
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Vet. Sci. , 15 June 2023
Sec. Veterinary clinical, anatomical, and comparative pathology
Volume 10 - 2023 | https://doi.org/10.3389/fvets.2023.1213150
 Anna Cutarelli1†
Anna Cutarelli1† Francesca De Falco2†
Francesca De Falco2† Bianca Cuccaro2
Bianca Cuccaro2 Vesna Milićević3
Vesna Milićević3 Branislav Kureljušić3
Branislav Kureljušić3 Jovan Bojkovski4
Jovan Bojkovski4 Pellegrino Cerino5
Pellegrino Cerino5 Antonella Perillo6
Antonella Perillo6 Raluca Marica7
Raluca Marica7 Cornel Catoi7
Cornel Catoi7 Sante Roperto2*
Sante Roperto2*Caprine papillomaviruses (ChPVs, Capra hircus papillomaviruses) were detected and quantified for the first time using droplet digital polymerase chain reaction (ddPCR) in blood samples of 374 clinically healthy goats from farms located in Italy, Romania, and Serbia. Overall, ddPCR revealed ChPV DNA in 78 of the 374 examined samples, indicating that ~21% of the goats harbored circulating papillomavirus DNA. In particular, in Italian goat farms, ChPV genotypes were detected and quantified in 58 of 157 blood samples (~37%), 11 of 117 samples from Serbian farms (~9.4%), and 9 of 100 from Romanian blood samples (9%). Blood samples from Italian goat farms showed a high prevalence of ChPV1, which was detected in 45 samples (28.6%). The ChPV2 genotype was detected in 13 samples (~8.3%). Therefore, significant differences in prevalence and genotype distributions were observed. On Serbian and Romanian farms, no significant differences were observed in the genotype prevalence of ChPVs. Molecular findings are consistent with ChPV prevalence, characterized by a territorial distribution similar to that of papillomaviruses in other mammalian species. Furthermore, this study showed that ddPCR is a very sensitive and accurate assay for ChPV detection and quantification. The ddPCR may be the molecular diagnostic tool of choice, ultimately providing useful insights into the molecular epidemiology and field surveillance of ChPV.
Papillomaviruses are a widespread family of pathogens that infect several mammalian and non-mammalian species (1). Overall, more than 400 papillomaviruses have been identified and sequenced including 50 genotypes of domestic ruminants (2). Papillomavirus infections have been observed in both large and small ruminants worldwide, and often have significant effects on livestock production (3). To date, 44 bovine papillomaviruses and four ovine papillomaviruses are known to cause infections in cattle and sheep, respectively (2). Bovine delta-papillomaviruses are highly pathogenic and are believed to be high-risk PV genotypes (4). Recent molecular surveys in large and small ruminants have been shown that there is a high prevalence of bovine and ovine papillomavirus infections in healthy cattle and sheep, respectively (5, 6). However, the extent of the caprine papillomavirus infection in goats remains unknown. Only two caprine papillomaviruses, Capra hircus papillomavirus 1 (ChPV1), which belongs to the genus Phipapillomavirus, and ChPV2, which are currently unclassified, are known to occur in domestic goats (2). ChPV1 was isolated from the skin of a healthy 7-year-old goat (7), its genome of ChPV2 was obtained and characterized from teat lesions of a Damascus goat, and it appears to be closely related to Xipapillomavirus1 species infecting cattle (8, 9).
Papillomavirus infection has been sporadically reported in goats (10), thereby very few cases of diseases associated with papillomavirus infection in goats have been documented. Although an infective agent has been proposed to be involved in goat papillomatosis of the mammary skin (11), the first papillomavirus-like sequences were reported in an outbreak of cutaneous fibropapillomas of the udder in Saanen goats (12). More recently, papillomavirus infection has been suggested to be a cause of caprine ocular squamous cell carcinoma (13). An immunohistochemical study performed on three goats with cutaneous papillomatosis suggested that benign neoplasia may be caused by papillomavirus infection (14).
This study aimed to investigate the prevalence and genotype distribution of ChPVs in whole blood of healthy goats from farms located in Italy, Romania, and Serbia, and to evaluate viral detection and load quantification using the droplet digital polymerase chain reaction (ddPCR) tool.
Blood samples from 374 apparently healthy 1- to 3-year-old goats (157 samples were obtained from goats living in Italy, 117 from Serbia, and 100 from Romania) were collected from the jugular vein in vacutainers containing ethylenediaminetetraacetic acid at public and private slaughterhouses after permission of medical authorities. Furthermore, the official veterinarians responsible for the health conditions of the flocks, where the goats belonged, provided us with the medical records of the animals showing that the examined goats did not have any diseases or therapeutic treatment. Total DNA was extracted using a DNeasy Blood & Tissue Kit (Qiagen, Wilmington, DE, USA) according to the manufacturer's instructions.
Positive controls were artificially created by inserting 270 and 400 base pairs of the ChPV1 and ChPV2 E7 sequences, respectively, into a plasmid (vector: pUCIDT-AMP) (IDT, Integrated DNA Technologies, IA, USA).
DNA extracted from 374 blood samples of healthy goats was analyzed using ddPCR. For ddPCR, the Bio-Rad QX100 ddPCR System was used according to the manufacturer's instructions as previously reported (15). The primer and probe sequences used to identify ChPV1 and ChPV2 are listed in Table 1. Finally, the results obtained by the ddPCR software QuantaSoft were converted directly into copy number/μL, multiplying the amount obtained by the instrument by 20 μL (the total volume of the reaction mixture) and then divided by 7 μL, that is, the volume of the DNA sample added at the beginning of the test. Each sample was analyzed in duplicate, and the samples were considered ChPV-positive if at least three droplets contained ChPV amplicons, as suggested for other papillomavirus infections (16, 17).
McNemar's test for two Related Binomial Proportions was used to evaluate the agreement between two tests performed on the same animals. The agreement between the two tests performed on different groups of animals from Italy, Romania, and Serbia (Italy vs. Romania and Italy vs. Serbia) was investigated using the Fisher's test. Statistical significance was set at P < 0.05. Statistical analyses were performed using R Studio.
Overall, ddPCR revealed ChPV DNA in 78 of the 374 examined blood samples, indicating that ~21% of the goats harbored circulating papillomavirus DNA (Supplementary Table 1). Rain plots of ddPCR for ChPV DNA are showed in Supplementary Figure 1. In particular, in Italian goat farms, ChPV genotypes were detected and quantified in 58 of 157 blood samples (~37%), 11 of 117 samples from Serbian farms (~9.4%), and 9 of 100 from Romanian blood samples (9%). The results are shown in Figure 1. Blood samples from Italian goat farms showed a high prevalence of ChPV1, which was detected in 45 samples (28.6%). The ChPV2 genotype was detected in 13 samples (~8.3%). McNemar's test showed significant differences in the prevalence of the two caprine genotypes (P < 0.05). In 117 samples from Serbian goat farms, eight samples were positive for ChPV1 and three for ChPV2, which were 6.8 and 2.6%, respectively. ChPV1 was observed in six of the 100 (6%) samples from Romania, whereas ChPV2 was detected in only three (3%) samples. On both Serbian and Romanian farms, no significant differences were observed in the genotype prevalence of ChPVs. Significant differences in both prevalence and genotype distribution were observed between Italian farms and Serbian and Romanian goat farms. Figure 2 shows the results for all the goat farms.
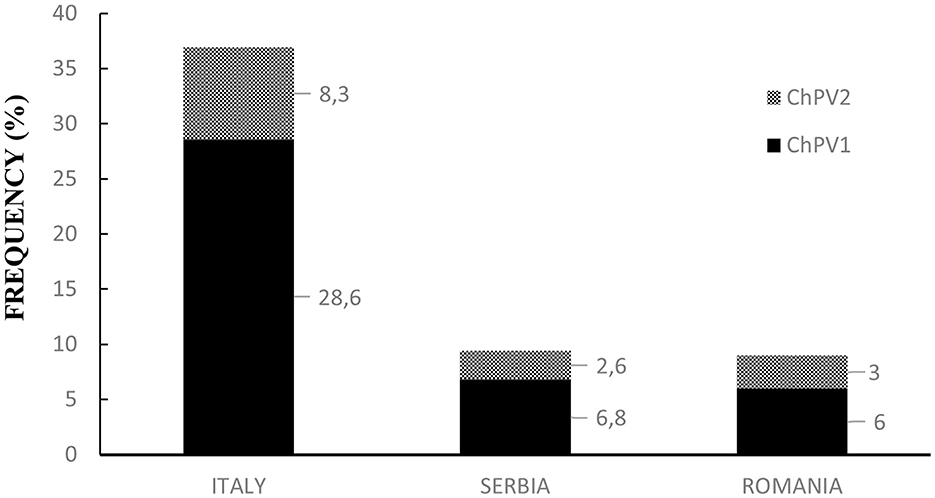
Figure 2. Prevalence and genotype distribution of ChPVs in Italian, Serbian, and Romanian goat farms.
Only isolated and sporadic ChPV DNA associated with neoplastic lesions has been reported in goats associated with neoplastic lesions (10). Therefore, the ecological epidemiology of ChPV genotypes remains unknown. To the best of our knowledge, this is the first systematic survey of the prevalence and genotype distribution of ChPVs among healthy goats from several farms in three European countries using ddPCR as a diagnostic assay, which has not yet been utilized for studying ChPV epidemiology. The current study shows that ddPCR is an advanced technology that can accurately diagnose ChPV infection with high specificity and sensitivity, thus representing a promising new tool for the accurate detection and quantification of ChPV nucleic acid load. This diagnostic approach quantifies a very small amount (<1 copy number/μL) of ChPV DNA, thus making it an important diagnostic procedure capable of detecting otherwise undetectable viral DNA 500 times (maximum) more sensitive than qPCR for low-level analyte (18). Notably, ddPCR has been shown to be the most accurate and sensitive method for quantifying the nucleic acids of bovine and ovine papillomaviruses in cattle and sheep, respectively (5, 6, 19).
On Italian goat farms, a high prevalence of ChPV1 and ChPV2 was detected in healthy goats, with the ChPV1 genotype being the most prevalent. There were no significant differences in the prevalence of ChPV1 and ChPV2 genotypes between Romanian and Serbian goat farms. Overall, both ChPV genotypes showed a significant geographical distribution, with a significantly higher prevalence in Italian goat farms than in Serbian and Romanian farms. We cannot exclude the fact that these data may be attributable to the limited number of blood samples examined from these countries.
As ChPVs have been found in healthy goats, it is conceivable that the peripheral blood represents an important primary route of infection and that ChPVs may spread through the bloodstream. However, the role of ChPVs in the pathogenesis of viral diseases in goats remains unknown. ChPV can cause persistent host infections, posing a threat to the goat industry. Therefore, new experimental therapeutic protocols against ChPV infections are being developed (20).
Finally, ddPCR can be used to improve diagnostic methods to allow researchers to accurately identify the genotypic distribution of ChPVs and better understand the territoriality of ChPV circulation. Gaining insight into the territorial prevalence and genotype distribution of ChPVs is important for improving the molecular and ecological epidemiology of ChPVs, as papillomavirus genotypes may be related to specific diseases, as has been reported in other animal species. Furthermore, the identification of the natural reservoirs of infectious pathogens, including viruses, plays a crucial role in controlling disease outbreaks in domestic animals, especially those diseases for which neither immunological nor therapeutic prophylaxis exists.
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
In this study, animal experiments were not performed. All the samples were collected from slaughterhouses, and therefore, no ethical approval was required.
SR, CC, and BK designed the experiments. AC, FD, BC, and VM carried out the experiments. SR, PC, AP, JB, and RM analyzed data. SR wrote the manuscript. All authors have read and approved the final manuscript.
This research was partially supported by Istituto Zooprofilattico Sperimentale del Mezzogiorno. The funders of the work did not influence study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The authors wish to thank S. Morace of the University of Catanzaro “Magna Graecia”, F. Di Domenico, from Azienda Sanitaria Locale (ASL) of Salerno, Antono Russillo from ASL of Potenza, and R.N. La Rizza from ASL of Vibo Valentia for their technical help.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2023.1213150/full#supplementary-material
1. Shah SD, Doorbar J, Goldstein RA. Analysis of host-parasite incongruence in Papillomavirus evolution using importance sampling. Mol Biol Evol. (2010) 27:1301–14. doi: 10.1093/molbev/msq015
2. The Papillomavirus Episteme. (2023). Available online at: pave.niaid.nih.gov/ (accessed June 3, 2023).
3. Medeiros-Fonseca B, Abreu-Silva AL, Medeiros R, Oliveira PA, Gil da Costa RM. Pteridium spp. and bovine papillomavirus: partners and cancer. Front Vet Sci. (2021) 8:758720. doi: 10.3389/fvets.2021.758720
4. Daudt C, Da Silva FRC, Lunardi M, Alves CBDT, Weber MN, Cibulski SP, et al. Papillomaviruses in ruminants: an update. Transbound Emerg Dis. (2018) 65:1381–95. doi: 10.1111/tbed.12868
5. De Falco F, Corrado F, Cutarelli A, Leonardi L, Roperto S. Digital droplet PCR for the detection and quantification of circulating bovine Deltapapillomavirus. Transbound Emerg Dis. (2021) 68:1345–52. doi: 10.1111/tbed.13795
6. De Falco F, Cutarelli A, D'Alessio N, Cerino P, Catoi C, Roperto S. Molecular epidemiology of ovine papillomavirus infections among sheep in southern Italy. Front Vet Sci. (2021) 8:790392. doi: 10.3389/fvets.2021.790392
7. Van Doorslaer K, Rector A, Vos P, Van Ranst M. Genetic characterization of the Capra hircus papillomavirus: a novel close-to-root artiodactyl papillomavirus. Virus Res. (2006) 118:164–9. doi: 10.1016/j.virusres.2005.12.007
8. Dogan F, Dorttas SD, Dagalp SB, Ataseven VS, Alkan F. A teat papillomatosis case in a Damascus goat (Shami goat) in Hatay Province, Turkey: a new putative papillomavirus? Arch Virol. (2018) 163:1635–42. doi: 10.1007/s00705-018-3781-2
9. Willemsen A, van der Boom A, Dietz J, Dagalp SB, Dogan F, Bravo IC, et al. Genomic and phylogenetic characterization of ChPV2, a novel goat closely related to the Xi-PV1 species infecting bovines. Virol J. (2020) 17:167. doi: 10.1186/s12985-020-01440-9
10. Odhah MNA, Garba B, Wen CX, Reduan MFH. A rare case of oral papillomatosis in a goat kid. Case Rep Vet Med. (2022) 2022:1598256. doi: 10.1155/2022/1598256
11. Theilen G, Wheeldon EB, East N, Madewell B, Lancaster WB, Munn R. Goat papillomatosis. Am J Vet Res. (1985) 46:2519–26.
12. Manni V, Roperto F, Di Guardo G, Galati D, Condoleo R, Venuti A. Presence of papillomavirus-like DNA sequences in cutaneous fibropapillomas of the goat udder. Vet Microbiol. (1998) 61:1–6. doi: 10.1016/S0378-1135(98)00168-0
13. Marà M, Di Guardo G, Venuti A, Marruchella G, Palmieri C, De Rugeriis M, et al. Spontaneous ocular squamous cell carcinoma in twin goats: pathological and biomolecular studies. J Comp Pathol. (2005) 132:96–100. doi: 10.1016/j.jcpa.2004.06.007
14. Al-Salihi KA, Al-Dabhawi AA, Ajeel AA, Erzuki IA, and Ali TAH. (2020). Clinico-histopathological and immunohistochemical study of ruminant's cutaneous papillomavirus in Iraq. Vet. Med. Int. 2020, 5691974, doi: 10.1155/2020/5691974
15. Cutarelli A, De Falco F, Uleri V, Buonavoglia C, Roperto S. The diagnostic value of the droplet digital PCR for the detection of bovine deltapapillomavirus in goats by liquid biopsy. Transbound Emerg Dis. (2021) 68:3624–30. doi: 10.1111/tbed.13971
16. Jeannot E, Latouche A, Bonneau C, Calméjane MA, Beaufort C, Ruigrok-Ritstier K, et al. Circulating HPV DNA as a marker for early detection of relapse in patients with cervical cancer. Clin Cancer Res. (2021) 27:5869–77. doi: 10.1158/1078-0432.CCR-21-0625
17. De Falco F, Cuccaro B, De Tullio R, Alberti A, Cutarelli A, De Carlo E, et al. (2023). Possible etiological association of ovine papillomaviruses with bladder tumors in cattle. Virus Res. 328, 199084. doi: 10.1016/j.virusres.2023.199084
18. Suo T, Liu X, Feng J, Gun M, Guo D, and Ullah H. (2020). ddPCR: a more accurate tool for SARS-CoV-2 detection in low viral load specimens. Emerg. Microbes Infect. 9, 1259–1268. doi: 10.1080/22221751.2020.1772678
19. Roperto S, Cutarelli A, Corrado F, De Falco F, and Buonavoglia C. (2021). Detection and quantification of bovine papillomavirus DNA by digital droplet PCR in sheep blood. Sci. Rep. 11, 10292. doi: 10.1038/s41598-021-89782-4
Keywords: blood, caprine papillomaviruses, ChPVs, droplet digital PCR, goat, liquid biopsy, molecular epidemiology
Citation: Cutarelli A, De Falco F, Cuccaro B, Milićević V, Kureljušić B, Bojkovski J, Cerino P, Perillo A, Marica R, Catoi C and Roperto S (2023) Prevalence and genotype distribution of caprine papillomavirus in peripheral blood of healthy goats in farms from three European countries. Front. Vet. Sci. 10:1213150. doi: 10.3389/fvets.2023.1213150
Received: 27 April 2023; Accepted: 01 June 2023;
Published: 15 June 2023.
Edited by:
Francisco José Pallarés, University of Cordoba, SpainReviewed by:
Tereza Cristina Cardoso, Universidade Estadual de São Paulo, BrazilCopyright © 2023 Cutarelli, De Falco, Cuccaro, Milićević, Kureljušić, Bojkovski, Cerino, Perillo, Marica, Catoi and Roperto. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sante Roperto, c2FudGUucm9wZXJ0b0B1bmluYS5pdA==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.