
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci., 23 May 2023
Sec. Animal Nutrition and Metabolism
Volume 10 - 2023 | https://doi.org/10.3389/fvets.2023.1183400
This article is part of the Research TopicGut Microbiota: Allied with Livestock Nutrition, Health, And WelfareView all 19 articles
 Seung-Won Yi1
Seung-Won Yi1 Han Gyu Lee1
Han Gyu Lee1 Eunju Kim1
Eunju Kim1 Young-Hun Jung1
Young-Hun Jung1 Eun-Yeong Bok1
Eun-Yeong Bok1 Ara Cho1
Ara Cho1 Yoon Jung Do1
Yoon Jung Do1 Tai-Young Hur1
Tai-Young Hur1 Sang-Ik Oh1,2*
Sang-Ik Oh1,2*Backgorund: Salmonella enterica serovar Typhimurium (ST) is one of the causative agents of gastroenteritis in pigs. Pigs fed a diet supplemented with raw potato starch (RPS) have improved gut health by the alteration of the microbiota composition and production of short-chain fatty acids (SCFAs). This study aimed to evaluate the effects of RPS supplementation in reducing infection severity and fecal shedding in ST-infected pigs.
Methods: The weaned experimental pigs were divided into two groups: CON (n = 6) fed a corn/soybean-based diet and TRT (n = 6) supplemented with 5% RPS. After 21 d, the pigs were inoculated with ST, and their body weight, clinical signs, and fecal shedding of ST were monitored for 14 d. At 14 d post-inoculation (dpi), the jejunum, cecum, ileum, and colon tissues were collected from euthanized pigs, and histopathological lesions and cytokine gene expression were compared. Additionally, blood samples at 2 dpi were analyzed for gene ontology enrichment. Moreover, the gutmicrobiome was analyzed using 16S rRNA metagenomic sequencing, and the SCFA concentration was measured using gas chromatography.
Results: The average daily weight gain was significantly higher in TRT than in CON during the ST infection period; however, histopathological lesion scores were significantly lower in TRT than in CON. The relative abundance of nine genera of butyrate- and acetate-producing bacteria significantly increased in TRT compared with that of only two acetate-producing bacteria in CON. Among the genes involved in the immune response, IL-18 expression level was significantly lower in the jejunum and colon in TRT than in CON. Furthermore, Reg3γ expression was significantly different in the cecum and colon of both groups.
Conclusion: The diet supplemented with RPS in weaned pigs could result in predominance of butyrate- and acetate-producing bacteria, reducing the severity of ST infection by improving the immune status.
Salmonella is an important foodborne pathogen that can cause human and animal infections (1). Among the various serovars of Salmonella, non-typhoidal Salmonella infections are a major public health problem worldwide (1). Pigs are an important non-typhoidal Salmonella infection source for humans, especially Salmonella enterica serovar Typhimurium (ST) (2). Intestinal inflammation caused by Salmonella infection can disrupt commensal microbiota and gut barriers, resulting in the bacteria colonizing the tissues of the host intestine (3, 4), leading to diarrhea, fibrinonecrotic enterocolitis, and dehydration in pigs (5).
Healthy gut microbiota can reduce the severity of Salmonella infection; therefore, a feed supplement diet that supports beneficial microbial populations is a potential on-farm strategy to control Salmonella infection in pigs (6, 7). Resistant starch (RS) is an important source of microbiota-accessible carbohydrates because it is digested in the large rather than the small intestines (8). RS feeding can increase short-chain fatty acids (SCFAs) in the intestinal tract, resulting in improved barrier functions, enhanced tolerance to commensal organisms, and reduced inflammation in the gut tissue (9, 10). Raw potato starch (RPS) is a common ingredient of RS that improves fermentation in the digestive tract and increases pro- and anti-inflammatory cytokine levels (11). Recently, studies have shown that RPS feeding could increase gene expression related to the cecal barrier function and improve the mucosal immune system in animals (8, 12).
Our previous study revealed that RPS consumption could promote the growth of beneficial microbes, promoting SCFA production in weaned pigs (13). Therefore, we hypothesized that feeding RPS as a supplement could reduce ST infection severity and fecal shedding in weaned pigs. This study aimed to determine the effect of feeding RPS in reducing ST infection severity in weaned pigs by comparing ST shedding and colonization, histopathological lesions, microbiota composition, and immunological responses in ST-inoculated pigs fed RS and non-RS diets. The findings could provide potential methods for preventing ST infection in pigs by altering microbiome composition and improving immune responses.
All experiments were approved by the Animal Ethics Committee of the National Institute of Animal Science, Republic of Korea (Approval No. NIAS 2021-503).
Twelve castrated male piglets (Landrace × Yorkshire, aged 25 d) were obtained from the same herd in a commercial farm. The average weight of the pigs was 5.00 ± 0.8 kg. All pigs were carefully monitored daily for 3 d before the diet experiment. During the adaptation period, all pigs were confirmed to be sero-negative for foot-and-mouth disease, porcine respiratory and reproductive syndrome, classical swine fever, Mycoplasma spp. infection, and Salmonella spp. infection. In addition, Salmonella spp. and Escherichia coli were not detected in the fecal samples of the experimental pigs. Piglets (aged 28 d) were randomly divided into two groups: the treatment (TRT, n = 6) and negative control (CON, n = 6) groups. The CON diet was formulated according to the nutritional requirements suggested by the Korean feeding standard for pigs, and the TRT pigs were fed the CON diet supplemented with 5% RPS for 21 d (Table 1). After 21 d, the TRT pigs' diet (aged 49 d) was changed to the CON diet until the end of the experiment. Subsequently, all experimental pigs aged 49 d were orally inoculated with 1 × 108 colony forming units of ST LT2 strain (ATCC 19585). The ST-infected pigs were then euthanized 14 d after bacterial inoculation and immediately necropsied to collect the tissue samples.
Fecal samples were collected at 0 and 21 d post-feeding (dpf) and 2, 5, 8, and 11 d post-ST inoculation (dpi). The fecal and intestinal tissues (jejunum, ileum, colon, and cecum) were collected from euthanized pigs at 14 dpi. The Rappaport–Vassiliadis R10 broth (BD, Sparks, MD, USA) containing the fecal and tissue samples (1 g) was incubated immediately at 42°C for 24 h, and one loop of the RV culture was streaked onto CHROMagar Salmonella Plus (CHROMagar, Paris, France). Lastly, the mauve colonies were identified as ST by polymerase chain reaction (PCR) using the AccuPower Salmonella spp. 3-Plex PCR Kit (Bioneer, Daejeon, Korea).
Tissue samples from the jejunum, ileum, cecum, and colon of necropsied pigs (at 14 dpi) were fixed in 10% neutral-buffered formalin and embedded in paraffin wax. Subsequently, 4-μm-thick sectioned tissues were stained with hematoxylin and eosin using a standard laboratory protocol and immunohistochemically stained with anti-Salmonella Typhimurium (BS-4801R; Thermo Fisher Scientific, Rockford, IL, USA). Lastly, the histopathological lesions were scored (from 0 to 5) using previously described parameters, including villus shortening and erosion, presence and concentration of ST, and inflammatory cell infiltration (14).
DNA was extracted using the DNeasy PowerSoil Kit (Qiagen, Hilden, Germany) following the manufacturer's instructions. Amplicons of the V3–V4 region were generated and sequenced following the Illumina 16S metagenomic sequencing library preparation protocol. Subsequently, paired-end sequencing of the amplicon was performed using the MiSeq platform (Illumina, San Diego, CA, USA). Afterward, bioinformatics analysis was performed as described in our previous study (13). Lastly, sequencing data were arranged according to the two experimental groups (TRT and CON) for analytical purposes.
