- 1Key Laboratory of Grassland Ecosystem (Ministry of Education), College of Grassland Science, Gansu Agricultural University, Lanzhou, China
- 2Gansu Agricultural University-Massey University Research Centre for Grassland Biodiversity, Gansu Agricultural University, Lanzhou, China
Introduction: Reproductive suppression is an adaptive strategy that affects the success rate and reproductive efficiency in animals, which in turn affects population continuation and evolution. However, no studies on the miRNAs in testicular development and spermatogenesis regulatory mechanisms under reproductive suppression have been reported.
Methods: In this study, the differentially expressed (DE) miRNAs, miRNA–mRNA interaction network and function of the plateau zokor testicular cells of non-breeders and breeders during the breeding season were comprehensively analyzed by transcriptomics.
Results: In total, 381 known and 94 novel miRNAs were determined. Compared with that in the breeders, 70 downregulated and 68 upregulated DE miRNAs were identified in the non-breeders. We predicted 1670 significant target mRNAs by analyzing the miRNA and mRNA expression profiles. According to the miRNA–mRNA interaction network, the target mRNAs of the DE miRNAs were related to testicular development and spermatogenesis. GO indicate that the target mRNAs were related to testicular development and spermatogenesis. KEGG indicate that pathways of target mRNAs enrichment related to testicular development, spermatogenesis, and energy metabolism. PROK2 was determined as the target mRNA of rno-miR-143-3p.
Discussion: Our study offers a basis for the regulatory mechanisms of miRNAs in testicular development and spermatogenesis in plateau zokor under reproductive suppression and offers a reference for reproductive regulation.
Introduction
Animals have evolved to display diverse reproductive strategies in response to both abiotic and biotic factors, and reproductive suppression is an adaptive strategy that affects the success and efficiency of reproduction, which in turn influence survival and evolution (1, 2). Reproductive suppression refers to the inhibition or damage of normal reproductive development, reproductive physiology, or reproductive behavior in animals due to the impact of specific environmental factors or internal conditions, resulting in reduced or lost reproductive capacity (1, 2). Reproductive suppression manifests as delayed puberty or inhibition of the development of secondary sexual signs in adolescence (3, 4), delayed gonadal and gametic development (5), and reduced serum testosterone or estradiol levels (5, 6). Transcriptome analysis of the testicular cells of non-breeding naked mole rats (Heterochephalus glaber) under reproductive suppression revealed downregulation of the genes that regulate testicular development and spermatogenesis, such as those related to postmeiotic spermatogenesis (e.g., PRM1, ODF3, and AKAP4) (7), lipid biosynthesis processes and steroid hormone biosynthetic pathways (e.g., CYP11A1, ABCG8, and SCARB1) (8), and endocrine signaling (e.g., SSTR3, TAC4, and CAMP) (9).
In addition to mRNAs, non-coding RNAs play a major role in reproductive regulation. Sperm cells house different RNA families (including miRNA, piRNA, snoRNA, and snRNA), 7% of which are miRNAs (10). miRNA is non-coding RNA that is 19–24 nt long, was first found in Cryptomeria elegans to regulate gene expression by forming a complementary structure with the 3’UTR of the target mRNA. miRNAs play a critical role in several stages of spermatogenesis, including self-renewal and differentiation of spermatogonia stem cells (SSC), spermatocyte meiosis, spermatid maturation, and proliferation of Sertoli cells (11). Increased expression of let-7 downregulates LIN28D expression, inhibits MYCN and CCDN1 expression, and promotes SSC differentiation (12). miR-224 regulates the self-renewal of mouse SSC by regulating PLZF and GFRα1 (13). miR-34c upregulates the expression of meiotic regulatory genes (STRA8, SCP3, and DAZL) through the target gene NANOS2 and promotes SSC differentiation and meiotic processes (14). The upregulation of miR-122 leads to the degradation of TNP2, which leads to sperm defects as TNP2 completes the compression and condensation of chromatin required for spermatogenesis (15). Additionally, miR-133b regulates the expression of CCNB1 and CCND1 through the target gene GLI3 and regulates the multiplication of Sertoli cells (16). Therefore, the function of miRNAs in testicular development and spermatogenesis must be investigated to increase our understanding of the regulatory mechanisms underlying reproductive suppression.
Plateau zokors (Eospalax baileyi) are subterranean rodents residing in alpine grasslands and meadows (17). They are regarded as “ecosystem engineers,” in the grassland ecosystem of the Qinghai–Tibet plateau because at a normal population density they promote the soil nutrient cycle, improve soil structure, and increase plant species diversity. However, at high population density, their activities such as digging the soil to build nests and competing with livestock for forage aggravates grassland degradation and soil erosion (18). Therefore, the population density of the plateau zokors needs to be controlled to maintained environmental balance. Recently, we observed reproductive suppression in plateau zokors and that spermatogenic arrest occurred at the spermatogonia stage in non-breeder (19), and the expression of the genes involved in spermatogenesis was downregulated during the meiotic and postmeiotic stages in non-breeders compared with that in breeders (19). Hence, the corresponding mRNAs may inhibit testicular development and spermatogenesis in non-breeders, resulting in spermatogenic arrest (19). Although miRNAs regulate mRNA expression and play a major role in spermatogenesis in plateau zokors under reproductive suppression, their specific role in testicular development and spermatogenesis under reproductive suppression has not been well studied, which limits our understanding of the regulatory mechanisms underlying reproductive suppression. In our study, we analyzed the miRNA expression profiles in the testicular cells of non-breeding and breeding plateau zokors during the breeding season and integrated the data with the mRNA expression profiles of testicular cells obtained in our previous studies (19). We aimed to predict the miRNA–mRNA interaction network and identify central miRNA and target mRNA that regulate testicular development and spermatogenesis in plateau zokors under reproductive suppression. Our results would provide perspectives on the molecular regulatory mechanisms underlying reproductive suppression in animals.
Materials and methods
Animals
Plateau zokors were caught alive using tube traps at the end of April 2020. We collected the testes for total RNA extraction (19). We previously found that based on testis weight, serum hormones, hematoxylin and eosin staining, and immunohistochemistry data, the plateau zokors were divided into non-breeders (BSA) during the breeding season and breeders (BSB) during the breeding season (19). Firstly, by observation and palpation, males in the breeding period had relatively large testicles, and the bulge was observed in the abdominal groin. Secondly, the testicular weight and testicular coefficient of reproductive inhibited individuals were significantly lower than those of normal reproductive individuals. Finally, testosterone level was decreased and spermatogenesis was blocked in reproductive inhibited individuals plateau zokors (19).
RNA sequencing
Three testicular samples each from the BSA and BSB groups were selected for transcriptome sequencing and sent to Beijing NovoGene (Beijing, China) (19).
Analysis and prediction of miRNA
The small RNA reads were aligned with the reference genome assembly (Nannospalax galili) using Bowtie (20). Known miRNAs were identified using the miRBase database and mirdeep2 (21). Novel miRNAs were predicted using mirdeep2 and miREvo (22). We selected the differentially expressed (DE) miRNAs based on the conditions |log2foldchange| > 1 and p-adj value <0.05.
Target mRNA prediction and enrichment analysis
To understand the underlying role of miRNAs in the testes, we predicted the target mRNAs of the DE miRNAs. The candidate target mRNA of the DE miRNA was predicted using miRanda (23) and RNAhybrid (24). To explore the underlying functions of the DE miRNAs, we performed Gene ontology (GO) functional analyses and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses of the significant target mRNAs of the DE miRNAs. GO and KEGG enrichment analysis was performed for candidate target mRNAs using DAVID (25).
miRNA–mRNA interactions network construction
We constructed an miRNA–mRNA interaction network by integrating the miRNA expression profile in the plateau zokor testicular cells with the mRNA expression profiles of testicular cells obtained in our previous studies (19). The miRNA–mRNA interaction network was integrated and visualized using Cytoscape 3.9.1 (26).
Validation using real-time quantitative PCR (RT-qPCR)
Nine DE miRNAs (miR-34b-5p, miR-148b-3p, miR-450b-5p, miR-9a-5p, miR-15b-5, miR-499-5p, miR-429, miR-128-3p, and miR-143-3p) were randomly screened for RT-qPCR analysis. Detection of changes in the expression level of PROK2. The RT-qPCR primer sequence information are shown in Supplementary Table 1. Method reference: Zhang et al. (27).
Dual-luciferase reporter assays
Fragments comprising PROK2-3′UTR (wild-type) of miR-143-3p and PROK2-3′UTR (mutant-type) of miR-143-3p and a positive control (miRNA inhibitor) were designed using Target Scan, and RNA22 and synthesized. The sequence information is listed in Table 1. Then, the PROK2-3′-UTR fragment containing the wild-type (WT-PROK2) and mutant (Mut-PROK2) sequences was cloned into the psiCHECK-2 vector. The miRNA (or miRNA inhibitor), recombinant vector, and Lipofectamine transfection reagent were mixed and transfected into HEK-293T cells. After 48 h of transfection, the activity of luciferase was determined.
