
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Vet. Sci. , 18 July 2023
Sec. Animal Nutrition and Metabolism
Volume 10 - 2023 | https://doi.org/10.3389/fvets.2023.1231072
This article is part of the Research Topic Gut Microbiota: Allied with Livestock Nutrition, Health, And Welfare View all 19 articles
 Sriniwas Pandey1
†
Sriniwas Pandey1
† Eun Sol Kim1†
Eun Sol Kim1† Jin Ho Cho2†
Jin Ho Cho2† Minho Song3†
Minho Song3† Hyunok Doo1
Hyunok Doo1 Sheena Kim1
Sheena Kim1 Gi Beom Keum1
Gi Beom Keum1 Jinok Kwak1
Jinok Kwak1 Sumin Ryu1
Sumin Ryu1 Yejin Choi1
Yejin Choi1 Juyoun Kang1
Juyoun Kang1 Jeong Jae Lee4
Jeong Jae Lee4 Hyeun Bum Kim1*
Hyeun Bum Kim1*Non-digestible carbohydrates are an unavoidable component in a pig’s diet, as all plant-based feeds contain different kinds of non-digestible carbohydrates. The major types of non-digestible carbohydrates include non-starch polysaccharides (such as cellulose, pectin, and hemicellulose), resistant starch, and non-digestible oligosaccharides (such as fructo-oligosaccharide and xylo-oligosaccharide). Non-digestible carbohydrates play a significant role in balancing the gut microbial ecology and overall health of the swine by promoting the production of short chain fatty acids. Although non-digestible carbohydrates are rich in energy, swine cannot extract this energy on their own due to the absence of enzymes required for their degradation. Instead, they rely on gut microbes to utilize these carbohydrates for energy production. Despite the importance of non-digestible carbohydrate degradation, limited studies have been conducted on the swine gut microbes involved in this process. While next-generation high-throughput sequencing has aided in understanding the microbial compositions of the swine gut, specific information regarding the bacteria involved in non-digestible carbohydrate degradation remains limited. Therefore, it is crucial to investigate and comprehend the bacteria responsible for the breakdown of non-digestible carbohydrates in the gut. In this mini review, we have discussed the major bacteria involved in the fermentation of different types of non-digestible carbohydrates in the large intestine of swine, shedding light on their potential roles and contributions to swine nutrition and health.
Dietary carbohydrates (DCs) are principal substrates for maintaining physiological health and serve as an energy source for animals. In the diets of pigs, carbohydrates contribute to the majority of feed energy, accounting for approximately 60–70% of overall energy intake. In addition, the digestion of carbohydrates has significant impacts on various aspects of colonic functions, including the metabolism, balance of commensal flora and the health of large intestine epithelial cells (1).
DCs encompass a group of chemical substances and can be classified based on molecular sizes, ranging from simple mono- and disaccharides to complex compounds with intricate structures (2). Moreover, carbohydrates can be divided into two nutritional categories based on chemical classification. The first category is digestible carbohydrates (DGCs), which are metabolized by the host’s endogenous enzymes and absorbed in the small intestine. This category includes monosaccharides, disaccharides, and polysaccharides such as starches. The second category is dietary fiber, which consists of non-digestible soluble and insoluble carbohydrates and lignin. These components have the potential to be degraded through microbial fermentation in the large intestine (3). Non-starch polysaccharides (NSPs) are a component of dietary fiber. American Association of Cereal Chemists (AACC) defined the dietary fiber in 2000 as edible plant parts or comparable carbohydrates that are resistant to digestion and absorption in the small intestine but can be completely or partially fermented in the large intestine (4). Dietary fiber contains a substantial amount of energy, but the majority of enzymes required for its breakdown are not encoded in the mammalian genome.
Starch is the principal source of energy for monogastric animals and cereals are the primary source of starch in animal feeds (5). Starch is a polysaccharide composed of polymers amylose and amylopectin. Resistant starch (RS), on the other hand, refers to starches that resist digestion in the small intestine by amylases and instead reach the large intestine, where they are available for bacterial fermentation (6).
NSPs (cellulose, pectin, and hemicellulose), RS and non-digestible oligosaccharides (NDOs) (Fructo-oligosaccharide and Xylo-oligosaccharide) are the major types of non-digestible carbohydrates (NDCs). The large intestine serves as an anaerobic digestive environment for complex molecules, such as NDCs. It is predominantly colonized by obligate anaerobic microorganisms, although a small number of aerobic and facultative microorganisms are also present (7, 8). These microorganisms within the large intestine utilize NDCs as their primary energy sources. As a result, they produce Short Chain Fatty Acids (SCFAs), vitamins, and participate in various metabolic processes. Moreover, these microorganisms engage in intricate interactions with host cells and the host immune system (9–11).
With the aid of next-generation high-throughput sequencing, researchers have been able to comprehend the gut microbial composition of swine. However, knowledge concerning the gastrointestinal tract microbiome that facilitates the fermentation of NDCs remains limited, despite several research endeavors aimed at understanding such bacterial species. Therefore, this mini review aims to consolidate information regarding the major bacterial species involved in the fermentation of different types of NDCs in the large intestine of swine.
DGCs are such carbs that can be digested by host’s enzymatic system (3), whereas NDCs are those carbs that resist the action of salivary and intestinal digestive enzymes and hence are fermented by microbes in the large intestine (12).
The carbohydrates in the swine feed like cereal grains, legumes, oil seeds, and potato are also composed of DGCs and NDCs (13). The legumes and oil seeds are source of protein however the cell wall of these crops contains NDCs (14). NDCs have a significant role in pig diets, and it is necessary to add a certain amount to ensure proper physiological functioning (15).
As summarized in Figure 1A, NDCs are a distinct group of carbohydrates found almost entirely in plants and is generally classified as NSPs (cellulose, hemicellulose, pectin), RS (potato starch) and NDOs (Fructo-oligosaccharide, Xylan-oligosaccharide, soybean Oligosaccharide) (16).
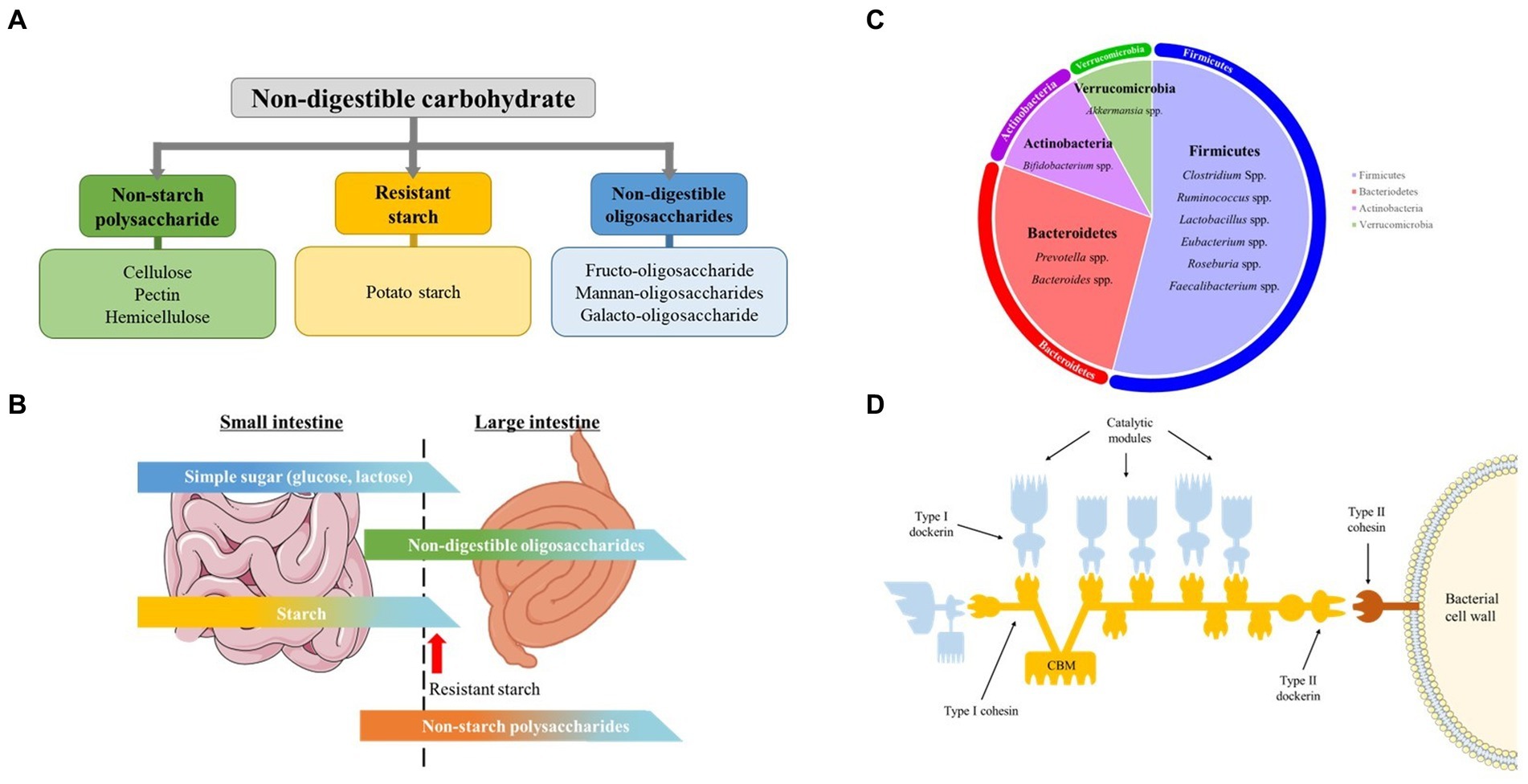
Figure 1. Non-digestible carbohydrates, pig gut microbiome, carbohydrate digestion in the intestine, and celluosome structure. (A) Classification and examples of non-digestible carbohydrate addressed in this mini review. (B) Schematic illustration of digestion of different carbohydrates in small and large Intestine. The figure gives a rough idea on the amount of carbohydrates digested in the small and large intestine. Modified from Bach Knudsen et al. (C) Predominant genus of swine gut microbiota. Each area of the circle signifies the domination of the respective phyla in the swine gut. (D) Cellulosome structure. The Type I dockerins, attached to the catalytic subunit (blue) interacts with the cohesin (yellow) of the primary CipA scaffoldin protein forming cellulosome complex. The cellulosome is attached to the bacterial surface through interaction of Type II dockerin in CipA with Type II cohesin module of a membrane-bound protein (red). The Cellulosome complex binds to cellulose through cellulose-binding module (CBM) of the CipA primary scaffoldin protein. Modified from Gilbert H.J.
NDCs are either water soluble or insoluble based on its solubility in water. Insoluble NDCs includes cellulose, hemicellulose, lignin, whereas soluble NDCs includes pectin, β-glucan, fructan, mucilage, gum, and psyllium fiber (17, 18). The most commonly present NDCs are cellulose, hemicellulose and pectic substances.
Digestion of simple carbs and starch occurs predominantly by enzymatic digestion, whereas the complex carbs that are resistant to host’s digestive enzymes are degraded by microbial fermentation after reaching large intestine (19).
Even though a host’s genome does not encode the enzymes required to break down the linkage between the monomers in NSPs and NDOs, 20 to 25% of NSPs and 40 to 95% of NDOs is found to be degraded while passing through the small intestine (Figure 1B). This breakdown is facilitated by the microbial enzymes of the microflora present in this part of the gut and not by the host’s enzymatic system (20). Nevertheless, the major types of carbs reach the large intestine, and those available for fermentation are plant cell wall polysaccharides, also known as NSPs, RS, and NDOs. Those carbohydrates are fermented by the swine gut microbiota.