Acetate, butyrate, and propionate were selected based on their specific differences reported in our previous study (13). Fecal concentrations of SCFAs were determined using an Agilent 6890 series gas chromatograph (Agilent Technologies, Santa Clara, CA, USA) coupled with mass spectrometry (13).
The jejunum, cecum, and colon tissues from pigs at 14 dpi were analyzed for cytokine quantification. First, total RNA was isolated using the RNeasy Mini Kit (Qiagen, Hilden, Germany) and reverse-transcribed into cDNA using a High-Capacity cDNA Synthesis Kit (Applied Biosystems, Foster City, CA, USA) following the manufacturer's instructions. Additionally, the relative expression of seven genes was quantified by reverse transcription (RT) PCR (RT-PCR), including four genes related to gut barrier function [claudin (CLDN), occludin (OCLN), zonula occludens-1 (ZO-1)], and regenerating islet-derived protein 3-gamma (Reg3γ), and three genes related to the immune response against Salmonella infection, including interleukin (IL)-10, IL-17A, and IL-18. Notably, RT-PCR was performed using the ABI 7500 Real-Time PCR System (Applied Biosystems) under the following conditions: 10 min at 95°C, 40 cycles at 95°C for 15 s, the annealing temperature of each primer for 30 min, and 72°C for 15 s. The primers and annealing temperatures are listed in Table 2. Lastly, the expression fold change was determined using the 2−ΔΔCt method with the beta-actin gene as the endogenous reference gene to normalize the level of target gene expression.
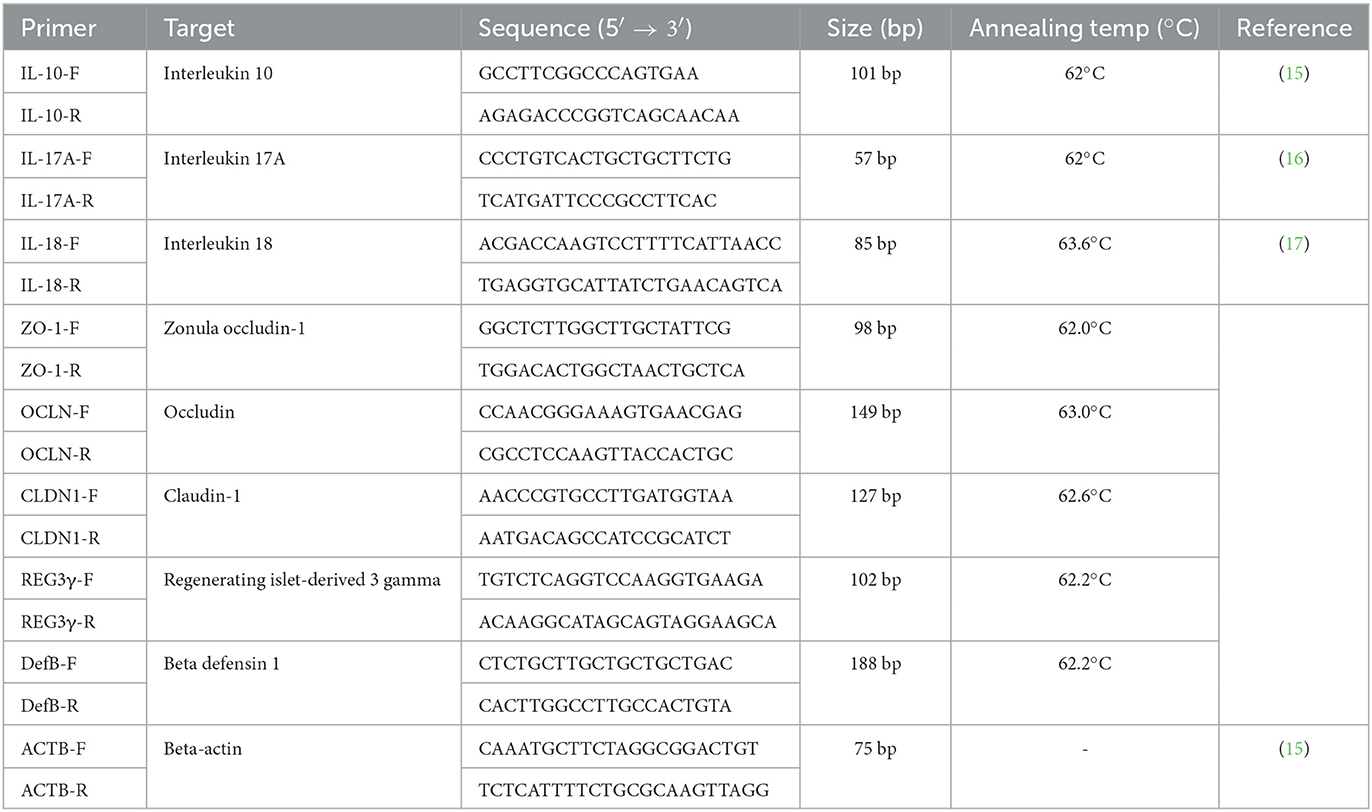
Table 2. The primer information and PCR condition for quantitative real-time polymer chain reaction.
Total RNA was obtained from all experimental pigs at 2 dpi using a Tempus Blood RNA Tube (Applied Biosystems, Seoul, Korea). To produce the transcriptome, TNT Research Corporation Limited (Anyang, Korea) conducted RNA and cDNA library construction and RNA sequencing as previously described (18). Lastly, the DEGs in TRT and CON groups' blood were analyzed based on the expression level of each transcript, as previously described (18).
The Kruskal–Wallis and unpaired Wilcoxon rank-sum tests were used for comparing the alpha diversities of the fecal microbiome composition in the TRT and CON groups at 14 dpi and three time points (0, 21 dpf, and 14 dpi), respectively. All statistical analyses adopted a P-value of 0.05 as the cut-off value and a linear discriminant analysis (LDA) score of 2.0 using the QIIME software version 2.0. Moreover, beta diversity was visualized by principal coordinate analysis (PCoA) matrix using Bray–Curtis distance and QIIME software version 2.0. Additionally, the LDA effect size (LEfSe) was used to determine the specific effect on the relative abundance of taxa in the RS and non-RS groups. Taxa with a significant difference (P < 0.05) between both groups were subjected to LEfSe analysis, and those with LDA score > 2.0 were considered to have been significantly altered after Salmonella infection in the TRT group compared with those in the CON group. Lastly, significant changes in average daily gain (ADG), histopathological lesion scores, and SCFA concentrations between the TRT and CON groups were compared by Student's t-test using the SPSS software (version 26.0; IBM, Armonk, NY, USA).
We compared the pigs in the TRT group (fed with RPS supplemented diet) with those in the CON group (Figure 1A). The ADG during the feeding period (from 0 to 21 dpf) was 0.15 and 0.12 kg/day in the TRT and CON groups, respectively; however, ADG during ST infection period (from 0 to 14 dpi) in the TRT group (0.27 kg/day) was significantly (P = 0.010) higher than that in the CON group (0.15 kg/day) (Figure 1B). Moreover, the TRT group showed marked reductions in ST shedding at 8 and 11 dpi compared with the CON group [8 dpi: 33.3% (TRT) vs. 66.7% (CON) and 11 dpi: 16.7% (TRT) vs. 50.0% (CON)] (Figure 1C).
