, such as 16S rDNA (Anaplasma/Ehrlichia spp.), groEL (Coxiella spp.), dsb (Ehrlichia spp.), 18S rDNA (Hepatozoon spp.), and gltA, ompA, and ompB (Rickettsia spp.). The nested PCR was used in the case of the groEL partial gene. Each PCR experiment contained a negative control (PCR water instead of the template DNA) and a positive control (the DNA of Rh. microplus for ticks, R. massiliae for bacterial species, and Theileria annulata for protozoal species). The primers used in the current study and their thermocycler conditions are given in Supplementary Table S1.
The PCR products were electrophoresed on 1.5% agarose gel, and the amplified samples were visualized under ultraviolet (UV) light using a gel documentation system (BioDoc-IT™ Imaging Systems UVP, LLC, Upland, United States). The PCR products that showed the expected size results were purified using a commercial PCR clean-up kit (Macherey-Nagel, Duren, Germany) following the manufacturer’s instructions.
DNA sequencing and phylogenetic analysis
PCR products of 16S rDNA and cox1 for ticks, as well as 16S rDNA, groEL, dsb, 18S rDNA, gltA, ompA, and ompB for associated tick-borne pathogens, were sent for capillary bidirectional sequencing (Macrogen Inc., Seoul, South Korea). The obtained sequences were trimmed and assembled using SeqMan v. 5 (DNASTAR, Inc., Madison/WI, United States) to remove poor reading sequences. The obtained multiple identical sequences for each gene were considered a consensus sequence and subjected to the basic local alignment search tool (BLAST) at the National Center for Biotechnology Information (NCBI). Sequences with maximum identities were downloaded in FASTA format from the NCBI. The obtained sequences were aligned with the downloaded sequences using ClustalW multiple alignments in BioEdit Sequence Alignment Editor v. 7.0.5 (36). The coding nucleotide sequences were aligned using MUSCLE (37). Phylogenetic trees were constructed individually for each partial sequence of tick and associated tick-borne pathogens, by employing the maximum likelihood (ML) method with Kimura 2-parameter in Molecular Evolutionary Genetics Analysis (MEGA-X), accompanied by a bootstrapping value of 1,000 (38).
Statistical analyzes
Tick data and associated pathogen information from all 14 districts were subjected to descriptive statistical analysis, and graphs were designed in Microsoft Excel v. 2016 (Microsoft Office 365®). The minimum infection rate (MIR) was used to estimate the prevalence of infected ticks. To accomplish this, the number of PCR product ticks was divided by the total number of collected ticks (39).
Results
A survey in the selected districts resulted in an overall collection of 575 ticks (392 and 183 from symptomatic and asymptomatic BASDs, respectively) from 117 BASDs (63 symptomatic and 54 asymptomatic), comprising four hard tick species. Among the identified ticks, Rh. sanguineus was the most prevalent tick species (n = 292/575, 50.78%), followed by Rh. haemaphysaloides (n = 127/575, 22.08%), Rh. turanicus counting (n = 87/575, 15.13%), and Rh. microplus clade C (n = 69/575, 12.00%). The spatial distribution of the collected ticks revealed that Mardan, Lakki Marwat, and Charsadda had the highest tick counts of 58, 53, and 51 ticks, respectively. The Malakand district (27) had the least number of collected ticks among the selected districts.
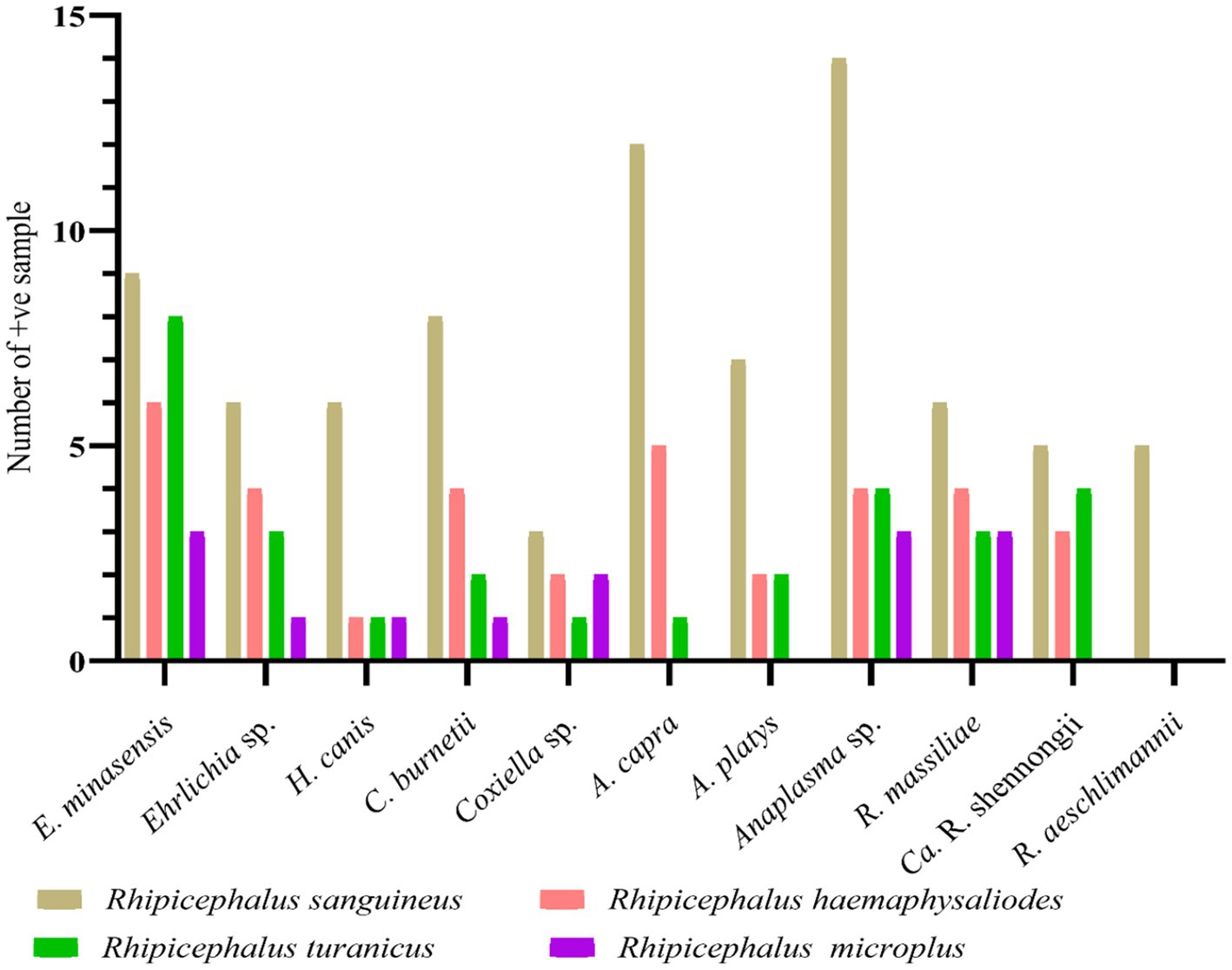
Of the 575 tick specimens, 271 (47.13%; 193 and 78 from symptomatic and asymptomatic BASDs, respectively) were subjected to PCR for the molecular screening of various tick-borne pathogens, including Ehrlichia, Anaplasma, Rickettsia, Coxiella and Hepatozoon species (Supplementary Table S1). According to the PCR-based screening and subsequent sequencing of targeted bacterial and protozoal agents, Anaplasma spp. were the most prevalent (n = 54/271, 19.92%), followed by Ehrlichia spp. (n = 40/271, 14.76%), Rickettsia spp. (n = 33/271, 12.17%), Coxiella spp. (n = 23/271, 8.49%), and Hepatozoon spp. (n = 9/271, 3.32%) in the ticks infesting BASDs (Table 1). None of the ticks was found positive for multiple pathogens.