Gastro-intestinal tract microbiota is defined as the ecological community made up of commensal, symbiotic and potentially pathogenic microorganisms that harbors the gut (17). The gut microbial composition of the swine is of great significance, as it affects the overall physiology and health, along with the feed conversion ratio. The swine gut microbiota are mainly made up of anaerobic & facultative anaerobic bacteria, and more than 90% of these bacteria belong to the phyla Firmicutes, Proteobacteria and Bacteroidetes (Figure 1C) (21–24). Several studies have shown ‘core’ genera consisting of Prevotella, Clostridium, Ruminococcus, Lactobacillus, Faecalibacterium, Bacteroides, Fusobacterium, and Alloprevotella in a larger portion of studied healthy pigs (25, 26). In this review, we will discuss major bacterial species involved in fermentation of different types of NDCs in the large intestine of swine.
NSPs comprise plant cell wall polysaccharides (Cellulose, hemicellulose, Pectin), structural non-polysaccharide (lignin) and non-structural polysaccharides. (7, 27). Numerous parameters, including the animal species, solubility, chemical composition, and consumption amount, influence the ease of digestion of NSPs. The order of microbial degradation in the large intestine is sugar residues = NDOs > Starch residues > Soluble NSP > RS=Insoluble NSP (3). Lignin, a component of plant cell wall, is however not digested by the enzymes of the small intestine and neither fermented by the gut bacteria. It is however supposed to impact the fermentability of other components in the diet (12, 28).
The degradation of complex fiber by the fibrolytic bacterial community is mainly carried out by several anaerobic gut microbes that possess the ability to produce enzymes. They belong to the dominant bacteria groups including Bacteroides, Roseburia, Ruminococcus or Bifidobacterium species (Table 1). Several specific as well as multi-carbohydrate degrading bacteria is found in swine gut. The anerobic bacteria mainly degrade cellulose through cell-bound organelle-like structure, cellulosome (46). Cellulosome is a large multi-enzyme complex bound to the bacterial cell wall, that helps degrade plant cell wall polysaccharides into usable sugars (47, 48). It basically consists of 2 major subunits: the enzymes and the noncatalytic subunit scaffoldin (Figure 1D). Scaffoldin possess 6 cohesin domains which bind with the dockerin module present in the enzymes and hence forms the functional cellulosomal-complex. Scaffoldin also has one another module, the cellulose-binding domain (CBM) that helps bind to the cellulosic substrates.
Ruminococcus flavefaciens (R. flavefaciens), one of the predominant and important cellulolytic bacteria, degrades cellulose and a variety of plant cell walls (30) using a high-molecular-weight enzyme complex, cellulosome (49). It is essential for the R. flavefaciens to bind to the cellulose in order to break it down. Another bacteria, Ruminococcus albus (R. albus), is also a well-known specialist cellulose-degrading bacterium found in rumen and gastro-intestinal tract of herbivores, but it has also been isolated from swine gut (8). It is also known to produce a cellulosome-like complex. However, it is supposed to employ another mechanism for adhesion to cellulose. It has been found that a cellulose-binding protein belonging to the Pil-protein family is involved in attachment (50). Ruminococcus champanellensis (R. champanellensis), a recently identified cellulolytic strain from human feces, ferments cellulose and xylase, while metabolizing cellobiose to acetate, succinate, ethanol, dihydrogen and small quantities of formate and lactate (31, 32).
Clostridium (C. leptum and C. herbivorans), which are normal inhabitants of the pig’s intestine, also possess multi-enzyme system including cellulosome and xylanosome, which aid in the breakdown of complex cellulosic polymers and several cellulosic by-products (29). However, it should be noted that not all polysaccharidase activities in R. flaveciens or the cellulolytic Clostrdia are linked to a cellulosome (51).
Bacteroides, another highly prevalent genus in swine gut, possess a high concentration of the carbohydrate-active enzyme (CAZyme) genes. These enzymes enable Bacteroides to degrade various components of plant cell wall, like glucronylxylans, xyloglucans and pectin (52). Moreover, a unique feature of Bacteroidetes is the presence of polysaccharide utilization locus (PUL), which consists of linked genes involved in the saccharification of complex carbohydrates, such as glycans (53, 54). Bacteroides ruminicola (B. ruminicola) and Bacteroides xylanisolvens (B. xylanisolvens) are significantly involved in the degradation of xylan (36, 37). Another species, Bacteroides succinogens (B. succinogens), isolated from the swine’s large intestine, also possesses the ability to degrade cellulose (34). Metagenome-assembled genomes (MAGs) have identified several members of the Bacteroidaceae family, including Bacteroides fragilis (B. fragilis), Bacteroides heparinolyticus (B. heparinolyticus), Bacteroides stercoris (B. stercoris), Bacteroides thetaiotaomicron (B. thetaiotaomicron), Bacteroides uniformis (B. uniformis), and Bacteroides xylanisolvens (B. xylanisolvens). These bacteria have been found to possess the carbohydrate-active enzyme (CAZyme) genes involved in the degradation of starch, pectin, fucose oligosaccharides, rhamnose oligosaccharides, and other complex carbohydrates. Additionally, using metagenome-assembled genomes (MAGs), it has been predicted that B. thetaiotaomicron and Bacteroides ovatus (B. ovatus) possess PULs.
Several species of Fibrobacter, including Fibrobacter intestinalis (F. intestinalis) and Fibrobacter succinogenes (F. succinogenes), possess the ability to ferment NSPs. These species, found in the caeca of pigs, have gained significant attention due to their relatively higher fibrolytic activity (33). F. succinogenes, in particular, is known for its efficient degradation of cellulose. The specific mechanism by which it utilizes cellulose is still not fully understood, but it has been proposed that F. succinogenes binds to cellulose through a protein present in its outer membrane, facilitating the degradation of cellulose (55, 56). Bacteroides, along with Prevotella, is known to contain carbohydrate-active enzymes (CAZymes) and play a major role in the degradation of pectin, a component of plant cell walls. However, it is important to note that while these bacterial genera contribute to the breakdown of various dietary carbohydrates, cellulose degradation is mainly carried out by other cellulolytic bacteria such as Ruminococcus, Fibrobacter, and certain species of Clostridium (38, 39). It is also widely known that Butyrivibrio spp., found in swine colon, can hydrolyze hemicellulose (8). Butyrivibrio fibrisolvens is a ruminal hemicellulose- degrading bacteria but also show modest cellulolytic activity (35).
Starch is the principal source of energy for monogastric animals, and cereals are the primary source of starch in animal feeds (5). Starch is a polysaccharide composed of polymers: amylose and amylopectin. RS, on the other hand, refers to starches that escape digestion in small intestine by the amylases and reach the large intestine, where they become available for bacterial fermentation (6).
Based on their physiochemical characteristics, resistant starch (RS) is classified into five different types. RS1 refers to starches that are physically inaccessible as they are located inside a fiber-protein matrix and are resistant to breakdown even with normal cooking. RS2 is a type of starch found in green bananas and raw potatoes, which can be reduced by thermal treatment. RS3 refers to retrograded starches that occur when starchy foods like bread or potatoes are gelatinized through heating and then undergo retrogradation upon cooling. RS4 is a group of resistant starches that are generated through chemical modifications such as esterification, etherification, and cross-linking. RS5 is predominantly associated with amylose-lipid V-type complexes, such as starch-monoglycerides and starch-fatty acids (57, 58). Given that starchy ingredients consumed by pigs typically contain significant quantities of RS1, RS2, and RS3, these three types of resistant starch are commonly regarded as the primary ones in the swine industry.
Several types of gut bacteria are involved in fermenting resistant starch in the hindgut, which leads to the production of SCFAs (such as acetate, butyrate, propionate, and valerate), gasses (including CO₂, H₂, and CH₄), as well as lesser amounts of organic acids (like lactate, succinate, and formate), branched SCFAs, and alcohols (such as methanol and ethanol) (57).
The main three phyla involved in starch fermentation are Firmicutes, Bacteroidetes, and Actinobacteria, which collectively account for 95% of the total gut bacteria in mammals. Numerous studies have indicated the interactions between resistant starch and microorganisms in the gut. It has been observed that as the levels of RS increase, there is an increase in the populations of the Bifidobacteria and Lactobacillus genera. Specifically, Lactobacillus sobrius and Lactobacillus amylovorus have been identified as the major amylolytic genera in the digestive system of swine. It is important to note that not all Bifidobacteria species are involved in the degradation of RS. However, Bifidobacterium breve, Bifidobacterium dentium, and Bifidobacterium pseudolongum have shown extracellular starch-degrading activities (59). A human study also evaluated the roles of four dominant amylolytic bacteria in the human colon, namely Bacteroides thetaiotaomicron (B. thetaiotaomicron), Ruminococcus bromii (R. bromii), Eubacterium rectale (E. rectale), and Bifidobacterium adolescentis (B. adolescentis), in the breakdown and utilization of RS. The findings of the study indicated that R. bromii exhibited a much greater capacity for RS degradation compared to the other three bacteria. However, all four bacteria demonstrated the ability to utilize RS (40, 60).
The starch-utilization system of B. thetaiotaomicron has been thoroughly investigated and found to possess a starch-utilization-structure (sus) gene clusters, which play a role in binding and utilization of starch (41, 61). On the other hand, the remarkable starch-degrading capacity of R. bromii is believed to be attributed to cohesion (Coh)-dockerin interactions, which are particularly significant in cellulosomal enzyme systems (62).
E. rectale depends on a large extracellular amylase attached to its cell wall, along with some membrane-associated binding proteins and hydrolases to use resistant starch, however it is not a primary degrader (63, 64) (Table 1).
NDOs are a type of oligosaccharides that cannot be broken down by mammalian endogenous enzymes (65). The terms “resistant oligosaccharides,” “NDOs” and “resistant short chain carbohydrates (RSCC)” are interchangeable and refer to the same compound. NDOs, such as fructo-oligosaccharides (FOS), transgalacto-oligosaccharides (TOS), xylo-oligosaccharides (XOS), and soybean oligosaccharides, occur naturally in legume seeds and cereals. They can also be artificially synthesized. These examples represent some common types of NDOs. Numerous studies have demonstrated that the presence of NDOs leads to an increase in Bifidobacterium species compared to other bacteria (42, 43). While Bifidobacterium shows a strong preference for fermenting NDOs, other bacteria such as Lactobacillus, Bacteroides, and Clostridium also have the ability to ferment NDOs, albeit at lower levels.
Lactic acid bacteria, such as Lactobacillus species can utilize simple carbohydrates broken down by other bacteria (66). However, their ability to utilize complex carbohydrates is generally limited, with only certain species such as Lactobacillus acidophilus and Lactobacillus plantarum having the capability to utilize NDOs (44, 45).
Bifidobacterium species are known for their production of glycolytic enzymes, which enable them to efficiently utilize NDOs. These enzymes allow Bifidobacterium strains to hydrolyze various monosaccharides and glycosidic linkages, providing them with a broader range of carbohydrate substrates. In contrast, other enteric bacteria such as Escherichia coli, Streptococcus, and Lactobacillus generally exhibit less diversified enzyme activities and lower levels of activity compared to Bifidobacterium (67) (Table 1).
NDCs play a crucial role in the overall health of pigs at all stages of life. When these carbohydrates undergo fermentation in the gut, they contribute to the production of SCFAs, gasses, and organic acids.