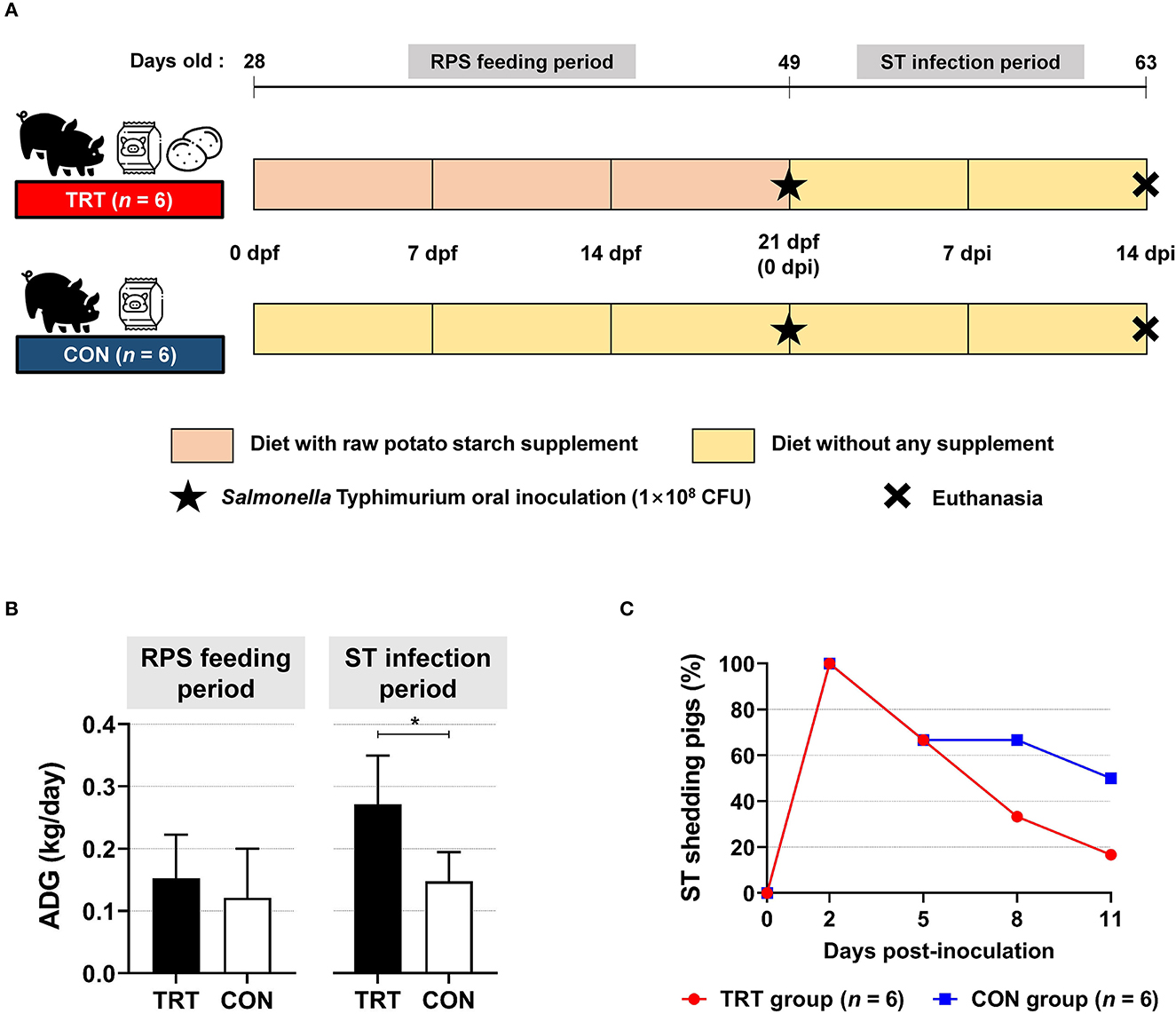
Figure 1. Effect of RPS diet supplement in Salmonella Typhimurium (ST)-infected weaned pigs. (A) Experimental scheme. (B) Average daily gains (ADG) in RS-fed (TRT) and non-RS-fed pigs (CON) during two experimental periods. *P < 0.05. (C) Isolation of ST in fecal samples from TRT and CON until 11 d post-inoculation.
Although histopathological lesions and ST were observed in both groups (Figure 2A), the CON pigs exhibited more severe organismal damage than the TRT pigs, indicated by the total average histopathological lesion scores in the jejunum, ileum, cecum, and colon [TRT (1.8 ± 0.9) vs. CON (3.6 ± 1.3), P < 0.001] (Figure 2B). Additionally, the average scores in the TRT pigs' ileum (1.2 ± 0.4), cecum (1.2 ± 0.4), and colon (2.5 ± 0.5) were significantly lower (P < 0.001, P = 0.001, and P < 0.001, respectively) than those in the CON pigs' ileum (3.5 ± 0.8), cecum (4.5 ± 0.5), and colon (4.3 ± 0.7).

Figure 2. Histopathological lesions in experimental pigs and Salmonella Typhimurium (ST) isolation from intestinal organ tissues. (A) Representative hematoxylin and eosin and immunohistochemical (anti-ST) staining of the ileum (a, e), cecum (b, f), jejunum (c, g), and colon (d, h). (B) Histopathological scores of the intestinal organs (jejunum, ileum, cecum, and colon). *P < 0.05. (C) Isolation of ST from the intestinal organs (jejunum, ileum, cecum, and colon). *P < 0.05.
For intestinal tissues, the estimated isolation rate of ST from the TRT pigs was lower than that from the CON pigs (Figure 2C). The isolation rate from the TRT pigs' jejunum (0%, zero of six pigs) was significantly (P = 0.046) lower than that for the CON pigs (50.0%, three of six pigs). Moreover, ST was detected in the ileum of only one CON pig. Lastly, one TRT pig (16.7%) and four CON pigs (66.7%) harbored ST in the cecum, and the bacteria were isolated from the colon tissue in all pigs, excluding one TRT pig.
Alpha diversity in fecal microbiota was compared between TRT and CON groups and time points (0, 21 dpf, and 14 dpi) (Figure 3A). Four indices were analyzed, including the number of amplicon sequence variants (ASV), Chao1 richness indices, and Shannon and Gini–Simpson diversity indices. The Kruskal–Wallis test showed no significant differences in the four alpha diversity indices: the number of ASV (P = 0.7) and Chao1 richness indices (P = 0.7), and Shannon (P = 0.59) and Gini–Simpson indices (P = 0.59). For the TRT group, the Gini–Simpson index (P = 0.016) was significantly higher at 14 dpi than at 0 and 21 dpf. Moreover, the Gini–Simpson (P = 0.0015) and Shannon indices (P = 0.0017) were significantly lower in the CON group than in the TRT group at 14 dpi. Furthermore, we performed beta diversity analysis to investigate the structure of the bacterial community at 14 dpi; the results are presented as a PCoA ordination plot based on Bray–Curtis distance matrices. Beta diversity differed between the TRT and CON groups at 14 dpi; however, no significant difference was observed between both groups (P = 0.529). The bacterial communities during ST infection in the TRT group shifted leftward along the PC1 axis in the opposite direction to those in the CON group (Figure 3B). Moreover, LEfSe analysis showed that 9 bacterial genera and 11 species of fecal microbes were significantly (P<0.05) increased in the TRT group; however, the abundance of only two genera and three species significantly increased in the CON group (Figure 3C).