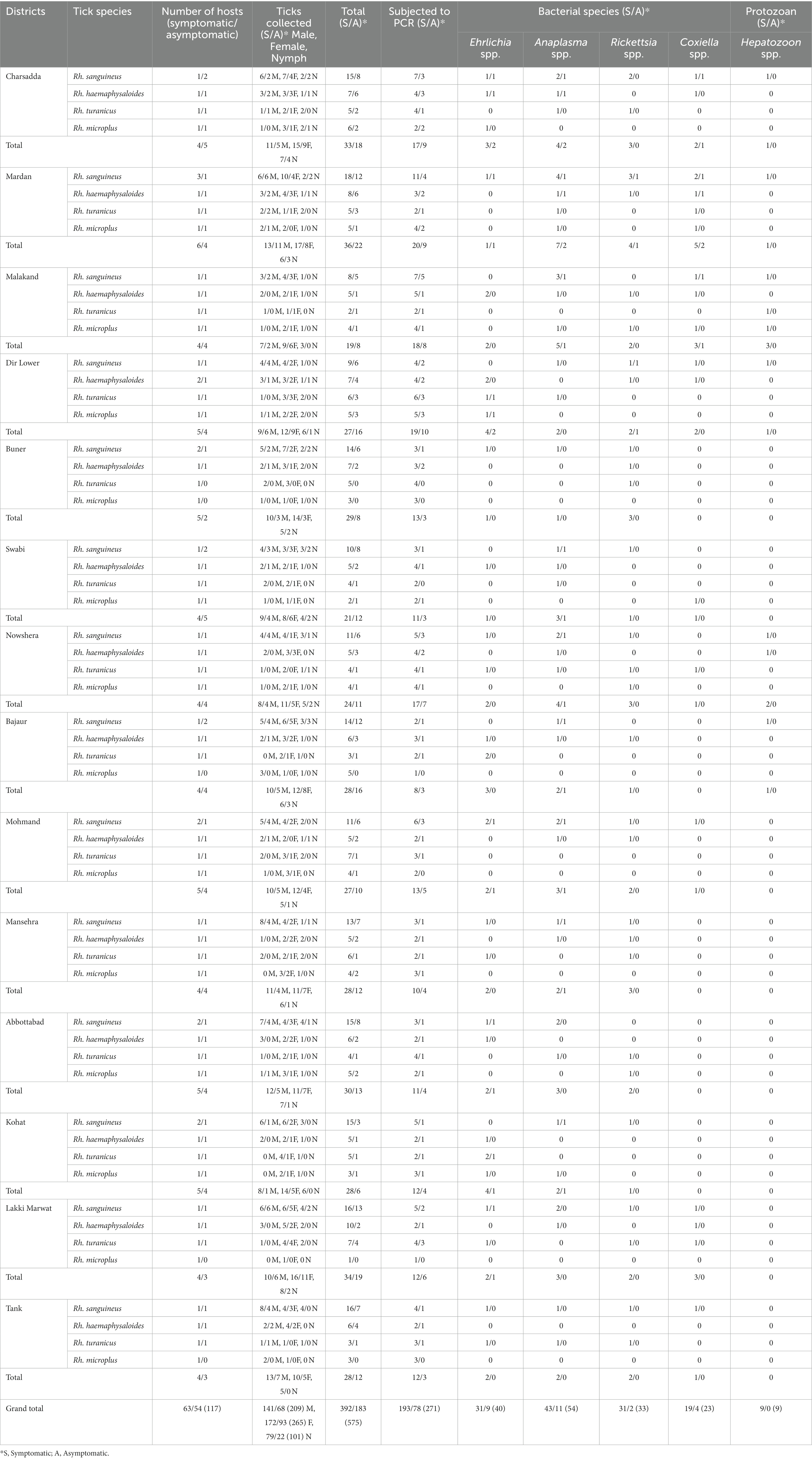
Table 1. Distribution pattern of butcher-associated stray dog ticks and the detection of bacterial and protozoal pathogens in these ticks.
The results of the prevalence of symptomatic and asymptomatic BASDs in ticks, as well as the molecular screening of tick-associated bacterial and protozoal pathogens, are presented in Table 1.
DNA sequences and phylogeny of ticks
Sequencing by BLAST analysis of the 16S rDNA for Rh. sanguineus, Rh. turanicus, Rh. microplus, and Rh. haemaphysaloides showed 99.24–100% maximum identities to the corresponding species. In the phylogenetic tree, the 16S rDNA sequences of Rh. sanguineus, Rh. turanicus, Rhipicephalus microplus (clade C), and Rhipicephalus haemaphysaloides were clustered with the corresponding species (Figure 3A).
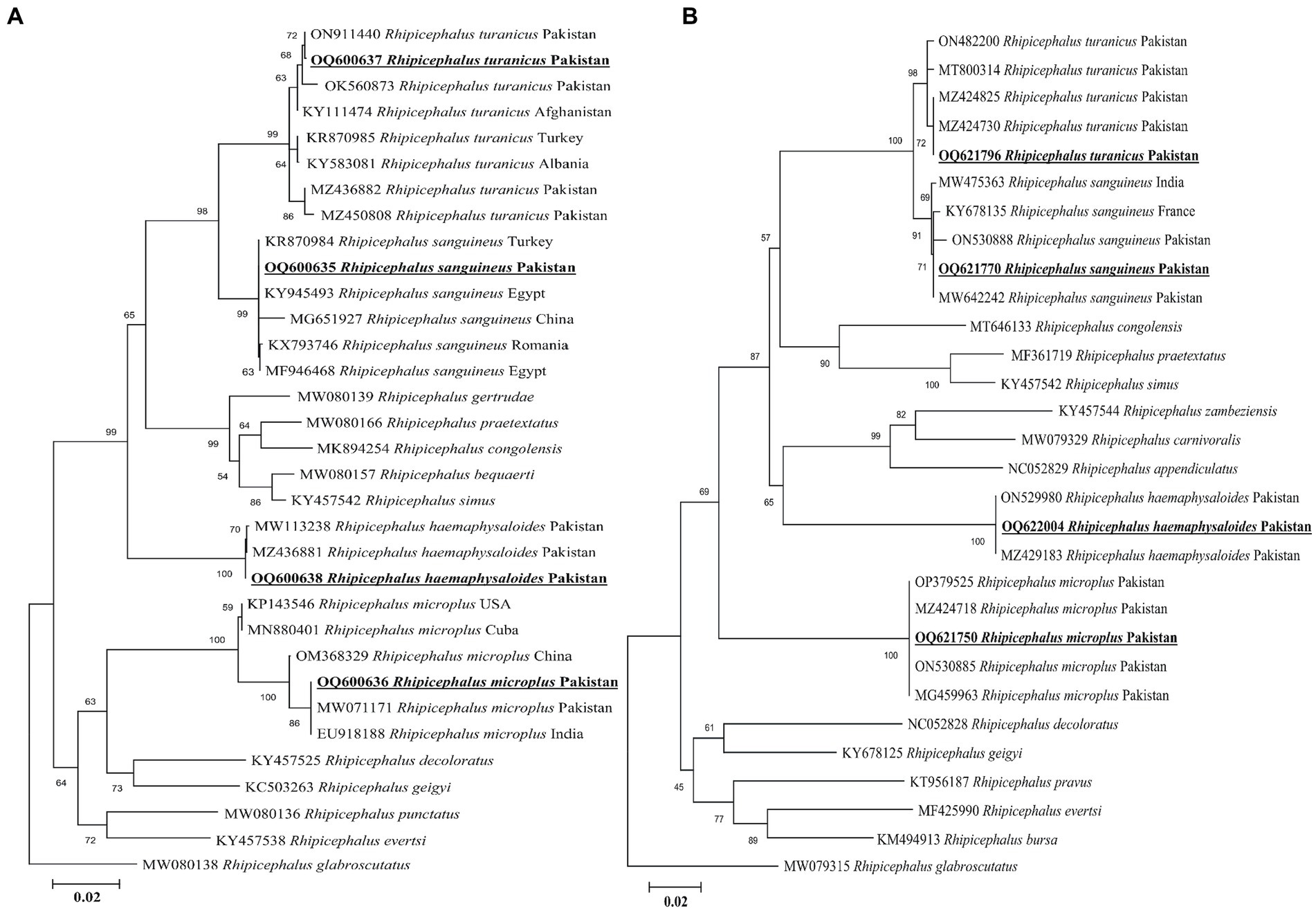
Figure 3. Maximum likelihood phylogenetic trees based on 16S rDNA (A) and cox1 (B) partial sequences of Rhipicephalus spp. The Rhipicephalus glabrosscutatus sequence was used as an outgroup. The obtained sequences are shown in bold underline font.
