- 1Department of Veterinary Medicine, College of Coastal Agricultural Sciences, Guangdong Ocean University, Zhanjiang, China
- 2Maoming Branch, Guangdong Laboratory for Lingnan Modern Agriculture, Maoming, China
Florfenicol is used worldwide for its low side effects and strong bactericidal effect. Florfenicol is physicochemically stable and can persist in natural water bodies and affect water denitrification. Indoor aquatic microcosm models were constructed and water samples were collected at different florfenicol concentrations (0.1, 1, 10, and 100 mg/L) on days 0, 7, 30, and 60 to extract the microbial genome DNA and determine the water properties. qPCR and amplicon sequencing were used to study the dynamic changes of nirS gene and nirS-type denitrifying communities structure, diversity and abundance, respectively. The results showed that higher florfenicol concentrations caused accumulation of nitrate and ammonium nitrogen in water. Florfenicol stress caused orders of magnitude changes in nirS gene abundance, showing a trend of increasing first and then decreasing. 100 mg/L florfenicol addition led to a sustained increase of nirS gene abundance in water bodies. The florfenicol addition altered denitrifying community structure and suppressed the richness and diversity index of denitrifying bacteria in water body. Over time, the richness and diversity index gradually recovered. Proteobacteria was always the dominant denitrifying phylum in water. The relative abundance of Pseudomonas and beta proteobacterium showed obvious positive correlation with nirS gene abundance and were the dominant genera under florfenicol stress. Our study provided a scientific basis for the rational use of florfenicol in aquaculture to maintain a healthy and stable microecological environment, and also provided a preliminary understanding of the response characteristics of water denitrifying microorganisms to florfenicol exposure.
1. Introduction
Aquaculture intensity requires the periodic use of antibiotics to treat and prevent bacterial disease outbreaks (1, 2). During farming, to ensure the proper growth of aquaculture animals, it also needs to be regularly replaced with an equal proportion of new water to treat the rapid accumulation of inorganic nitrogen compounds in the water (3, 4). According to the FAO 2020 report, the scale of worldwide aquaculture is increasing and is expected to reach 150 Mt by 2030 (5, 6). The rapid growth of intensive industrial aquaculture will produce large amounts of farm wastewater, causing natural water bodies to become eutrophic. Unused antibiotics can also eventually go back into the aquatic environment, making water bodies an important reservoir for antibiotics. The rapid growth of intensive industrial aquaculture will produce large amounts of farm wastewater, causing natural water bodies to become eutrophic. Unutilized antibiotics will also eventually enter the aquatic environment, causing water bodies to become important storage tanks for antibiotics (7). In addition, antibiotics can enter surface water, groundwater, and even possibly drinking water, which can pose a serious threat to human health and safety (8, 9).
Florfenicol shows low side effects and strong bactericidal activity and has been one of the antibiotics allowed to be used in aquaculture by the Food and Agriculture Organization of the United Nations (10). Since 2012, FLO has become the main antibiotic ingredient in fish feed premixes and is commonly recommended for fish bacterial diseases (11, 12), and the amount of florfenicol used in China is as high as 10,000 tons (13). However, researchers have shown that 45–60% of florfenicol cannot be absorbed by animals and is excreted in manure and urine (14). Furthermore, florfenicol is stable physicochemically and persists in aquatic systems under ambient temperature and pH conditions typical of natural waters (15). Different concentrations of florfenicol residues have been detected in coastal waters, sediments, and organisms (16). The residual concentration of florfenicol was as high as 11 mg/L in the waters around the Dalian Bay aquacultural farms in China (17). Six months after antibiotic treatment was stopped, the detected concentration of florfenicol was as high as 23.1 ng/L in salmon farms in Chile (18). Florfenicol was also detected in human urine and judged to be one of the antibiotics with a high health risk (19). Despite the high use of Florfenicol in farming all over the world, it is still a challenge to evaluate its negative environmental impacts, mainly on water bodies (20).
Denitrification, as the important nitrogen purification mechanism in lake waters (21), is the key part of the nitrogen cycle and the most economical approach to solving nitrate pollution in wastewater, which can purify water quality and alleviate water eutrophication (22). Denitrification is a series of enzymatic reactions mediated by microorganisms and involves four key enzymes: nitrate reductase, nitrite reductase, NO reductase and N2O reductase. Nitrite reductase, encoded by the nirS and nirK genes, is the rate-limiting enzyme for denitrification (23). The nirS and nirK genes have been used as functional markers for the presence of denitrification (24). The nirS gene is more broadly found among bacteria than nirK gene, and is more widely present in the physiological groups of bacteria (25). The nirS gene was used as a molecular marker to construct a clone library, which is an effective means to understand the composition and diversity of the denitrifying microbial community structure. Research has also reported varying degrees of adverse or inhibitory effects of antibiotics on denitrification (26, 27). Antibiotic residues can inhibit key enzyme activities of denitrifying microorganisms (27). However, it has also been reported that the negative effects of antibiotics decrease to some extent over time, and overall a gradual adaptation to the stress can be observed (26, 28). There is a lack of research regarding the effects of florfenicol on denitrifying microorganisms in the water body. In this study, an aquatic microcosm model was constructed to simulate an aquatic ecosystem with florfenicol added exogenously to explore the dynamic changes in water properties, nirS gene abundance, and the nirS-type denitrifying communities structure, abundance and diversity after different florfenicol concentrations were added to the aquatic environment. This study may provide theoretical data for the scientific use of florfenicol in aquaculture and the assessment of the ecological risk of florfenicol residues.
2. Materials and methods
2.1. Aquatic mesocosm experiment design and sampling
The mesocosm experiment was constructed to simulate the aquatic ecosystem. Surface water (1–10 cm) and surface sediments (1–10 cm) were collected from lakes at the Guangdong Ocean University (Zhanjiang, China), removing larger stones and other debris, and divided into transparent plastic boxes (50 × 40 × 30 cm). A plastic box contained 30 L of lake water and 10 cm of sediment. Keep at ambient temperature (25 ± 3°C) for 3 days until the aquatic mesocosm systems have stabilized. Florfenicol solution was added so that the concentration of florfenicol in the water column was 0, 0.1, 1, 10, and 100 mg/L. Water samples (numbered W0 to W4) were collected on days 0, 7, 30, and 60 (D0, D7, D30, and D60). Among them, the 0-day water samples (D0) were collected before the addition of florfenicol. The experiment had 5 groups with 3 replicates each. Three replicate samples from each group were mixed and immediately split into two parts for the determination of physicochemical parameters and DNA extraction.
2.2. Analysis of water sample properties
Several experiments were performed to assess the physiochemical characteristics of the samples. The pH and conductivity of the water samples were measured using a pH meter and conductivity meter, and nitrate nitrogen (colorimetric method) and ammonium nitrogen (indigo method) were estimated using an ultraviolet-visible spectrophotometer. Each sample was analyzed in triplicate.