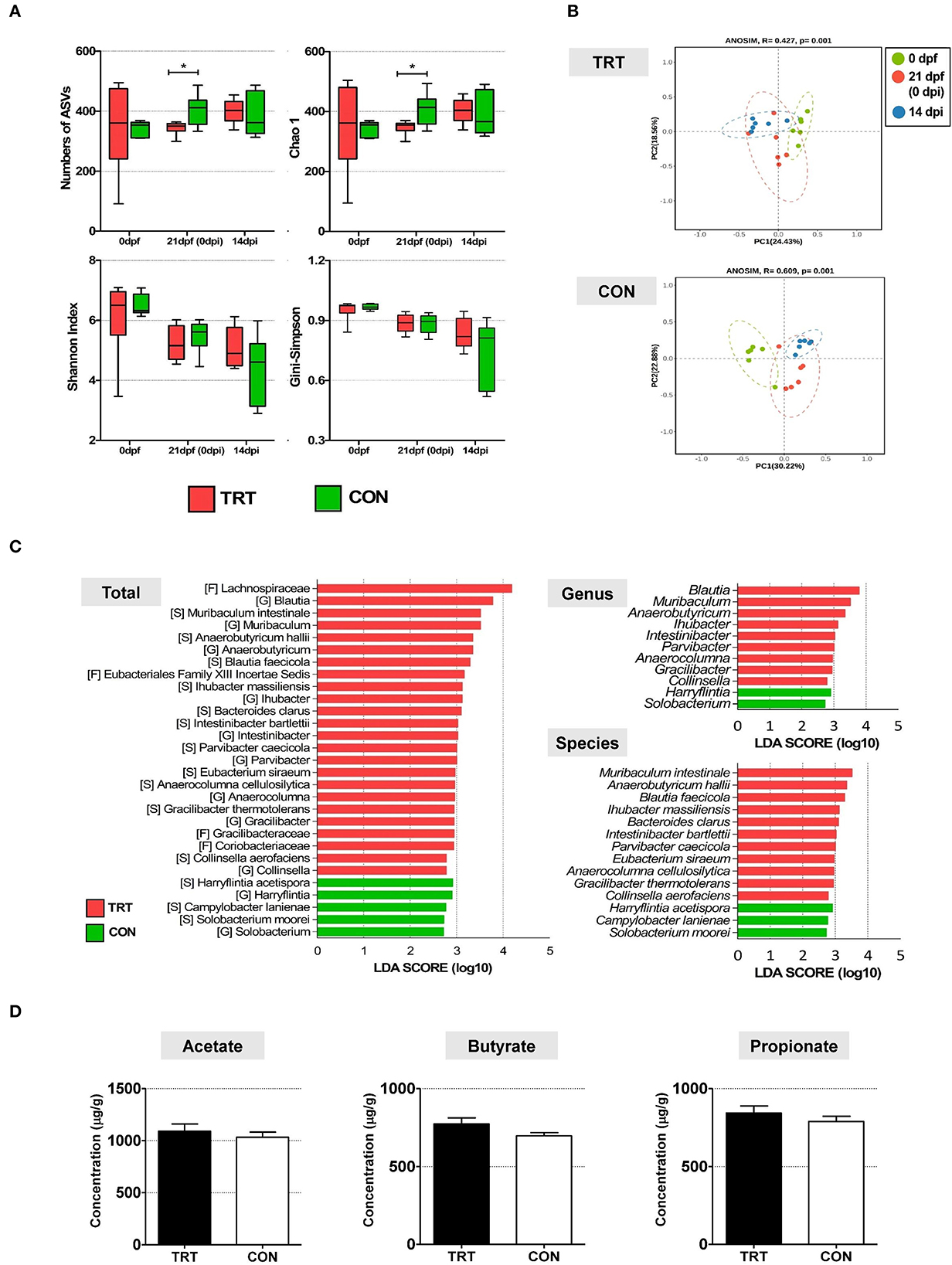
Figure 3. The comparison of alteration of microbiota diversity and composition and SCFA concentration in RPS-fed pigs (TRT) and non-RPS-fed pigs (CON) after Salmonella Typhimurium inoculation (at 14 dpi). (A) Comparison of alpha diversity of microbiome from feces between TRT and CON. The four indices included the number of ASV, Chao1 richness indices, and Shannon and Gini –Simpson diversity indices. (B) PCoA of beta diversity analysis based on the Bray–Curtis dissimilarity matrix. (C) LEfSe revealed predicted biological effect sizes of differential taxa in fecal microbiota between TRT and CON. The LDA scores show a significant difference in the abundance and consistency of the detected bacterial taxa at the genus and species levels. (D) Concentrations of three SCFAs (acetate, propionate, and butyrate) (μg/g) in TRT and CON fecal samples at 14 dpi. *P < 0.05.
The concentrations of three SCFAs (acetate, butyrate, and propionate) were evaluated in all experimental pig fecal samples (14 dpi) to investigate the effect of altered bacterial communities in the TRT and CON groups (Figure 3D). The levels of the three SCFAs were higher in the TRT group than in the CON group. Additionally, the concentrations of acetate, butyrate, and propionate were 1,090.8 ± 170.3 μg/g, 843.3 ± 110.6 μg/g, and 773.6 ± 93.8 μg/g, respectively, in the TRT group, and 1,033.4 ± 120.4 μg/g, 789.3 ± 81.6 μg/g, and 697.3 ± 49.6 μg/g, respectively, in the CON group.
The relative mRNA expression of the proinflammatory cytokine IL-18 was significantly lower in the jejunum [0.00010 ± 0.00014 (TRT) vs. 0.41 ± 0.11 (CON); P = 0.0139] and colon of the TRT group [0.00003 ± 0.00002 (TRT) vs. 0.73 ± 0.05 (CON); P = 0.00001] than in those of the CON group (Figure 4A). Additionally, the anti-inflammatory cytokine IL-10 was less expressed in the jejunum, cecum, and colon. Moreover, the expression of the proinflammatory cytokine IL-17A was higher in the TRT group's colon than in the CON group's colon; however, the differences were insignificant due to the large deviations between samples.