According to the BLAST results, the obtained cox1 sequences of Rh. sanguineus, Rh. turanicus, Rh. microplus, and Rh. haemaphysaloides shared 99.54–100% maximum identities with the corresponding species. In the phylogenetic tree, the cox1 sequences of Rh. sanguineus, Rh. turanicus, Rhipicephalus microplus, and Rhipicephalus haemephysalides were clustered with the corresponding species (Figure 3B). All the obtained 16S rDNA and cox1 sequences of Rh. sanguineus, Rh. turanicus, Rh. microplus, and Rh. haemaphysaloides were deposited to the GenBank (Supplementary Table S2).
Prevalence of pathogens in ticks
Altogether, 271 out of 575 collected ticks were screened for detecting bacterial and protozoal pathogens. Among the four tested tick species, Rh. sanguineus was the most infected tick comprising 81 positive samples with a minimum infection rate of 29.88% (n = 81/271), followed by Rh. haemaphysaloides with a minimum infection rate of 12.91% (n = 35/271), Rh. turanicus with a minimum infection rate of 10.70% (n = 29/271), and Rh. microplus with a minimum infection rate of 5.16% (n = 14/271). All these minimum infection rates were statistically significant. Sequencing of the PCR-based amplified DNA resulted in the detection of eight pathogens–E. minasensis, H. canis, C. burnetii, Anaplasma capra, A. platys, R. massiliae, “Candidatus Rickettsia shennongii” and R. aeschlimannii. Three undetermined species, Ehrlichia sp., Coxiella sp., and Anaplasma sp. were also detected. All pathogens and undetermined species were detected in the collected four tested tick species, except R. aeschlimannii, which was detected only in Rh. sanguineus, whereas A. capra, A. platys, and “Ca. R. shennongii” were detected in Rh. sanguineus, Rh. haemaphysaloides, and Rh. turanicus (Figure 4).
Sequence and phylogenetic analyzes of tick-borne pathogens
Ehrlichia spp.
According to the BLAST results, the 16S rDNA sequences of Ehrlichia sp. and E. minasensis showed 100% identity with the corresponding species. In the phylogenetic tree, the 16S rDNA sequences of Ehrlichia sp. and E. minasensis were clustered with the corresponding sequences (Figure 5A). In the case of the dsb sequence, the BLAST results of the dsb sequence of Ehrlichia sp. showed 100% identity with the E. minasensis sequence. In the phylogenetic tree, the dsb sequence of E. minasensis was clustered with the same species (Figure 5B). The obtained 16S rDNA and dsb sequences of Ehrlichia sp. and E. minasensis were deposited to the GenBank (Supplementary Table S2).
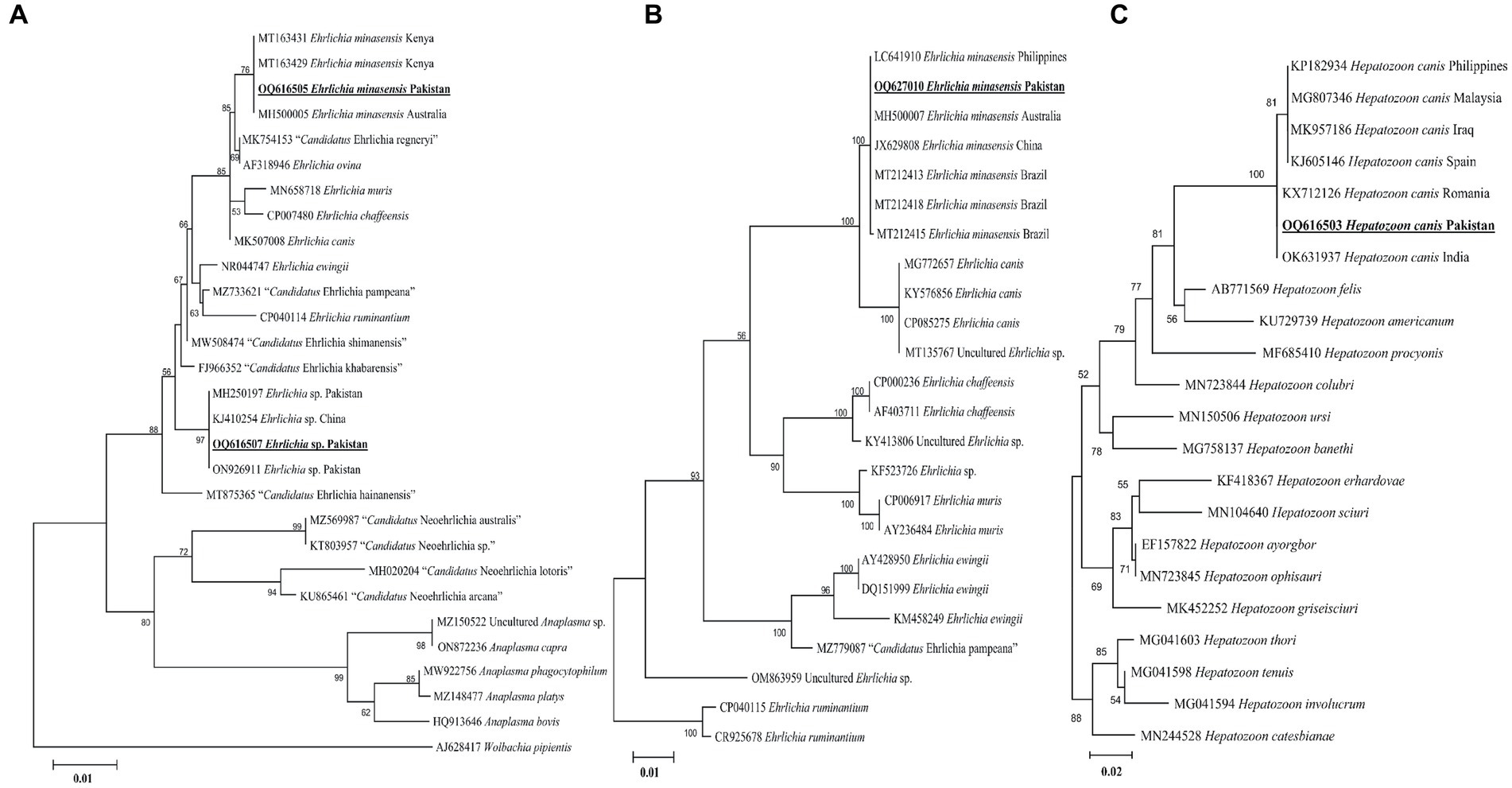
Figure 5. Maximum likelihood phylogenetic trees based on 16S rDNA (A) and dsb (B) for Ehrlichia spp. and 18S rDNA (C) partial sequences for Hepatozoon canis. The Wolbachia pipientis 16S rDNA, Ehrlichia ruminantium dsb, and Hepatozoon catesbianae, Hepatozoon thori, Hepatozoon tenuis, and Hepatozoon involucrum 18S rDNA sequences were used as an outgroup. The obtained sequences are shown in bold underline font.