The anaerobic fermentation of complex carbohydrates in the large intestine primarily produces SCFAs, which are small organic monocarboxylic acids (68, 69). SCFAs have several beneficial effects on the host’s gut health. They contribute to the maintenance of intestinal barrier integrity, promoting a healthy gut lining and preventing the entry of harmful substances into the bloodstream. SCFAs also stimulate the production of mucus, which forms a protective layer in the gut and aids in the proper functioning of the digestive system. Furthermore, SCFAs have anti-inflammatory properties and help regulate the immune response in the gut, reducing the risk of inflammation-related conditions. Overall, the production of SCFAs through the fermentation of complex carbohydrates plays a crucial role in supporting gut health and protecting against intestinal inflammation (70). The major SCFAs produced in the gut are acetate, propionate, and butyrate (71). Among these, butyrate plays a crucial role as it serves as the primary energy source for colonocytes (72). Propionate, on the other hand, is transported to the liver where it contributes to gluconeogenesis. Acetate, being the most abundant SCFA, has various important functions including improving cholesterol metabolism and lipogenesis, regulating the immune system, and exhibiting anti-inflammatory responses (73–75). SCFAs cause a decrease in the pH of the large intestine, which helps inhibit the growth of pathogenic microorganisms and facilitates the absorption of calcium and magnesium (71). Furthermore, SCFAs can function as signaling molecules by binding to G protein-coupled receptors (GPCRs) present in gut epithelial cells and immune cells (76). In both anaerobic ecosystems and the alimentary canal, an inevitable byproduct of microbial fermentation is gas, with representative gasses being H₂ and CO₂.
In addition to the energy production through fermentation of NDCs, they also play a crucial role in maintaining gut health by producing a diverse range of metabolites, such as SCFAs. Gut microbes contribute to host health through SCFA production, lowering pH, and synthesizing vitamins.
The specific gut anaerobes involved in the breakdown of complex carbohydrates in the large intestine are still not fully understood. Advancing our knowledge in this area is crucial to better understand the microbial ecosystem and its impact on swine health. Prioritizing research efforts to identify the bacterial species responsible for fermenting various dietary fibers will not only contribute to maintaining overall health but also promote optimal growth and well-being of swine. This knowledge can aid in developing targeted interventions and strategies to optimize gut health and maximize the benefits of complex carbohydrate fermentation in swine. Identifying and understanding the functional roles of different bacterial species involved in the fermentation of dietary fibers can greatly enhance swine production. By optimizing diets and developing targeted interventions based on this knowledge, we can promote efficient fermentation and maximize the utilization of dietary fibers by beneficial gut bacteria. This can lead to improved nutrient absorption, enhanced gut health, and ultimately, better swine production outcomes. Additionally, such advancements can contribute to more sustainable and efficient farming practices by reducing feed waste and improving the overall utilization of available resources.
JC, MS, and HK: conceptualization. SP, EK, HD, SK, GK, JiK, SR, YC, JuK, and JL: resources. HK: supervision. SP, EK, and HK: writing – original draft. JC, MS, and HK: writing – review & editing. All authors contributed to the article and approved the submitted version.
This work was supported by National Research Foundation of Korea (the Ministry of Education: 2021R1I1A3059910 and the Ministry of Science & ICT: 2019M3A9F3065227).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
DCs, Dietary carbohydrates; DGCs, Digestible carbohydrates; NSPs, Non-starch polysaccharides; RS, Resistant starch; NDOs, Non-digestible oligosaccharides; NDCs, Non-digestible carbohydrates; SCFAs, Short Chain Fatty Acids.
1. Knudsen, KEB, Hedemann, MS, and Laerke, HN. The role of carbohydrates in intestinal health of pigs. Anim Feed Sci Tech. (2012) 173:41–53. doi: 10.1016/j.anifeedsci.2011.12.020
2. Cummings, JH, and Stephen, AM. Carbohydrate terminology and classification. Eur J Clin Nutr. (2007) 61:S5–S18. doi: 10.1038/sj.ejcn.1602936
3. Erik, K, Knudsen, B, Lærke, HN, and Jørgensen, H. Carbohydrates and carbohydrate utilization in swine In: LI Chiba, editor. Sustainable Swine Nutrition. Hoboken, NJ: John Wiley & Sons (2013). 109–37.
4. Devries, J, Camire, M, Cho, S, Craig, S, Gordon, D, Jones, JM, et al. The definition of dietary fiber. Cereal food world (2001) 46:112–29.
5. Stevnebo, A, Sahlstrom, S, and Svihus, B. Starch structure and degree of starch hydrolysis of small and large starch granules from barley varieties with varying amylose content. Anim Feed Sci Tech. (2006) 130:23–38. doi: 10.1016/j.anifeedsci.2006.01.015
6. Walsh, SK, Lucey, A, Walter, J, Zannini, E, and Arendt, EK. Resistant starch-an accessible fiber ingredient acceptable to the Western palate. Compr Rev Food Sci F. (2022) 21:2930–55. doi: 10.1111/1541-4337.12955
7. Chassard, C, and Lacroix, C. Carbohydrates and the human gut microbiota. Curr Opin Clin Nutr. (2013) 16:453–60. doi: 10.1097/MCO.0b013e3283619e63
8. Varel, VH, and Yen, JT. Microbial perspective on fiber utilization by swine. J Anim Sci. (1997) 75:2715–22. doi: 10.2527/1997.75102715x
9. Rowland, I, Gibson, G, Heinken, A, Scott, K, Swann, J, Thiele, I, et al. Gut microbiota functions: metabolism of nutrients and other food components. Eur J Nutr. (2018) 57:1–24. doi: 10.1007/s00394-017-1445-8
10. Flint, HJ, Duncan, SH, Scott, KP, and Louis, P. Interactions and competition within the microbial community of the human colon: links between diet and health. Environ Microbiol. (2007) 9:1101–11. doi: 10.1111/j.1462-2920.2007.01281.x
11. Hooper, LV, Wong, MH, Thelin, A, Hansson, L, Falk, PG, and Gordon, JI. Molecular analysis of commensal host-microbial relationships in the intestine. Science. (2001) 291:881–4. doi: 10.1126/science.291.5505.881
12. Chanmuang, S, Nguyen, QA, and Kim, HJ. Current research on the effects of non-digestible carbohydrates on metabolic disease. Appl Sci-Basel. (2022) 12:3768. doi: 10.3390/app12083768
13. Navarro, D, Abelilla, JJ, and Stein, HH. Structures and characteristics of carbohydrates in diets fed to pigs: a review. J Anim Sci Biotechnol. (2019) 10:39. doi: 10.1186/s40104-019-0345-6
14. Knudsen, KE. Fiber and nonstarch polysaccharide content and variation in common crops used in broiler diets. Poult Sci. (2014) 93:2380–93. doi: 10.3382/ps.2014-03902
15. Wenk, C. The role of dietary fibre in the digestive physiology of the pig. Anim Feed Sci Tech. (2001) 90:21–33. doi: 10.1016/S0377-8401(01)00194-8
16. Jha, SK, Singh, HR, and Prakash, P. Chapter 1 - dietary Fiber and human health: an introduction In: RA Samaan, editor. Dietary Fiber for the Prevention of Cardiovascular Disease. Cambridge, MA: Academic Press (2017). 1–22.
17. Kalyani Nair, K, Kharb, S, and Thompkinson, DK. Inulin dietary Fiber with functional and health attributes—a review. Food Rev Intl. (2010) 26:189–203. doi: 10.1080/87559121003590664
18. Mudgil, D, and Barak, S. Composition, properties and health benefits of indigestible carbohydrate polymers as dietary fiber: a review. Int J Biol Macromol. (2013) 61:1–6. doi: 10.1016/j.ijbiomac.2013.06.044
19. Drochner, W. Digestion of carbohydrates in the pig. EAAP Public. (1991) 54:367–88. doi: 10.1080/17450399309386027
20. Knudsen, KEB, Laerke, HN, Ingerslev, AK, Hedemann, MS, Nielsen, TS, and Theil, PK. Carbohydrates in pig nutrition - recent advances. J Anim Sci. (2016) 94:1–11. doi: 10.2527/jas.2015-9785
21. Isaacson, R, and Kim, HB. The intestinal microbiome of the pig. Anim Health Res Rev. (2012) 13:100–9. doi: 10.1017/S1466252312000084
22. Ma, J, Chen, J, Gan, M, Chen, L, Zhao, Y, Zhu, Y, et al. Gut microbiota composition and diversity in different commercial swine breeds in early and finishing growth stages. Animals (Basel). (2022) 12:1607. doi: 10.3390/ani12131607
23. Kim, HB, and Isaacson, RE. The pig gut microbial diversity: understanding the pig gut microbial ecology through the next generation high throughput sequencing. Vet Microbiol. (2015) 177:242–51. doi: 10.1016/j.vetmic.2015.03.014
24. Kim, HB, Borewicz, K, White, BA, Singer, RS, Sreevatsan, S, Tu, ZJ, et al. Longitudinal investigation of the age-related bacterial diversity in the feces of commercial pigs. Vet Microbiol. (2011) 153:124–33. doi: 10.1016/j.vetmic.2011.05.021
25. Luo, YH, Ren, W, Smidt, H, Wright, ADG, Yu, B, Schyns, G, et al. Dynamic distribution of gut microbiota in pigs at different growth stages: composition and contribution. Microbiol Spectr. (2022) 10:e0068821. doi: 10.1128/spectrum.00688-21
26. Holman, DB, Brunelle, BW, Trachsel, J, and Allen, HK. Meta-analysis to define a Core microbiota in the swine gut. Msystems. (2017) 2:e00004–17. doi: 10.1128/mSystems.00004-17
27. Selvendran, RR. The plant cell wall as a source of dietary fiber: chemistry and structure. Am J Clin Nutr. (1984) 39:320–37. doi: 10.1093/ajcn/39.2.320
28. Metzler, BU, and Mosenthin, R. A review of interactions between dietary fiber and the gastrointestinal microbiota and their consequences on intestinal phosphorus metabolism in growing pigs. Asian Austral J Anim. (2008) 21:603–15. doi: 10.5713/ajas.2008.r.03
29. Thomas, L, Joseph, A, and Gottumukkala, LD. Xylanase and cellulase systems of clostridium sp.: an insight on molecular approaches for strain improvement. Bioresour Technol. (2014) 158:343–50. doi: 10.1016/j.biortech.2014.01.140
30. Krause, DO, Bunch, RJ, Smith, WJM, and McSweeney, CS. Diversity of Ruminococcus strains: a survey of genetic polymorphisms and plant digestibility. J Appl Microbiol. (1999) 86:487–95. doi: 10.1046/j.1365-2672.1999.00688.x
31. Chassard, C, Delmas, E, Robert, C, Lawson, PA, and Bernalier-Donadille, A. Ruminococcus champanellensis sp nov., a cellulose-degrading bacterium from human gut microbiota. Int J Syst Evol Micr. (2012) 62:138–43. doi: 10.1099/ijs.0.027375-0
32. Froidurot, A, and Julliand, V. Cellulolytic bacteria in the large intestine of mammals. Gut Microbes. (2022) 14:2031694. doi: 10.1080/19490976.2022.2031694
33. Miron, J, and Ben-Ghedalia, D. Digestion of cell-wall monosaccharides of ryegrass and alfalfa hays by the ruminal bacteria Fibrobacter succinogenes and Butyrivibrio fibrisolvens. Can J Microbiol. (1993) 39:780–6. doi: 10.1139/m93-115
34. Varel, VH, Fryda, SJ, and Robinson, IM. Cellulolytic bacteria from pig large intestine. Appl Environ Microbiol. (1984) 47:219–21. doi: 10.1128/aem.47.1.219-221.1984
36. Chassard, C, Delmas, E, Lawson, PA, and Bernalier-Donadille, A. Bacteroides xylanisolvens sp. nov., a xylan-degrading bacterium isolated from human faeces. Int J Syst Evol Microbiol. (2008) 58:1008–13. doi: 10.1099/ijs.0.65504-0
37. Varel, VH, Robinson, IM, and Jung, HJ. Influence of dietary fiber on xylanolytic and cellulolytic bacteria of adult pigs. Appl Environ Microbiol. (1987) 53:22–6. doi: 10.1128/aem.53.1.22-26.1987
38. Martens, EC, Lowe, EC, Chiang, H, Pudlo, NA, Wu, M, McNulty, NP, et al. Recognition and degradation of plant Cell Wall polysaccharides by two human gut symbionts. PLoS Biol. (2011) 9:e1001221. doi: 10.1371/journal.pbio.1001221
39. Flint, HJ, Scott, KP, Duncan, SH, Louis, P, and Forano, E. Microbial degradation of complex carbohydrates in the gut. Gut Microbes. (2012) 3:289–306. doi: 10.4161/gmic.19897
40. Ze, XL, Duncan, SH, Louis, P, and Flint, HJ. Ruminococcus bromii is a keystone species for the degradation of resistant starch in the human colon. ISME J. (2012) 6:1535–43. doi: 10.1038/ismej.2012.4
41. Reeves, AR, Wang, GR, and Salyers, AA. Characterization of four outer membrane proteins that play a role in utilization of starch by Bacteroides thetaiotaomicron. J Bacteriol. (1997) 179:643–9. doi: 10.1128/jb.179.3.643-649.1997
42. Gibson, GR, Beatty, ER, Wang, X, and Cummings, JH. Selective stimulation of bifidobacteria in the human colon by oligofructose and inulin. Gastroenterology. (1995) 108:975–82. doi: 10.1016/0016-5085(95)90192-2
43. Tanaka, R, Takayama, H, Morotomi, M, Kuroshima, T, Ueyama, S, Matsumoto, K, et al. Effects of administration of TOS and Bifidobacterium breve 4006 on the human Fecal Flora. Bifidobacteria Microflora. (1983) 2:17–24. doi: 10.12938/bifidus1982.2.1_17
44. Barrangou, R, Azcarate-Peril, MA, Duong, T, Conners, SB, Kelly, RM, and Klaenhammer, TR. Global analysis of carbohydrate utilization by Lactobacillus acidophilus using cDNA microarrays. Proc Natl Acad Sci U S A. (2006) 103:3816–21. doi: 10.1073/pnas.0511287103
45. Saulnier, DAA, Molenaar, D, de Vos, WA, Gibson, GR, and Kolida, S. Identification of prebiotic fructooligosaccharide metabolism in Lactobacillus plantarum WCFS1 through microarrays. Appl Environ Microb. (2007) 73:1753–65. doi: 10.1128/AEM.01151-06
46. Poulsen, HV, Willink, FW, and Ingvorsen, K. Aerobic and anaerobic cellulase production by Cellulomonas uda. Arch Microbiol. (2016) 198:725–35. doi: 10.1007/s00203-016-1230-8
47. Bayer, EA, Shimon, LJ, Shoham, Y, and Lamed, R. Cellulosomes-structure and ultrastructure. J Struct Biol. (1998) 124:221–34. doi: 10.1006/jsbi.1998.4065
48. Himmel, ME, Xu, Q, Luo, Y, Ding, S-Y, Lamed, R, and Bayer, EA. Microbial enzyme systems for biomass conversion: emerging paradigms. Biofuels. (2010) 1:323–41. doi: 10.4155/bfs.09.25
49. Rincon, MT, Cepeljnik, T, Martin, JC, Barak, Y, Lamed, R, Bayer, EA, et al. A novel cell surface-anchored cellulose-binding protein encoded by the sca gene cluster of Ruminococcus flavefaciens. J Bacteriol. (2007) 189:4774–83. doi: 10.1128/JB.00143-07
50. Morrison, M, and Miron, J. Adhesion to cellulose by Ruminococcus albus: a combination of cellulosomes and Pil-proteins? FEMS Microbiol Lett. (2000) 185:109–15. doi: 10.1111/j.1574-6968.2000.tb09047.x
51. Flint, HJ, Bayer, EA, Rincon, MT, Lamed, R, and White, BA. Polysaccharide utilization by gut bacteria: potential for new insights from genomic analysis. Nat Rev Microbiol. (2008) 6:121–31. doi: 10.1038/nrmicro1817
52. Liu, GS, Li, PH, Hou, LM, Niu, Q, Pu, G, Wang, BB, et al. Metagenomic analysis reveals new microbiota related to Fiber digestion in pigs. Front Microbiol. (2021) 12:12. doi: 10.3389/fmicb.2021.746717
53. Grondin, JM, Tamura, K, Dejean, G, Abbott, DW, and Brumer, H. Polysaccharide Utilization Loci: Fueling Microbial Communities. J Bacteriol. (2017) 199:e00860-16. doi: 10.1128/JB.00860-16
54. Terrapon, N, Lombard, V, Gilbert, HJ, and Henrissat, B. Automatic prediction of polysaccharide utilization loci in Bacteroidetes species. Bioinformatics. (2015) 31:647–55. doi: 10.1093/bioinformatics/btu716
55. Raut, MP, Karunakaran, E, Mukherjee, J, Biggs, CA, and Wright, PC. Influence of substrates on the surface characteristics and membrane proteome of Fibrobacter succinogenes S85. PLoS One. (2015) 10:e0141197. doi: 10.1371/journal.pone.0141197
56. Jun, HS, Qi, M, Gong, J, Egbosimba, EE, and Forsberg, CW. Outer membrane proteins of Fibrobacter succinogenes with potential roles in adhesion to cellulose and in cellulose digestion. J Bacteriol. (2007) 189:6806–15. doi: 10.1128/JB.00560-07
57. Birt, DF, Boylston, T, Hendrich, S, Jane, JL, Hollis, J, Li, L, et al. Resistant starch: promise for improving human health. Adv Nutr. (2013) 4:587–601. doi: 10.3945/an.113.004325
58. Gutierrez, TJ, and Tovar, J. Update of the concept of type 5 resistant starch (RS5): self-assembled starch V-type complexes. Trends Food Sci Tech. (2021) 109:711–24. doi: 10.1016/j.tifs.2021.01.078
59. Giuberti, G, Gallo, A, Moschini, M, and Masoero, F. New insight into the role of resistant starch in pig nutrition. Anim Feed Sci Tech. (2015) 201:1–13. doi: 10.1016/j.anifeedsci.2015.01.004
60. Macfarlane, GT, and Englyst, HN. Starch utilization by the human large intestinal microflora. J Appl Bacteriol. (1986) 60:195–201. doi: 10.1111/j.1365-2672.1986.tb01073.x
61. Reeves, AR, D'Elia, JN, Frias, J, and Salyers, AA. A Bacteroides thetaiotaomicron outer membrane protein that is essential for utilization of maltooligosaccharides and starch. J Bacteriol. (1996) 178:823–30. doi: 10.1128/jb.178.3.823-830.1996
62. Ze, XL, Ben David, Y, Laverde-Gomez, JA, Dassa, B, Sheridan, PO, Duncan, SH, et al. Unique Organization of Extracellular Amylases into Amylosomes in the resistant starch-utilizing human colonic firmicutes bacterium Ruminococcus bromii. MBio. (2015) 6:e01058–15. doi: 10.1128/mBio.01058-15
63. Cockburn, DW, Orlovsky, NI, Foley, MH, Kwiatkowski, KJ, Bahr, CM, Maynard, M, et al. Molecular details of a starch utilization pathway in the human gut symbiont Eubacterium rectale. Mol Microbiol. (2015) 95:209–30. doi: 10.1111/mmi.12859
64. Ramsay, AG, Scott, KP, Martin, JC, Rincon, MT, and Flint, HJ. Cell-associated alpha-amylases of butyrate-producing firmicute bacteria from the human colon. Microbiology (Reading). (2006) 152:3281–90. doi: 10.1099/mic.0.29233-0
65. Englyst, KN, Liu, S, and Englyst, HN. Nutritional characterization and measurement of dietary carbohydrates. Eur J Clin Nutr. (2007) 61:S19–39. doi: 10.1038/sj.ejcn.1602937
66. Walter, J. Ecological role of lactobacilli in the gastrointestinal tract: implications for fundamental and biomedical research. Appl Environ Microb. (2008) 74:4985–96. doi: 10.1128/AEM.00753-08
67. Sako, T, Matsumoto, K, and Tanaka, R. Recent progress on research and applications of non-digestible galacto-oligosaccharides. Int Dairy J. (1999) 9:69–80. doi: 10.1016/S0958-6946(99)00046-1
68. Louis, P, and Flint, HJ. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol Lett. (2009) 294:1–8. doi: 10.1111/j.1574-6968.2009.01514.x
69. Miller, TL, and Wolin, MJ. Pathways of acetate, propionate, and butyrate formation by the human fecal microbial flora. Appl Environ Microbiol. (1996) 62:1589–92. doi: 10.1128/aem.62.5.1589-1592.1996
70. Silva, YP, Bernardi, A, and Frozza, RL. The role of short-chain fatty acids from gut microbiota in gut-brain communication. Front Endocrinol. (2020) 11:11. doi: 10.3389/fendo.2020.00025
71. Topping, DL, and Clifton, PM. Short-chain fatty acids and human colonic function: roles of resistant starch and nonstarch polysaccharides. Physiol Rev. (2001) 81:1031–64. doi: 10.1152/physrev.2001.81.3.1031
72. Gardiner, GE, Metzler-Zebeli, BU, and Lawlor, PG. Impact of intestinal microbiota on growth and feed efficiency in pigs. Review Microorganisms. (2020) 8:1886. doi: 10.3390/microorganisms8121886
73. Duncan, SH, Holtrop, G, Lobley, GE, Calder, AG, Stewart, CS, and Flint, HJ. Contribution of acetate to butyrate formation by human faecal bacteria. Br J Nutr. (2004) 91:915–23. doi: 10.1079/BJN20041150
74. De Vadder, F, Kovatcheva-Datchary, P, Goncalves, D, Vinera, J, Zitoun, C, Duchampt, A, et al. Microbiota-generated metabolites promote metabolic benefits via gut-brain neural circuits. Cells. (2014) 156:84–96. doi: 10.1016/j.cell.2013.12.016
75. Nogal, A, Louca, P, Zhang, X, Wells, PM, Steves, CJ, Spector, TD, et al. Circulating levels of the short-chain fatty acid acetate mediate the effect of the gut microbiome on visceral fat. Front Microbiol. (2021) 12:711359. doi: 10.3389/fmicb.2021.711359
Keywords: pigs, non-digestible carbohydrates, bacteria, fermentation, nutrition
Citation: Pandey S, Kim ES, Cho JH, Song M, Doo H, Kim S, Keum GB, Kwak J, Ryu S, Choi Y, Kang J, Lee JJ and Kim HB (2023) Swine gut microbiome associated with non-digestible carbohydrate utilization. Front. Vet. Sci. 10:1231072. doi: 10.3389/fvets.2023.1231072
Received: 30 May 2023; Accepted: 04 July 2023;
Published: 18 July 2023.
Edited by:
Wen-Chao Liu, Guangdong Ocean University, ChinaReviewed by:
Muhammad Akbar Shahid, Bahauddin Zakariya University, PakistanCopyright © 2023 Pandey, Kim, Cho, Song, Doo, Kim, Keum, Kwak, Ryu, Choi, Kang, Lee and Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hyeun Bum Kim, aGJraW1AZGFua29vay5hYy5rcg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.