Results
Summary of sRNA libraries
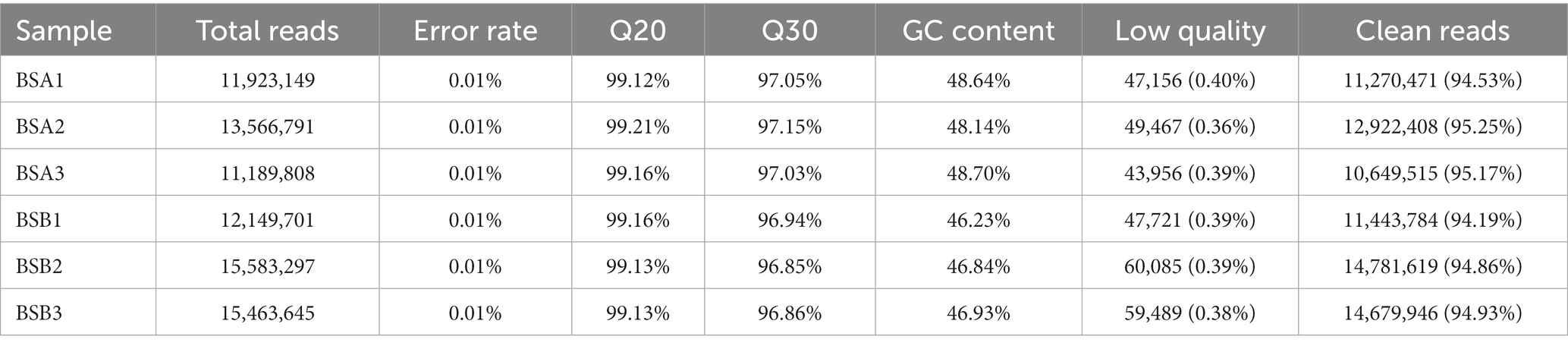
To identify miRNAs in plateau zokor testicular cells, we constructed and sequenced six small RNA (sRNA) libraries designated as BSA1, BSA2, BSA3, BSB1, BSB2, and BSB3 libraries containing 11,923,149, 13,566,791, 11,189,808, 12,149,701, 15,583,297, and 15,463,645 raw reads, respectively. The low-quality sequences, erroneous reads, and reads with N > 10% were removed, and sequences shorter than 18 nt were discarded. In total, 11,270,471, 12,922,408, 10,649,515, 11,443,784, 14,781,619, and 14,679,946 clean reads were generated in the BSA1, BSA2, BSA3, BSB1, BSB2, and BSB3 libraries, respectively, and were used for further analysis (Table 2).
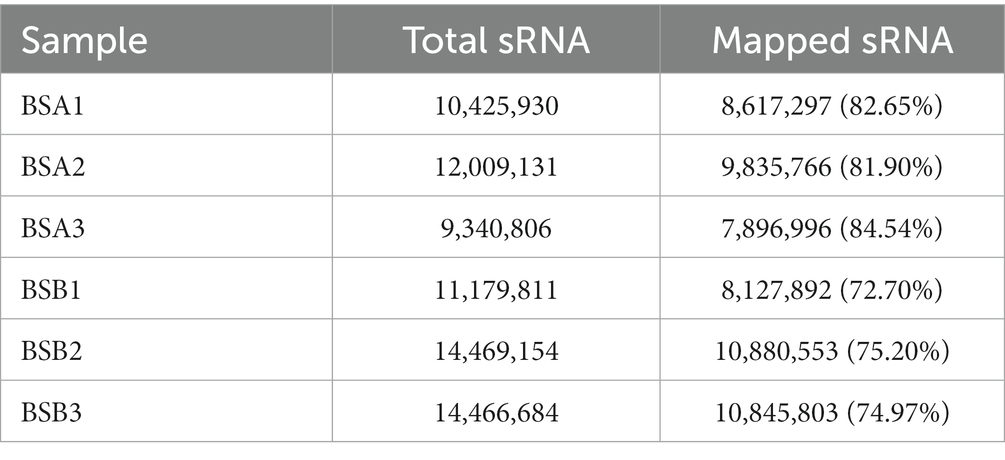
Mapping the length-filtered sRNAs to the reference sequences (N. galili) covered 72.70–84.54% of the reads from six libraries (Table 3). Pearson’s correlation analysis showed that the correlation between each group of samples was strong, providing reliable data for the next step (Supplementary Table 2).
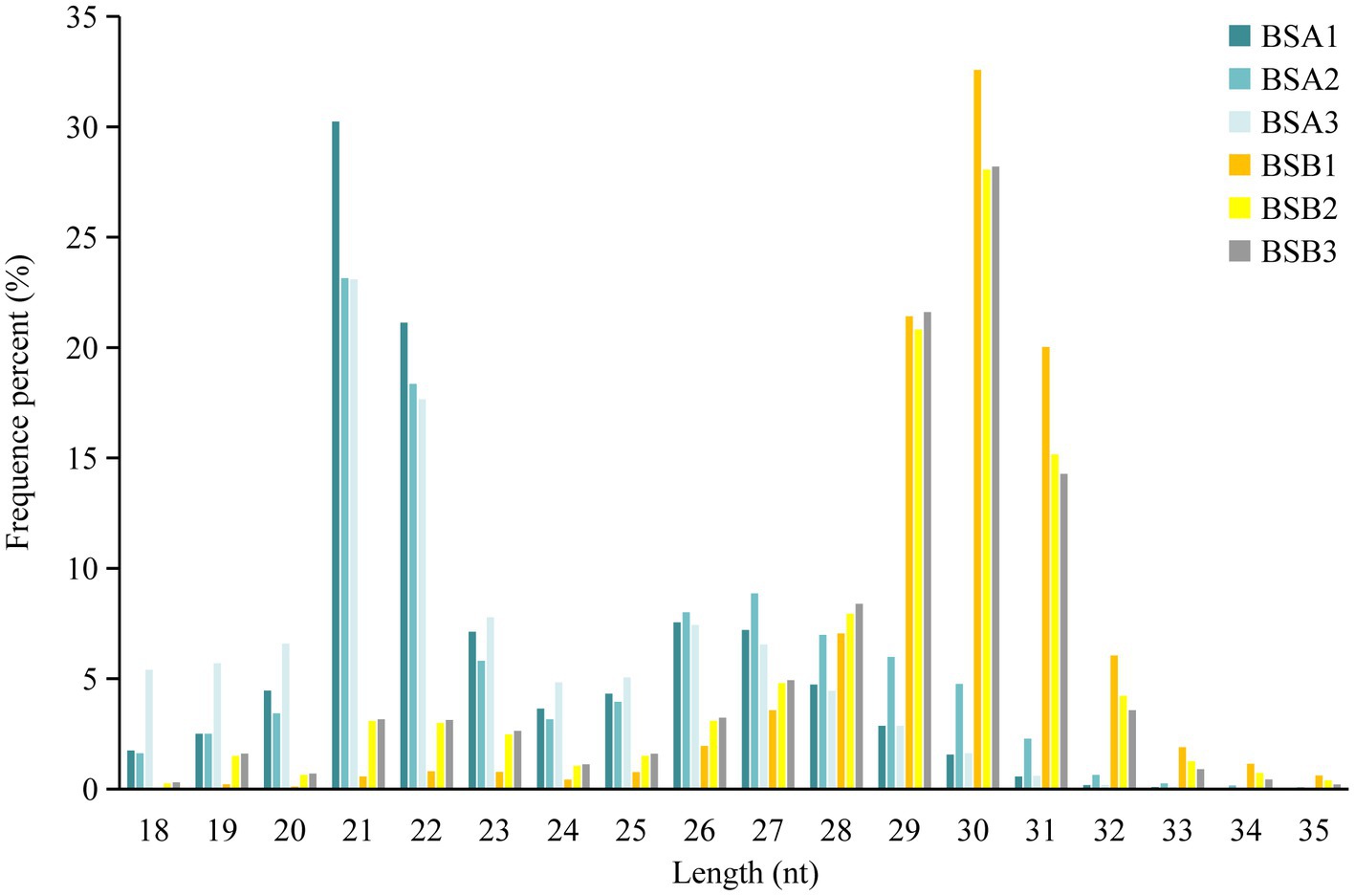
Comparative analysis of the reading size distribution and abundance of six sRNA libraries revealed that the sRNA peaks for BSA1, BSA2, and BSA3 were 21–22 nt, whereas that for BSB1, BSB2, and BSB3 were 29–31 nt. sRNAs longer than 25 nt were Piwi-interacting RNAs (piRNAs), which have been reported to be abundant in the mature testicular cells in animals. The sRNA size distribution and abundance of BSA and BSB groups differed (Figure 1).
Differentially expressed miRNAs in plateau zokor testicular cells
The reads mapped to the reference sequence reads were aligned to the range of the sequences indicated in miRBase to obtain details of the known miRNAs. Novel miRNAs were predicted using miREvo and mirdeep2. Statistical analysis indicated that among all the sRNAs annotated in our study, the known miRNAs and other sRNA accounted for 46.29 and 42.20% of the total number of sRNA reads in BSA, respectively, whereas other sRNAs were the most abundant type in BSB, accounting for 88.04% of the small sRNA reads (Figure 2A). The other sRNAs in the BSB group may be piRNAs. Statistical results indicate that in total 475 miRNAs comprising 381 known miRNAs and 94 novel miRNAs from both BSA and BSB plateau zokor testicular cells were predicted in the six libraries (Supplementary Table 3).
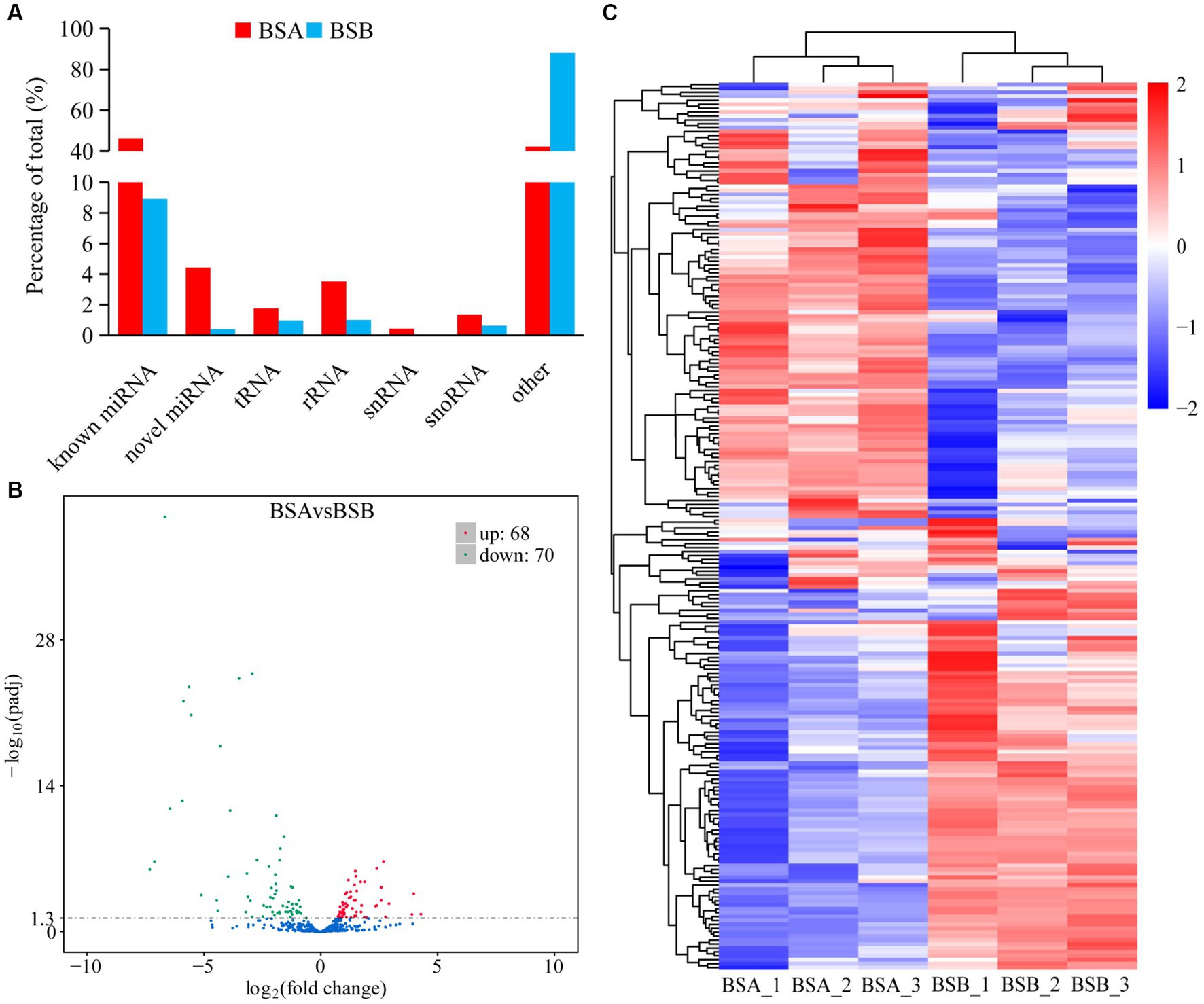
Figure 2. Differentially expressed miRNA in plateau zokor testes. (A) the percentage of small RNA types in plateau zokor testes; (B) The plot of the number of differentially expressed miRNA; (C) differentially expressed miRNA clustering graph.
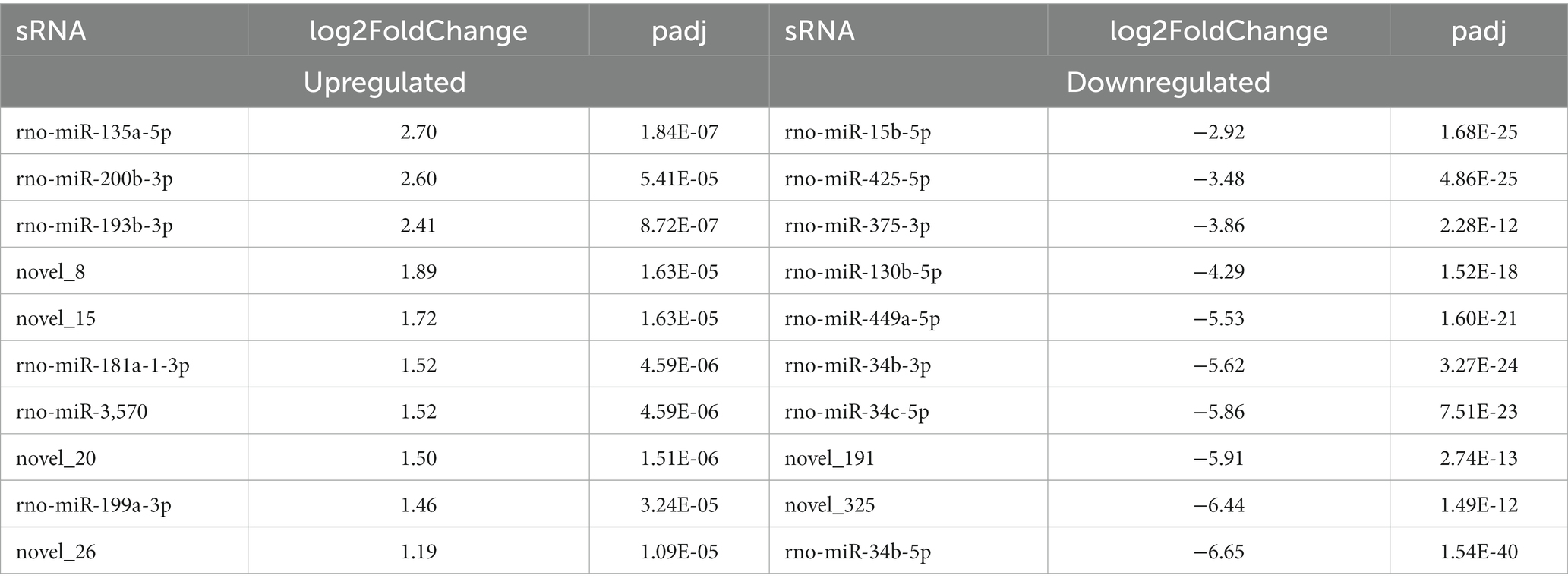
We further analyzed the DE miRNAs derived from both the BSA and BSB testicular cells. The volcano plot shows 138 DE miRNAs (including 105 known and 33 novel miRNAs) from BSA and BSB testicular cells, of which 68 were upregulated and 70 were downregulated in the BSA compared with that in the BSB (Figure 2B; Supplementary Table 4). The top 10 DE miRNAs are listed in Table 4. The miRNA expression patterns in BSA and BSB were different from those in the cluster diagram (Figure 2C).
Target mRNA prediction and enrichment analysis
In total, 4,860 miRNA–mRNA target pairs were predicted, and all 138 DE miRNAs were associated with the target mRNAs previously screened in the BSA and BSB groups (Supplementary Table 5). In total, 1,670 significant target mRNAs were identified (Supplementary Table 6).
Gene ontology analysis indicated that the top ten biological processes in which the downregulated target mRNAs in BSA were significantly enriched compared with that in BSB included spermatogenesis, cell differentiation, cell projection organization, flagellated sperm motility, multicellular organism development, and lipid metabolic process (p < 0.05), and the upregulated target mRNAs were significantly enriched in positive regulation of apoptotic process, extracellular matrix organization, response to drug, and negative regulation of smoothened signaling pathway (p < 0.05). The top 10 cellular components in which the downregulated target mRNAs were significantly enriched include motile cilium, sperm midpiece, sperm flagellum, acrosomal vesicle, and sperm principal piece (p < 0.05), whereas the top 10 molecular functions in which the downregulated target mRNAs were significantly enriched include phosphoric diester hydrolase activity, anaphase-promoting complex binding, calcium ion binding, and ubiquitin ligase activator activity (p < 0.05; Figure 3).
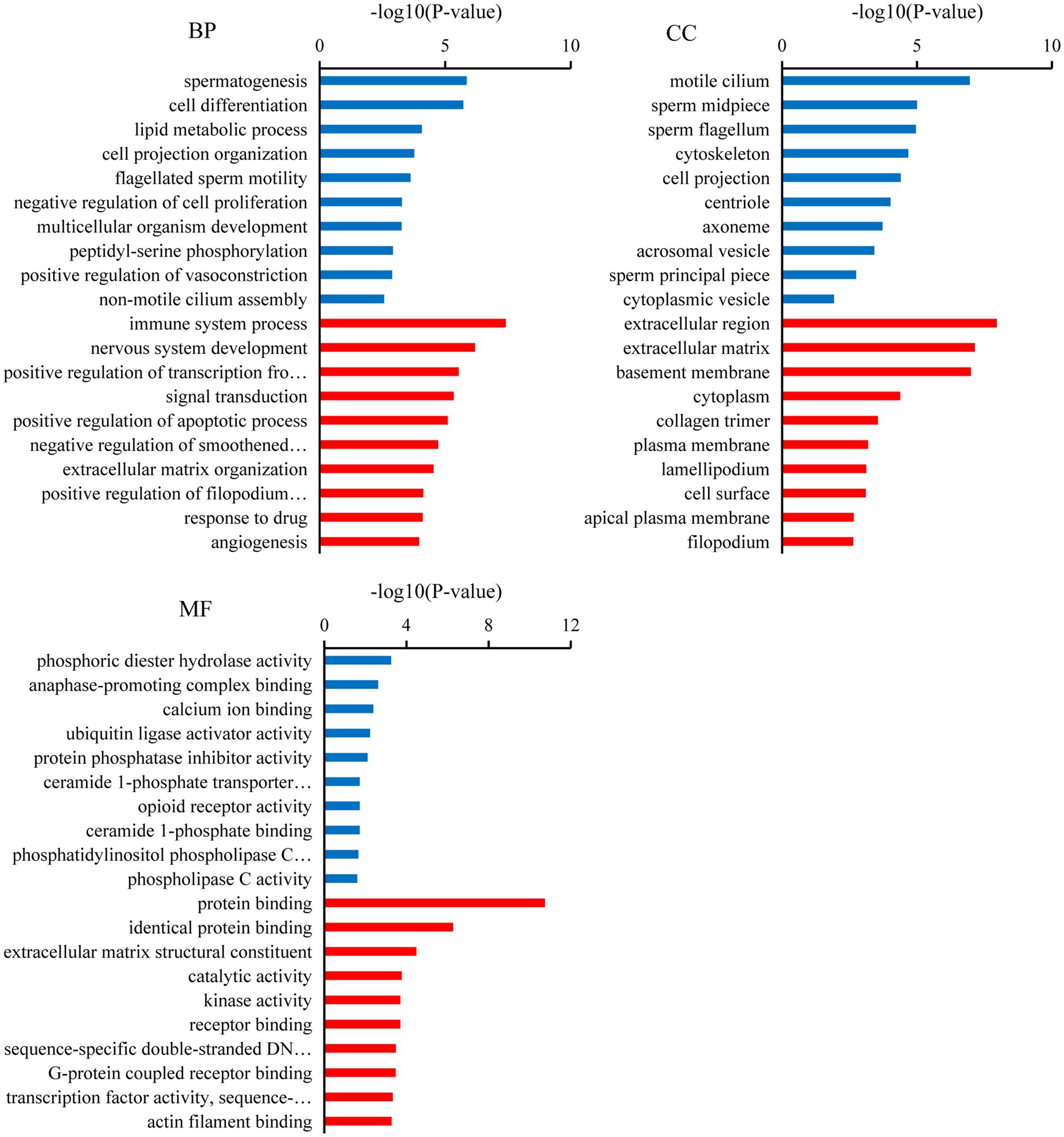
Figure 3. Gene ontology (GO) analysis of the candidate target mRNAs. A color code indicates overrepresentation (red) and underrepresentation (blue).
Kyoto Encyclopedia of Genes and Genomes analysis showed that the downregulated target mRNAs in BSA were significantly enriched in oocyte meiosis, AMPK signaling pathway, cell cycle, progesterone-mediated oocyte maturation, metabolic pathways, thyroid hormone signaling pathway, inositol phosphate metabolism, fructose and mannose metabolism, and phosphatidylinositol signaling system (p < 0.05). The upregulated target mRNAs were significantly enriched in ECM-receptor interaction, focal adhesion, and chemokine signaling pathway, Rap1 signaling pathway (p < 0.05; Figure 4).
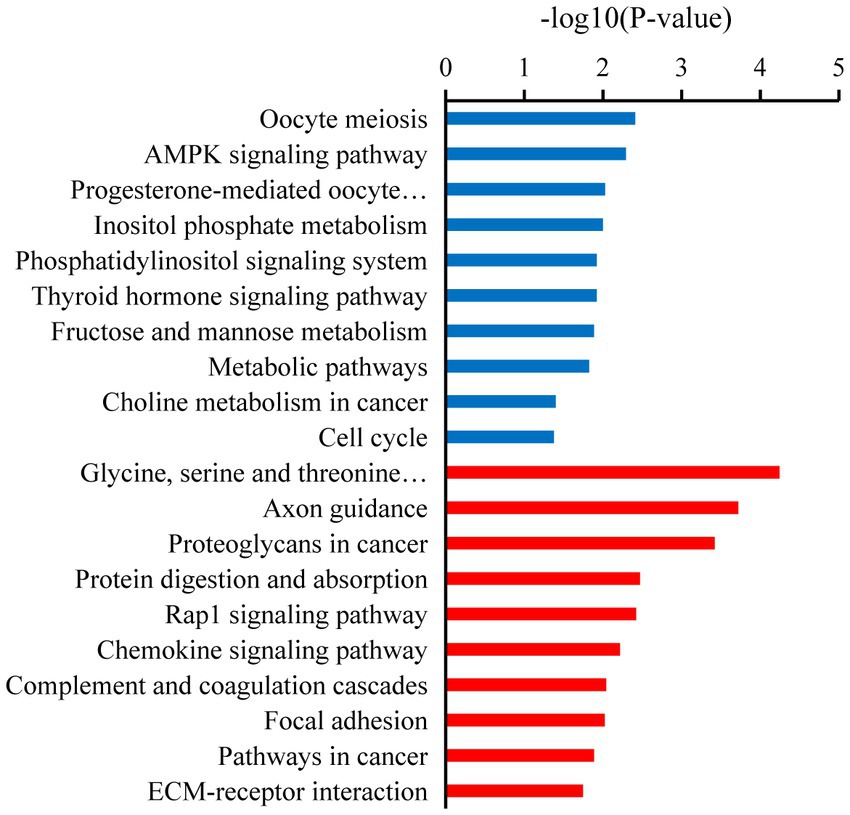
Figure 4. Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment pathways of the candidate target mRNAs. A color code indicates overrepresentation (red) and underrepresentation (blue).
miRNA–mRNA interactions network construction
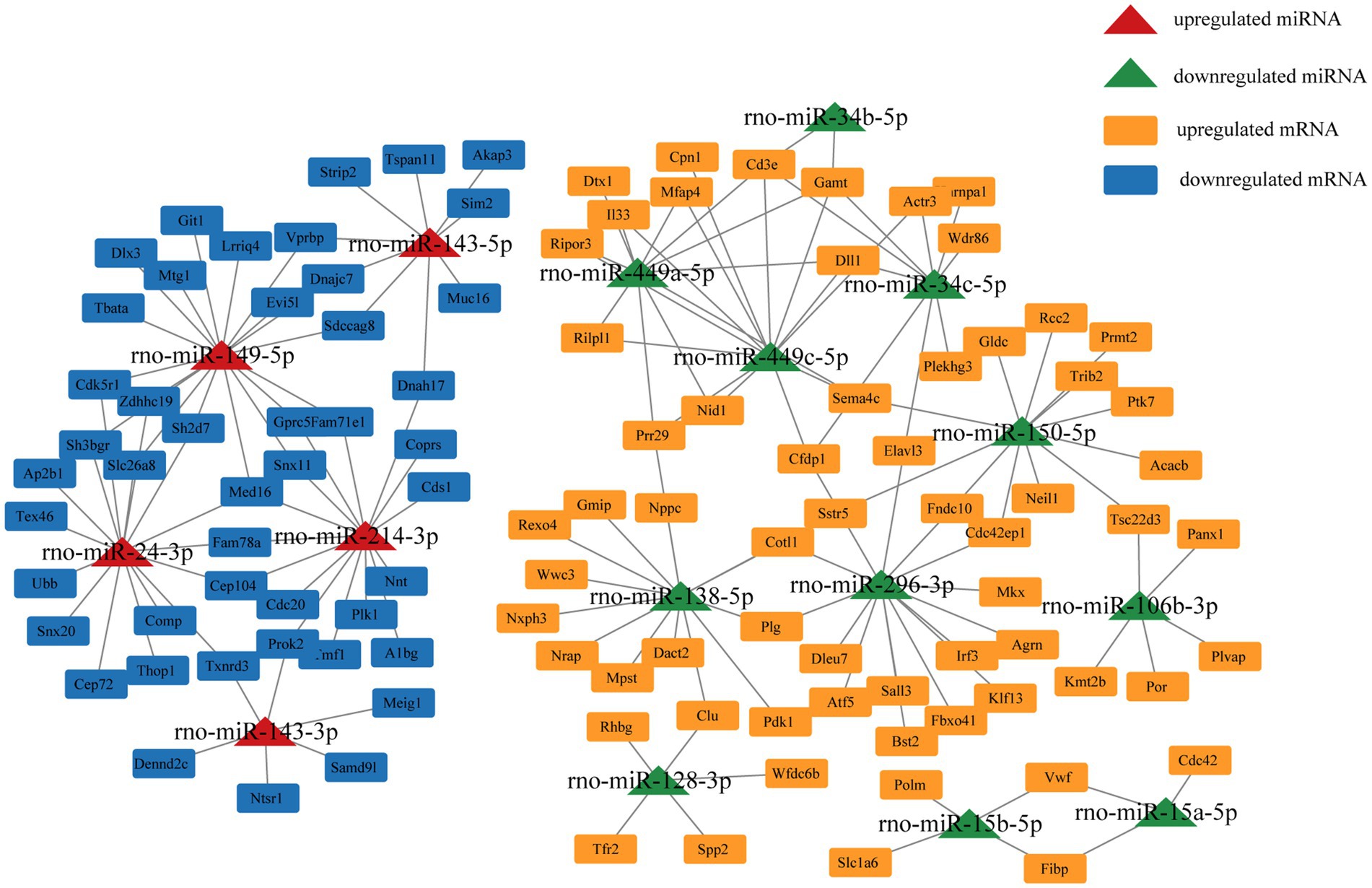
We selected miRNA–mRNA target pairs related to testicular development and spermatogenesis from 30,229 miRNA–mRNA target pairs and visualized them using Cytoscape (Figure 5). These key DE miRNAs (rno-miR-449a/c-5p, rno-miR-149-5p, rno-miR-296-3p, rno-miR-138-5p, rno-miR-143, and rno-miR-214-3p) significantly regulate mRNA expression in plateau zokor testicular cells. They also play a central role in spermatogenesis in plateau zokors.
Validation using RT-qPCR
We randomly selected nine DE miRNAs (miR-34b-5p, miR-450b-5p, miR-9a-5p, miR-15b-5, miR-499-5p, miR-429, miR-148b-3p, miR-128-3p, and miR-143-3p) for RT-qPCR analysis. The variation trend of qRT-PCR results was consistent with that of RNA-seq data. Our results prove that RNA-seq data was accurate (Figure 6).
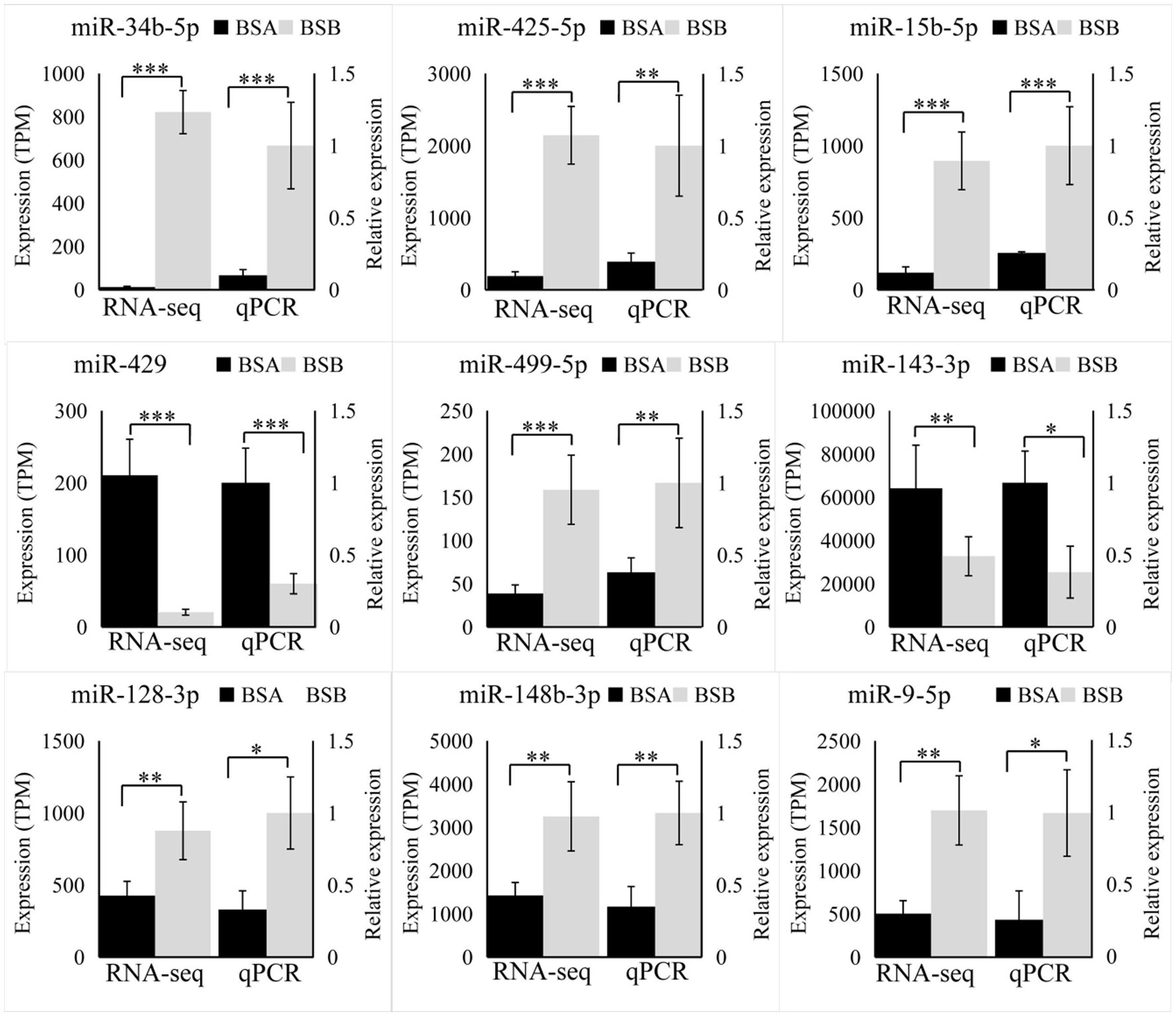
Figure 6. Validation by RT-qPCR. Data were shown as mean ± SD. The right axis represents gene expression levels verified by qRT-PCR, the left axis represents the expression levels in TPM units of RNA-seq. *p < 0.05, **p < 0.01, ***p < 0.001.
rno-miR-143-3p targets PROK2
According to GO and KEGG pathway analyses, the biological process and signaling pathway of target mRNA enrichment are associated with testicular development and spermatogenesis in plateau zokors. PROK2 is closely related to spermatogenesis. The expression level of PROK2 in BSB is significantly higher than that in BSA (Figure 7A). According to a bioinformatics database, PROK2 is a major target pair of miR-143-3p. Thus, we chose miR-143-3p and PROK2 to validate the miRNA–mRNA regulatory relationship. We found that co-transfection of rno-miR-143-3p mimics for 48 h reduced the luciferase activity of PROK2 receptors by 24.4% compared to the negative control (p < 0.05). The results show that rno-miR-143-3p directly targets PROK2-3′UTR (Figure 7B).
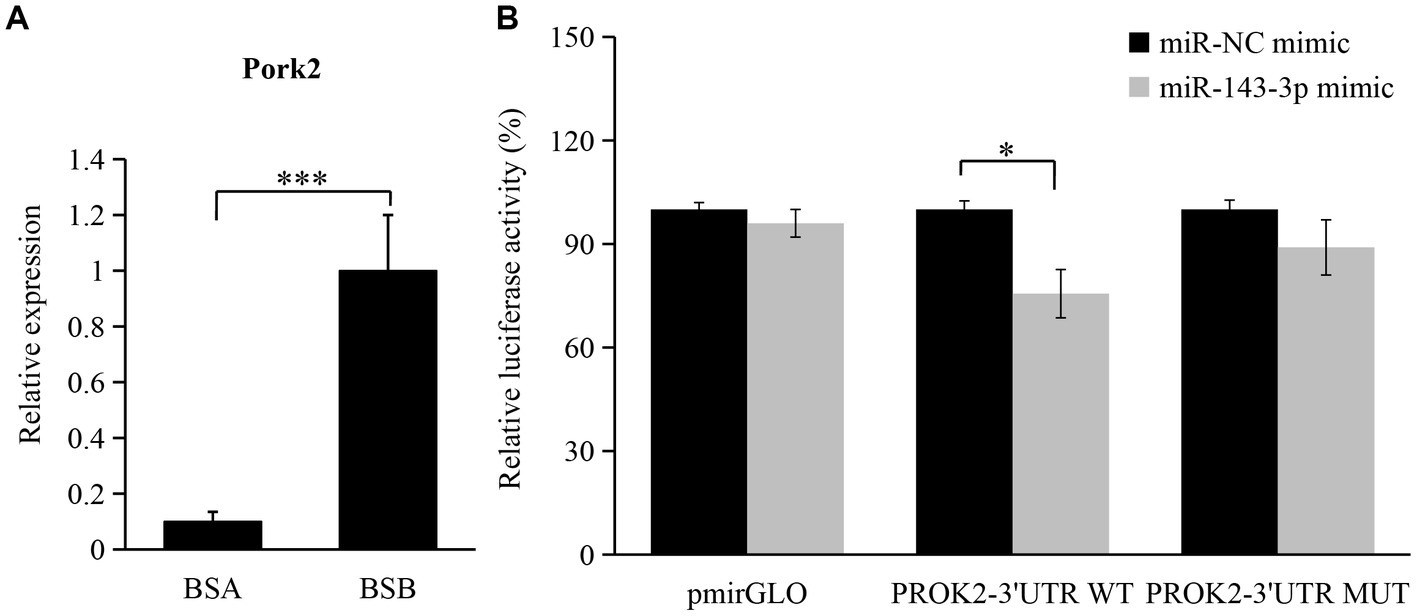
Figure 7. Validation of regulatory relationship between rno-miR-143-3p and PROK2. (A) Changes in the expression level of PROK2 in BSA and BSB; (B) miR-143-3p mimic was cotransfected with PROK2-3 ‘UTR WT or PROK2-3 ‘UTR MUT to detect the luciferase activity for 48 h. *p < 0.05, ***p < 0.001.
Discussion
Studying the regulatory mechanisms of reproductive suppression is helpful in understanding the reproductive strategies in animals. In our study, we comparatively analyzed the miRNA expression profiles of plateau zokor testicular cells in both non-breeders and breeders and integrated them with the mRNA expression profiles obtained in previous studies. We determined 138 DE miRNAs, of which 68 were upregulated and 70 were downregulated in BSA and BSB testes. The top 10 DE miRNAs were mostly related to spermatogenesis and testicular development. GO and KEGG enrichment analyses suggested that the target mRNAs of the DE miRNAs were associated with spermatogenesis and testicular development. We predicted the miRNA–mRNA interaction network and identified the key miRNAs (rno-miR-449a/c-5p, rno-miR-149-5p, rno-miR-296-3p, rno-miR-138-5p, rno-miR-143, and rno-miR-214-3p) that may regulate testicular development and spermatogenesis in plateau zokors under reproductive suppression. We confirmed that PROK2 is a binding site for miR-143-3p.
Based on sRNA length distribution, we found that sRNAs from non-breeder testes were mainly distributed between 21 and 22 nt, which was consistent with the result that miRNA expression levels in the immature testes were higher than that in the mature testes (28). However, most sRNAs in the testes of the breeders were piRNAs with a length of 29–31 nt (29), and piRNAs were most abundantly expressed in male spermatogenesis. piRNAs are abundantly expressed in germ cells at meiosis I stage and are lost before the production of mature sperm (29, 30). Our RNA-seq data included piRNAs from the plateau zokor testicular cells and did not ignore the differences in miRNA and piRNA length distribution. This suggests that piRNA may be the key factor in suppressing spermatogenesis in plateau zokors under reproductive suppression.
In total, 138 (68 upregulated and 70 downregulated) DE miRNAs were determined in the six libraries. Among the top 10 DE miRNAs, miR-34b/c-5p, miR-34b-3p, miR-449a-5p, miR-130b-5p, miR-375-3p, miR-425-5p, and miR-15b-5p were significantly downregulated. Previous studies state that miR-34b/c is preferentially expressed in germ cells during the meiosis stage in mouse testes. Germ cells carry miRNA-34c as it is vital during the first meiosis stage of spermatogenesis (31). miR-449a is upregulated at the beginning of meiosis and is preferentially expressed in spermatocytes and haploid spermatids. miR-449 promotes proliferation and inhibits apoptosis of Sertoli cells by suppressing PTEN expression (32). Thus, reduced expression of miR-34/449 in the testes of non-breeders may reflect impaired spermatogenesis during meiosis. miR-130a activates SMAD5 through the TGF-β-PI3K/AKT signaling pathway and promotes Sertoli cell growth in immature porcine (Sus scrofa) testes (33). The expression level of miR-375 in mature porcine testes was 5.4 times higher than that in immature porcine testes (28), leading to speculation that miR-375 affects porcine testicular development. miR-15a is significantly upregulated during the processes of spermatogonia to spermatocyte transformation (34), and its expression is significantly downregulated in dysspermia and asthenospermia (35). Thus, we conclude that the low expression of these miRNAs would affect testicular development and spermatogenesis in plateau zokors under reproductive suppression.
In our study, several GO terms associated with the male reproductive process were enriched, such as spermatogenesis, flagellated sperm motility, sperm midpiece, sperm flagellum, and sperm principal piece (36). Upregulated candidate target mRNA were significantly enriched in the apoptotic process. A dramatic increase in germ cell apoptosis occurs in some pathological conditions, which include idiopathic infertility in males. It has now been shown that the expression of apoptotic markers increases in the cryptorchidism testis which compromises fertility (37). Furthermore, the mRNAs that play a key role in male reproduction were significantly upregulated in the testicular cells of breeders, whereas these mRNAs were expressed at low levels in non-breeders. This reflects the limiting non-breeder testicular development and spermatogenesis. Similarly, KEGG analysis shows that most of the downregulated target mRNAs were enriched in spermatogenesis pathways, such as the oocyte meiosis, AMPK signaling pathway, progesterone-mediated oocyte maturation, thyroid hormone signaling pathway, fructose and mannose metabolism, and phosphatidylinositol signaling system. In non-breeders, a large number of pathways related to spermatogenesis were downregulated, which would limit testicular development and spermatogenesis, whereas in breeder, these pathways were upregulated, and testicular development and spermatogenesis were normal. Male meiotic stages I and II were significantly regulated by the oocyte meiotic pathway, which was crucial for spermatogenesis (38). Progestogen stimulates spermatogenesis through the progesterone-mediated oocyte maturation pathway, plays a key role in spermatogonial differentiation (39), and activates CatSper channels to accelerate sperm maturation in rhesus monkeys (Macaca mulatta) (40). In Sertoli cells, the AMPK signaling pathway regulates energy metabolism, junction complex stability, and cell proliferation (41) and is a key regulatory factor in the energy metabolism of germ cells, providing lactic acid and maintain spermatogenesis (42). Spermatogenesis was a process highly dependent on glycolytic metabolism for energy production, and endogenous synthetic fructose was the main source of energy for spermatozoa and may be important for fertility (43). Phosphatidylinositol signaling system play a central role in spermatogenesis, including the maintenance and proliferation and survival of germ cells and dynamic remodeling of cell adhesion in Sertoli cells (44). The thyroid promotes the eventual maturation of Sertoli cells by inhibiting their proliferation (45). Upregulated candidate target mRNA were significantly enriched in the chemokine signaling pathway and Rap1 signaling pathway. Studies have shown that some chemokines may have some effect on the germ cells involved in spermatogenesis. CXCL12 (C–C motif ligand 12) and its receptor type 4 (C–C receptor type 4) signal transduction regulate SSC activity, because failure of this axis leads to SSC loss and SSC in vivo (46). Interfering with Rap1 specifically in haploid cells results in an anomalous release of immature spermatids within the lumen of seminiferous tubuli and in low sperm counts; the loss of nondifferentiated cells correlated with impaired spermatid–Sertoli cell adhesion (47). Therefore, we hypothesized that these miRNAs participate in pathway through target mRNAs to regulate testicular development, spermatogenesis, and energy metabolism in plateau zokor testicular cells. The downregulation and upregulation of these signaling pathway in non-breeder testicular cells suggesting that testicular development, spermatogenesis, and energy metabolism in non-breeders are abnormal.
The core miRNA–target mRNA interaction network may reveal a regulatory relationship between testicular development and spermatogenesis in plateau zokors. We found that rno-miR-296-3p, rno-miR-138-5p, rno-miR-449a/c-5p, rno-miR-149-5p, rno-miR-143, and rno-miR-214-3p may be key miRNAs that regulate testicular functional development in the plateau zokors. Previous research have indicated that miR-296 as a tumor suppressor and cell motion suppressor, was upregulated in mature rhesus monkey testes (48). miR-138-5p targeted CASPASE3 through the BCL2 signaling pathway and inhibited testicular cell apoptosis (49). miR-138-5p, miR-449, and miR-296-3p were down-regulated in the testes of non-breeders, which may damage spermatogenesis. The upregulation of miR-214 suppressed the target TCP11, which is a testes-specific gene that regulates haploid spermatids in inducing proper vitality in mature sperm (50). miR-149-5p regulates the multiplication and apoptosis of ovarian cancer cell (51). Nevertheless, the potential mechanism of action of rno-miR-149-5p in the testis remains unclear. miR-143-3p plays a tumor suppressive role in multiple of malignancies (e.g., liver cancer, bladder cancer, and ovarian cancer) (52). miR-143-3p suppressed cancer cell proliferation and migration in the ovary by regulating TAK1 in ovarian cancer cells (53). miR-143-3p was expressed in porcine testes at various developmental stages, suggesting it was a key regulator of spermatogenesis (54). Compared with that in patients with obstructive azoospermia, miR-143-3p was upexpressed in human spermatogonia of patients with non-obstructive azoospermia, which led to spermatogenesis failure. SMAD3 was the predicted target gene of miRNA-143-3p (55). SMAD3 belongs to the SMAD superfamily and is transformed during TGF-β activation, which is crucial for the proliferation and differentiation of spermatogonia (56). These findings support our inference that upregulated miR-149-5p, miR-143, and miR-214-3p in the testes of non-breeders may impair spermatogenesis.
Additionally, PROK2 was a secreted protein involved in cell migration, proliferation, and apoptosis. Under normal physiological conditions, PROK2 was upexpressed in the testes and was located in primary spermatocytes. PROK2 knockout mice showed impaired male sexual development and infertility, as evidenced by the absence of spermatocytes and spermatids in the seminiferous tubule (57, 58). Therefore, PROK2 is essential for regulating testicular development and spermatogenesis in animals. Bioinformatics and dual luciferase reporter analyses show that the 3′UTR of PROK2 matched the seed sequence of miR-143-3p, which downregulates PROK2 expression. However, the hypothesis that miR-143-3p regulates testicular development and spermatogenesis in plateau zokors by targeting PROK2 remains to be elucidated.
Conclusion
Our study provides the first comprehensively analyze miRNAs and mRNAs expression profiles during testicular development and spermatogenesis in plateau zokors. We identified central miRNA and target mRNA associated with testicular development and spermatogenesis in plateau zokors under reproductive suppression. Our results provide new perspectives on testicular development and spermatogenesis and help understand and illustrate the molecular regulatory mechanisms underlying reproductive suppression in plateau zokors. In particular, we confirmed the binding site of miR-143-3p on PROK2. In future, we plan to study the function of miRNAs in testicular development under reproductive suppression and elucidate the mechanism underlying spermatogenesis in plateau zokors to provide additional information for understanding the regulatory mechanisms of reproductive suppression in testicular development and spermatogenesis. We hope our findings will advance the overall understanding of reproductive suppression in mammals.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://ngdc.cncb.ac.cn/gsa/browse/CRA008119, PRJCA010603.
Ethics statement
The animal study was reviewed and approved by Animal Ethics Committee of Gansu Agricultural University (approval number GAU-LC-2020-014).
Author contributions
BY and JS did the data analysis and wrote the manuscript. BY, YK, KA, YT, QH, and DZ performed investigation and collected the samples. BY, YK, KA, YT, and QH performed the formal analysis, methodology, and software. JS did the project administration and revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by the National Natural Science Foundation of China (32272566), the “Fuxi Talent” Plan (Gaufx-02J03) of Gansu Agricultural University, and the Industrial Support Program Project (2022CYZC-47) of Gansu Provincial Education Department.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2023.1184120/full#supplementary-material
References
1. Beehner, JC, and Lu, A. Reproductive suppression in female primates: a review. Evol Anthropol. (2013) 22:226–38. doi: 10.1002/evan.21369
2. Zizzari, ZV, Jessen, A, and Koene, JM. Male reproductive suppression: not a social affair. Curr Zool. (2017) 63:573–9. doi: 10.1093/cz/zow089
3. Maggioncalda, AN, Czekala, NM, and Sapolsky, RM. Male orangutan subadulthood: a new twist on the relationship between chronic stress and developmental arrest. Am J Phys Anthropol. (2002) 118:25–32. doi: 10.1002/ajpa.10074
4. Charpentier, MJ, Tung, J, Altmann, J, and Alberts, SC. Age at maturity in wild baboons: genetic, environmental and demographic influences. Mol Ecol. (2008) 17:2026–40. doi: 10.1111/j.1365-294X.2008.03724.x
5. Freeman, AR. Female-female reproductive suppression: impacts on signals and behavior. Integr Comp Biol. (2021) 61:1827–40. doi: 10.1093/icb/icab027
6. Faulkes, CG, Abbott, DH, and Jarvis, JU. Social suppression of reproduction in male naked mole-rats, Heterocephalus glaber. J Reprod Fertil. (1991) 91:593–604. doi: 10.1530/jrf.0.0910593
7. Mulugeta, E, Marion-Poll, L, Gentien, D, Ganswindt, S B, Ganswindt, A, Bennett, NC, et al. Molecular insights into the pathways underlying naked mole-rat eusociality. BioRxiv [Epub ahead of preprint] (2017); 209932. doi: 10.1101/209932
8. Sahm, A, Platzer, M, Koch, P, Henning, Y, Bens, M, Groth, M, et al. Increased longevity due to sexual activity in mole-rats is associated with transcriptional changes in the HPA stress axis. eLife. (2021) 10:e57843. doi: 10.7554/eLife.57843
9. Bens, M, Szafranski, K, Holtze, S, Sahm, A, Groth, M, Kestler, HA, et al. Naked mole-rat transcriptome signatures of socially suppressed sexual maturation and links of reproduction to aging. BMC Biol. (2018) 16:77. doi: 10.1186/s12915-018-0546-z
10. Burgos, CF, Cikutovic, R, and Alarcón, M. MicroRNA expression in male infertility. Reprod Fertil Dev. (2022) 34:805–18. doi: 10.1071/RD21131
11. Walker, WH. Regulation of mammalian spermatogenesis by miRNAs. Semin Cell Dev Biol. (2022) 121:24–31. doi: 10.1016/j.semcdb.2021.05.009
12. Tong, MH, Mitchell, D, Evanoff, R, and Griswold, MD. Expression of Mirlet7 family microRNAs in response to retinoic acid-induced spermatogonial differentiation in mice. Biol Reprod. (2011) 85:189–97. doi: 10.1095/biolreprod.110.089458
13. Cui, N, Hao, G, Zhao, Z, Wang, F, Cao, J, and Yang, A. MicroRNA-224 regulates self-renewal of mouse spermatogonial stem cells via targeting DMRT1. J Cell Mol Med. (2016) 20:1503–12. doi: 10.1111/jcmm.12838
14. Yu, M, Mu, H, Niu, Z, Chu, Z, Zhu, H, and Hua, J. miR-34c enhances mouse spermatogonial stem cells differentiation by targeting Nanos2. J Cell Biochem. (2014) 115:232–42. doi: 10.1002/jcb.24655
15. Yu, Z, Raabe, T, and Hecht, NB. MicroRNA Mirn122a reduces expression of the posttranscriptionally regulated germ cell transition protein 2 (Tnp2) messenger RNA (mRNA) by mRNA cleavage. Biol Reprod. (2005) 73:427–33. doi: 10.1095/biolreprod.105.040998
16. Yao, C, Sun, M, Yuan, Q, Niu, M, Chen, Z, Hou, J, et al. MiRNA-133b promotes the proliferation of human Sertoli cells through targeting GLI3. Oncotarget. (2016) 7:2201–19. doi: 10.18632/oncotarget.6876
17. Yao, B, Hegab, IM, Kang, Y, Tan, Y, Zhang, D, and Su, J. Underground environment increases the differentiation of personality traits between male and female plateau zokors (Eospalax baileyi). Acta Ethol. (2023) 26:21–30. doi: 10.1007/s10211-023-00414-8
18. Kang, Y, Tan, Y, Wang, C, Yao, B, An, K, Liu, M, et al. Antifertility effects of levonorgestrel, quinestrol, and their mixture (EP-1) on plateau zokor in the Qinghai-Tibetan plateau. Integr Zool. (2022) 17:1002–16. doi: 10.1111/1749-4877.12642
19. Yao, B, An, K, Kang, Y, Tan, Y, Zhang, D, and Su, J. Reproductive suppression caused by spermatogenic arrest: transcriptomic evidence from a non-social animal. Int J Mol Sci. (2023) 24:4611. doi: 10.3390/ijms24054611
20. Langmead, B, and Salzberg, SL. Fast gapped-read alignment with bowtie 2. Nat Methods. (2012) 9:357–9. doi: 10.1038/nmeth.1923
21. Friedländer, MR, Mackowiak, SD, Li, N, Chen, W, and Rajewsky, N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. (2012) 40:37–52. doi: 10.1093/nar/gkr688
22. Wen, M, Shen, Y, Shi, S, and Tang, T. miREvo: an integrative microRNA evolutionary analysis platform for next-generation sequencing experiments. BMC Bioinf. (2012) 13:140. doi: 10.1186/1471-2105-13-140
23. Betel, D, Koppal, A, Agius, P, Sander, C, and Leslie, C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. (2010) 11:R90. doi: 10.1186/gb-2010-11-8-r90
24. Krüger, J, and Rehmsmeier, M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. (2006) 34:W451–4. doi: 10.1093/nar/gkl243
25. Sherman, BT, Hao, M, Qiu, J, Jiao, X, Baseler, MW, Lane, HC, et al. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. (2022) 50:W216–21. doi: 10.1093/nar/gkac194