Hepatozoon canis
The obtained 18S rDNA sequence of Hepatozoon sp. showed 99–100% identity with the H. canis sequence. In the phylogenetic tree, the 18S rDNA sequence of H. canis was clustered with the same species (Figure 5C). The obtained 18S rDNA sequence of H. canis was deposited to the GenBank (Supplementary Table S2).
Coxiella spp.
The present groEL sequence of Coxiella sp. showed 97–98% maximum identity with the Coxiella sp. sequence, followed by 96.55% identity with C. burnetii. Other Coxiella sp. groEL sequences showed 100% identity with C. burnetii. In the phylogenetic tree, the groEL sequence of Coxiella sp. was clustered with the Coxiella sp. endosymbiont sequences, while the sequence of C. burnetii was clustered with the corresponding species (Figure 6A). The obtained groEL sequences of Coxiella sp. and C. burnetii were deposited to the GenBank (Supplementary Table S2).
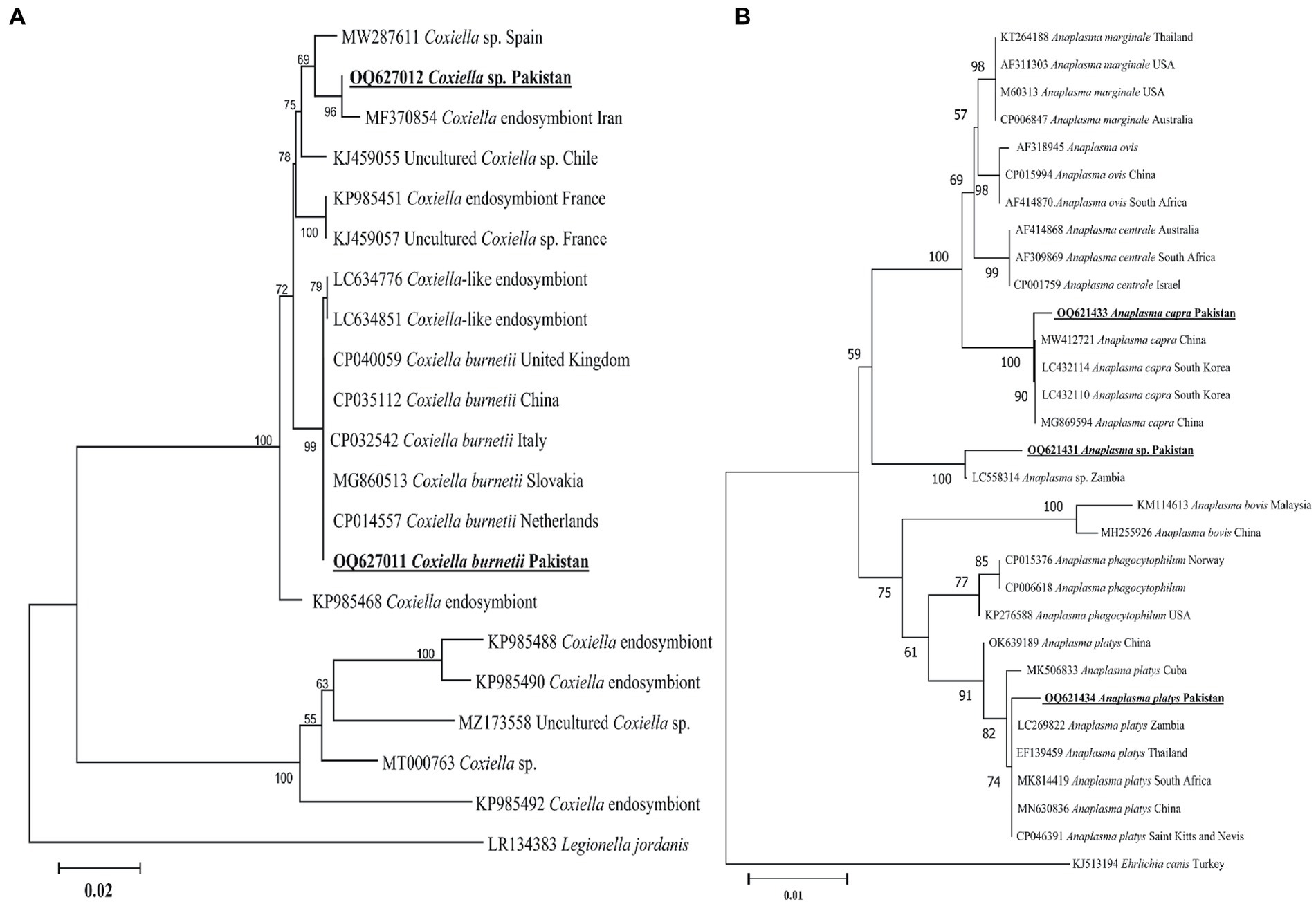
Figure 6. Maximum likelihood phylogenetic trees based on groEL (A) and 16S rDNA (B) partial sequences of Coxiella spp. and Anaplasma spp., respectively. The Legionella jordanis groEL and Ehrlichia canis 16S rDNA sequences were used as an outgroup. The obtained sequences are shown in bold underline font.
Anaplasma spp.
According to the BLAST results, 16S rDNA Anaplasma spp. sequences showed 99.12% maximum identity with A. capra, 100% maximum identity with A. platys sequences, and 99.13% maximum identity with an undermined Anaplasma sp. In the phylogenetic tree, the 16S rDNA sequences of A. capra, A. platys, and undetermined Anaplasma sp. were clustered with the corresponding species (Figure 6B). The obtained 16S rDNA sequences of A. capra, A. platys, and Anaplasma sp. were deposited to the GenBank (Supplementary Table S2).
Rickettsia spp.
A rickettsial gltA sequence showed 100% identity with R. massiliae, and in the phylogenetic tree, it was clustered with the corresponding species (Figure 7A). Another obtained gltA sequence showed 100% identity with “Ca. R. shennongii,” and it was phylogenetically clustered with the corresponding species (Figure 7A). A third rickettsial gltA sequence showed 100% identity with R. aeschlimannii, and in the phylogenetic tree, it was clustered with the corresponding species (Figure 7A).
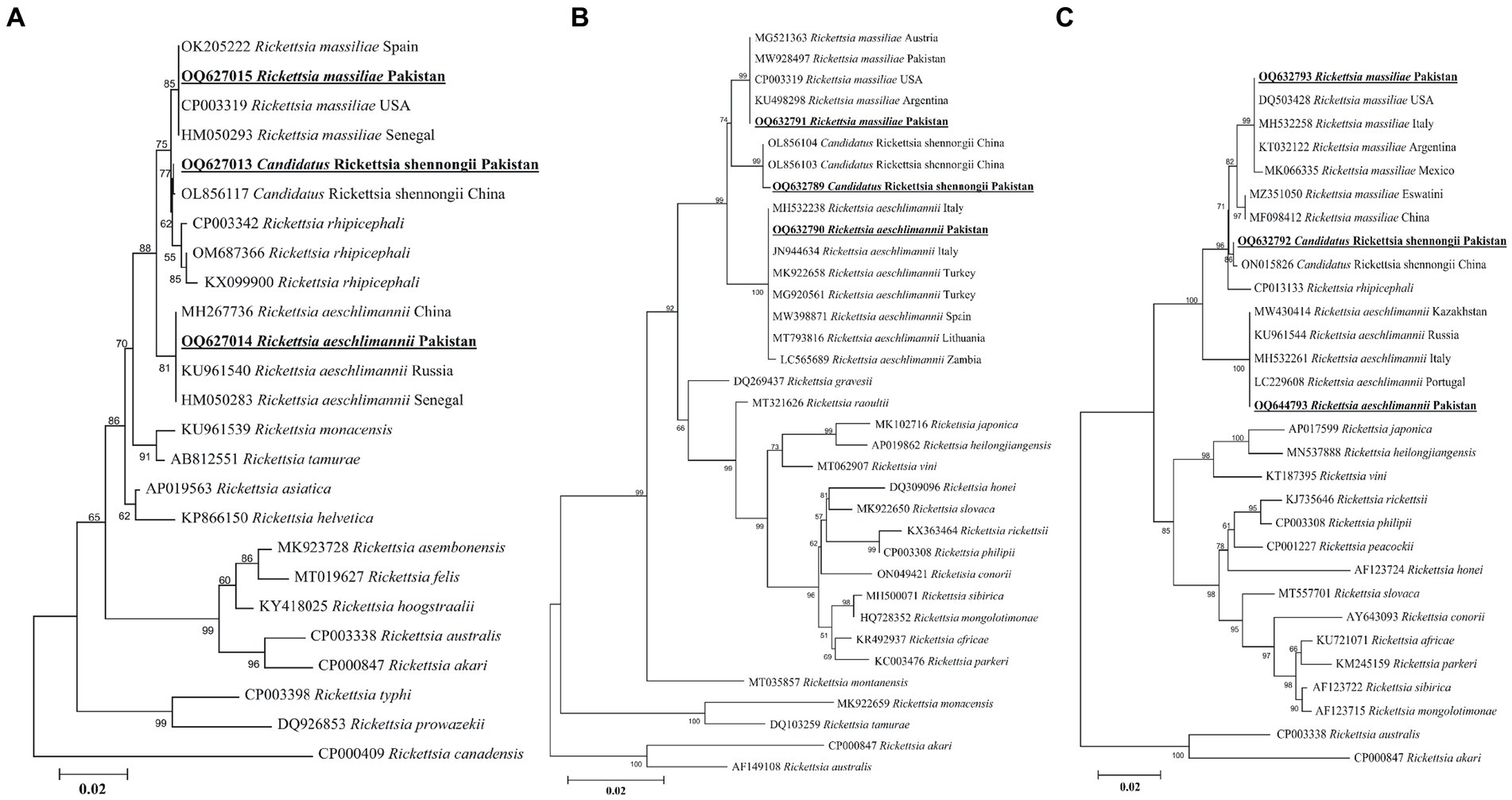
Figure 7. Maximum likelihood phylogenetic trees based on gltA (A), ompA (B), and ompB (C) partial sequences of Rickettsia spp. The Rickettsia canadensis gltA, Rickettsia akari and Rickettsia australis ompA, and Rickettsia akari and Rickettsia australis ompB sequences were used as an outgroup. The obtained sequences are shown in bold underline font.
The obtained ompA sequence showed 100% identity with R. massiliae and was phylogenetically clustered with the same species (Figure 7B). Another ompA sequence showed 100% identity with “Ca. R. shennongii” and in the phylogenetic tree, it was clustered with the same species (Figure 7B). A third ompA fragment showed 99.8–100% identity with R. aeschlimannii sequences, and in the phylogenetic tree, it was clustered with the same species (Figure 7B).
In the case of ompB, the obtained sequences showed 100% identity with R. massiliae. In the phylogenetic tree, the obtained R. massiliae sequence was clustered with the same species. Another ompB sequence showed 100% identity with “Ca. R. shennongii” and in the phylogenetic tree, this sequence was clustered with “Ca. R. shennongii” (Figure 7C). A third ompB sequence showed 100% identity with R. aeschlimannii, and it was phylogenetically clustered with the corresponding species (Figure 7C). The obtained gltA, ompA, and ompB sequences of R. massiliae, “Ca. R. shennongii” and R. aeschlimannii were deposited to the GenBank (Supplementary Table S2).
Discussion
BASDs are commonly exposed to vector-borne pathogens due to the lack of adequate protection against ectoparasites in different regions. Different bacterial and protozoal infections have been reported in ticks infesting dogs, depending on the status of reservoir hosts, geographical variations in the exposure to tick vectors, detection methods, and the time of testing (2). The epidemiological situations of ticks and tick-borne pathogens associated with BASDs near butcher shops have not been previously investigated. A combination of morphological and molecular characterization resulted in the identification of several tick species infesting BASDs, including Rh. sanguineus, Rh. haemaphysaloides, Rh. turanicus, and Rh. microplus. The examined ticks were found positive for a variety of pathogenic and undetermined microorganisms, including species of the genera Ehrlichia, Coxiella, Hepatozoon, Anaplasma, and Rickettsia, which can be transmitted and pose health threats to humans, as well as wild and domestic animals. The different detected pathogenic and undermined species included E. minasensis, Ehrlichia sp., H. canis, Coxiella sp., C. burnetii, A. capra, A. platys, Anaplasma sp., R. massiliae, “Ca. R. shennongii” and R. aeschlimannii. This is the first investigation of Ehrlichia spp., Hepatozoon spp., Coxiella spp., Anaplasma spp., and Rickettsia spp. in ticks infesting BASDs. In Pakistan, E. minasensis, H. canis, C. burnetii, A. capra, A. platys, “Ca. R. shennongii” and R. aeschlimannii were detected for the first time, and they pose zoonotic threats to public health.
Abandoned stray dogs are frequently exposed and particularly vulnerable to the high risks of tick infestation and dispersal (5). Several tick species infest stray dogs in urban and peri-urban regions, and various pathogens are transmitted from dogs to other animals and humans (28). Surveillance requires a thorough understanding of tick dentification to reduce losses associated with tick-borne disease-causing agents (9, 16, 40). Morphological identification notwithstanding, the best way to determine the taxonomic status of the tick species is through molecular characterization utilizing 16S rDNA and cox1 genetic markers (10, 15, 41, 42). The 16S rDNA and cox1 sequences for Rh. sanguineus, Rh. turanicus, Rh. microplus, and Rh. haemaphysaloides in the present study revealed the closest identities of these ticks to the corresponding species from the Oriental and Palearctic regions.
BASDs freely roam outdoors, which increases their exposure to ticks and leads to uncontrolled breeding. They are mostly spotted in urban and rural localities. In general, stray dogs tend to carry a number of ticks, most commonly the dog tick Rh. sanguineus, which is a vector for several pathogens (43). Two Ehrlichia species (E. minasensis and Ehrlichia sp.) were detected in ticks in BASDs. In previous studies, E. minasensis has been detected in ticks infesting different hosts, such as cattle, horses, goats, and dogs (44, 45). For the genetic characterization of Ehrlichia spp., the 16S rDNA and dsb partial fragments have been identified as accurate genetic markers at the species level (46, 47). The 16S rDNA fragments of Ehrlichia spp. were closely related to the Ehrlichia sp. and E. minasensis species. In the 16S rDNA-based phylogeny, Ehrlichia sp. formed a sister clustered with E. ewingii and E. ruminantium, suggesting that this species may be of zoonotic concern. In the case of dsb fragments, the present E. minasensis sequences were phylogenetically clustered with the same species reported from the Palearctic, Neotropical, Australian, and Oriental regions. In both phylogenetic trees based on the 16S rDNA and dsb, the appearance of E. minasensis as a sister clade to E. canis might suggest that both are potentially canine pathogens (45, 47). Considering the detection of Ehrlichia spp. in ticks infesting free-roaming BASDs, further investigations are critical to disclose its association between stray dogs (BASDs) and humans.
Hepatozoon canis is a tick-borne apicomplexan parasite infecting canids and has long been known to be transmitted by ingesting infected Rh. sanguineus and Rh. turanicus ticks (48). The available phylogeny of H. canis is based on the 18S rDNA sequence, which has been demonstrated to be useful for inferring phylogenetic analyzes at the species level (48). Phylogenetic analysis based on the 18S rDNA sequence of this species suggests its close resemblance with the same species circulating in the Palearctic and Oriental regions.