2.3. DNA extraction and qPCR
According to the manufacturer's instructions, genomic DNA was extracted from water samples using the water DNA kit (Omega Bio-Tek, Norcross, GA, USA). The concentration and purity of the extracted DNA were assessed using a nanodrop UV-vis spectrophotometer. Three DNA of samples were extracted from each group for analysis. Based on previous work, the SYBR-primer method was employed with the CFX Connect Real-Time System instrument (BIO-RAD) to determine the abundance of the nirS gene. The denitrifying bacterial nirS gene was amplified using the primer pair CD3AF/R3CDR. The qPCR reaction system was: 10 μL SYBR, 1 μL DNA template, 1 μL upstream and downstream primers (10 mol/L) each, and 8 μL ddH2O. The qPCR reaction program was: 94°C, 5 min; 94°C for 30 s, 58°C for 30 s, 72°C for 30 s, 39 cycles.
2.4. Amplicon sequencing of nirS gene and bioinformatics processing
The DNA samples from each of the three replicates were combined into the mixed sample and sent to Jinweizhi (Suzhou, China) for Illumina MiSeq paired-end sequencing of the nirS gene amplicon regions. The nirS gene was amplified for sequencing analysis using the same primer pairs as qPCR. The high-throughput sequencing raw data was optimized using QIIME 1.9.1 by splicing overlapping regions at the ends of the sequences, removing sequences that were shorter than 200 bp, and removing chimeric sequences to produce valid data. An operational taxonomic unit (OTU) cluster analysis was carried out based on 97% similarity. The species taxonomic annotation was completed using the NCBI database. The 20 water samples were divided into different groups according to the sampling time and florfenicol addition concentration, and then OTU clustering analysis was performed. According to the OTU cluster analysis results, a Venn diagram was drawn. Based on OTU analysis results are obtained, using the method of random sampling sample sequences is flat, calculate Shannon, Chao1 alpha diversity index, community species abundance and diversity. Through the (UN) weighted unifrac analysis and comparison between samples whether there are significant differences in the microbial community. PCoA is display beta diversity visualization, PCoA is based on the distance between the matrix Brary - Curtis. Through the weighted clustering hierarchy and the group average method construct UPGMA (Unweighted pair group method with arithmetic mean) clustering tree. And diversity index was examined using QIIME 1.9.1 software and R language. The water properties and nirS gene abundance were used as independent variables, and denitrifying bacteria relative abundance was used as the response variable. By establishing a multiple linear regression model, the response variables were explained by the independent variables, and the correlation between them was explored.
2.5. Data statistic analyses
Excel 2016 was used to analyze the nirS gene abundance, R 3.3.1 was used to analyze alpha diversity, and the correlation between environmental factors and community structure was examined using CANOCO 5.0 software. The two-factor variance analysis was applied to test the effects of florfenicol concentration, time, and their interactions on water properties and nirS gene abundance. Statistical analyses were performed with IBM SPSS Statistics 24. All significant differences were at P ≤ 0.05.
3. Results
3.1. Effect of florfenicol on water properties
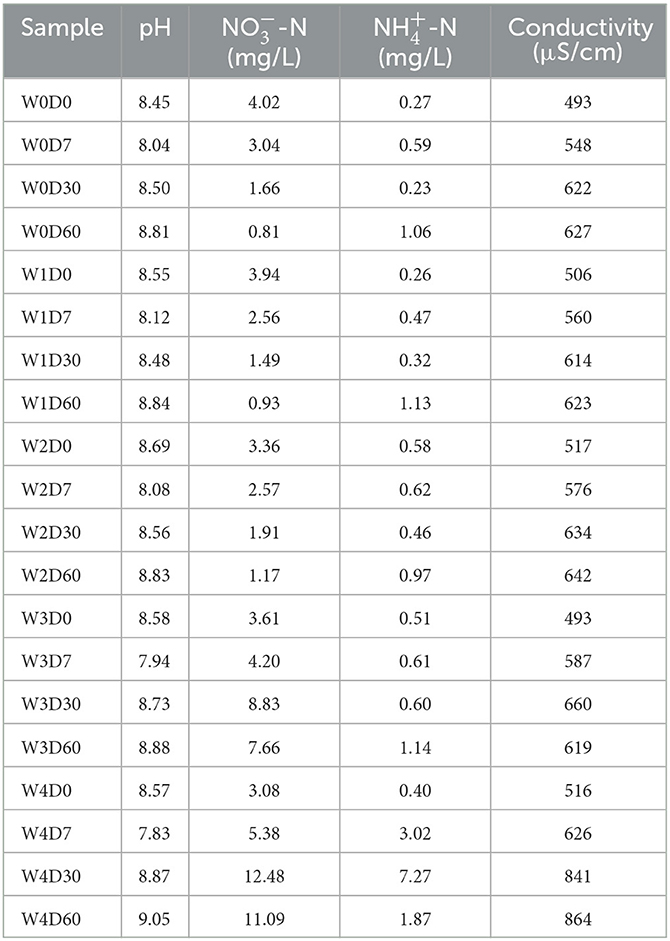
As shown in Table 1, florfenicol concentration and sampling time both affected clearly water properties. A higher effect on water properties was observed for sampling time than florfenicol addition concentrations. This trend was not broken by the lower florfenicol additions, but by the higher florfenicol additions. The pH of all water samples was alkaline and weakly alkaline, with a significant decrease in pH on day 7 and a gradual increase in water pH on days 30 and 60. The treatment groups with higher florfenicol additions showed a larger variation amplitude of pH. Higher concentrations of florfenicol caused an accumulation of nitrate nitrogen in the water compared to the control group. In addition, 100 mg/L florfenicol caused a marked accumulation of ammonium nitrogen and a marked rise in water conductivity at day 30. According to the statistical analysis results, concentrations of florfenicol and time significantly affected the water properties in an aquatic microcosm model (P < 0.05).
3.2. Effect of florfenicol on nirS gene abundance
As shown in Figure 1, the absolute abundance of nirS genes in the water bodies was 2.53 × 104-2.02 × 107 copies/μL. The nirS denitrifying community showed two response patterns to the florfenicol concentration in the water body when stressed by florfenicol. On Day 7, after the addition of florfenicol, the abundance of the nirS gene was 1–2 orders of magnitude higher. Moreover, the abundance of nirS gene increased more with increasing florfenicol addition concentration. On 30 and 60 days, the abundance of nirS gene decreased continuously over time, with a slower decrease in 10 mg/L florfenicol treatment groups. However, with 100 mg/L florfenicol, the abundance of nirS gene kept increasing over time until 60 days, when the abundance of nirS gene reached maximum value and increased by 3 orders of magnitude compared with no florfenicol addition. According to the statistical analysis results, concentrations of florfenicol and sampling time significantly affected the abundance of nirS genes in an aquatic microcosm model (P < 0.05).
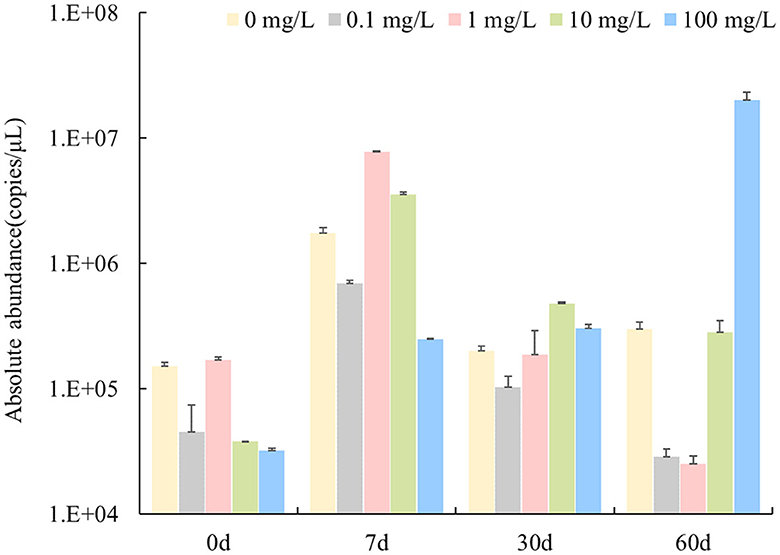
Figure 1. Effects of different concentrations of florfenicol on the abundance of nirS genes in an aquatic microcosm model.
3.3. Effect of florfenicol on nirS-type denitrifying communities structure
After the quality optimization of the original data, the number of valid sequences obtained in water samples was 63,042–186,410. After OTU cluster analysis, the number of OTUs in each sample ranged from 285 to 2,607, with an average of 1,135 OTUs per sample, of which the W4D7 group had the fewest OTUs. The results of the cluster analysis showed that sampling time had an obvious influence on OTU composition in the water column without florfenicol. As shown in Figure 2, the 0-day samples had 512 common OTUs; the four samples without added florfenicol but collected at different times had 82 common OTUs. 1 mg/L florfenicol could markedly reduce the number of common OTUs. Florfenicol concentrations affect its degradation rate and produce different effects on microbial communities in the water body.
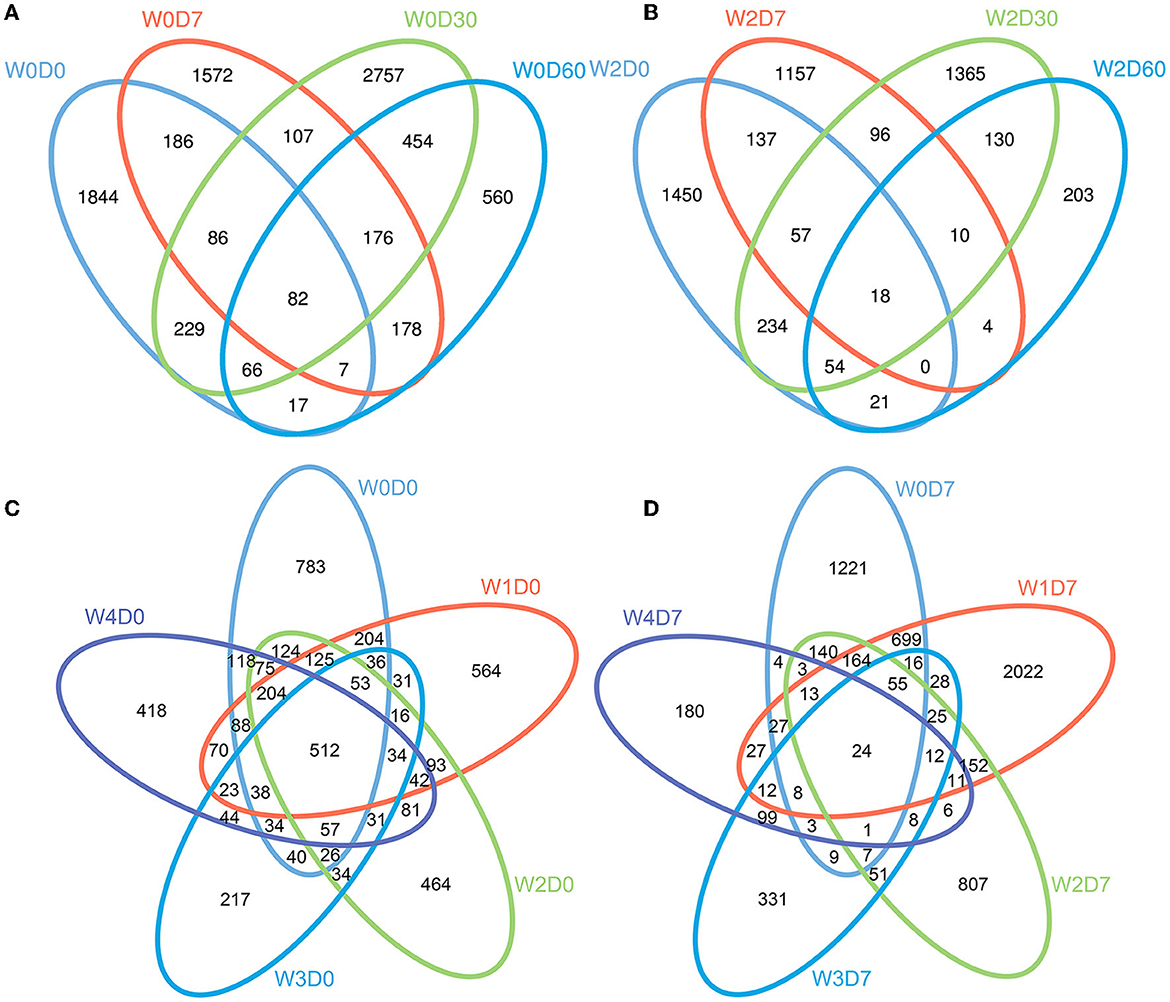
Figure 2. Effects of different concentrations of florfenicol on operational taxonomic units (OTUs) of bacteria (Venn diagram) in an aquatic microcosm model. (A) Effect of sampling time on the OUTs in group W0 samples. (B) Effect of sampling time on OUTs in group W2 samples. (C) OTUs differences between the 5 groups at 0 days. (D) Effects of different concentrations of florfenicol on OTUs at 7 days. Differently colored circles represent different samples, and the numbers represent the number of unique OTUs in each sample or the number of common OTUs in all samples.
The number of valid sequences and OTUs in each sample was obtained from the OTU analysis results. The random sampling method was used to extract a different number of sequences from each sample and calculate the number of OTUs after extraction. The dilution curve between OTU and the number of sample sequences was constructed by using the number of valid sequences extracted from the samples as the abscissa and the number of observed OTUs as the ordinate. As presented in Figure 3, with the increased number of extracted sequences, the number of detected OTUs increases and gradually tends to be flat, indicating that the sequencing is saturated, the sampling is essentially reasonable and the most abundant microbiome is included in the sequencing results. This provided a more realistic representation of the microbial community structure.