26. Shannon, P, Markiel, A, Ozier, O, Baliga, NS, Wang, JT, Ramage, D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. (2003) 13:2498–504. doi: 10.1101/gr.1239303
27. Zhang, B, Yan, Z, Gao, Y, Li, J, Wang, Z, Wang, P, et al. Integrated analysis of miRNA and mRNA expression profiles in testes of landrace and Hezuo boars. Front Vet Sci. (2022) 9:942669. doi: 10.3389/fvets.2022.942669
28. Lian, C, Sun, B, Niu, S, Yang, R, Liu, B, Lu, C, et al. A comparative profile of the microRNA transcriptome in immature and mature porcine testes using Solexa deep sequencing. FEBS J. (2012) 279:964–75. doi: 10.1111/j.1742-4658.2012.08480.x
29. Lau, NC, Seto, AG, Kim, J, Kuramochi-Miyagawa, S, Nakano, T, Bartel, DP, et al. Characterization of the piRNA complex from rat testes. Science. (2006) 313:363–7. doi: 10.1126/science.1130164
30. Hirano, T, Iwasaki, YW, Lin, ZY, Imamura, M, Seki, NM, Sasaki, E, et al. Small RNA profiling and characterization of piRNA clusters in the adult testes of the common marmoset, a model primate. RNA. (2014) 20:1223–37. doi: 10.1261/rna.045310.114
31. Liu, WM, Pang, RT, Chiu, PC, Wong, BP, Lao, K, Lee, KF, et al. Sperm-borne microRNA-34c is required for the first cleavage division in mouse. Proc Natl Acad Sci U S A. (2012) 109:490–4. doi: 10.1073/pnas.1110368109
32. Gao, H, Chen, B, Luo, H, Weng, B, Tang, X, Chen, Y, et al. miR-499 promotes immature porcine Sertoli cell growth through the PI3K/AKT pathway by targeting the PTEN gene. Reproduction. (2020) 159:145–57. doi: 10.1530/REP-19-0303
33. Luo, H, Chen, B, Weng, B, Tang, X, Chen, Y, Yang, A, et al. miR-130a promotes immature porcine Sertoli cell growth by activating SMAD5 through the TGF-β-PI3K/AKT signaling pathway. FASEB J. (2020) 34:15164–79. doi: 10.1096/fj.202001384R
34. Teng, Y, Wang, Y, Fu, J, Cheng, X, Miao, S, and Wang, L. Cyclin T2: a novel miR-15a target gene involved in early spermatogenesis. FEBS Lett. (2011) 585:2493–500. doi: 10.1016/j.febslet.2011.06.031
35. Curry, E, Safranski, TJ, and Pratt, SL. Differential expression of porcine sperm microRNAs and their association with sperm morphology and motility. Theriogenology. (2011) 76:1532–9. doi: 10.1016/j.theriogenology.2011.06.025
36. Cannarella, R, Condorelli, RA, Mongioì, LM, La Vignera, S, and Calogero, AE. Molecular biology of spermatogenesis: novel targets of apparently idiopathic male infertility. Int J Mol Sci. (2020) 21:1728. doi: 10.3390/ijms21051728
37. Shukla, KK, Mahdi, AA, and Rajender, S. Apoptosis, spermatogenesis and male infertility. Front Biosci. (2012) 4:746–54. doi: 10.2741/415
38. Li, B, He, X, Zhao, Y, Bai, D, du, M, Song, L, et al. Transcriptome profiling of developing testes and spermatogenesis in the Mongolian horse. BMC Genet. (2020) 21:46. doi: 10.1186/s12863-020-00843-5
39. Skaftnesmo, KO, Edvardsen, RB, Furmanek, T, Crespo, D, Andersson, E, Kleppe, L, et al. Integrative testis transcriptome analysis reveals differentially expressed miRNAs and their mRNA targets during early puberty in Atlantic salmon. BMC Genomics. (2017) 18:801. doi: 10.1186/s12864-017-4205-5
40. Zhu, Z, Li, C, Yang, S, Tian, R, Wang, J, Yuan, Q, et al. Dynamics of the transcriptome during human spermatogenesis: predicting the potential key genes regulating male gametes generation. Sci Rep. (2016) 6:19069. doi: 10.1038/srep19069
41. Ni, FD, Hao, SL, and Yang, WX. Multiple signaling pathways in Sertoli cells: recent findings in spermatogenesis. Cell Death Dis. (2019) 10:541. doi: 10.1038/s41419-019-1782-z
42. Galardo, MN, Riera, MF, Pellizzari, EH, Sobarzo, C, Scarcelli, R, Denduchis, B, et al. Adenosine regulates Sertoli cell function by activating AMPK. Mol Cell Endocrinol. (2010) 330:49–58. doi: 10.1016/j.mce.2010.08.007
43. Hannou, SA, Haslam, DE, McKeown, NM, and Herman, MA. Fructose metabolism and metabolic disease. J Clin Invest. (2018) 128:545–55. doi: 10.1172/JCI96702
44. Brill, JA, Yildirim, S, and Fabian, L. Phosphoinositide signaling in sperm development. Semin Cell Dev Biol. (2016) 59:2–9. doi: 10.1016/j.semcdb.2016.06.010
45. Shah, W, Khan, R, Shah, B, Khan, A, Dil, S, Liu, W, et al. The molecular mechanism of sex hormones on Sertoli cell development and proliferation. Front Endocrinol. (2021) 12:648141. doi: 10.3389/fendo.2021.648141
46. Yang, QE, Kim, D, Kaucher, A, Oatley, MJ, and Oatley, JM. CXCL12-CXCR4 signaling is required for the maintenance of mouse spermatogonial stem cells. J Cell Sci. (2013) 126:1009–20. doi: 10.1242/jcs.119826
47. Aivatiadou, E, Mattei, E, Ceriani, M, Tilia, L, and Berruti, G. Impaired fertility and spermiogenetic disorders with loss of cell adhesion in male mice expressing an interfering Rap1 mutant. Mol Biol Cell. (2007) 18:1530–42. doi: 10.1091/mbc.e06-10-0902
48. Vaira, V, Faversani, A, Dohi, T, Montorsi, M, Augello, C, Gatti, S, et al. miR-296 regulation of a cell polarity-cell plasticity module controls tumor progression. Oncogene. (2012) 31:27–38. doi: 10.1038/onc.2011.209
49. He, L, Gong, H, You, S, Zhang, C, Zhong, C, and Li, L. miRNA-138-5p suppresses cigarette smoke-induced apoptosis in testicular cells by targeting Caspase-3 through the Bcl-2 signaling pathway. J Biochem Mol Toxicol. (2021) 35:e22783. doi: 10.1002/jbt.22783
50. Castaneda, JM, Miyata, H, Archambeault, DR, Satouh, Y, Yu, Z, Ikawa, M, et al. Mouse t-complex protein 11 is important for progressive motility in sperm. Biol Reprod. (2020) 102:852–62. doi: 10.1093/biolre/ioz226
51. Sun, L, Zhai, R, Zhang, L, and Zhao, S. MicroRNA-149 suppresses the proliferation and increases the sensitivity of ovarian cancer cells to cisplatin by targeting X-linked inhibitor of apoptosis. Oncol Lett. (2018) 15:7328–34. doi: 10.3892/ol.2018.8240
52. Tan, X, Shao, Y, Teng, Y, Liu, S, Li, W, Xue, L, et al. The Cancer-testis long non-coding RNA PCAT6 facilitates the malignant phenotype of ovarian cancer by sponging miR-143-3p. Front Cell Dev Biol. (2021) 9:593677. doi: 10.3389/fcell.2021.593677
53. Shi, H, Shen, H, Xu, J, Zhao, S, Yao, S, and Jiang, N. MiR-143-3p suppresses the progression of ovarian cancer. Am J Transl Res (2018) 10:866–74. PMCID: PMC5883127
54. Weng, B, Ran, M, Chen, B, Wu, M, Peng, F, Dong, L, et al. Systematic identification and characterization of miRNAs and piRNAs from porcine testes. Genes Genom. (2017) 39:1047–57. doi: 10.1007/s13258-017-0573-0
55. Yao, C, Yuan, Q, Niu, M, Fu, H, Zhou, F, Zhang, W, et al. Distinct expression profiles and novel targets of microRNAs in human spermatogonia, pachytene spermatocytes, and round spermatids between OA patients and NOA patients. Mol Therapy Nucleic Acids. (2017) 9:182–94. doi: 10.1016/j.omtn.2017.09.007
56. Young, JC, Wakitani, S, and Loveland, KL. TGF-β superfamily signaling in testis formation and early male germline development. Semin Cell Dev Biol. (2015) 45:94–103. doi: 10.1016/j.semcdb.2015.10.029
57. Pitteloud, N, Zhang, C, Pignatelli, D, Li, JD, Raivio, T, Cole, LW, et al. Loss-of-function mutation in the prokineticin 2 gene causes Kallmann syndrome and normosmic idiopathic hypogonadotropic hypogonadism. Proc Natl Acad Sci U S A. (2007) 104:17447–52. doi: 10.1073/pnas.0707173104
Keywords: microRNA, spermatogenesis, testicular development, reproductive suppression, plateau zokor
Citation: Yao B, Kang Y, An K, Tan Y, Hou Q, Zhang D and Su J (2023) Comparative analysis of microRNA and messengerRNA expression profiles in plateau zokor testicular cells under reproductive suppression. Front. Vet. Sci. 10:1184120. doi: 10.3389/fvets.2023.1184120
Edited by:
Hsiuying Wang, National Yang Ming Chiao Tung University, TaiwanCopyright © 2023 Yao, Kang, An, Tan, Hou, Zhang and Su. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Junhu Su, c3VqaEBnc2F1LmVkdS5jbg==
 Baohui Yao
Baohui Yao Yukun Kang1,2
Yukun Kang1,2 Junhu Su
Junhu Su