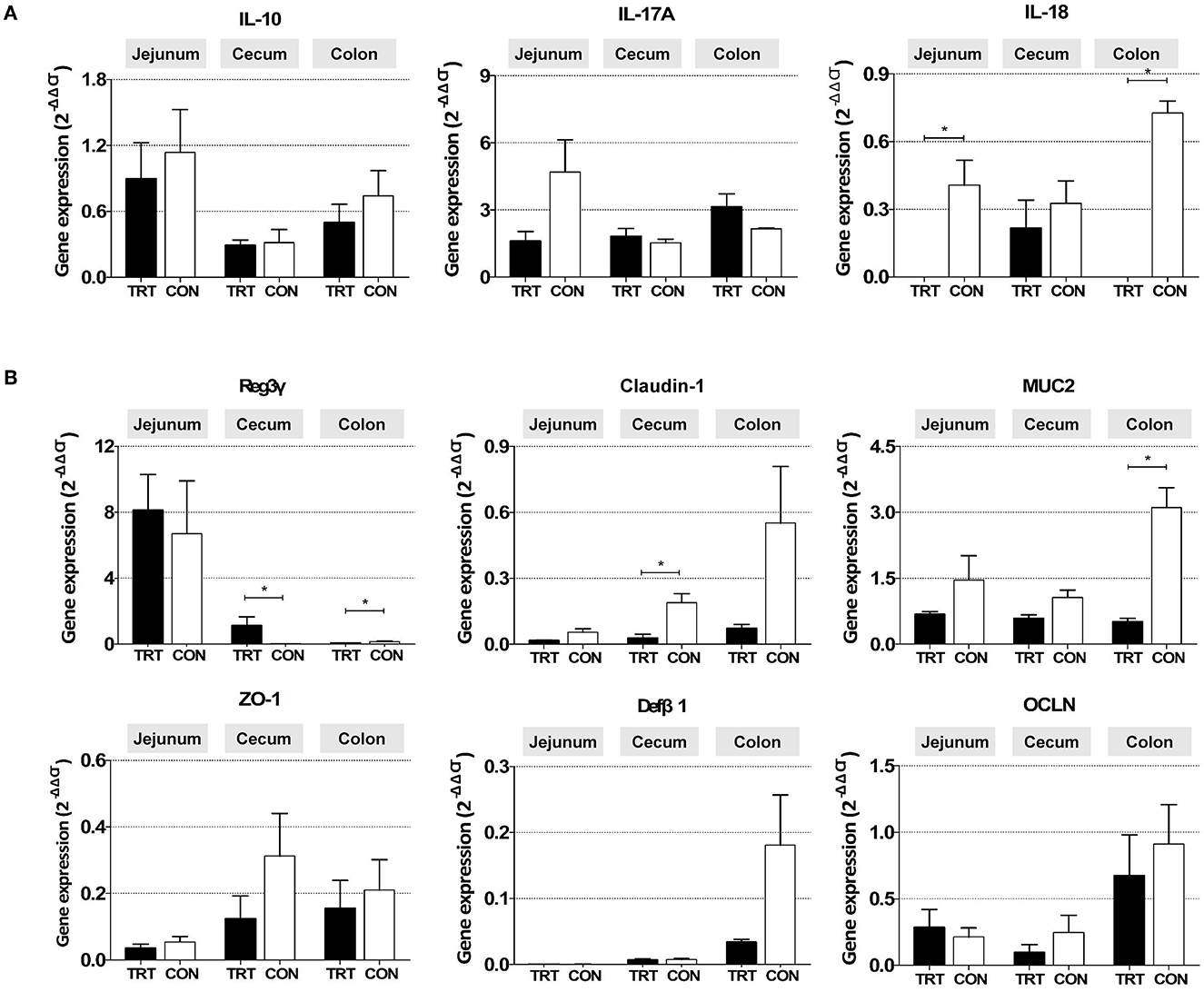
Figure 4. Gene mRNA expression in three intestinal organs (jejunum, cecum, and colon) from RPS-fed (TRT) and non-RPS-fed pigs (CON) after Salmonella Typhimurium inoculation (at 14 dpi). (A) Genes associated with inflammatory response (IL-10, IL-17A, and IL-18). (B) Genes associated with gut barrier function (Reg3γ, Claudin-1, MUC2, Zo-1, Defβ1, and OCLN). *P < 0.05.
Among the genes related to the gut barrier, the expression of the antimicrobial peptide gene Reg3γ was significantly higher in the cecum [1.120 ± 0.535 (TRT) vs. 0.023 ± 0.004 (CON); P = 0.0309] but lower in the colon of the TRT group [0.073 ± 0.012 (TRT) vs. 0.148 ± 0.036 (CON); P = 0.0190] than in those of the CON group (Figure 4B). Furthermore, the relative mRNA expression levels of CLDN-1 in the cecum [0.216 ± 0.124 (TRT) vs. 0.327 ± 0.099 (CON); P = 0.0254] and MUC2 in the colon [0.51 ± 0.08 (TRT) vs. 3.11 ± 0.45 (CON); P = 0.0095] were significantly lower in the TRT group than in the CON group. However, insignificant difference was observed in CLDN-1 expression in the jejunum and colon, MUC2 in the jejunum and cecum, and the other three genes (ZO-1, OCLN, and DefB1) in the jejunum, cecum, and colon between the two feeding groups.
An average of 6.7 Gb raw data for each sample were collected from paired-end transcriptome sequencing using the Illumina NovaSeq 6000 platform. Raw data were subjected to quality control using Trimmomatic (ver. 0.38), and the trimmed data were mapped using HISAT2 ver. 2.1.0 (Bowtie2 aligner). Next, the mapped reads were assembled using the StringTie-e option ver. 1.3.4d. Afterward, the obtained genes were filtered by excluding those with at least one zero count, leaving 12,189 genes for DEG analysis. Overall, 424 genes were considered differentially expressed based on the threshold level (fold change [log2] ≥ 2 and P < 0.05) (Figure 5A). Lastly, the biological function of 424 DEGs was determined by gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes pathways using the DAVID 6.8 tool. Figure 5B shows the GO functional analysis of the biological process (top 20), cellular component (top 12), and molecular function (top 6).
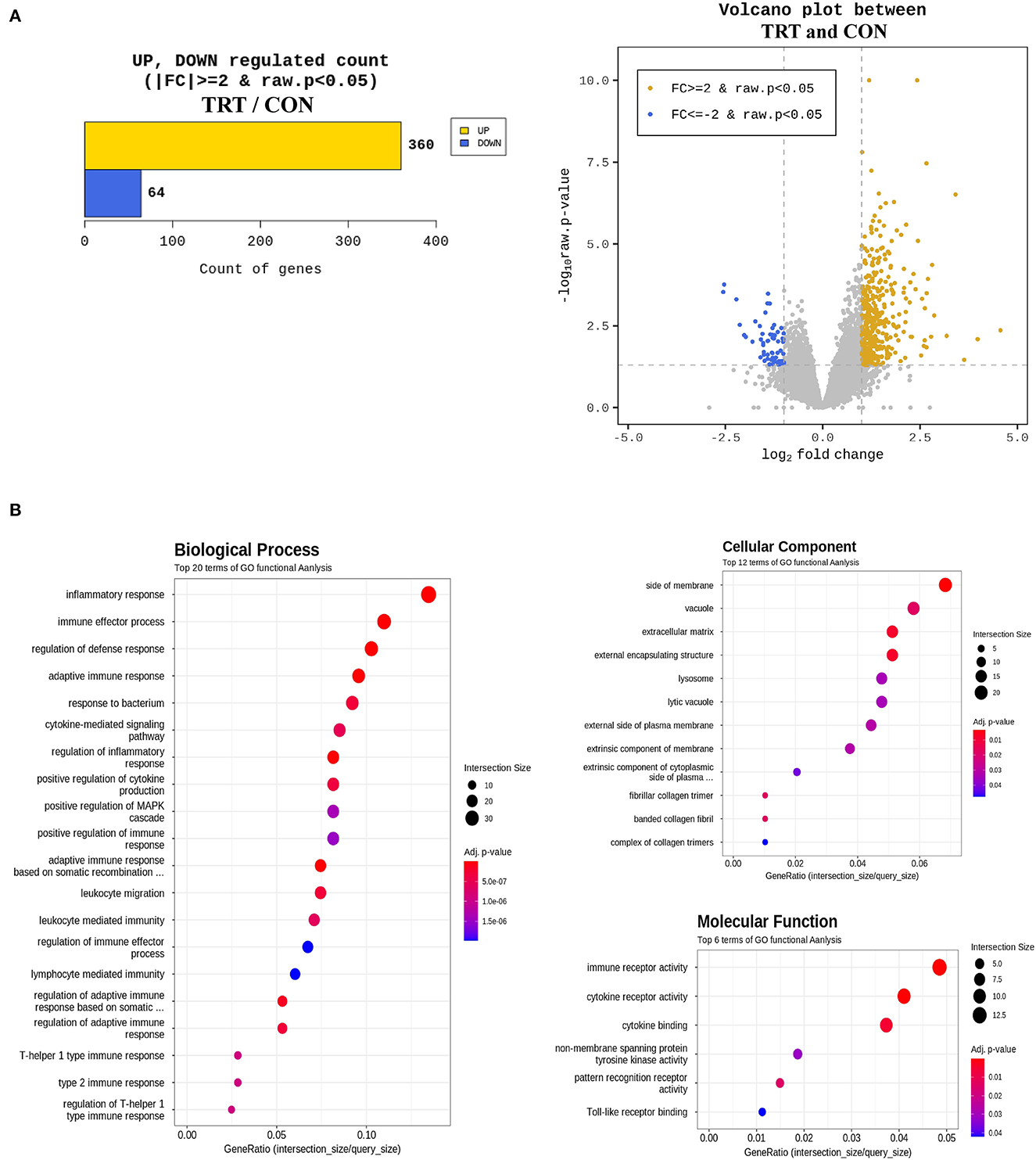
Figure 5. Differentially expressed genes (DEG) and gene ontology (GO) enrichment analysis in blood samples from RPS-fed (TRT) and non-RPS-fed pigs (CON) at 2 d after Salmonella Typhimurium inoculation. (A) Numbers of up- and down-regulated genes after comparison of normalized values using the DESeq2 package. (B) GOTERM_Biological process, GOTERM_Cellular Component, and GOTERM_Molecular Function.
Salmonella is a major causative agent of diarrhea in pigs, threatening food safety and human health. Local inflammation in Salmonella-infected pigs can reportedly cause changes in the gut microbiome, favoring the survival of Salmonella (4, 19). We previously revealed that RPS feeding of weaned pigs could improve gut health by maintaining the balance of beneficial bacteria and promoting SCFA production (13). Therefore, we investigated the gut microbiota composition and immunological response for preventing ST infection in RPS-fed pigs.
RPS is a type II RS that can decrease body weight (BW) in humans and animals. However, the ADG of TRT pigs was not significantly different from that of CON pigs during the RPS feeding period (until 21 dpf), consistent with the finding in our previous study (13). Although Salmonella infection in pigs reduces BW and ADG (20), the ADG of TRT pigs was significantly higher than that of CON pigs during the ST infection period. These findings could explain the gut health-promoting effect in RPS-fed pigs because ADG in pigs is strongly related to intestinal morphology (21). Additionally, healthy gut microbiota and its derived SCFAs could prevent the colonization of pathogenic bacteria by decreasing gut mucosal permeability (13, 22). The histopathological lesions in the intestinal organ tissues of TRT pigs were significantly milder than those of CON pigs. Moreover, ST fecal shedding was reduced in RPS-fed pigs (TRT) at 8 dpi. These results suggest improved gut health after the post-weaning diet supplemented with RPS.
The abundance of nine bacterial genera significantly increased in TRT pigs, among which Blautia (P = 0.0374), Muribaculum (P = 0.0104), Anaerobutyricum (P = 0.0374), and Anaerocolumna (P = 0.0247) were the main butyrate-producing bacteria (23–26). Blautia is considered as a novel potential probiotic due to its ability to produce bacteriocin (sactipeptide and lanthipeptide), inhibit pathogenic bacterial colonization, and regulate inflammatory responses (25). Thus, we speculated that the increased abundance of Blautia might have contributed to the mild histopathological lesions and reduced ST colonization in the TRT pigs' intestinal organ tissues. Yuan et al. (27) showed that metabolites from Muribaculum could improve gut barrier function and integrity, preventing leakage of inflammatory mediators into the systemic circulation. Moreover, herein, the genus Anaerobutyricum and its subtaxon A. hallii (P = 0.0374) increased more in the TRT group than in the CON group. A. hallii is a potential next-generation probiotic bacterium because of its capacity to produce propionate and butyrate (28). Additionally, the four main acetate-producing bacteria in TRT, Intestinibacter (P = 0.0250), Anaerocolumna (P = 0.0247), Gracilibacter (P = 0.0374), and Collinsella (P = 0.0250), were more prevalent in the TRT group than in the CON group. Anaerocolumna reportedly decomposes cellulose, oligosaccharides, polysaccharides, and organic acids into energy sources (29). Moreover, Gracilibacter can degrade glucose, and Intestinibacter is involved in mucin consumption by degrading fucose (30). Lastly, Collinsella is significantly and positively correlated with most bile acids and is related to lipid metabolism (31). Conversely, the abundance of only two genera, Harryflintia (P = 0.0463) and Solobacterium (P = 0.0278) increased in the CON group, which can only produce acetate (32, 33). Lawhon et al. (34) reported that unbalanced SCFA ratio (e.g., high acetate and low butyrate/propionate concentration) could cause a more invasive ST infection. Further, Harryflintia abundance reduced when mice were fed high concentrations of RPS (0–10%) (35). Although the abundant species Campylobacter lanienae (P = 0.0222) and Solobacterium moorei (P = 0.0278) are common in the gastrointestinal tract of pigs, they have emerged as a potential cause of human gastroenteritis (32, 36).
Our previous study showed that feeding 5% RPS resulted in significantly higher concentrations of acetate, butyrate, and total SCFAs in healthy pigs (13). In the present study, however, the three SCFAs in the TRT and CON groups showed no significant difference, despite the increased numbers of butyrate-producing bacteria. ST reportedly uses and decreases microbiota-derived butyrate by altering the gut microbiota composition; consequently, the intestinal epithelium shifts to lactate fermentation (37, 38). In addition, sufficient concentrations of butyrate and propionate reportedly enabled the abrogation of ST-induced gut inflammation by regulating the expression of genes responsible for ST invasion and pathogenesis; moreover, they increased the sensitivity of the pathogens to butyrate-mediated repression of invasion-related gene expression (34, 37). Herein, butyrate consumption by ST might not significantly increase butyrate and propionate concentrations in the gut of TRT pigs; however, this may affect the severity and bacterial colonization results in RS-fed pigs.
In the present study, the mRNA expression of Reg3γ (antimicrobial peptide gene) markedly increased in the cecum and colon of the TRT pigs. Reg3γ restricts bacterial colonization of the intestinal mucosal surface and maintains spatial segregation between bacteria and intestinal epithelium (39). Therefore, the poor colonization of ST in TRT pigs could result from the enhanced Reg3γ expression in the cecum and colon. Furthermore, IL-18 mRNA expression levels and histopathological scores were significantly reduced in TRT pigs' colons. Previous studies have suggested that IL-18 is necessary to initiate mucosal inflammation (40, 41). Moreover, the upregulation of IL-18 is central to the pathogenesis of tissue destruction and the severity of gastroenteritis in humans and mice (41, 42). Therefore, the high Reg3γ expression and low IL-18 expression in TRT are related to the reduced ST colonization and histopathology results.
In this study, GO analysis revealed that upregulated DEGs were primarily involved in immune and inflammatory responses at 2 dpi. The top five biological processes were the inflammatory response, immune effector process, defense response regulation, adaptive immune response, and response to bacteria. Furthermore, GO enrichment analysis of the upregulated DEG included immune receptor activity, cytokine receptor activity, and cytokine binding. These results are consistent with previous findings that inflammatory features peaked, and inflammatory infiltration significantly increased in the small intestinal tissues of ST-infected piglets at 2 dpi (43, 44). Considering these results together with qPCR results, the GO analysis revealed that ST infection triggered immune responses in the early phase. At 2 dpi, transcript levels of the tight junction proteins claudin and occludin (CLDND1 and OCEL1) in blood increased by 22.33 and 1.21 folds, respectively. However, at 14 dpi, the genes were less expressed in the TRT group than in the CON group. This result suggests that CLDN and OCLN might have been expressed earlier, making it unnecessary by 14 dpi. Additionally, at 2 dpi, the cytokine genes related to inflammation, IL-10 subunit alpha and subunit beta (IL10Rα and IL10RB) showed 1.62- and 2.23-fold increases, respectively, and the IL-17 gene (IL17B) showed a 41-fold increase. Moreover, we observed 4.84-, 2.49-, and 1.91-fold increases in IL-18 receptor (IL18R1), binding protein (IL18BP), and IL18 genes, respectively. However, excluding IL-17A in the colon, IL-10 and IL-17A were lower in all tissues examined in this study, and IL-18 was significantly lower in the jejunum and colon of the TRT group than in those of the CON group. Reg3γ overexpression reportedly induces high immunosuppression (45). Therefore, the dramatic reduction in cytokine genes in our study might be caused by a significant increase in Reg3γ at 14 dpi.
Overall, the results demonstrate that the RS-supplement diet could prevent ST infection in weaned pigs and provide insights into the mechanisms underlying the immune responses of RS-fed pigs against ST infection. However, there are some limitations to providing a general conclusion of using RS feeding as a preventative measure for ST in pigs. First, this study was conducted on a limited number of pigs; therefore, the results may not represent the general pig population. Further studies with larger sample sizes and different breeds of pigs are required to validate these findings. Second, the animal experiment was conducted under controlled conditions; therefore, the results may be difficult to replicate immediately on a pig farm. An application experiment in actual farm units is required in the future.
The study results suggest that feeding weaned pigs with RPS—a type II RS—could improve gut health and reduce ST infection. The TRT group showed higher ADG during the infection period and milder histopathological lesions in the intestinal organs than the CON group. In addition, TRT pigs exhibited marked reduction in ST shedding compared with that in CON pigs. These results suggested that RPS feeding in weaned pigs could reduce economic losses in farm due to ST infection. The gut microbiota of the TRT group showed an increased abundance of four main butyrate-producing bacteria and four main acetate-producing bacteria. The increased levels of these beneficial bacteria could have contributed to promoting gut health and reducing ST colonization in the TRT group. Moreover, Reg3γ expression was markedly increased in the TRT pigs, preventing ST colonization in RPS-fed pigs. Overall, our findings highlight the potential use of RPS as a dietary intervention to improve gut health and reduce Salmonella infections in pigs.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/bioproject; PRJNA881483, PRJNA952690.
All the experiments were approved by the Animal Ethics Committee of the National Institute of Animal Science, Republic of Korea (Approval No. NIAS 2021-503).
S-IO made substantial contributions to the conception and design of the work and revised the manuscript prior to the submission. S-WY was responsible for laboratory analyses, data curation, and interpretation of experimental data. S-WY, HL, EK, and S-IO were responsible for animal experiments and investigations. S-WY, Y-HJ, E-YB, AC, YD, T-YH, and S-IO were responsible for data validation and resources. S-WY and S-IO wrote the original draft. All authors read and approved the final manuscript.
This study was supported by the 2021 RDA Fellowship Program of the National Institute of Animal Science, Rural Development Administration, the “Cooperative Research Program for Agriculture Science and Technology Development (Project Title: Development of gut microbiota for preventing intestinal diseases and its impact on host immunity in pigs, Project No. PJ01564401),” and Rural Development Administration, Republic of Korea.
The authors thank Hyung Joon Lee for his technical support in collecting samples from the pigs.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Callaway TR, Edrington TS, Anderson RC, Byrd JA, Nisbet DJ. Gastrointestinal microbial ecology and the safety of our food supply as related to Salmonella. J Anim Sci. (2008) 86:E163–72. doi: 10.2527/jas.2007-0457
2. Oh SI, Kim JW, Chae M, Jung JA, So B, Kim B, et al. Characterization and antimicrobial resistance of Salmonella Typhimurium isolates from clinically diseased pigs in Korea. J Food Prot. (2016) 79:1884–90. doi: 10.4315/0362-028X.JFP-16-131
3. Bescucci DM, Moote PE, Ortega Polo R, Uwiera RRE, Inglis GD. Salmonella enterica serovar typhimurium temporally modulates the enteric microbiota and host responses to overcome colonization resistance in swine. Appl Environ Microbiol. (2020) 86:e01569–20. doi: 10.1128/AEM.01569-20
4. Drumo R, Pesciaroli M, Ruggeri J, Tarantino M, Chirullo B, Pistoia C, et al. Salmonella enterica serovar Typhimurium exploits inflammation to modify swine intestinal microbiota. Front Cell Infect Microbiol. (2016) 5:106. doi: 10.3389/fcimb.2015.00106
5. Costa MO, Fouhse J, Silva APP, Willing B, Harding JCS. Putting the microbiota to work: Epigenetic effects of early life antibiotic treatment are associated with immune-related pathways and reduced epithelial necrosis following Salmonella Typhimurium challenge in vitro. PLoS ONE. (2020) 15:e0231942. doi: 10.1371/journal.pone.0231942
6. Argüello H, Estellé J, Leonard FC, Crispie F, Cotter PD, O'Sullivan O, et al. Influence of the intestinal microbiota on colonization resistance to salmonella and the shedding pattern of naturally exposed pigs. mSystems. (2019) 4:e00021–19. doi: 10.1128/mSystems.00021-19
7. Doyle MP, Erickson MC. Opportunities for mitigating pathogen contamination during on-farm food production. Int J Food Microbiol. (2012) 152:54–74. doi: 10.1016/j.ijfoodmicro.2011.02.037
8. Trachsel J, Briggs C, Gabler NK, Allen HK, Loving CL. Dietary resistant potato starch alters intestinal microbial communities and their metabolites, and markers of immune regulation and barrier function in swine. Front Immunol. (2019) 10:1381. doi: 10.3389/fimmu.2019.01381
9. Trachsel JM, Bearson BL, Kerr BJ, Shippy DC, Byrne KA, Loving CL, et al. Short chain fatty acids and bacterial taxa associated with reduced Salmonella enterica serovar I 4, (5), 12: i: Shedding in Swine Fed a Diet Supplemented with Resistant Potato Starch. Microbiol Spec. (2022) 3:e0220221. doi: 10.1128/spectrum.02202-21
10. Van Der Hee B, Wells JM. Microbial regulation of host physiology by short-chain fatty acids. Trends Microbiol. (2021) 29:700–12. doi: 10.1016/j.tim.2021.02.001
11. Regmi PR, Metzler-Zebeli BU, Gänzle MG, Van Kempen TATG, Zijlstra RT. Starch with high amylose content and low in vitro digestibility increases intestinal nutrient flow and microbial fermentation and selectively promotes bifidobacteria in pigs. J Nutr. (2011) 141:1273–80. doi: 10.3945/jn.111.140509
12. Qin SM, Zhang KY, Ding XM, Bai SP, Wang JP, Zeng QF. Effect of dietary graded resistant potato starch levels on growth performance, plasma cytokines concentration, and intestinal health in meat ducks. Poult Sci. (2019) 98:3523–32. doi: 10.3382/ps/pez186
13. Yi SW, Lee HG, So KM, Kim E, Jung YH, Kim M, et al. Effect of feeding raw potato starch on the composition dynamics of the piglet intestinal microbiome. Anim Biosci. (2022) 35:1698–710. doi: 10.5713/ab.22.0045
14. Argüello H, Estellé J, Zaldívar-López S, Jiménez-Marín Á, Carvajal A, López-Bascón MA, et al. Early Salmonella Typhimurium infection in pigs disrupts microbiome composition and functionality principally at the ileum mucosa. Sci Rep. (2018) 8:7788. doi: 10.1038/s41598-018-26083-3
15. Walsh AM, Sweeney T, Bahar B, Flynn B, O'Doherty JV. The effect of chitooligosaccharide supplementation on intestinal morphology, selected microbial populations, volatile fatty acid concentrations and immune gene expression in the weaned pig. Animal. (2012) 6:1620–6. doi: 10.1017/S1751731112000481
16. Ryan MT, O'Shea CJ, Collins CB, O'Doherty JV, Sweeney T. Effects of dietary supplementation with Laminaria hyperborea, Laminaria digitata, and Saccharomyces cerevisiae on the IL-17 pathway in the porcine colon. J Anim Sci. (2012) 90:263–5. doi: 10.2527/jas.53802
17. Bouwhuis MA, Mcdonnell MJ, Sweeney T, Mukhopadhya A, O'Shea CJ, O'Doherty JV. Seaweed extracts and galacto-oligosaccharides improve intestinal health in pigs following Salmonella Typhimurium challenge. Animal. (2017) 11:1488–96. doi: 10.1017/S1751731117000118
18. Lopez BI, Santiago KG, Lee D, Ha S, Seo K, RNA. sequencing (RNA-seq) based transcriptome analysis in immune response of Holstein cattle to killed vaccine against bovine viral diarrhea virus type I. Animals. (2020) 10:344. doi: 10.3390/ani10020344
19. Diaz-Ochoa VE, Lam D, Lee CS, Klaus S, Behnsen J, Liu JZ, et al. Salmonella mitigates oxidative stress and thrives in the inflamed gut by evading calprotectin-mediated manganese sequestration. Cell Host Microbe. (2016) 19:814–25. doi: 10.1016/j.chom.2016.05.005
20. Price KL, Totty HR, Lee HB, Utt MD, Fitzner GE, Yoon I, et al. Use of Saccharomyces cerevisiae fermentation product on growth performance and microbiota of weaned pigs during Salmonella infection. J Anim Sci. (2010) 88:3896–908. doi: 10.2527/jas.2009-2728
21. Park S, Lee JJ, Yang BM, Cho JH, Kim S, Kang J, et al. Dietary protease improves growth performance, nutrient digestibility, and intestinal morphology of weaned pigs. J Anim Sci Technol. (2020) 62:21–30. doi: 10.5187/jast.2020.62.1.21
22. Jeong YD, Ko HS, Hosseindoust A, Choi YH, Chae BJ Yu DJ, et al. Lactobacillus-based fermentation product and lactose level in the feed for weanling pigs: effects on intestinal morphology, microbiota, gas emission, and targeted intestinal coliforms. Livest Sci. (2019) 227:90–6. doi: 10.1016/j.livsci.2019.06.018
23. Berni Canani R, Sangwan N, Stefka AT, Nocerino R, Paparo L, Aitoro R, et al. Lactobacillus rhamnosus gg-supplemented formula expands butyrate-producing bacterial strains in food allergic infants. ISME J. (2016) 10:742–50. doi: 10.1038/ismej.2015.151
24. Lagkouvardos I, Lesker TR, Hitch TCA, Gálvez EJC, Smit N, Neuhaus K, et al. Sequence and cultivation study of Muribaculaceae reveals novel species, host preference, and functional potential of this yet undescribed family. Microbiome. (2019) 7:28. doi: 10.1186/s40168-019-0637-2
25. Liu X, Mao B, Gu J, Wu J, Cui S, Wang G, et al. Blautia—a new functional genus with potential probiotic properties? Gut Microbes. (2021) 13:1–21. doi: 10.1080/19490976.2021.1875796
26. Shetty SA, Zuffa S, Bui TPN, Aalvink S, Smidt H, De Vos WM. Reclassification of Eubacterium hallii as Anaerobutyricum hallii gen.nov, comb nov, and description of Anaerobutyricum soehngenii sp nov, a butyrate and propionate-producing bacterium from infant faeces. Int J Syst Evol Microbiol. (2018) 68:3741–6. doi: 10.1099/ijsem.0.003041
27. Yuan Y, Zhou J, Zheng Y, Xu Z, Li Y, Zhou S, et al. Beneficial effects of polysaccharide-rich extracts from Apocynum venetum leaves on hypoglycemic and gut microbiota in type 2 diabetic mice. Biomed Pharmacother. (2020) 127:110182. doi: 10.1016/j.biopha.2020.110182
28. Engels C, Ruscheweyh HJ, Beerenwinkel N, Lacroix C, Schwab C. The common gut microbe Eubacterium hallii also contributes to intestinal propionate formation. Front Microbiol. (2016) 7:713. doi: 10.3389/fmicb.2016.00713
29. Ueki A, Ohtaki Y, Kaku N, Ueki K. Descriptions of Anaerotaenia torta gen. nov, sp nov and Anaerocolumna cellulosilytica gen nov, sp nov isolated from a methanogenic reactor of cattle waste and reclassification of Clostridium aminovalericum, Clostridium jejuense and Clostridium xylanovorans as Anaerocolumna species. Int J Syst Evol Microbiol. (2016) 66:2936–43. doi: 10.1099/ijsem.0.001123
30. Forslund K, Hildebrand F, Nielsen T, Falony G, Le Chatelier E, Sunagawa S, et al. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature. (2015) 528:262–6. doi: 10.1038/nature15766
31. Wang J, Li Y, Cao C, Yang R, He M, Yan J, et al. The periparturient Gut microbiota's modifications in Shaziling Sows concerning bile acids. Metabolites. (2023) 13:68. doi: 10.3390/metabo13010068
32. Alauzet C, Aujoulat F, Lozniewski A, Ben Brahim S, Domenjod C, Enault C, et al. A new look at the genus Solobacterium: a retrospective analysis of twenty-seven cases of infection involving S. moorei and a review of sequence databases and the literature. Microorganisms. (2021) 9:1229. doi: 10.3390/microorganisms9061229
33. Petzoldt D, Breves G, Rautenschlein S, Taras D. Harryflintia acetispora gen. nov, sp nov, isolated from chicken caecum. Int J Syst Evol Microbiol. (2016) 66:4099–104. doi: 10.1099/ijsem.0.001317
34. Lawhon SD, Maurer R, Suyemoto M, Altier C. Intestinal short-chain fatty acids alter Salmonella Typhimurium invasion gene expression and virulence through bara/sira. Mol Microbiol. (2002) 46:1451–64. doi: 10.1046/j.1365-2958.2002.03268.x
35. Smith AD, Chen C, Cheung L, Ward R, Hintze KJ, Dawson HD. Resistant potato starch alters the cecal microbiome and gene expression in mice fed a Western diet based on NHANES data. Front Nutr. (2022) 9:782667. doi: 10.3389/fnut.2022.782667
36. Fornefett J, Busch A, Döpping S, Hotzel H, Rimek D. Bacterial gastroenteritis caused by the putative zoonotic pathogen Campylobacter lanienae: First reported case in Germany. Access Microbiol. (2021) 3:000199. doi: 10.1099/acmi.0.000199
37. Bronner DN, Faber F, Olsan EE, Byndloss MX, Sayed NA, Xu G, et al. Genetic ablation of butyrate utilization attenuates gastrointestinal Salmonella disease. Cell Host Microbe. (2018) 23:266–73. doi: 10.1016/j.chom.2018.01.004
38. Rivera-Chávez F, Zhang L, Faber F, Lopez CA, Byndloss MX, Olsan EE, et al. