- 1State Key Laboratory for Managing Biotic and Chemical Threats to the Quality and Safety of Agro-Products, Ningbo University, Ningbo, Zhejiang, China
- 2Laboratory for Marine Fisheries Science and Food Production Processes, Qingdao National Laboratory for Marine Science and Technology, Qingdao, China
Vibrio splendidus is an opportunistic pathogen that causes skin ulcer syndrome and results in huge losses to the Apostichopus japonicus breeding industry. Ferric uptake regulator (Fur) is a global transcription factor that affects varieties of virulence-related functions in pathogenic bacteria. However, the role of the V. splendidus fur (Vsfur) gene in the pathogenesis of V. splendidus remains unclear. Hence, we constructed a Vsfur knock-down mutant of the V. splendidus strain (MTVs) to investigate the role of the gene in the effect of biofilm, swarming motility, and virulence on A. japonicus. The result showed that the growth curves of the wild-type V. splendidus strain (WTVs) and MTVs were almost consistent. Compared with WTVs, the significant increases in the transcription of the virulence-related gene Vshppd mRNA were 3.54- and 7.33-fold in MTVs at the OD600 of 1.0 and 1.5, respectively. Similarly, compared with WTVs, the significant increases in the transcription of Vsm mRNA were 2.10- and 15.92-fold in MTVs at the OD600 of 1.0 and 1.5, respectively. On the contrary, the mRNA level of the flagellum assembly gene Vsflic was downregulated 0.56-fold in MTVs at the OD600 of 1.0 compared with the WTVs. MTVs caused delayed disease onset time and reduced A. japonicus mortality. The median lethal doses of WTVs and MTVs were 9.116 × 106 and 1.658 × 1011 CFU·ml−1, respectively. Compared with WTVs, the colonization abilities of MTVs to the muscle, intestine, tentacle, and coelomic fluid of A. japonicus were significantly reduced. Correspondingly, the swarming motility and biofilm formation in normal and iron-replete conditions were remarkably decreased compared with those of WTVs. Overall, these results demonstrate that Vsfur contributes to the pathogenesis of V. splendidus by regulating virulence-related gene expression and affecting its swarming and biofilm formation abilities.
Introduction
The gram-negative rod-shaped bacterium Vibrio splendidus can cause severe vibriosis in numerous aquatic animals (1, 2). In particular, it is the main pathogen of “skin ulcer syndrome” (SUS), which occurs in sea cucumbers, Apostichopus japonicus (Echinodermata, Holothuroidea) (3, 4). Sea cucumber farming frequently suffers from SUS, resulting in serious economic losses (5). In recent years, research on the immune mechanism of sea cucumbers has made great progress. Different pattern recognition receptors that recognize pathogens have been identified (6, 7), complex immune regulatory networks have been depicted, and the relationship between metabolism and immunity has been further explored (8). Unfortunately, there is still a lack of effective means to prevent SUS. The fundamental reason is that research on the pathogenic mechanism is relatively scarce. It mainly focuses on the exploration of V. splendidus virulence factors such as hemolysin and extracellular metalloprotease (9, 10), lacking the cross integration of host immunity, pathogenic pathogenicity, and environmental factors.
Environmental factors have an important impact on the virulence and survival of pathogenic bacteria. One important environmental signal is iron, which is an essential micro-nutrient in almost all living organisms (11). However, excessive intracellular iron concentration is toxic to bacteria due to the reactive oxygen species produced by Fenton and Haber-Weiss reactions (12, 13). Therefore, many pathogens must control the iron concentration within a certain range (14). Ferric uptake regulator (Fur), a global transcription regulator factor, is a key regulator for bacteria to respond to changing iron availability in the environment, thereby regulating bacterial gene expression related to iron uptake, utilization, and storage (15, 16). Significantly, the fur gene also plays a critical role in the expression of virulence-related genes, such as oxidative stress response, quorum sensing, swarming motility, and biofilm formation in numerous pathogens during host invasion (17–20). Under normal and iron-replete conditions, Fur binds to Fe2+ as an inhibitor and then combines with the promoter region. It inhibits the transcription of the target gene by preventing RNA polymerase from binding to the target gene. The typical representative of this regulation mode is the siderophore transportation system (21, 22). Under iron-poor conditions, Fur combines with a conserved DNA sequence called the “fur box,” which is located near promoter regions and subsequently activates or inhibits transcription of the target genes (23). Moreover, the regulation of Fur can occur indirectly via small RNAs (24).
Our previous study confirmed that the iron uptake pathway of V. splendidus depends on hydroxamate siderophore-IutA (25). Alignment of the amino acid sequence demonstrated that V. splendidus fur (Vsfur) showed high similarities to that of the Furs from other Vibrio sp. (26). However, the regulation of bacterial pathogenicity caused by Vsfur is not known. In the present study, the Vsfur gene was knocked down by antisense RNA interference. The Vsfur knock-down mutant of V. splendidus (MTVs) was assessed for growth curve and expression of virulence-related genes compared with wild-type V. splendidus (WTVs). The WTVs and MTVs cells were collected to analyze swarming motility and biofilm formation in normal, iron-replete, and iron-starved conditions. Furthermore, the colonization ability and pathogenicity of WTVs and MTVs in A. japonicus were analyzed and compared.
Materials and methods
Bacterial strains, culture conditions, and chemicals
The wild-type V. splendidus strain was dissociated from the focus of sea cucumbers with SUS and preserved in our laboratory. Zhang et al. tested the strain’s pathogenicity in sea cucumbers by means of a reinfection experiment (27). The bacterial cultures of the wild-type strain and indicated mutant were shaken (180 rpm) at 28°C in Zobell’s 2216E medium (5 g·L−1 tryptone, 1 g·L−1 yeast extract in filtered natural seawater). Escherichia coli S17λpir and DH5α (Takara, China) were grown in Luria–Bertani (LB) broth or agar at 37°C. The antimicrobial agent was purchased from a commercial source Sangon (Shanghai, China). Antibiotics were added to the medium at the following concentrations: kanamycin (Kn) 50 μg·ml−1, gentamicin (Gm) 100 μg·ml−1, and ampicillin (Ap) 100 μg·ml−1. The vectors used in the experiment were pMD19-T, which was purchased from Takara (Beijing, China); vector pBBR1MCS-5, which was purchased from Fenghui Biotechnology (Changsha, China); and vectors pET28a and pBT3, which were kept in our laboratory. Restriction endonucleases were purchased from New England Biolabs and used according to the manufacturer’s instructions.
Mutant of Vibrio splendidus construction
The primers used in the present study were designed according to the genomic DNA of the V. splendidus strain LGP32 with accession number FM954973.2 and listed in Table 1. MTVs was constructed as described previously (28, 29). Plasmid pET28aVsfur containing constitutive promoter PTrc was generated to construct the MTVs with the reverse Vsfur sequence. The antisense RNA fragment of Vsfur was amplified with the forward primer VsfurF (Xho I) and reverse primer VsfurR (BamH I) and ligated into pMD19-T for subsequent restriction digestion. Plasmid pBT3Vsfur was constructed by ligating Vsfur into pBT3, between the BamH I/Xho I sites. The reverse Vsfur fragment fusing Trc promoter was obtained after pBT3Vsfur was digested with Swa I. The PTrc-Vsfur fragment was inserted into the blunt end site EcoR V of pET28a and then transformed into S17λπ to construct S17λπ/pET28aVsfur. To transform the recombinant plasmid pET28aVsfur into wild-type V. splendidus, bacterial conjugation was conducted as described previously (30). The conjugants were placed on 2216E agar together with Kn and Ap, and the positive conjugants were picked out and confirmed by PCR (T7/T7ter) and sequencing.
Complementation of the fur deletion mutant of Vibrio splendidus
To complement the fur deletion mutant of V. splendidus, we designed a pair of primers, furCF/R, to amplify the promoter region and complete ORF of the Vsfur gene. The fragment was ligated into pMD19-T and digested with BamH I and Xho I. Subsequently, fragments with BamH I and Xho I cleavage sites were cloned into the wide-host vector pBBR1MCS-5, which had different replicon and antibiotic resistance from pET28a. The recombinant vector pBBR1MCS-5Vsfur was transferred into MTVs by bacterial conjugation. The positive conjugants were selected on 2216E agar plates amended with Kn and Gm. Finally, the complemented strain furC was confirmed by PCR (pBBR1MCS-5F/R) and used for further research.
Growth curve measurement
Vibrio splendidus strains, including WTVs, MTVs and furC were coated to 2216E agar at 28°C for 24 h. Single colonies were transferred into 10 ml of fresh 2216E broth and allowed to reach the optical density at 600 nm (OD600) of 1.0. The culture was diluted proportionally to the same concentration. One hundred microlitre aliquots of WTVs, MTVs, and furC were reinoculated into flasks with 100 ml of fresh 2216E broth, 2216E broth supplemented with 100 μM iron chelator 2,2′-dipyridyl (DP), and 2216E broth supplemented with 50 μM FeCl3 and grown at 28°C with agitation at 180 rpm. The OD600 was recorded at different time points with an ultraviolet spectrophotometer (Mapada Instruments Co. Ltd., Shanghai, China). The experiment was repeated three times for each sample.
Quantification of mRNA expression
Total RNA was isolated from WTVs and MTVs cells at different OD600 values. Data were standardized with the endogenous reference gene 16S rDNA (933F and 16SRTR1). The PrimeScript RT kit (Takara, Japan) and the reagent for removing genomic DNA were used for reverse transcription. Real-time polymerase chain reaction (RT-PCR) was performed in an ABI 7500 RT-PCR detection system (Applied Biosystems) with a total volume of 20 μl containing 0.4 μl of Dye-II (ROX), 0.8 μl of each of the forward and reverse primers (10 μM), 8 μl of diluted cDNA template, and 10 μl of 2 × SYBR Green Mix. The experimental procedure was set as follows: 95°C for 5 min to activate the polymerase, 40 cycles of 95°C for 15 s, 60°C for 20 s, and 72°C for 20 s. Fluorescent signals were collected at the extension stage of each cycle. The primers for RT-PCR are listed in Table 1. The fold changes of gene expression were determined using the 2−ΔΔCT method (31).
Artificial infection
The sea cucumber artificial infection experiments proceeded as described by Liang et al. (32). Healthy sea cucumbers (weight, 3 ± 1 g) were temporarily reared for 2–3 days before the experiment. Then, 105 sea cucumbers were randomly divided into seven groups. Each group was placed in an aquarium containing 10 ml of aerated natural seawater (salinity, 28 psu), and the temperature was maintained at approximately 16°C. For microbial infection experiments, the WTVs and MTVs strains were prepared as described in culture conditions. The cells were collected when OD600 reached 1.0 by centrifugation at 8000 × g, washed three times with PBS, and resuspended in seawater. Three groups were continuously immersed in 1 × 107, 1 × 106, and 1 × 105 CFU·ml−1 WTVs and the other three groups were immersed in 1 × 107, 1 × 106, and 1 × 105 CFU·ml−1 MTVs. At the same time, the negative control group was inoculated with sterile PBS. The symptoms of infected A. japonicus were observed and the daily mortality was recorded.
Colonization quantities of Vibrio splendidus in Apostichopus japonicus tissues
The distributions of V. splendidus in the different A. japonicus tissues were determined as described previously (32). Sea cucumbers in the same growth state were equally divided into two groups, and each group was infected by WTVs and MTVs in a final concentration of 107 CFU·ml−1. After A. japonicus was subjected to immersion infection for 48 h, the body wall, muscle, respiratory tree, intestine, and tentacle of A. japonicus were collected using sterilized scissors and tweezers. Three tissue samples from three sea cucumbers were collected and mixed together; this was repeated three times. The specimens were supplemented with sterile PBS to 2 ml. Subsequently, the mixtures were homogenized by a homogenizer. One hundred microlitre gradient diluted mixtures were spread on 2216E agar plates supplemented with Ap (100 μg·ml−1). Meanwhile, the coelomic fluids were filtered through a 200-mesh and spread on 2216E agar plates supplemented with Ap. Single colonies from each plate were identified by 16S rDNA sequencing analysis and the number of colonies was counted after incubation at 28°C for 24 h.
Motility analysis
Swarming motility was assayed as described previously (33). WTVs, MTVs, and furC strains were separately grown in fresh 2216E broth at 28°C with agitation at 180 rpm, and OD600 was adjusted to 1.0. Then, 2 μl bacterial cell suspension was dropped on the center of 2216E, 2216E supplemented with 200 μM iron chelator DP, and 2216E supplemented with 100 μM FeCl3 plates containing 0.3% agar. The 2216E agar plates were then incubated at 28°C for 3 days to observe the diameters of the swarming halos. The iron treatment with different concentrations was repeated three times.
Biofilm formation assay
Biofilm formation assays for WTVs, MTVs, and furC were conducted as described by Luo et al. (34). Briefly, the WTVs, MTVs, and furC strains were prepared as described in culture conditions. The cell suspensions were adjusted to OD600 = 1.0 with fresh 2216E broth. Then, 2 μl of WTVs, MTVs, and furC strains were transferred into 198 μl of 2216E broth, 2216E broth supplemented with 200 μM DP, and 2216E broth supplemented with 100 μM FeCl3 in a 96-well plate and allowed to grow at 28°C for 48 h. Subsequently, the plate was washed three times with sterile PBS. Biofilms were stained with 200 μl of crystal violet (1%) for 30 min and washed with sterile PBS. The culture plate was then placed upside down on the absorbent paper. The adsorbed biofilm was dissolved with 200 μl of ethanol and the OD570 was measured. The iron treatment with different concentrations was conducted in five independent biological replicates.
Statistical analyses
Promoter prediction analyses were conducted with online prediction tools at http://www.softberry.com. The 500-bp upstream sequences of virulence-related genes from the initial codon site were used to search the Fur binding box. All data are expressed as the mean ± SD of at least three sets of independent experiments. Statistical significance was determined by one-way Analysis of Variance (ANOVA) with a Dunnett’s test. The significance level was defined as *p < 0.05, **p < 0.01.
Results
The role of Vsfur in vitro Vibrio splendidus growth
The growth curves of WTVs, MTVs, and furC were measured under different iron concentration culture conditions. The OD600 was measured at 3, 6, 9, 12, 21, 24, 45, and 48 h. Although iron chelator DP inhibited the growth of three strains within 24 h, the growth curves of the WTVs, MTVs, and furC strains were almost the same in 2216E broth (Figure 1A), 2216E broth supplemented with 50 μM FeCl3 (Figure 1B), and 2216E broth supplemented with 100 μM DP (Figure 1C). In this study, Vsfur was knocked down by antisense RNA interference to construct MTVs. Our previous research demonstrated that V. splendidus showed resistance to ampicillin, which is a β-lactam antibiotic, but V. splendidus was sensitive to kanamycin (35). Thus, plasmid pET28a with kanamycin resistance, which can replicate in V. splendidus, was used as a carrier vector to introduce an exogenous DNA fragment containing PTrc-Vsfur. Meanwhile, constitutive promoter PTrc could regulate the mRNA expression constant to a certain extent, improving the interference efficiency. Single colonies of WTVs and MTVs were inoculated into tubes with 10 ml of fresh 2216E medium supplemented with Ap and Kn cultured at 28°C overnight. No bacteria grew in the blank control and WTVs group, and the bacteria of MTVs could grow in the 2216E medium supplemented with Ap and Kn (Figure 2A). These results suggest that the recombinant plasmid pET28aVsfur was successfully transformed into V. splendidus (MTVs) and had no obvious influence on the growth rate of V. splendidus.
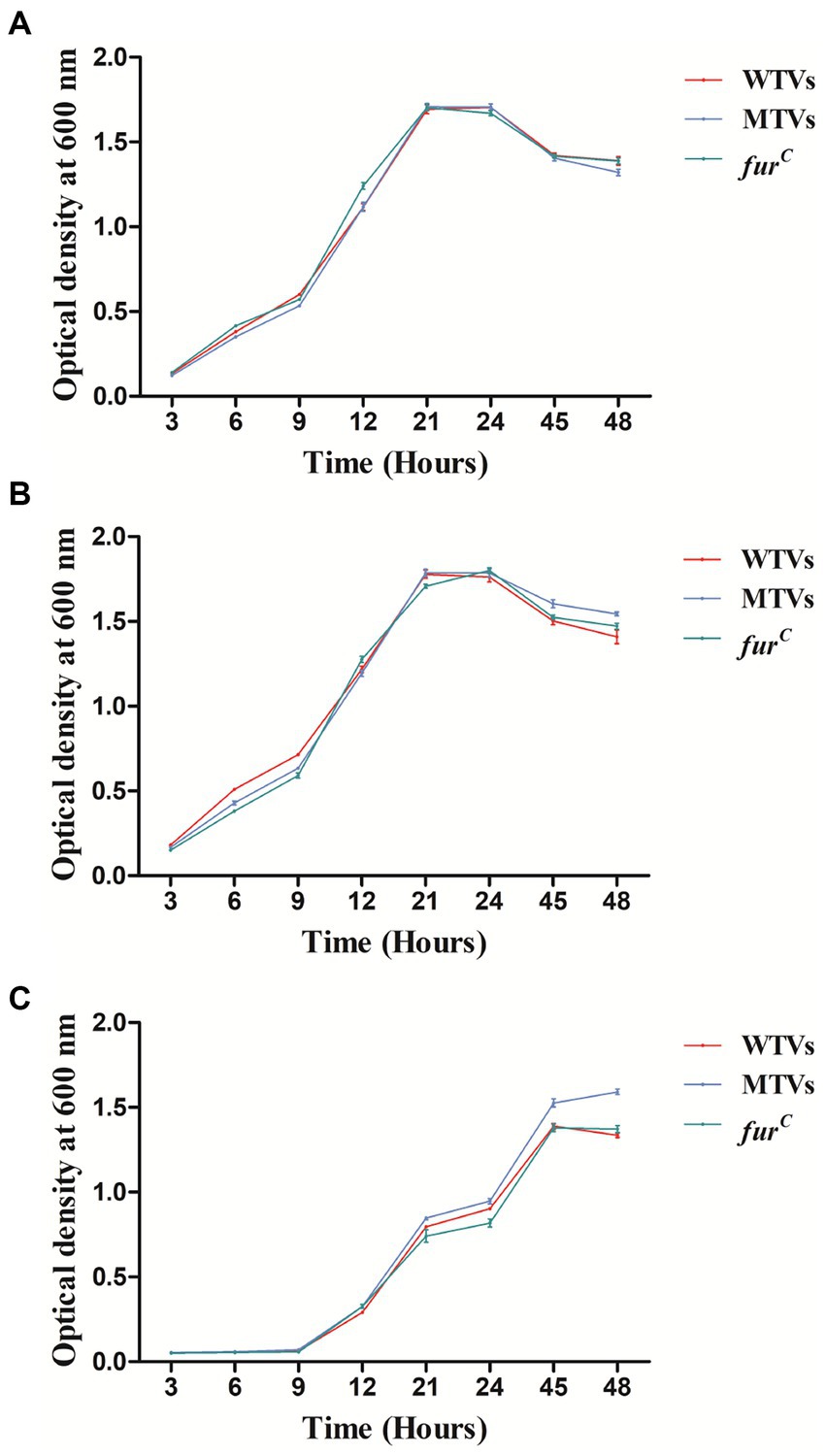
Figure 1. Growth curves of WTVs, MTVs, and furC. WTVs, MTVs, and furC were spread onto 2216E solid plates at 28°C overnight. Three single colonies were inoculated into flasks with 100 ml of fresh 2216E medium and incubated at 28°C with shaking at 180 rpm. Overnight cultures were diluted to the same concentration, and 200 μl aliquots of WTVs, MTVs, and furC were transferred into flasks with 100 ml of fresh 2216E broth (A), 2216E broth supplemented with 50 μM FeCl3 (B), and 2216E broth supplemented with 100 μM iron chelator DP (C). OD600 values were measured at different time points.
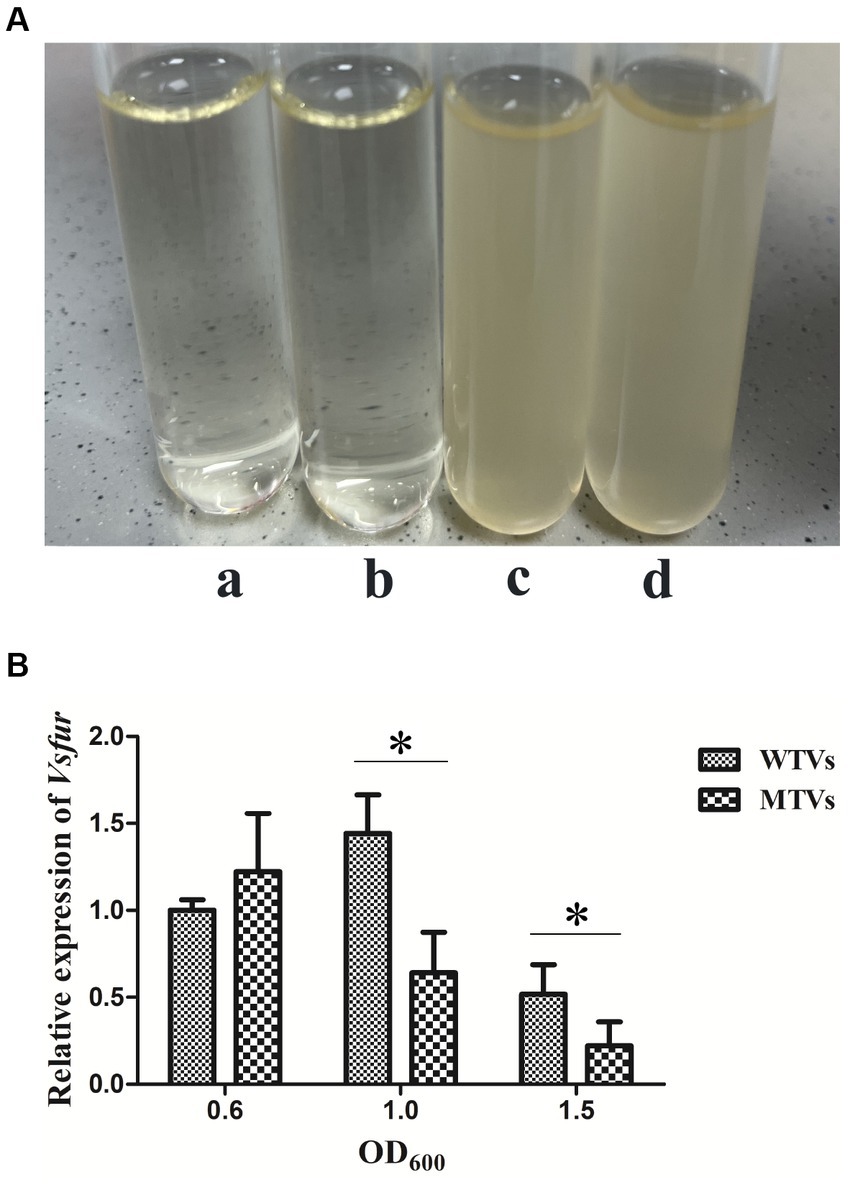
Figure 2. (A) Antibiotic resistance test of WTVs, MTVs, and furC. WTVs and MTVs were spread onto 2216E solid plates at 28°C overnight. Single colonies were inoculated into tubes with 5 ml of fresh 2216E medium supplemented with Ap and Kn and incubated at 28°C with shaking at 180 rpm. (a) Negative control group, (b) WTVs group, and (c) MTVs group. furC was spread onto 2216E solid plates at 28°C overnight. Single colonies were inoculated into tubes with 5 ml of fresh 2216E medium supplemented with Ap, Kn and Gm and incubated at 28°C with shaking at 180 rpm. (d) furC group. (B) Temporal expression analyses of Vsfur in WTVs or MTVs at the OD600 of 0.6, 1.0, and 1.5. Values are presented as mean ± SD (n = 5). Asterisks indicate significant differences: *p < 0.05 and **p < 0.01.
The differential expression of virulence-related genes between WTVs and MTVs
The WTVs and MTVs cells were collected at the OD600 of 0.6, 1.0, and 1.5 to determine the temporal expressions of Vsfur and virulence-related genes. The expression levels of Vsfur, Vshppd, Vsm, and Vsflic from MTVs were not significantly affected in the early log phase at the OD600 of 0.6. The expression of the Vsfur mRNA level was significantly downregulated 0.44- and 0.43-fold (p < 0.05) in MTVs at the OD600 of 1.0 and 1.5 compared with the WTVs (Figure 2B). Compared with the WTVs, the significant increases in the transcription of Vshppd mRNA were 3.54- and 7.33-fold (p < 0.01) in MTVs at the OD600 of 1.0 and 1.5 (Figure 3A). Meanwhile, the expression of Vsm mRNA was extremely significantly upregulated at the OD600 of 1.0 and 1.5 compared with the OD600 of 0.6. Compared with the WTVs, the significant increases in the transcription of Vsm mRNA were 2.10- and 15.92-fold (p < 0.01) in MTVs at the OD600 of 1.0 and 1.5 (Figure 3B). Under the same conditions, the mRNA level of Vsflic was downregulated 0.56-fold (p < 0.05) in MTVs at the OD600 of 1.0 compared with the WTVs (Figure 3C). These results suggest that the expression of the Vsfur gene was knocked down successfully in the mid and stationary log phase. Moreover, the downregulation of Vsfur, a global transcription factor that affects a number of virulence-related genes in V. splendidus, including upregulated genes in the MTVs for iron acquisition Vshppd (hemolysin) and Vsm (metalloprotease) and downregulated genes such as Vsflic (flagella C) related to flagellum biosynthesis. In summary, Vsfur has both positive and negative regulatory functions.
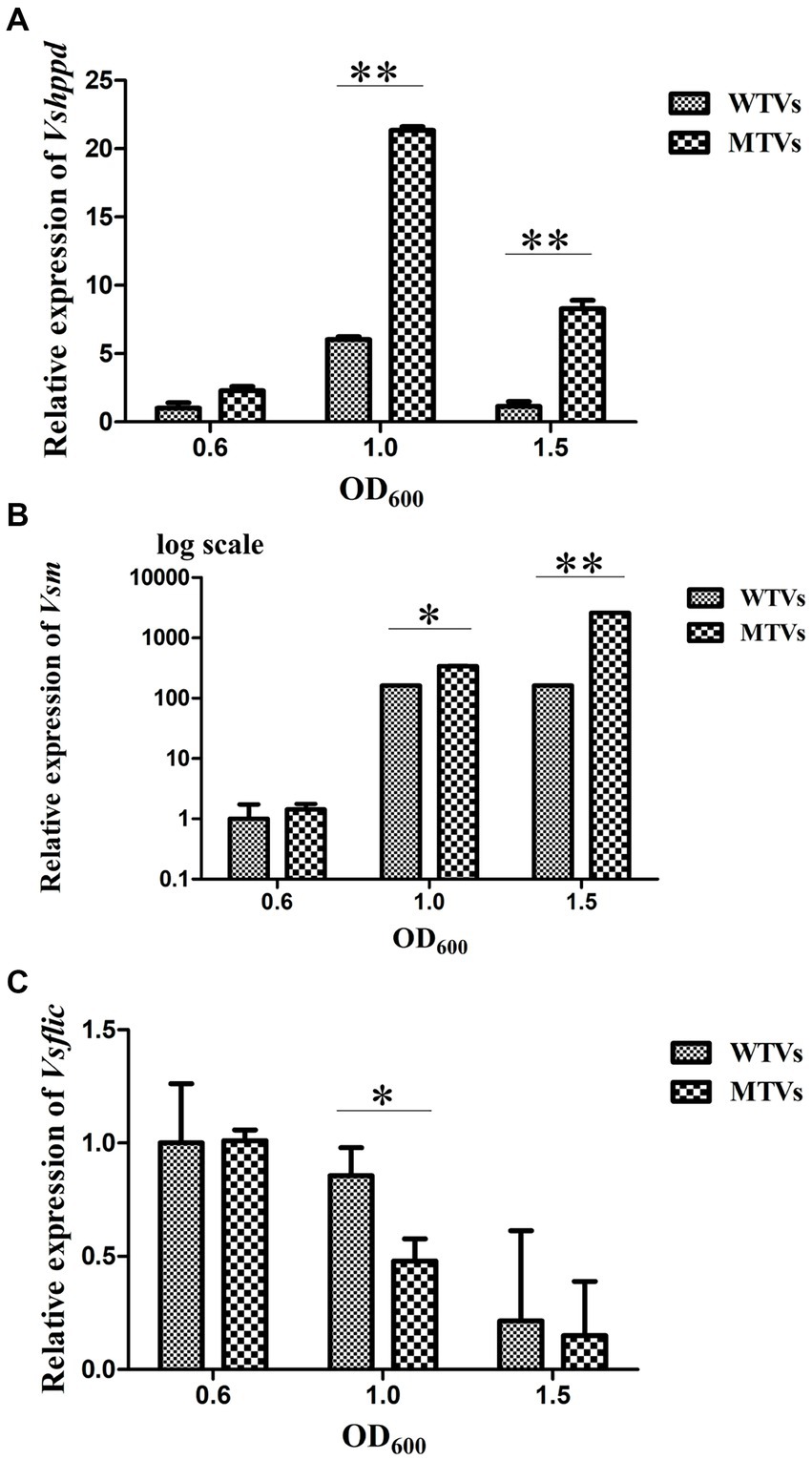
Figure 3. Temporal expression analyses of (A) Vshppd, (B) Vsm (The Y-axes were shown as a log scale), and (C) Vsflic in WTVs or MTVs at the OD600 of 0.6, 1.0, and 1.5. Values are presented as mean ± SD (n = 5). Asterisks indicate significant differences: *p < 0.05 and **p < 0.01.
The promoter regions from the functional genes showed Fur binding sites
Promoter sequence analyses from the virulence-related genes of Vshppd, Vsm, and Vsflic were performed to predict the putative Fur binding boxes in the V. splendidus strain. The promoter regions before the ATG initial cordon of the three virulence-related genes were searched in the genomic DNA database. BPROM1 prediction proved that the upstream of each virulence-related gene contained a typical promoter containing-35 and-10 regions (Figure 4). The Fur binding site was also searched and predicted near the-35 and-10 regions. These putative Fur binding boxes of Vshppd, Vsm, and Vsflic showed 52.63, 63.16, and 52.63% homology to the consensus Fur binding site sequence (5′-GATAATGATAATGATTATC-3′), respectively (Figure 4D). This sequence alignment suggests that Fur might bind directly to the promoter and regulate the expression of Vshppd, Vsm, and Vsflic.
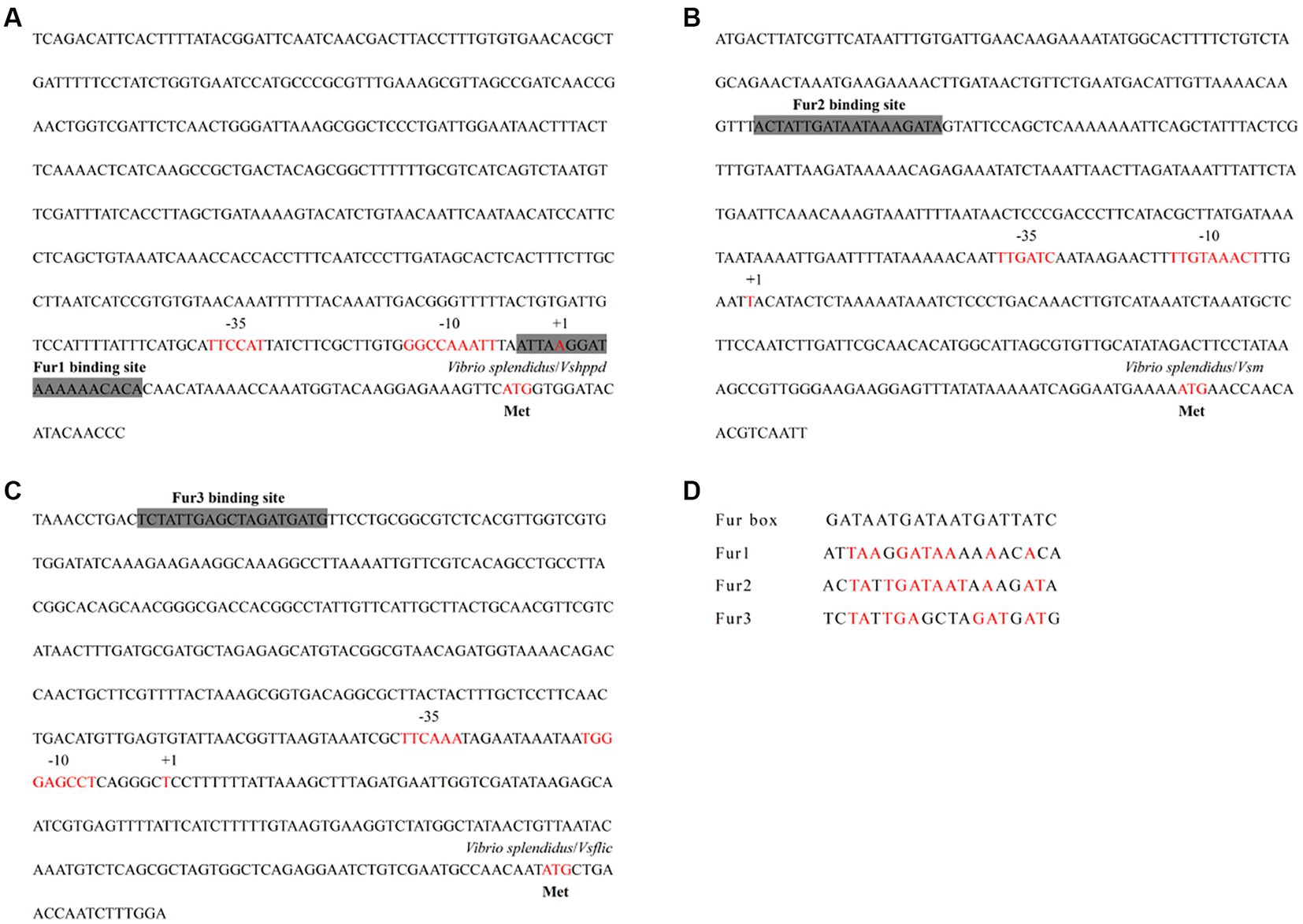
Figure 4. The analysis for Fur binding box prediction in the (A) Vshppd, (B) Vsm, and (C) Vsflic promoter region. The putative Fur binding boxes are indicated as gray boxes. The transcriptional initiation site, the corresponding −10 and −35 boxes, and the translational start site are indicated in red letters. (D) Sequence alignment of these putative Fur binding boxes with the Fur box consensus sequence. Bases identical to the consensus are shown in red.
Mutant of Vibrio splendidus showed lower pathogenicity in Apostichopus japonicus
In the artificial infection experiment, WTVs showed stronger pathogenicity than MTVs. Compared with the WTVs group, the A. japonicus infected with the MTVs strain showed a decrease in mortality and an obvious delay in the time of death (Figure 5A). The first mortalities of WTVs were observed 4 days post-infection with 1 × 107 CFU·ml−1 WTVs, and the first mortalities of MTVs were observed 4 days post-infection with 1 × 106 CFU·ml−1 MTVs. The total number of deaths in the WTVs group in 18 days were 7, 6, and 2 post-infection with 1 × 107, 1 × 106, and 1 × 105 CFU·ml−1 WTVs, respectively. The total number of deaths in the MTVs group in 18 days were 1, 2, and 0 post-infection with 1 × 107, 1 × 106, and 1 × 105 CFU·ml−1 MTVs, respectively. The A. japonicus infected with WTVs showed obvious characteristics of SUS, such as head shaking and skin ulceration. However, the A. japonicus group infected with MTVs did not show obvious characteristics of SUS (Figure 5B). No death occurred in the negative control group during the whole experiment. The median lethal doses (LD50) of WTVs and MTVs calculated by SPSS were 9.116 × 106 and 1.658 × 1011 CFU·ml−1, respectively. These results indicate that Vsfur is crucial to the pathogenicity of V. splendidus.
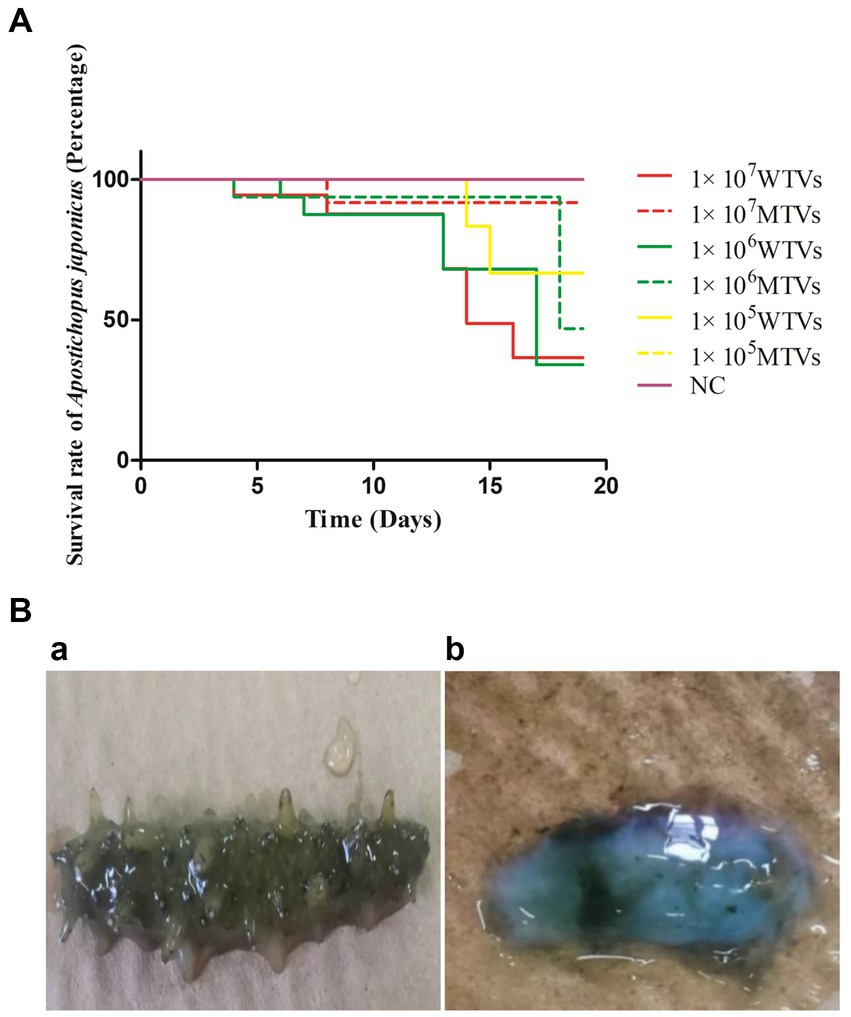
Figure 5. (A) The lethality of WTVs and MTVs. Apostichopus japonicus was randomly divided into seven tanks with 15 individuals each. The WTVs and MTVs strains used for infection were cultured in 2216E medium (24 h, 28°C) until OD600 was approximately 1.0. The strains were then washed and re-suspended in PBS (28°C). For survival assays, weight-matched A. japonicus individuals were infected with 1 × 107, 1 × 106, and 1 × 105 CFU·ml−1 V. splendidus (WTVs or MTVs). Apostichopus japonicus infected with PBS was used as the negative control. The water temperature during infection was 16°C. The daily mortality of infected A. japonicus was recorded. (B) The observed symptoms of (a) MTVs and (b) WTVs. Dead A. japonicus were removed in a timely manner and photographed to observe symptoms.
Mutant of Vibrio splendidus showed lower colonization ability compared to WTVs
The quantitative detection of cell colonization in various tissues of A. japonicus after immersion infection with WTVs and MTVs is shown in Figure 6. The colonization quantities of WTVs to the body wall, muscle, respiratory tree, intestine, and tentacle tissues were 9.53 × 107, 5.51 × 108, 7.77 × 107, 1.24 × 108, and 2.42 × 108 CFU·g−1, respectively. The colonization amount of WTVs to the coelomic fluid was 2.95 × 107 CFU·ml−1. The colonization quantities of MTVs to all tissues except the body wall and respiratory tree decreased. The colonization quantities of MTVs to the body wall, muscle, respiratory tree, intestine, and tentacle tissues were 1.71 × 107, 8.92 × 107, 4.78 × 107, 2.70 × 107, and 6.71 × 107 CFU·g−1, respectively. The colonization amount of MTVs to the coelomic fluid was 1.02 × 107 CFU·ml−1.
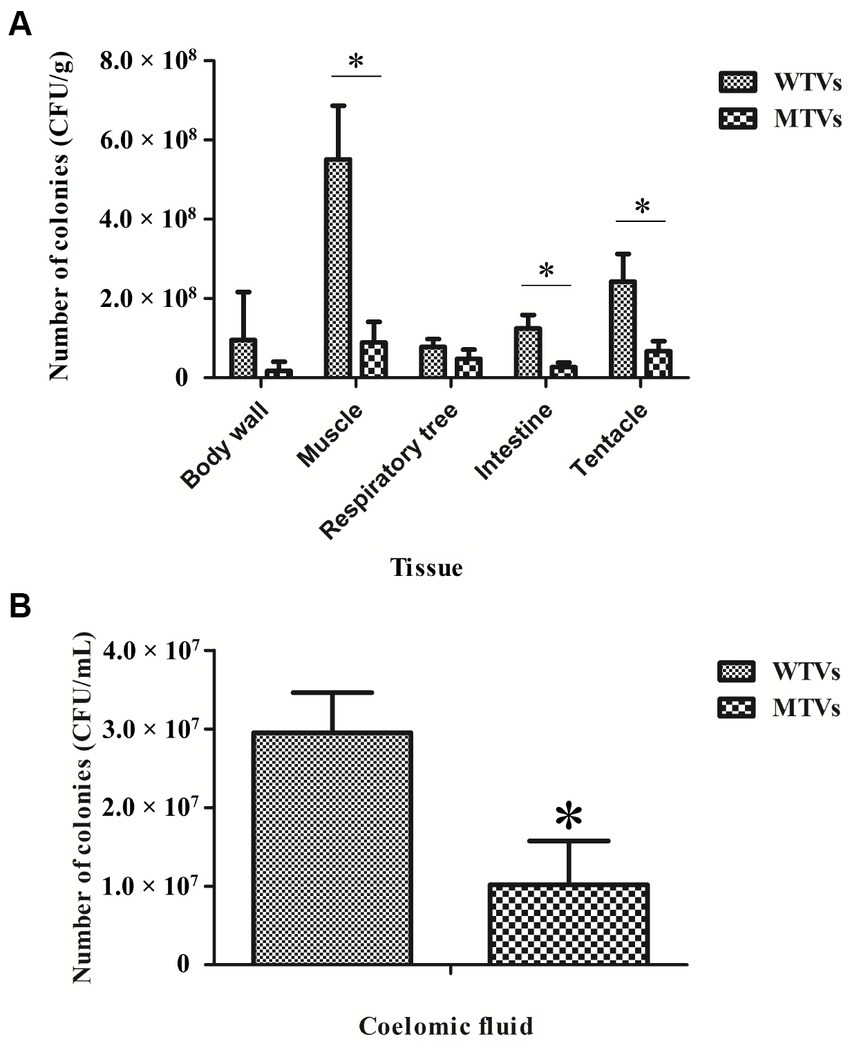
Figure 6. The colonization abilities of WTVs and MTVs to different A. japonicus tissues (A) and coelomic fluids (B) were demonstrated by colony counting. A. japonicus was soaked in WTVs and MTVs (1.0 × 107 CFU·ml−1) for 48 h infection. The A. japonicus tissues were weighed and homogenized. The coelomic fluids were filtered through a 200-mesh. The homogenized solution was diluted in gradients and then coated on 2216E plates.
Mutant of Vibrio splendidus showed lower swarming motility to WTVs
The motility of WTVs, MTVs, and furC strains was measured in 2216E agar plates, and the results demonstrated an obvious difference in the swarming motility of WTVs and MTVs (Figure 7A). The swarming halo diameter of WTVs was 3–4 mm more than that of MTVs at each time point and the diameter of MTVs was 79–81% (p < 0.01) of that of WTVs in normal conditions (Figure 7B). Meanwhile, the swarming motility of MTVs was reduced by approximately 0.68-fold compared with that of WTVs in iron-replete conditions (Figure 7C). Under the iron-starved conditions, the motility of WTVs and MTVs was restricted and could not be spread normally (Figure 7D). These results suggest that Vsfur contributes to the swarming motility of V. splendidus.
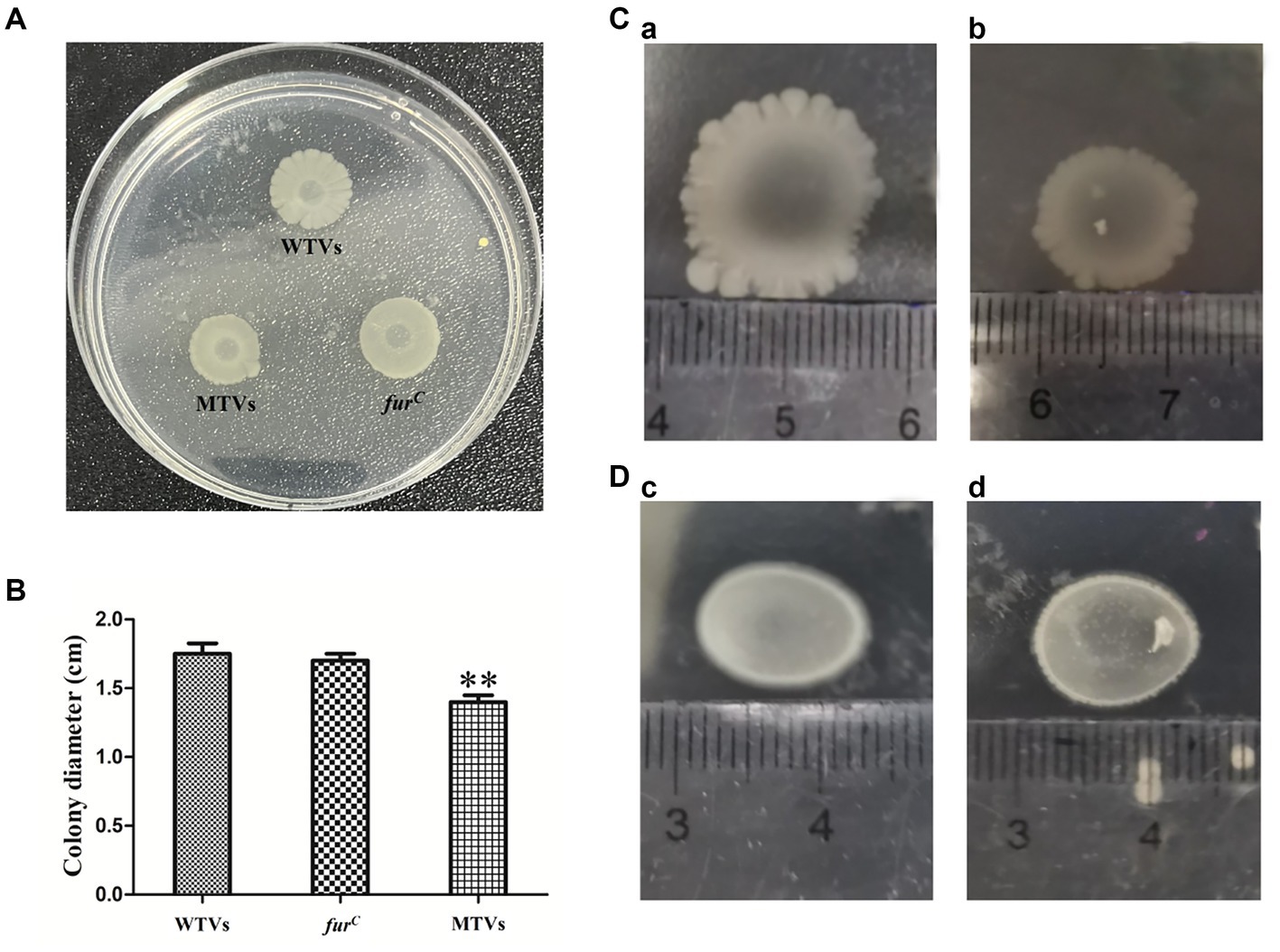
Figure 7. (A) Swarming motility of WTVs, MTVs, and furC in normal conditions. (B) Bar graph of the swimming motility of WTVs, MTVs, and furC. (C) The same concentration of WTVs (a) and MTVs (b) (2 μl) were dropped on low-agar 2216E plates supplemented with 100 μM FeCl3 and cultured for 48 h at 28°C. (D) The same concentration of WTVs (c) and MTVs (d) (2 μl) was dropped on low-agar 2216E plates supplemented with 200 μM DP and cultured for 48 h at 28°C. The error line represents the SD (n = 5). Asterisks represent the significant difference (*p<0.05, **p<0.01).
Mutant of Vibrio splendidus showed lower biofilm formation to WTVs
The biofilm formation was determined using a crystal violet (CV) staining assay. The biofilms of WTVs, MTVs, and furC that attached to 96-well plates were analyzed in normal 2216E medium. Consistent with the results of swarming motility, the biofilm formation of MTVs on the surface of a 96-well plate was 24% (p < 0.01) and 22% of WTVs and furC, respectively (Figure 8A). The biofilm formation of MTVs was 81% (p < 0.05) of WTVs in iron-replete conditions. Under the iron-starved conditions, the biofilm formation of WTVs and MTVs showed no significant difference (Figure 8B). Taken together, these results indicate that deletion of Vsfur results in decreased biofilm formation, which depends on the presence of Fe3+.
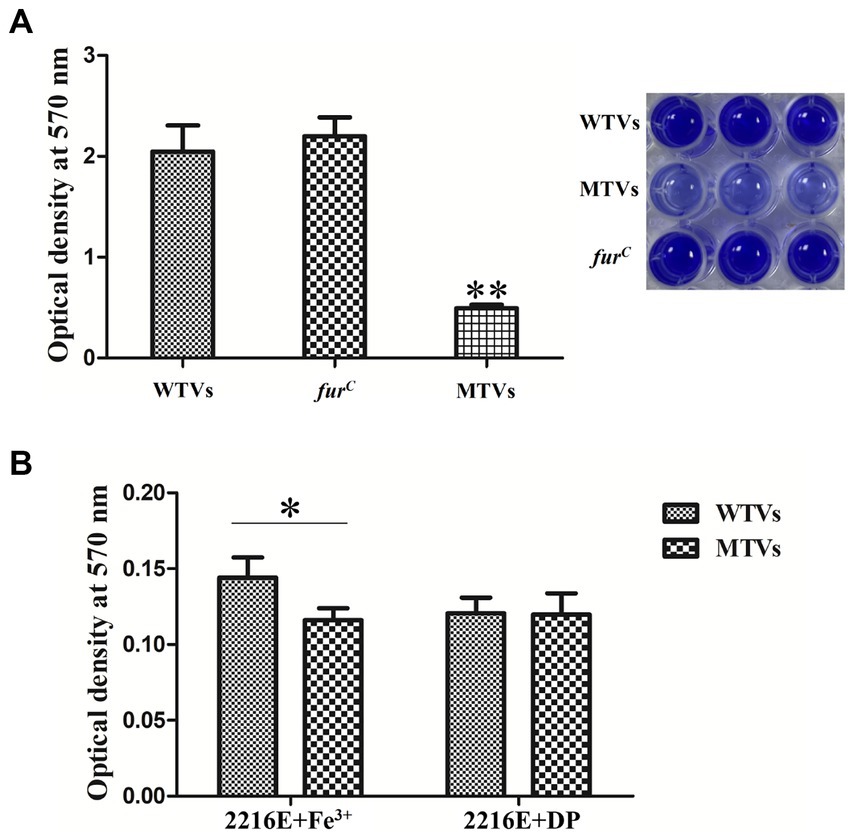
Figure 8. (A) Biofilm formation of WTVs, MTVs, and furC in normal conditions. (B) Biofilm formation of WTVs and MTVs in iron-replete conditions and iron-starved conditions. The error line represents the SD (n = 5). Asterisks represent the significant difference (*p < 0.05, **p < 0.01).
Discussion
Many genes have been proven to be involved in regulating the virulence of aquatic pathogenic bacteria by knock-out technology (36, 37). Differently from other bacteria, attempts to obtain fur deletion mutants of V. splendidus failed, leading to the assumption that Fur is an essential protein in V. splendidus. While several attempts to obtain Vsfur in-frame deletion mutants by suicide vector failed, we constructed a Vsfur knock-down mutant of V. splendidus through antisense RNA interference. The recombinant plasmid pET28aVsfur containing promoter PTrc could efficiently express the interference sequence in V. splendidus (28, 35). In previous studies, fur mutants of other Pseudomonas aeruginosa and Acidovorax citrulli have shown significantly reduced growth rates in low-and high-iron environments (18, 38). In this study, the growth rate of the MTVs strain was the same as that of the WTVs strain in iron normal conditions.
Bacteria depend on Fur to regulate the absorption and utilization of iron, which has a critical impact on bacterial survival and virulence (39). Moreover, recent studies have demonstrated that Fur can function as both an activator and a repressor on a global level through both direct and indirect mechanisms to affect a variety of virulence-related traits (14). However, no studies on the contribution of the knock-out of the fur gene to the pathogenesis of V. splendidus have been reported. In this study, the expression of the Vsfur mRNA level was not significantly different in MTVs at the OD600 of 0.6 compared with the WTVs. After that, the expression level was significantly downregulated and maintained a low level at the OD600 of 1.0 and 1.5. It might be that the interference had not occurred effectively in the early log phase of bacteria. With the continuous copy of the vector, a large number of interfering sequences were expressed, leading to the effective blocking of the Vsfur gene. Meanwhile, the result showed that the expression of virulence-related genes Vshppd and Vsm upregulated significantly with the decrease of the Vsfur gene. We speculate that it was the high-iron concentration inhibition phenomenon dependent on Fur. When the concentration of iron reaches a stable state, Fur binds and forms a complex with Fe2+, and the resulting Fur-Fe2+ complex binds to the promoter region of the target gene (40). This binding results in the transcriptional repression of genes involved in iron acquisition and storage (21). The siderophore synthesis pathway of E. coli, Vibrio cholerae, and Bacillus subtilis showed a high-iron concentration inhibition mode (23, 41). In addition to iron metabolism-related genes, the expression of the hemolysin synthesis gene hly (42), the outer membrane protein synthesis gene irgA of V. cholerae (43), and the catalase gene catC of Streptomyces coelicolor (44) was inhibited by Fur. Our previous research demonstrated that HGA-melanin produced by Vshppd with the ability to reduce Fe3+ to Fe2+, contributes to the ferrous iron uptake pathway in V. splendidus (45). In addition, BPROM prediction suggested that the upstream of the Vshppd and Vsm had apparent Fur binding sites. Overall, these results demonstrated that Fur may regulate the expression of Vshppd and Vsm through a high-iron concentration inhibition mode in V. splendidus. However, the detailed regulation mechanism of Fur needs to be investigated in future studies.
Motility is an important virulence determinant during the adhesion and invasion stages in many pathogenic bacteria and enables bacterial cells to swim to nutrient-rich niches or avoid environmental stresses (46). Previous research has suggested that Fur directly activates the transcription of toxin coregulated pilus tcpA, which was necessary for colonization by binding to their promoters in V. cholerae (47). In Shigella flexneri, Fur could indirectly activate the icsA gene that facilitates bacterial movement by activating VirF (48). Our results demonstrate that the swarming motility of the MTVs was significantly reduced compared to the WTVs. Consistent with these findings, gene expression levels of Vsflic were significantly reduced in the MTVs. Our previous research demonstrated that FliC could contribute to swarming motility in V. splendidus (49). Combining these results, Fur could positively regulate the expression of Vsflic to contribute to swarming motility in V. splendidus, whereas the repressive mechanism of Fur has been well understood; emerging research on direct and indirect Fur-mediated activation mechanisms will reveal novel global regulatory circuits (22).
In consideration of the complex interaction relationship between motility and biofilm formation (50), we further compared the biofilm formation of MTVs and WTVs at different iron concentrations. The results showed that the biofilm formation of MTVs was decreased compared to the WTVs in normal and iron-replete conditions. Considering that low-iron concentration limits the growth of bacteria, the biofilm formation of WTVs and MTVs showed no significant difference. Moreover, despite the increased expression of virulence-related genes in MTVs, motility and adhesion are prerequisites for pathogen infection (51). We speculated that the pathogenicity of MTVs in A. japonicus was lower than that of WTVs. The results confirmed that A. japonicus infected with the MTVs strain exhibited a significant reduction in mortality, a remarkable delay in the time of death, and alleviated SUS symptoms compared with the WTVs group. All these findings support that the knock-down of Vsfur leads to the decrease of swarming motility and biofilm formation in Vibrio splendidus, affecting its virulence in Apostichopus japonicus.
In summary, the knock-down of the Vsfur gene in V. splendidus showed no effect on growth, but MTVs infection resulted in a reduced death rate, delayed time of death, and alleviated SUS symptoms in A. japonicus. Vsfur regulates biofilm formation and swarming motility, which are responsible for the virulence of V. splendidus. These findings will provide a theoretical basis for the prevention and control of SUS.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Ethics statement
The animal study was reviewed and approved by the Experimental Animal Ethics Committee of Ningbo University, China. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
YS performed the experiments and interpreted the data. CYL, FD, and YZ performed the partial experiment. CHL contributed new reagents, analytic tools, and revised the manuscript. WL participated in the experimental design, interpreted the data, and wrote the manuscript. All authors read and approved the manuscript.
Funding
This work was financially supported by the National Natural Science Foundation of China (32102843 and 32073003), Zhejiang Provincial Natural Science Foundation (LY23C190003), and the K. C. Wong Magna Fund at Ningbo University.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
1. Lacoste, A, Jalabert, F, Malham, S, Cueff, A, Gelebart, F, Cordevant, C, et al. A Vibrio splendidus strain is associated with summer mortality of juvenile oysters Crassostrea gigas in the bay of Morlaix (North Brittany, France). Dis Aquat Org. (2001) 46:139–45. doi: 10.3354/dao046139
2. Thomson, R, Macpherson, HL, Riaza, A, and Birkbeck, TH. Vibrio splendidus biotype 1 as a cause of mortalities in hatchery-reared larval turbot, Scophthalmus maximus (L.). J Appl Microbiol. (2005) 99:243–50. doi: 10.1111/j.1365-2672.2005.02602.x
3. Deng, H, He, CB, Zhou, ZC, Liu, C, Tan, KF, Wang, NB, et al. Isolation and pathogenicity of pathogens from skin ulceration disease and viscera ejection syndrome of the sea cucumber Apostichopus japonicus. Aquaculture. (2009) 287:18–27. doi: 10.1016/j.aquaculture.2008.10.015
4. Liu, HZ, Zheng, FR, Sun, XQ, Hong, XG, Dong, SL, Wang, B, et al. Identification of the pathogens associated with skin ulceration and peristome tumescence in cultured sea cucumbers Apostichopus japonicus (Selenka). J Invertebr Pathol. (2010) 105:236–42. doi: 10.1016/j.jip.2010.05.016
5. Wang, YG, Zhang, FP, Li, SZ, Chen, X, and Cui, YL. Detection of Vibrio splendidus causing skin ulcer syndrome of sea cucumber (Apostichopus japonicus) with DNA probes. J Fish China. (2009) 33:119–25. doi: 10.3321/j.issn:1000-0615.2009.01.016 (in Chinese)
6. Chen, KY, Zhang, SY, Shao, YN, Guo, M, Zhang, WW, and Li, CH. A unique NLRC4 receptor from echinoderms mediates Vibrio phagocytosis via rearrangement of the cytoskeleton and polymerization of F-actin. PLoS Pathog. (2021) 17:e1010145. doi: 10.1371/journal.ppat.1010145
7. Dai, F, Guo, M, Shao, YN, and Li, CH. Vibrio splendidus flagellin C binds tropomodulin to induce p38 MAPK-mediated p53-dependent coelomocyte apoptosis in Echinodermata. J Biol Chem. (2022) 298:102091. doi: 10.1016/j.jbc.2022.102091
8. Sun, LL, Zhou, FY, Shao, YN, Lv, ZM, and Li, CH. Sedoheptulose kinase bridges the pentose phosphate pathway and immune responses in pathogen-challenged sea cucumber Apostichopus japonicus. Dev Comp Immunol. (2020) 109:103694. doi: 10.1016/j.dci.2020.103694
9. Liang, WK, Zhang, C, Liu, NN, Zhang, WW, Han, QX, and Li, CH. Cloning and characterization of Vshppd, a gene inducing haemolysis and immune response of Apostichopus japonicus. Aquaculture. (2016) 464:246–52. doi: 10.1016/j.aquaculture.2016.06.039
10. Liang, WK, Zhang, WW, Shao, YN, Zhao, XL, and Li, CH. Dual functions of a 4-hydroxyphenylpyruvate dioxygenase for Vibrio splendidus survival and infection. Microb Pathog. (2018) 120:47–54. doi: 10.1016/j.micpath.2018.04.055
11. Chen, P, Meulenaere, ED, Deheyn, DD, and Bandaru, PR. Iron redox pathway revealed in ferritin via electron transfer analysis. Sci Rep. (2020) 10:4033. doi: 10.1038/s41598-020-60640-z
12. Kobayashi, M, Suhara, T, Baba, Y, Kawasaki, NK, Higa, JK, and Matsui, T. Pathological roles of iron in cardiovascular disease. Curr Drug Targets. (2018) 19:1068–76. doi: 10.2174/1389450119666180605112235
13. Morales, M, and Xue, X. Targeting iron metabolism in cancer therapy. Theranostics. (2021) 11:8412–29. doi: 10.7150/thno.59092
14. Porcheron, G, and Dozois, CM. Interplay between iron homeostasis and virulence: Fur and RyhB as major regulators of bacterial pathogenicity. Vet Microbiol. (2015) 179:2–14. doi: 10.1016/j.vetmic.2015.03.024
15. Hantke, K. Iron and metal regulation in bacteria. Curr Opin Microbiol. (2001) 4:172–7. doi: 10.1016/s1369-5274(00)00184-3
16. Pohl, E, Haller, JC, Mijovilovich, A, Meyer, KW, Garman, E, and Vasil, ML. Architecture of a protein central to iron homeostasis: crystal structure and spectroscopic analysis of the ferric uptake regulator. Mol Microbiol. (2003) 47:903–15. doi: 10.1046/j.1365-2958.2003.03337.x
17. Shao, S, Li, CL, Zhao, LY, Zhang, YX, Yin, KY, and Wang, QY. Interplay between ferric uptake regulator Fur and horizontally acquired virulence regulator Esr B coordinates virulence gene expression in Edwardsiella piscicida. Microbiol Res. (2021) 253:126892. doi: 10.1016/j.micres.2021.126892
18. Liu, J, Tian, YL, Zhao, YQ, Zeng, R, Chen, BH, Hu, BS, et al. Ferric uptake regulator (FurA) is required for Acidovorax citrulli virulence on watermelon. Phytopathology. (2019) 109:1997–2008. doi: 10.1094/PHYTO-05-19-0172-R
19. Cha, JY, Lee, JS, Oh, JI, Choi, JW, and Baik, HS. Functional analysis of the role of Fur in the virulence of Pseudomonas syringae pv. Tabaci 11528: Fur controls expression of genes involved in quorum-sensing. Biochem Biophys Res Commun. (2008) 366:281–7. doi: 10.1016/j.bbrc.2007.11.021
20. Gao, H, Ma, LZ, Qin, Q, Qiu, Y, Zhang, JY, Li, J, et al. Fur represses Vibrio cholerae biofilm formation via direct regulation of vieSAB, cdgD, vpsU, and vpsA-K transcription. Front Microbiol. (2020) 11:587159. doi: 10.3389/fmicb.2020.587159
21. Carpenter, BM, Whitmire, JM, and Merrell, DS. This is not your mother's repressor: the complex role of Fur in pathogenesis. Infect Immun. (2009) 77:2590–601. doi: 10.1128/IAI.00116-09
22. Yu, CX, and Genco, CA. Fur-mediated global regulatory circuits in pathogenic Neisseria species. J Bacteriol. (2012) 194:6372–81. doi: 10.1128/JB.00262-12
23. Troxell, B, and Hassan, HM. Transcriptional regulation by ferric uptake regulator (Fur) in pathogenic bacteria. Front Cell Infect Microbiol. (2013) 3:59. doi: 10.3389/fcimb.2013.00059
24. Hoe, CH, Raabe, CA, Rozhdestvensky, TS, and Tang, TH. Bacterial sRNAs: regulation in stress. Int J Med Microbiol. (2013) 303:217–29. doi: 10.1016/j.ijmm.2013.04.002
25. Song, TX, Liu, HJ, Lv, TT, Zhao, XL, Shao, YN, Han, QX, et al. Characteristics of the iron uptake-related process of a pathogenic Vibrio splendidus strain associated with massive mortalities of the sea cucumber Apostichopus japonicus. J Invertebr Pathol. (2018) 155:25–31. doi: 10.1016/j.jip.2018.05.001
26. Liang, WK, Zhang, SS, Zhang, WW, Li, CH, and Han, QX. Characterization of functional furVs as a biomarker for detection of aquatic pathogen Vibrio splendidus. J World Aquacult Soc. (2016) 47:577–86. doi: 10.1111/jwas.12288
27. Zhang, WW, Liang, WK, and Li, CH. Inhibition of marine Vibrio sp. by pyoverdine from Pseudomonas aeruginosa PA1. J Hazard Mater. (2016) 302:217–24. doi: 10.1016/j.jhazmat.2015.10.003
28. Zhang, M, Sun, K, and Sun, L. Regulation of autoinducer 2 production and lux S expression in a pathogenic Edwardsiella tarda strain. Microbiology. (2008) 154:2060–9. doi: 10.1099/mic.0.2008/017343-0
29. Hu, YH, Liu, CS, Hou, JH, and Sun, L. Identification, characterization, and molecular application of a virulence-associated autotransporter from a pathogenic Pseudomonas fluorescens strain. Appl Environ Microbiol. (2009) 75:4333–40. doi: 10.1128/AEM.00159-09
30. Sawabe, T, Fukui, Y, and Stabb, EV. Simple conjugation and outgrowth procedures for tagging vibrios with GFP, and factors affecting the stable expression of the gfp tag. Lett Appl Microbiol. (2006) 43:514–22. doi: 10.1111/j.1472-765X.2006.01992.x
31. Livak, KJ, and Schmittgen, TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. (2001) 25:402–8. doi: 10.1006/meth.2001.1262
32. Liang, WK, Zhang, WW, and Li, CH. Vibrio splendidus virulence to Apostichopus japonicus is mediated by hppD through glutamate metabolism and flagellum assembly. Virulence. (2022) 13:458–70. doi: 10.1080/21505594.2022.2046949
33. Wang, Z, Lin, BC, Mostaghim, A, Rubin, RA, Glaser, ER, Mittraparp-Arthorn, P, et al. Vibrio campbellii hmgA-mediated pyomelanization impairs quorum sensing, virulence, and cellular fitness. Front Microbiol. (2013) 4:379. doi: 10.3389/fmicb.2013.00379
34. Luo, G, Huang, LX, Su, YQ, Qin, YX, Xu, XJ, Zhao, LM, et al. flrA, flr B and flrC regulate adhesion by controlling the expression of critical virulence genes in Vibrio alginolyticus. Emerg Microbes Infect. (2016) 5:e85. doi: 10.1038/emi.2016.82
35. Dai, F, Zhuang, QT, Zhao, XL, Shao, YN, Guo, M, Lv, ZM, et al. Green fluorescent protein-tagged Vibrio splendidus for monitoring bacterial infection in the sea cucumber Apostichopus japonicus. Aquaculture. (2020) 523:735169. doi: 10.1016/j.aquaculture.2020.735169
36. Guo, LN, Huang, LX, Su, YQ, Qin, YX, Zhao, LM, and Yan, QP. secA, secD, secF, yajC, and yidC contribute to the adhesion regulation of Vibrio alginolyticus. Microbiology. (2018) 7:00551. doi: 10.1002/mbo3.551
37. Luo, G, Sun, YJ, Huang, LX, Su, YQ, Zhao, LM, Qin, YX, et al. Time-resolved dual RNA-seq of tissue uncovers Pseudomonas plecoglossicida key virulence genes in host-pathogen interaction with Epinephelus coioides. Environ Microbiol. (2020) 22:677–93. doi: 10.1111/1462-2920.14884
38. Pasqua, M, Visaggio, D, Sciuto, AL, Genah, S, Banin, E, Visca, P, et al. Ferric uptake regulator Fur is conditionally essential in Pseudomonas aeruginosa. J Bacteriol. (2017) 199:e00472–17. doi: 10.1128/JB.00472-17
39. Pajuelo, D, Hernandez-Cabanyero, C, Sanjuan, E, Lee, CT, Silva-Hernandez, FX, Hor, LI, et al. Iron and Fur in the life cycle of the zoonotic pathogen Vibrio vulnificus. Environ Microbiol. (2016) 18:4005–22. doi: 10.1111/1462-2920.13424
40. Crosa, JH. Signal transduction and transcriptional and posttranscriptional control of iron-regulated genes in bacteria. Microbiol Mol Biol Rev. (1997) 61:319–36. doi: 10.1128/mmbr.61.3.319-336.1997
41. Lorenzo, MD, and Stork, M. Plasmid-encoded iron uptake systems. Microbiol Spectr. (2014) 2:26104436. doi: 10.1128/microbiolspec.PLAS-0030-2014
42. Stoebner, JA, and Payhe, SM. Iron-regulated hemolysin production and utilization of heme and hemoglobin by Vibrio cholerae. Infect Immun. (1988) 56:2891–5. doi: 10.1128/iai.56.11.2891-2895.1988
43. Mey, AR, Wyckoff, EE, Oglesby, AG, Rab, E, Taylor, RK, and Payne, SM. Identification of the Vibrio cholerae enterobactin receptors VctA and IrgA: IrgA is not required for virulence. Infect Immun. (2002) 70:3419–26. doi: 10.1128/IAI.70.7.3419-3426.2002
44. Hahn, JS, Oh, SY, and Roe, JH. Regulation of the furA and catC operon, encoding a ferric uptake regulator homologue and catalase-peroxidase, respectively, in Streptomyces coelicolor A3(2). J Bacteriol. (2000) 182:3767–74. doi: 10.1128/JB.182.13.3767-3774.2000
45. Zhang, WW, and Li, CH. Virulence mechanisms of vibrios belonging to the Splendidus clade as aquaculture pathogens, from case studies and genome data. Rev Aquac. (2021) 13:2004–26. doi: 10.1111/raq.12555
46. Hossain, MM, Shibata, S, Aizawa, SI, and Tsuyumu, S. Motility is an important determinant for pathogenesis of Erwinia carotovora subsp. carotovora. Physiol Mol Plant Pathol. (2005) 66:134–43. doi: 10.1016/j.pmpp.2005.06.001
47. Gao, H, Zhang, JY, Lou, J, Li, J, Qin, Q, Shi, QN, et al. Direct binding and regulation by Fur and HapR of the intermediate regulator and virulence factor genes within the ToxR virulence regulon in Vibrio cholerae. Front Microbiol. (2020) 11:709. doi: 10.3389/fmicb.2020.00709
48. Wing, HJ, Yan, AW, Goldman, SR, and Goldberg, MB. Regulation of IcsP, the outer membrane protease of the Shigella actin tail assembly protein IcsA, by virulence plasmid regulators VirF and VirB. J Bacteriol. (2004) 186:699–705. doi: 10.1128/JB.186.3.699-705.2004
49. Dai, F, Li, Y, Shao, YN, Li, CH, and Zhang, WW. FliC of Vibrio splendidus-related strain involved in adhesion to Apostichopus japonicus. Microb Pathog. (2020) 149:104503. doi: 10.1016/j.micpath.2020.104503
50. Ushijima, B, and Hase, CC. Influence of chemotaxis and swimming patterns on the virulence of the coral pathogen Vibrio coralliilyticus. J Bacteriol. (2018) 200:e00791–17. doi: 10.1128/JB.00791-17
Keywords: Vibrio splendidus , Apostichopus japonicus , ferric uptake regulator, biofilm, swarming motility
Citation: Shi Y, Liao C, Dai F, Zhang Y, Li C and Liang W (2023) Vibrio splendidus Fur regulates virulence gene expression, swarming motility, and biofilm formation, affecting its pathogenicity in Apostichopus japonicus. Front. Vet. Sci. 10:1207831. doi: 10.3389/fvets.2023.1207831
Edited by:
Lixing Huang, Jimei University, ChinaReviewed by:
Jingfeng Sun, Tianjin Agricultural University, ChinaHassan Al-Tameemi, University of Basrah, Iraq
Copyright © 2023 Shi, Liao, Dai, Zhang, Li and Liang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Weikang Liang, bGlhbmd3ZWlrYW5nQG5idS5lZHUuY24=
 Yue Shi
Yue Shi Changyu Liao1
Changyu Liao1 Chenghua Li
Chenghua Li Weikang Liang
Weikang Liang