More than 40 Ixodid (Rhipicephalus, Dermacentor, Haemaphysalis, Amblyomma, and Ixodes) and Argasid (Ornithodoros and Argas) tick species are known to harbor bacterial microbes related to C. burnetii and other Coxiella-like endosymbionts, which have been commonly detected in Rhipicephalus spp., particularly Rh. sanguineus (25, 49). Rhipicephalus spp. may play an important role in the transmission of C. burnetii among the hosts. Except for C. burnetii, there is no evidence that Coxiella spp. cause disease in the host; however, the pathogenic roles of these species have remained undetermined (49). Q fever is a vector-borne zoonotic disease caused by C. burnetii that affects a wide range of hosts globally. Coxiella spp. have been detected in Rh. sanguineus (s.l.) based on the groEL sequence (26). Since its successful detection in various ticks, the groEL sequences are potentially specific for Coxiella-like endosymbionts (18, 25). Likewise, the groEL partial sequence of Coxiella sp. and C. burnetii was detected in Rhipicephalus ticks of BASDs, which were closely related to the corresponding species.
Several tick species have been reported as vectors of Anaplasma, particularly in the genus Rhipicephalus (14, 50). Although Anaplasma spp. are prevalent in ixodid ticks, relatively limited studies have been conducted to investigate its detection, especially in BASD ticks (2). Anaplasma spp. were detected in high abundance in BASD ticks, which is nearly comparable to prior studies (2). Anaplasma platys and A. capra have mostly been considered to be transmitted by the dog tick Rh. sanguineus, and dogs have been confirmed as the hosts for A. capra in China (27). For the genetic characterization of Anaplasma spp., the highly conserved 16S rDNA marker has been historically employed (14, 27, 50). Likewise, the 16S rDNA fragments of Anaplasma spp., such as A. capra and A. platys, and an undermined Anaplasma sp. detected in Rhipicephalus spp. align with previous reports (14, 50). The detection of Anaplasma spp. in ticks infesting BASDs in Pakistan might suggest its geographic expansion, highlighting the need for further comprehensive studies on its pathogenicity to screen for the epidemiological and evolutionary status, which was previously underestimated due to the lack of sufficient data.
In Pakistan, earlier epidemiological investigations on the detection of Rickettsia spp. were mostly performed on ticks infesting small and large ruminants, equine, and wild hosts (11, 15, 16, 29, 51). Despite this gap, we used molecular screening of Rickettsia spp. in ticks infesting BASDs. Genetic markers, such as gltA, ompA, and ompB, have been utilized to determine a high degree of intraspecific variation and are extensively used for reliable characterization at the species level (42). We detected and characterized for the first time three rickettsial agents, Rickettsia massiliae, “Ca. Rickettsia shennongii” and Rickettsia aeschlimannii in ticks infesting BASDs. Rickettsia massiliae and Rickettsia aeschlimannii have been detected in ticks of multiple hosts in several districts of Pakistan (19, 52). The detection of “Ca. R. shennongii” represents the first record of any host or tick in Pakistan. Recently, this species was detected in Rh. haemaphysaloides ticks in China as a novel spotted fever group (SFGR) (25). The detection of “Ca. R. shennongii” in Rh. sanguineus, Rh. haemaphysaloides, and Rh. turanicus suggests its diverse host and distribution range and enforces the need to investigate its zoonotic threat to other hosts, including humans. These findings might advance our knowledge of the diversity of circulating tick-borne pathogens in the region, highlighting the need for further comprehensive surveillance studies to properly monitor ticks for potential zoonotic threats.
Conclusion
To effectively manage ticks and tick-borne infections, the community must be educated regarding the associated risks and prevention strategies. The current endeavor highlights the risks of ticks infesting BASDs to public health. A comprehensive analysis was undertaken to determine which bacterial and protozoal pathogens might be carried by ticks infesting BASDs that inhabit butcher shops. This study will provide fundamental knowledge about the risks associated with these free-roaming BASDs, convincing health policymakers to ensure the control of any zoonotic consequences associated with these ticks infesting dogs near butcher shops. To minimize the risks of ticks and tick-borne diseases related to BASDs, proper disposal of meat and butcher waste should be adopted, and open-street butcher shops should be properly managed. Further surveillance studies are essential to understand the status of ticks and tick-borne pathogens in BASDs in different regions.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Ethics statement
The animal studies were approved by the current study was approved by the Advance Studies Research Board (ASRB: Dir/A&R/AWKUM/2022/9396) committee members of the Faculty of Chemical and Life Sciences, Abdul Wali Khan University, Mardan Khyber Pakhtunkhwa (KP), Pakistan. Butchers were informed verbally about their observations and collection of ticks. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
AiA designed the study. AiA, SU, and MN collected the ticks and carried out phylogenetic and statistical analysis. AiA, AdA, MA, and TT performed the experiments. All authors contributed to the article and approved the submitted version.
Funding
The researchers supporting project number (RSP2023R494), King Saud University, Riyadh, Saudi Arabia. This work was supported by JSPS KAKENHI Grant Numbers JP20KK0154 and JP22H02522, and JSPS Bilateral Program (Grant number JPJSBP120239937).
Acknowledgments
The authors acknowledge the financial support provided by the Higher Education Commission (HEC), Pakistan, and the Pakistan Science Foundation (PSF). The researchers supporting project number (RSP2023R494), King Saud University, Riyadh, Saudi Arabia.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2023.1246871/full#supplementary-material
Abbreviations
BASDs, Butcher-associated stray dogs; BLAST, Basic local alignment search too; gltA, Citrate synthase; SFGR, Spotted fever group Rickettsia; VBDs, Vector-borne diseases; KP, Khyber Pakhtunkhwa; MEGA, Molecular evolutionary genetics analysis; NCBI, National Center for Biotechnology Information; CME, Canine monocytic ehrlichiosis; Q fever, Query fever; ompA, Outer membrane protein; PCR, Polymerase chain reaction; rDNA, Ribosomal DNA.
References
1. Butler, JRA, Du Toit, JT, and Bingham, J. Free-ranging domestic dogs (Canis familiaris) as predators and prey in rural Zimbabwe: threats of competition and disease to large wild carnivores. Biol Conserv. (2004) 115:369–78. doi: 10.1016/S0006-3207(03)00152-6
2. Maksimović, Z, Dervišević, M, Zahirović, A, and Rifatbegović, M. Seroprevalence of Anaplasma spp. and Ehrlichia spp. and molecular detection of Anaplasma phagocytophilum and Anaplasma platys in stray dogs in Bosnia and Herzegovina. Ticks Tick Borne Dis. (2022) 13:101875. doi: 10.1016/j.ttbdis.2021.101875
3. Smith, LM, Hartmann, S, Munteanu, AM, Dalla Villa, P, Quinnell, RJ, and Collins, LM. The effectiveness of dog population management: a systematic review. Animals. (2019) 9:1020. doi: 10.3390/ani9121020
4. Larson, G, Karlsson, EK, Perri, A, Webster, MT, Ho, SY, Peters, J, et al. Rethinking dog domestication by integrating genetics, archeology, and biogeography. Proc Natl Acad Sci U S A. (2012) 109:8878–83. doi: 10.1073/pnas.1203005109
5. Koh, FX, Panchadcharam, C, and Tay, ST. Vector-borne diseases in stray dogs in peninsular Malaysia and molecular detection of Anaplasma and Ehrlichia spp. from Rhipicephalus sanguineus (Acari: Ixodidae) ticks. J Med Entomol. (2016) 53:183–7. doi: 10.1093/jme/tjv153
6. Murawat, F. Countering the stray dog crisis using CNVR in Sindh, Pakistan. (2022). Available at: https://ssrn.com/abstract=4138427.
7. Do, T, Phoosangwalthong, P, Kamyingkird, K, Kengradomkij, C, Chimnoi, W, and Inpankaew, T. Molecular detection of tick-borne pathogens in stray dogs and Rhipicephalus sanguineus sensu lato ticks from Bangkok. Thailand Pathog. (2021) 10:561. doi: 10.3390/pathogens10050561
8. Wyk, CLV, Mtshali, K, Taioe, MO, Terera, S, Bakkes, D, Ramatla, T, et al. Detection of ticks and tick-borne pathogens of urban stray dogs in South Africa. Pathogens. (2022) 11:862. doi: 10.3390/pathogens11080862
9. Gondard, M, Cabezas-Cruz, A, Charles, RA, Vayssier-Taussat, M, Albina, E, and Moutailler, S. Ticks and tick-borne pathogens of the Caribbean: current understanding and future directions for more comprehensive surveillance. Front Cell Infect Microbiol. (2017) 7:490. doi: 10.3389/fcimb.2017.00490
10. Lu, X, Lin, XD, Wang, JB, Qin, XC, Tian, JH, Guo, WP, et al. Molecular survey of hard ticks in endemic areas of tick-borne diseases in China. Ticks Tick Borne Dis. (2013) 4:288–96. doi: 10.1016/j.ttbdis.2013.01.003
11. Ali, A, Numan, M, Khan, M, Aiman, O, Muñoz-Leal, S, Chitimia-Dobler, L, et al. Ornithodoros (Pavlovskyella) ticks associated with a Rickettsia sp. in Pakistan. Parasit Vectors. (2022) 15:138. doi: 10.1186/s13071-022-05248-0
12. Ali, A, Numan, M, Ullah, S, Khan, M, and Kamran, K. Genetic characterization of Haemaphysalis (Rhipistoma) indica and Haemaphysalis (Segalia) montgomeryi ticks (Ixodoidea: Ixodidae). Ticks Tick Borne Dis. (2023) 14:102105. doi: 10.1016/j.ttbdis.2022.102105
13. Aiman, O, Ullah, S, Chitimia-Dobler, L, Nijhof, AM, and Ali, A. First report of Nosomma monstrosum ticks infesting Asian water buffaloes (Bubalus bubalis) in Pakistan. Ticks Tick Borne Dis. (2022) 13:101899. doi: 10.1016/j.ttbdis.2022.101899
14. Alam, S, Khan, M, Alouffi, A, Almutairi, MM, Ullah, S, Numan, M, et al. Spatio-temporal patterns of ticks and molecular survey of Anaplasma marginale, with notes on their phylogeny. Microorganisms. (2022) 10:1663. doi: 10.3390/microorganisms10081663
15. Numan, M, Islam, N, Adnan, M, Zaman Safi, S, Chitimia-Dobler, L, Labruna, MB, et al. First genetic report of Ixodes kashmiricus and associated Rickettsia sp. Parasit Vectors. (2022) 15:378–2. doi: 10.1186/s13071-022-05509-y
16. Khan, SM, Khan, M, Alouffi, A, Almutairi, MM, Numan, M, Ullah, S, et al. Phylogenetic position of Haemaphysalis kashmirensis and Haemaphysalis cornupunctata, with notes on Rickettsia spp. Genes. (2023) 14:360. doi: 10.3390/genes14020360
17. Dantas-Torres, F, Chomel, BB, and Otranto, D. Ticks and tick-borne diseases: a one health perspective. Trends Parasitol. (2012) 28:437–46. doi: 10.1016/j.pt.2012.07.003
18. Bogunović, D, Stević, N, Sidi-Boumedine, K, Misić, D, Tomanović, S, Kulisić, Z, et al. Molecular evidence of Q fever agent Coxiella burnetii in Ixodid ticks collected from stray dogs in Belgrade (Serbia). Acta Vet. (2018) 68:257–68. doi: 10.2478/acve-2018-0023
19. Beall, MJ, Chandrashekar, R, Eberts, MD, Cyr, KE, Diniz, PP, Mainville, C, et al. Serological and molecular prevalence of Borrelia burgdorferi, Anaplasma phagocytophilum, and Ehrlichia species in dogs from Minnesota. Vector Borne Zoonotic Dis. (2008) 8:455–64. doi: 10.1089/vbz.2007.0236
20. Parola, P, Paddock, CD, Socolovschi, C, Labruna, MB, Mediannikov, O, Kernif, T, et al. Update on tick-borne rickettsioses around the world: a geographic approach. Clin Microbiol Rev. (2013) 26:657–702. doi: 10.1128/CMR.00032-13
21. Li, J, Zhang, C, Lu, M, Wang, Y, Liu, F, Wang, W, et al. Infection by a previously uncharacterized Ehrlichia species in rodents from Inner Mongolia, Northern China. Ticks Tick Borne Dis. (2023) 14:102116. doi: 10.1016/j.ttbdis.2022.102116
22. Choubdar, N, Karimian, F, Koosha, M, Nejati, J, and Oshaghi, MA. Hyalomma spp. ticks and associated Anaplasma spp. and Ehrlichia spp. on the Iran-Pakistan border. Parasit Vectors. (2021) 14:1–8. doi: 10.1186/s13071-021-04956-3
23. Nicholson, WL, Allen, KE, McQuiston, JH, Breitschwerdt, EB, and Little, SE. The increasing recognition of rickettsial pathogens in dogs and people. Trends Parasitol. (2010) 26:205–12. doi: 10.1016/j.pt.2010.01.007
24. Allen, KE, Li, Y, Kaltenboeck, B, Johnson, EM, Reichard, MV, Panciera, RJ, et al. Diversity of Hepatozoon species in naturally infected dogs in the southern United States. Vet Parasitol. (2008) 154:220–5. doi: 10.1016/j.vetpar.2008.03.027
25. Duron, O, Noël, V, McCoy, KD, Bonazzi, M, Sidi-Boumedine, K, Morel, O, et al. The recent evolution of a maternally-inherited endosymbiont of ticks led to the emergence of the Q fever pathogen, Coxiella burnetii. PLoS Pathog. (2015) 11:e1004892. doi: 10.1371/journal.ppat.1004892
26. Rahal, M, Medkour, H, Diarra, AZ, Bitam, I, Parola, P, and Mediannikov, O. Molecular identification and evaluation of Coxiella-like endosymbionts genetic diversity carried by cattle ticks in Algeria. Ticks Tick Borne Dis. (2020) 11:101493. doi: 10.1016/j.ttbdis.2020.101493
27. Rar, V, and Golovljova, I. Anaplasma, Ehrlichia, and “Candidatus Neoehrlichia” bacteria: pathogenicity, biodiversity, and molecular genetic characteristics, a review. Infect Genet Evol. (2011) 11:1842–61. doi: 10.1016/j.meegid.2011.09.019
28. Jongejan, F, and Uilenberg, G. The global importance of ticks. Parasitology. (2004) 129:S3–S14. doi: 10.1017/S0031182004005967
29. Ali, A, Shehla, S, Zahid, H, Ullah, F, Zeb, I, Ahmed, H, et al. Molecular survey and spatial distribution of Rickettsia spp. in ticks infesting free-ranging wild animals in Pakistan (2017–2021). Pathogens. (2022) 11:11162. doi: 10.3390/pathogens11020162
30. Laušević, D, Ilić, T, Nenadović, K, Bacić, D, and Obrenović, S. Seroprevalences of Rickettsia conorii, Ehrlichia canis and Coxiella burnetii in dogs from Montenegro. Acta Parasitol. (2019) 64:769–78. doi: 10.2478/s11686-019-00098-w
31. Otranto, D, Dantas-Torres, F, Mihalca, AD, Traub, RJ, Lappin, M, and Baneth, G. Zoonotic parasites of sheltered and stray dogs in the era of the global economic and political crisis. Trends Parasitol. (2017) 33:813–25. doi: 10.1016/j.pt.2017.05.013
32. Chan, EY, Sham, TS, Shahzada, TS, Dubois, C, Huang, Z, Liu, S, et al. Narrative review on health-edrm primary prevention measures for vector-borne diseases. Int J Environ Res Public Health. (2020) 17:5981. doi: 10.3390/ijerph17165981
33. Beck, AM. The ecology of stray dogs: A study of free-ranging urban animals. West Lafayette, Indiana: Purdue University Press (2002).
34. Walker, JB, Keirans, JE, and Horak, IG. The genus Rhipicephalus (Acari, Ixodidae): A guide to the brown ticks of the world. Cambridge: Cambridge University Press (2000).
35. Sambrook, J. Molecular cloning: a laboratory manual. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press (1989).
36. Hall, T, Biosciences, I, and Carlsbad, CJ. BioEdit: an important software for molecular biology. GERF Bull Biosci. (2011) 2:60–1.
37. Edgar, RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. (2004) 32:1792–7. doi: 10.1093/nar/gkh340
38. Kumar, S, Stecher, G, Li, M, Knyaz, C, and Tamura, K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. (2018) 35:1547–9. doi: 10.1093/molbev/msy096
39. Springer, A, Shuaib, YA, Isaa, MH, Ezz-Eldin, MI, Osman, AY, Yagoub, IA, et al. Tick fauna and associated Rickettsia, Theileria, and Babesia spp. in domestic animals in Sudan (North Kordofan and Kassala states). Microorganisms. (2020) 8:81969. doi: 10.3390/microorganisms8121969
40. Ali, A, Khan, MA, Zahid, H, Yaseen, PM, Qayash Khan, M, Nawab, J, et al. Seasonal dynamics, record of ticks infesting humans, wild and domestic animals and molecular phylogeny of Rhipicephalus microplus in Khyber Pakhtunkhwa Pakistan. Front Physiol. (2019) 10:793. doi: 10.3389/fphys.2019.00793
41. Ahmad, I, Ullah, S, Alouffi, A, Almutairi, MM, Khan, M, Numan, M, et al. Description of male, redescription of female, host record, and phylogenetic position of Haemaphysalis danieli. Pathogens. (2022) 11:1495. doi: 10.3390/pathogens11121495
42. Khan, M, Islam, N, Khan, A, Islam, ZU, Muñoz-Leal, S, Labruna, MB, et al. New records of Amblyomma gervaisi from Pakistan, with detection of a reptile-associated Borrelia sp. Ticks Tick Borne Dis. (2022) 13:102047. doi: 10.1016/j.ttbdis.2022.102047
43. Asgarali, Z, Pargass, I, Adam, J, Mutani, A, and Ezeokoli, C. Haematological parameters in stray dogs seropositive and seronegative to Ehrlichia canis in North Trinidad. Ticks Tick Borne Dis. (2012) 3:207–11. doi: 10.1016/j.ttbdis.2012.03.006
44. Muraro, LS, Souza, AD, Leite, TN, Cândido, SL, Melo, AL, Toma, HS, et al. First evidence of Ehrlichia minasensis infection in horses from Brazil. Pathogens. (2021) 10:265. doi: 10.3390/pathogens10030265
45. Hu, H, Liu, Z, Fu, R, Liu, Y, Ma, H, and Zheng, W. Detection and phylogenetic analysis of tick-borne bacterial and protozoan pathogens in a forest province of eastern China. Acta Trop. (2022) 235:106634. doi: 10.1016/j.actatropica.2022.106634
46. Almeida, AP, Souza, TD, Marcili, A, and Labruna, MB. Novel Ehrlichia and Hepatozoon agents infecting the crab-eating fox (Cerdocyon thous) in southeastern Brazil. J Med Entomol. (2013) 50:640–6. doi: 10.1603/ME12272
47. Moura de Aguiar, D, Pessoa Araújo Junior, J, Nakazato, L, Bard, E, Aguilar-Bultet, L, Vorimore, F, et al. Isolation and characterization of a novel pathogenic strain of Ehrlichia minasensis. Microorganisms. (2019) 7:528. doi: 10.3390/microorganisms7110528
48. Helm, CS, Von Samson-Himmelstjerna, G, Liesner, JM, Kohn, B, Müller, E, Schaper, R, et al. Identical 18S rRNA haplotypes of Hepatozoon canis in dogs and foxes in Brandenburg, Germany. Ticks Tick Borne Dis. (2020) 11:101520. doi: 10.1016/j.ttbdis.2020.101520
49. Machado-Ferreira, E, Dietrich, G, Hojgaard, A, Levin, M, Piesman, J, Zeidner, NS, et al. Coxiella symbionts in the Cayenne tick Amblyomma cajennense. Microb Ecol. (2011) 62:134–42. doi: 10.1007/s00248-011-9868-x
50. Khan, Z, Shehla, S, Alouffi, A, Kashif Obaid, M, Zeb Khan, A, Almutairi, MM, et al. Molecular survey and genetic characterization of Anaplasma marginale in ticks collected from livestock hosts in Pakistan. Animals. (2022) 12:121708. doi: 10.3390/ani12131708
51. Ali, A, Zahid, H, Zeb, I, Tufail, M, Khan, S, Haroon, M, et al. Risk factors associated with tick infestations on equids in Khyber Pakhtunkhwa, Pakistan, with notes on Rickettsia massiliae detection. Parasit Vectors. (2021) 14:363–2. doi: 10.1186/s13071-021-04836-w
52. Chitimia, L, Lin, RQ, Cosoroaba, I, Wu, XY, Song, HQ, Yuan, ZG, et al. Genetic characterization of ticks from southwestern Romania by sequences of mitochondrial cox 1 and nad 5 genes. Exp Appl Acarol. (2010) 52:305–11. doi: 10.1007/s10493-010-9365-9
53. Mangold, AJ, Bargues, MD, and Mas-Coma, S. Mitochondrial 16S rDNA sequences and phylogenetic relationships of species of Rhipicephalus and other tick genera among Metastriata (Acari: Ixodidae). Parasitol Res. (1998) 84:478–84. doi: 10.1007/s004360050433
54. Inokuma, H, Raoult, D, and Brouqui, P. Detection of Ehrlichia platys DNA in brown dog ticks (Rhipicephalus sanguineus) in Okinawa Island, Japan. J Clin Microbiol. (2000) 38:4219–21. doi: 10.1128/JCM.38.11.4219-4221.2000
55. Labruna, MB, Whitworth, T, Horta, MC, Bouyer, DH, McBride, JW, Pinter, A, et al. Rickettsia species infecting Amblyomma cooperi ticks from an area in the state of São Paulo, Brazil, where Brazilian spotted fever is endemic. J Clin Microbiol. (2004) 42:90–8. doi: 10.1128/JCM.42.1.90-98.2004
56. Roux, V, Fournier, PE, and Raoult, D. Differentiation of spotted fever group rickettsiae by sequencing and analysis of restriction fragment length polymorphism of PCR-amplified DNA of the gene encoding the protein rOmpA. J Clin Microbiol. (1996) 34:2058–65. doi: 10.1128/jcm.34.9.2058-2065.1996
Keywords: ticks, pathogens, stray dogs, butcher, one-health
Citation: Ali A, Ullah S, Numan M, Almutairi MM, Alouffi A and Tanaka T (2023) First report on tick-borne pathogens detected in ticks infesting stray dogs near butcher shops. Front. Vet. Sci. 10:1246871. doi: 10.3389/fvets.2023.1246871
Edited by:
Calin Mircea Gherman, University of Agricultural Sciences and Veterinary Medicine of Cluj-Napoca, RomaniaReviewed by:
Zsuzsa Kalmar, University of Agricultural Sciences and Veterinary Medicine of Cluj-Napoca, RomaniaFrans Kooyman, Utrecht University, Netherlands
Copyright © 2023 Ali, Ullah, Numan, Almutairi, Alouffi and Tanaka. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Abid Ali, dW9wX2FsaUB5YWhvby5jb20=; Tetsuya Tanaka, azYxOTk0MzFAa2FkYWkuanA=
 Abid Ali
Abid Ali Shafi Ullah
Shafi Ullah Muhammad Numan
Muhammad Numan Mashal M. Almutairi
Mashal M. Almutairi Abdulaziz Alouffi
Abdulaziz Alouffi Tetsuya Tanaka
Tetsuya Tanaka