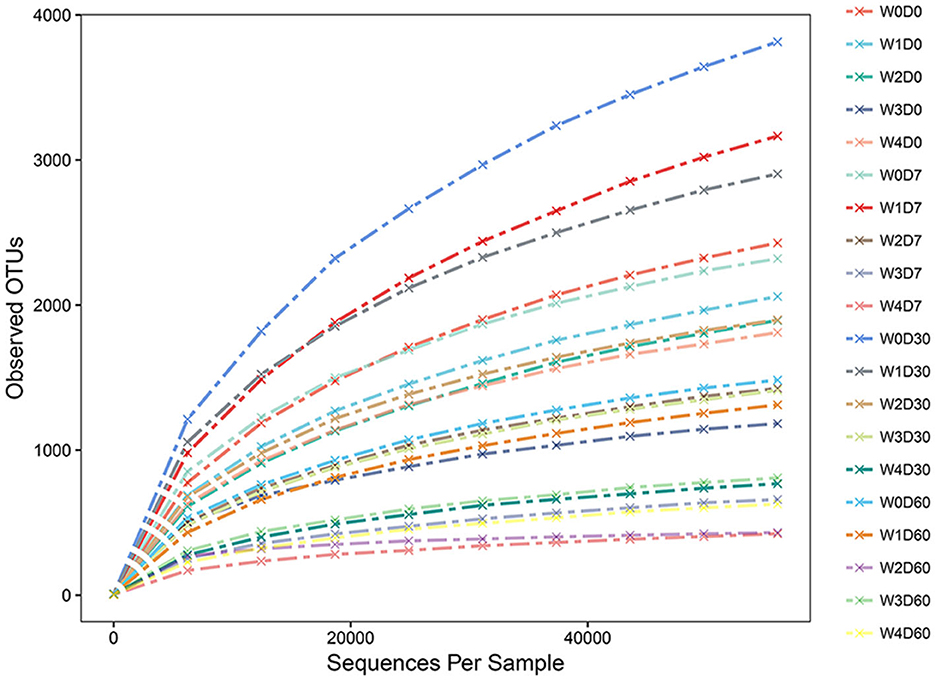
Figure 3. Effects of different concentrations of florfenicol on dilution curve of operational taxonomic units (OTUs) in an aquatic microcosm model.
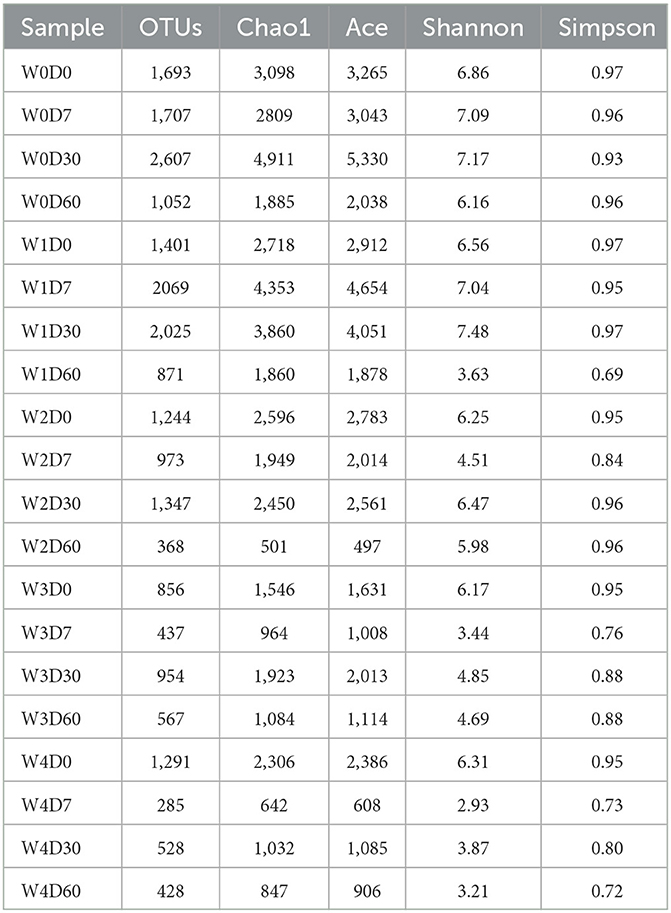
The denitrifying microbial community diversity indices mainly include the Shannon and Simpson indices, and the richness indices mainly include the Chao1 and ACE indices. Larger indices represent a greater total species number and higher community diversity. As shown in Table 2, denitrifying microbial community richness and diversity showed a decreasing trend in the water body as the concentration of florfenicol increased; with the change of time, the community richness and diversity were first suppressed and then gradually recovered. Its recovery rate was bounded by the florfenicol concentration. As shown in Figure 4, the Chao1 and Shannon indices of the denitrifying microbial community decreased with increased florfenicol concentrations. The most inhibition of denitrifying microbial community richness and diversity index was observed in the 100 mg/L florfenicol treatment group on day 7.
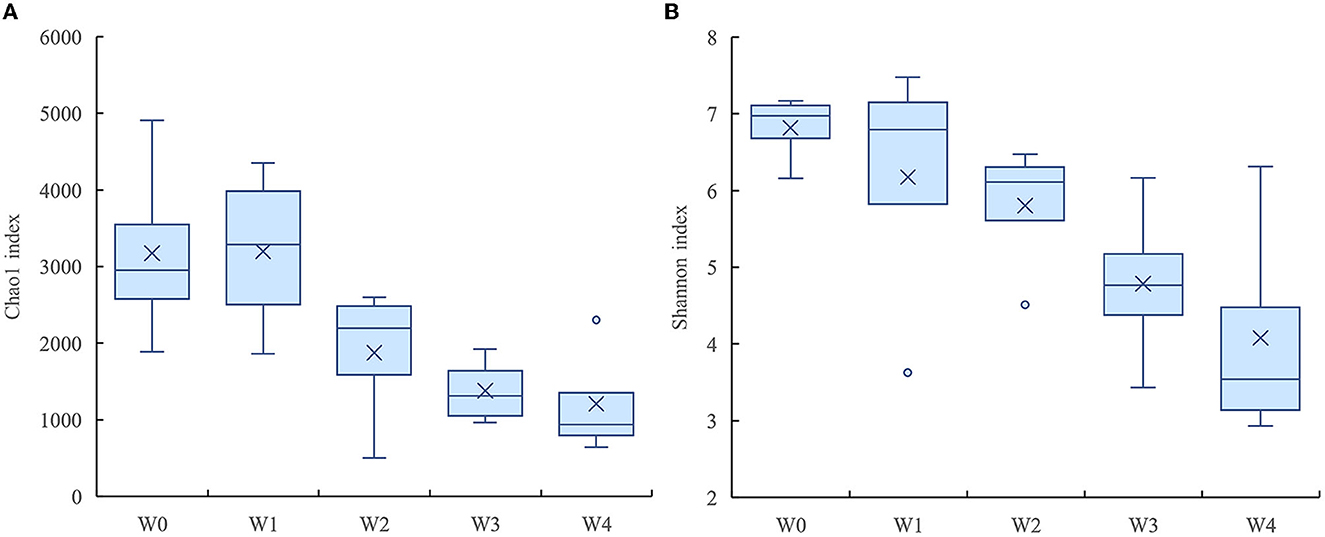
Figure 4. Effects of different concentrations of florfenicol on the Shannon index (A) and chao1 index (B) of denitrifying microorganisms in an aquatic microcosm model.
The denitrifying microbial community composition of each sample was statistical under different species classification levels, as shown in Figure 5. The identified species were classified into 7 phylum, 12 classes, 2 orders, 23 families, 52 genera, and 106 species. The relative abundance of each species in the denitrifying communities was calculated, with the relative abundance of unclassified species ranging from 4.57% to 87.83%. At the phylum level, the relative abundance of Proteobacteria ranged from 8.43% to 95.37% and mainly included Betaproteobacteria, Alphaproteobacteria, and Gammaproteobacteria, which were the absolute dominant bacteria. The relative abundance of Betaproteobacteria was the highest and the relative abundance of Gammaproteobacteria was the lowest, but the relative abundance of Gammaproteobacteria in the W4D60 treatment group was 91.22%. However, the relative abundance of Gammaproteobacteria was 91.22% in 60-day water sample with 100 mg/L florfenicol. At 7 days, the relative abundances of Gemmobacter and Comamonas were increased by lower florfenicol concentrations while higher florfenicol concentrations decreased the abundance of both. However, Comamonas was more sensitive to florfenicol than Gemmobacter, and 1 mg/L florfenicol reduced its abundance. The relative abundance of Pseudomonas and Pelomonas was <0.2% on day 0, but they showed different responses to the florfenicol concentration on day 60. The relative abundance of Pelomonas was suppressed in groups with florfenicol added, except for 0.1 mg/L, where the relative abundance of Pelomonas reached 71.79%. The relative abundance of Acidovorax was not markedly changed at 7 days and markedly suppressed at 30 days, whereas higher florfenicol concentrations increased the relative abundance of Acidovorax at 60 days and returned to the 0-day level. In addition, the relative abundance of Azovibrio showed a steep increase to 29.84% in the 30-day water sample with 10 mg/L florfenicol. Higher florfenicol concentrations also caused enrichment of some unclassified species, for example, the relative abundance of OTU5, OTU13 and OTU14 could reach 41.83%, 25.77% and 37.55% respectively.
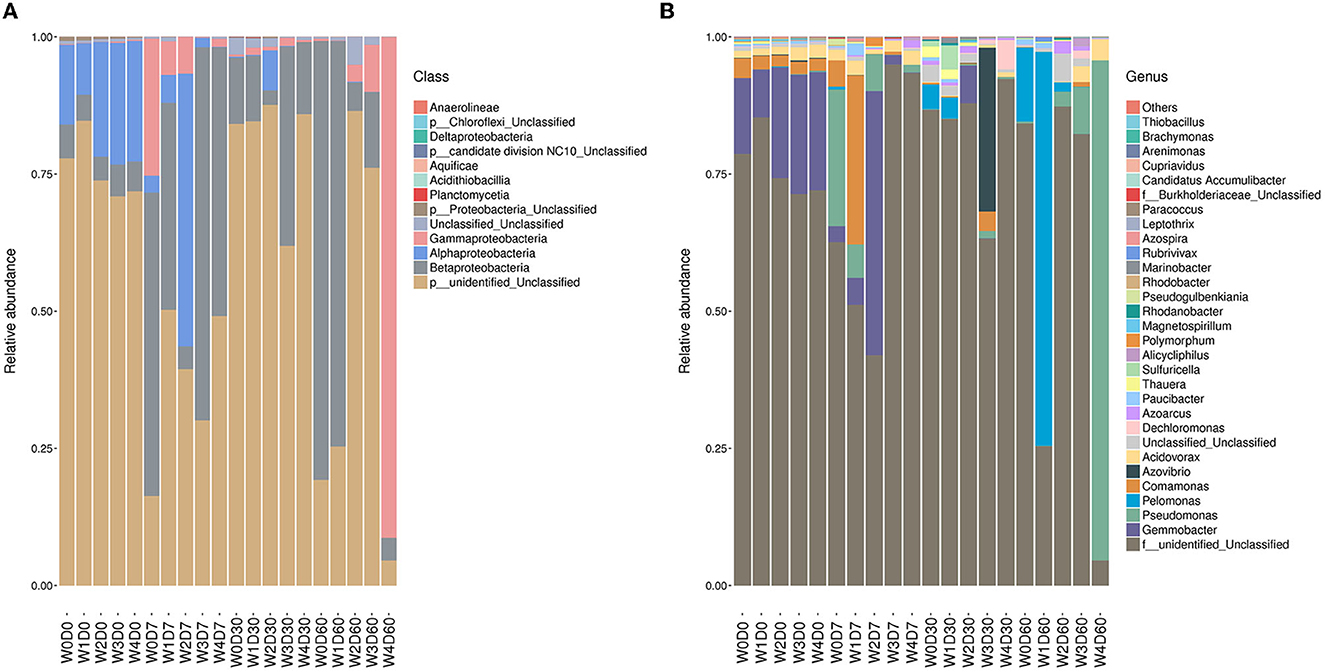
Figure 5. Effects of different concentrations of florfenicol on class-level (A) and genus-level (B) denitrifying microorganisms abundance in an aquatic microcosm model.
3.4. Beta diversity
PCoA (Principal Co-ordinates Analysis) and UPGMA (Unweighted pair group method with arithmetic mean) clustering methods were used to express the similarity and differences degree between different samples. As shown in Figure 6A, the contribution of PC1 and PC2 principal coordinates to sample differences were 22.11% and 13.96% respectively in PCoA. The 0-day samples were completely separated from others samples collected at other times, indicating that sampling time had a greater effect on the degree of variation between samples. Samples with higher florfenicol concentrations added were closer together, where they had a higher similarity of denitrifying microbial communities. The similarity of denitrifying microbial communities in samples with lower florfenicol concentrations added was mainly influenced by time. Samples with similar denitrifying microbial community structure were clustered together in the clustering analysis. As shown in Figure 6B, 0-day samples were clustered together; 30- and 60-day samples with lower florfenicol concentrations were clustered with control (W0D30, W1D30, W0D60, W1D60 and W2D60); samples with higher florfenicol concentrations (W3D7, W4D7, W4D30W, W3D30, and W3D60) were clustered together.
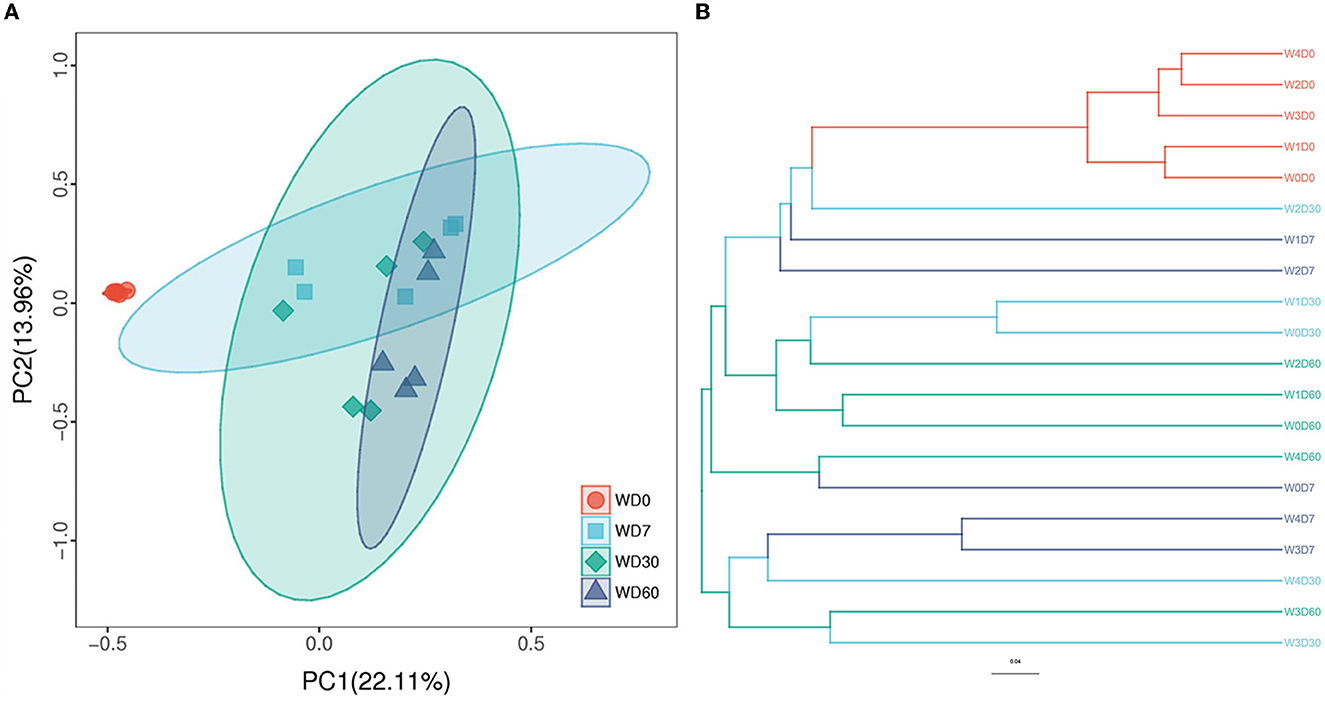
Figure 6. Principal Co-ordinates Analysis (PCoA) and unweighted pair group method with arithmetic mean (UPGMA) reveal the degree of similarity and difference of denitrifying microorganisms between different samples in an aquatic microcosm model. (A) The results of the PCoA analysis; (B) The results of the UPGMA analysis.
3.5. Linkage among water properties, nirS gene abundance, and denitrifying communities
The top 10 denitrifying bacteria with higher relative abundance in each sample were selected, and redundancy analysis (RAD) was used to investigate its correlation with water properties and nirS gene abundance. The results are shown in Figure 7, beta proteobacterium and Pseudomonas were strongly correlated with nirS gene abundance and showed a positive correlation, while Gemmobacter, Pelomonas and Paucibacter showed a clearly negative correction. Pseudomonas, beta proteobacterium, Azovibrio and Dechloromonas showed positive correlations with NO and NH; while Gemmobacter, Comamonas and Paucibacter showed negative correlations. Acidovorax was sensitive to variations in pH and showed a significant negative correlation.
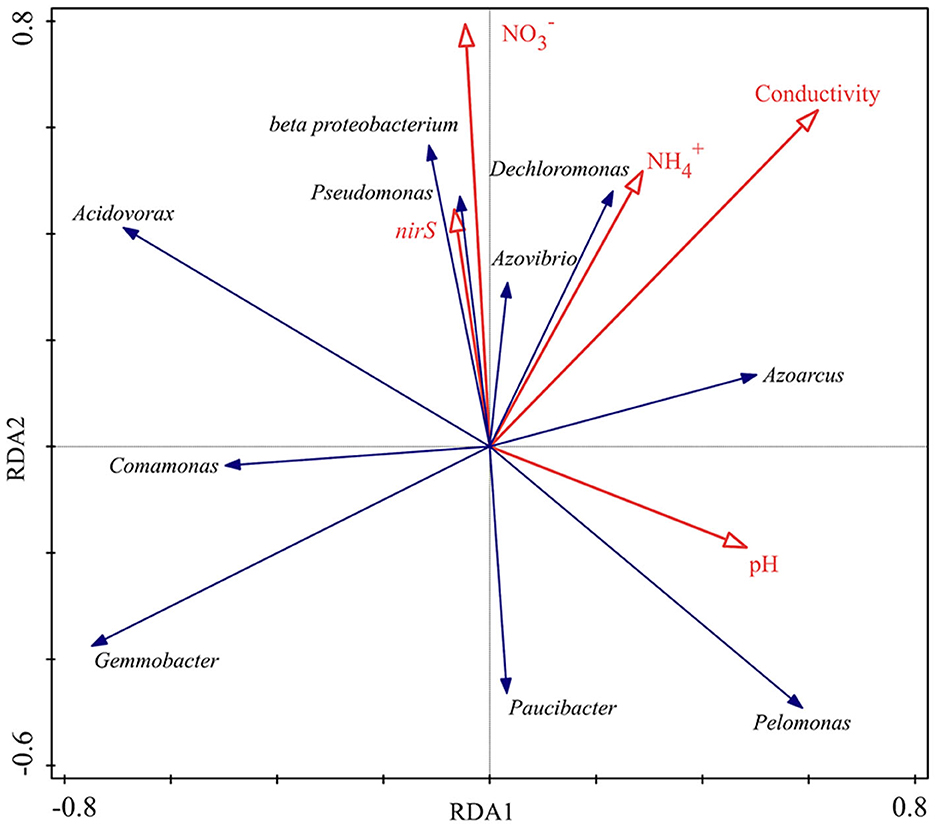
Figure 7. Correlation analysis between denitrifying communities and water properties, nirS gene abundance in an aquatic microcosm model.
4. Discussion
Florfenicol inhibits the production of bacterial proteins by affecting the transpeptidase reaction of peptidyl transferase for bacterial inhibition. In aquaculture, florfenicol was fed in therapeutic doses together with feed. Bulk quantities of unabsorbed florfenicol persist in the water body until degradation, with the sediment carrying low florfenicol levels (29). The degradation products of florfenicol may still be biologically active in water-borne environments (15). The metabolic processes of microorganisms are also inevitably affected by the residual antibiotics in the water body. This study showed that 100 mg/L florfenicol caused the obvious change in water properties, massive accumulation of nitrate and ammonium nitrogen, increase in conductivity, and a dramatic change in water pH. The denitrifying enzyme activities are closely linked to substrate concentrations (30). Dynamic changes in substrate concentrations lead to variation in denitrifying enzyme activities, and the sensitivities of denitrifying enzymes activities to environmental conditions are different with different substrate concentrations (31). It has been shown that nitrate reduction might be the rate-limiting step in the denitrification process (32), so it was confirmed that high florfenicol concentrations hindered the water denitrification process and inhibited nitrate reductase activity in our study. Many studies have also reported the inhibitory effect of antibiotics on denitrification. For example, sulfamethoxazole, ciprofloxacin, amoxicillin and aureomycin showed various inhibition on the denitrification performance of microorganisms in short-term acute stress experiments and led to a reduction in nitrate removal (32). Chen et al. (31) reported that norfloxacin inhibited nitrate reduction and nitrate reductase activity, and enhanced nitrite reductase activity to some extent.
The inhibition of denitrification of antibiotics may be achieved by affecting bacterial growth, gene expression, denitrifying enzyme activity, and electron transport system activity (31). The dynamics of nirS genes, which encode key enzymes for denitrification, were quantified by qPCR. The results showed that florfenicol addition stimulated different increases of nirS gene abundance in water body. The abundance of nirS genes showed two response patterns to different florfenicol concentrations in aquatic microcosm models. Apart from 100 mg/L florfenicol, nirS genes abundance in water tended to increase and then decrease over time; it showed orders of magnitude increase at day 7; and the higher florfenicol concentration was used, the more obvious gene abundance increase compared with the control. While 100 mg/L florfenicol addition led to a sustained increase of nirS gene abundance throughout all experimental phases. It has been shown that florfenicol slightly promoted the nitrite reductase-coding gene in a short time (25, 32). As shown by Xie and Wang (33, 34), florfenicol promoted the nirK gene in sediment. However, the richness and diversity of denitrifying microorganisms showed an opposite trend to nirS genes in our experiment. Florfenicol inhibited the richness and diversity of denitrifying microorganisms while stimulating the abundance of nirS genes to increase, but the inhibition recovered slowly with time. Higher florfenicol concentrations resulted in greater inhibition of richness and diversity indices and a slower recovery process. This is in line with our previous findings regarding the effects of florfenicol on the richness and diversity of bacteria in water bodies (35). The inhibition of denitrifying microorganisms caused by florfenicol may lead to reduced denitrification capacity in water bodies. An et al. reported that sulfamethoxazole inhibited the denitrifying process by effectively suppressing the expression of denitrifying genes, rather than a reduce in total denitrifying microorganism abundance (36). This differs from our experimental results.
Effects of antibiotics on the denitrification process are mainly attributed to influences of denitrifying microorganisms. Florfenicol significantly affected the bacterial community structure and even led to a complete reversal of the microbial community (35). In this study florfenicol also had a significant effect on the water denitrifying microbial community structure, with higher and lower concentrations of florfenicol showed two developmental trends in the water denitrifying microbial community evolution over time. Pseudomonas is the dominant bacterium under florfenicol stress and considered as a potential host bacterium for florfenicol resistance genes. In this study, we found that Pseudomonas was also an important denitrifying bacterium under high florfenicol concentrations. This may be due to its ability to carry both the florfenicol resistance gene and the denitrification function gene, which could become the superior bacteria under florfenicol stress.
The studies have been reported about the effects and mechanisms of types, addition times and concentrations of antibiotics on denitrification in different environmental media such as sediment, soil, groundwater and wastewater (37). Different conclusions have also been drawn from the effects of antibiotics on denitrification, including promotion, no significant effect and inhibition. However, there are limited studies on the effects of florfenicol on denitrification, and its effects on water denitrification have not been reported. Our study filled this gap and provided preliminary understanding of the response characteristics of water denitrifying microorganisms to florfenicol exposure. Our study provided a preliminary understanding of the response characteristics of water denitrifying microorganisms to florfenicol exposure.
5. Conclusion
In this study, florfenicol additions changed the water properties and disrupted the original community structure. High florfenicol concentrations caused a massive accumulation of nitrate and ammonium nitrogen. nirS gene abundance showed orders of magnitude changes. Two trends in nirS gene abundance were observed, increased then decreased and increased continuously, depending on florfenicol concentrations. Florfenicol addition reduced the diversity and richness of nirS-type denitrifying microbial communities, which gradually recovered over time.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA943538.
Author contributions
Funding acquisition and supervision: YM. Investigation and writing-original draft: TZ. Methodology: TZ and JP. Project administration and data curation: YDi and YDa. Software and visualization: SL and XX. Writing-review and editing: JP and YM. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Natural Science Foundation of Guangdong Province, China (2023A1515012181); the Zhanjiang Science and Technology Bureau 2021 Provincial Science and Technology Special Funds (Big Special + Task List) Competitive Allocation Project (2021A05231); and the Start-Up Research Project of Maoming Laboratory, Guangdong Laboratory for Lingnan Modern Agriculture (2021TDQD002).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Lulijwa R, Rupia EJ, Alfaro AC. Antibiotic use in aquaculture, policies and regulation, health and environmental risks: a review of the top 15 major producers. Rev Aquacult. (2020) 12:640–63. doi: 10.1111/raq.12344
2. Miranda CD, Godoy FA, Lee MR. Current status of the use of antibiotics and the antimicrobial resistance in the Chilean salmon farms. Front. Microbiol. (2018) 9:389. doi: 10.3389/fmicb.2018.01284
3. Dong D, Sun HY Qi ZL, Liu XL. Improving microbial bioremediation efficiency of intensive aquacultural wastewater based on bacterial pollutant metabolism kinetics analysis. Chemosphere. (2021) 265:129151. doi: 10.1016/j.chemosphere.2020.129151
4. Mishra SS, Markande AR, Keluskar RP, Karunasagar I, Nayak BB. Simultaneous nitrification and denitrification by novel heterotrophs in remediation of fish processing effluent. J Basic microb. (2015) 55:772–9. doi: 10.1002/jobm.201400783
5. FAO. The state of world Fisheries and aquaculture 2020. Sustainability in action. Rome: FAO. (2020). doi: 10.4060/ca9229en
6. Wang R, Xu Q, Chen CL Li XK, Zhang CF, Zhang DD. Microbial nitrogen removal in synthetic aquaculture wastewater by fixed-bed baffled reactors packed with different biofilm carrier materials. Bioresource Technol. (2021) 331:125045. doi: 10.1016/j.biortech.2021.125045
7. Danner MC, Robertson A, Behrends V, Reiss J. Antibiotic pollution in surface fresh waters: occurrence and effects. Sci Total Environ. (2019) 664:793–804. doi: 10.1016/j.scitotenv.2019.01.406
8. Carvalho IT, Santos L. Antibiotics in the aquatic environments: a review of the European scenario. Environ Int. (2016) 94:736–57. doi: 10.1016/j.envint.2016.06.025
9. Segura PA, François M, Gagnon C, Sauvé S. Review of the occurrence of anti-infectives in contaminated wastewaters and natural and drinking waters. Environ Health Persp. (2009) 117:675–84. doi: 10.1289/ehp.11776
10. Rozas M, Enríquez R. Piscirickettsiosis and Piscirickettsia salmonis in fish: a review. J Fish Dis. (2014) 37:163–88. doi: 10.1111/jfd.12211
11. Gaikowski MP, Whitsel MK, Charles S, Schleis SM, Crouch LS, Endris RG. Depletion of florfenicol amine in tilapia (Oreochromis sp) maintained in a recirculating aquaculture system following Aquaflor®-medicated feed therapy. Aquac Res. (2015) 46:1842. doi: 10.1111/are.12340
12. Zhang Q, Guo R, Zhang YQ, Zhang XY, Mostafizur RMD, Liu PW, et al. Study on florfenicol-contained feeds delivery in Dicentrarchus labrax seawater recirculating and flowing aquaculture systems. Aquaculture. (2020) 526:735326. doi: 10.1016/j.aquaculture.2020.735326
13. Qiao M, Ying GG, Singer AC, Zhu YG. Review of antibiotic resistance in China and its environment. Environ Int. (2018) 110:160–72. doi: 10.1016/j.envint.2017.10.016
14. Liu JZ, Fung KF, Chen ZL, Zeng ZL, Zhang J. Pharmacokinetics of florfenicol in healthy pigs and in pigs experimentally infected with Actinobacillus pleuropneumoniae. Antimicrob Agents Ch. (2003) 47:820–3. doi: 10.1128/AAC.47.2.820-823.2003
15. Mitchell SM, Ullman JL, Teel AL, Watts RJ. Hydrolysis of amphenicol and macrolide antibiotics: Chloramphenicol, florfenicol, spiramycin, and tylosin. Chemosphere. (2015) 134:504–11. doi: 10.1016/j.chemosphere.2014.08.050
16. Zong HM, Ma DY, Wang JY, Hu JT. Research on florfenicol residue in coastal area of Dalian (Northern China) and analysis of functional diversity of the microbial community in marine sediment. B Environ Contami Tox. (2010) 84:245–9. doi: 10.1007/s00128-009-9923-1
17. Na GS, Fang XD, Cai YQ, Ge LK, Zong HM, Yuan XT, et al. Occurrence, distribution, and bioaccumulation of antibiotics in coastal environment of Dalian, China. Mar Pollut Bull. (2013) 69:233–7. doi: 10.1016/j.marpolbul.2012.12.028
18. Jara B, Tucca F, Srain B M, Méjanelle L, Aranda M, Fernández C, et al. Antibiotics florfenicol and flumequine in the water column and sediments of Puyuhuapi Fjord, Chilean Patagonia[J]. Chemosphere. (2021) 275:130029. doi: 10.1016/j.chemosphere.2021.130029
19. Hu Y, Zhu QQ, Wang YW, Liao CY, Jiang GB. A short review of human exposure to antibiotics based on urinary biomonitoring. Sci Total Environ. (2022) 830:154775. doi: 10.1016/j.scitotenv.2022.154775
20. Barreto FM, da Silva MR, Braga PAC, Bragotto APA, Hisano H, Reyes FGR. Evaluation of the leaching of florfenicol from coated medicated fish feed into water. Environ Pollut. (2018) 242:1245–52. doi: 10.1016/j.envpol.2018.08.017
21. Pang M, Huang ZL, Zhang L. Experimental study on the effect of nitrate nitrogen content on denitrification in the typical area of the west of Taihu Lake. Acta Scientiae Circumstantiae. (2023) 43:223–30. doi: 10.13671/j.hjkxxb.2022.0240
22. Huang C, Liu Q, Li ZL, Ma Xd, Hou YN, Ren NQ, et al. Relationship between functional bacteria in a denitrification desulfurization system under autotrophic, heterotrophic, and mixotrophic conditions. Water Res. (2021) 188:116526. doi: 10.1016/j.watres.2020.116526
23. Wang M, Xie Xy, Wang MZ, Wu J, Zhou Q, Sun YX. The bacterial microbiota in florfenicol contaminated soils: The antibiotic resistome and the nitrogen cycle. Environ Pollut. (2020) 259:113901. doi: 10.1016/j.envpol.2019.113901
24. Cui PY, Fan FL, Yin C, Song AL, Huang PR, Tang YJ, et al. Long-term organic and inorganic fertilization alters temperature sensitivity of potential N[[sb]]2[[/s]]O emissions and associated microbes. Soil Biol Biochem. (2016) 93:131–41. doi: 10.1016/j.soilbio.2015.11.005
25. Cydzik-Kwiatkowska A, Rusanowska P, Zielińska M, Bernat K, Wojnowska-Baryła I. Structure of nitrogen-converting communities induced by hydraulic retention time and COD/N ratio in constantly aerated granular sludge reactors treating digester supernatant. Bioresource Technol. (2014) 154:162–70. doi: 10.1016/j.biortech.2013.11.099
26. Chen QQ, Wu WD, Zhang ZZ, Xu JJ, Jin RC. Inhibitory effects of sulfamethoxazole on denitrifying granule properties: short- and long-term tests. Bioresource Technol. (2017) 233:391–8. doi: 10.1016/j.biortech.2017.02.102
27. Guo HX, Chen Z, Lu CC, Guo JB Li HB, Song YY, et al. Effect and ameliorative mechanisms of polyoxometalates on the denitrification under sulfonamide antibiotics stress. Bioresource Technol. (2020) 305:123073. doi: 10.1016/j.biortech.2020.123073
28. Li ZL, Cheng R, Chen F, Lin XQ, Yao XJ, Liang B, et al. Selective stress of antibiotics on microbial denitrification: inhibitory effects, dynamics of microbial community structure and function. J Hazard Mater. (2021) 405:124366. doi: 10.1016/j.jhazmat.2020.124366
29. Liu CW, Meng Y, Zhu XH Li BP, Xia LP, Xu ZH, et al. Elimination and migration of florfenicol and its metabolic residues in Letaurus punetaus and aquaculture environment under pond conditions. Chinese Fishery Quality and Standards. (2019) 9:30–40. doi: 10.3969/j.issn.2095-1833.2019.05.005
30. Li MJ, Qian WJ, Gao YQ, Shi L, Liu CX. Functional enzyme-based approach for linking microbial community functions with biogeochemical process kinetics. Environ Sci Technol. (2017) 51:11848–57. doi: 10.1021/acs.est.7b03158
31. Chen LP, Huang FY, Zhang C, Zhang J, Liu F, Guan XY. Effects of norfloxacin on nitrate reduction and dynamic denitrifying enzymes activities in groundwater. Environ Pollut. (2021) 273:116492. doi: 10.1016/j.envpol.2021.116492
32. Guo GX, Deng H, Qiao M, Yao HY, Zhu YG. Effect of long-term wastewater irrigation on potential denitrification and denitrifying communities in soils at the watershed scale. Environ Sci Technol. (2013) 47:3105–13. doi: 10.1021/es304714a
33. Xie XY. The Regulation of Acyl-Homoserine Lactones Signal Molecules on the Response of Sediment Nitrogen Cycle Under Florfenicol Stress. Guangzhou: South China Agricultural University. (2020) doi: 10.27152/d.cnki.ghanu.2020.000986
34. Wang M. The Response Characteristics of Nitrogen Cycle and ARGs in Soilsand Aquatic Sediment Under The Stress of FLorfenicol and Sulfadimidine. Guangzhou: South China Agricultural University. (2019) doi: 10.27152/d.cnki.ghanu.2019.001308
35. Zhang TY, Ding YX, Peng JJ Dai Y, Luo SS, Liu WC, et al. Effects of broad-spectrum antibiotic (florfenicol) on resistance genes and bacterial community structure of water and sediments in an aquatic microcosm model. Antibiotics. (2022) 11:1299. doi: 10.3390/antibiotics11101299
36. An YL, Qin XM. Effects of sulfamethoxazole on the denitrifying process in anoxic activated sludge and the responses of denitrifying microorganisms. Water Sci Technol. (2018) 78:1228–36. doi: 10.2166/wst.2018.394
Keywords: florfenicol, nirS gene, denitrifying microbial community, water body microorganisms, aquatic microcosm model
Citation: Zhang T, Peng J, Dai Y, Xie X, Luo S, Ding Y and Ma Y (2023) Effect of florfenicol on nirS-type denitrifying communities structure of water in an aquatic microcosm model. Front. Vet. Sci. 10:1205394. doi: 10.3389/fvets.2023.1205394
Received: 13 April 2023; Accepted: 03 July 2023;
Published: 17 July 2023.
Edited by:
Nora Mestorino, National University of La Plata, ArgentinaReviewed by:
Alejandro Perretta, University of the Republic, UruguayAyhan Filazi, Ankara University, Türkiye
Copyright © 2023 Zhang, Peng, Dai, Xie, Luo, Ding and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yi Ma, bWF5aTc2MUBnZG91LmVkdS5jbg==
†These authors have contributed equally to this work and share first authorship
 Tengyue Zhang
Tengyue Zhang Jinju Peng1†
Jinju Peng1†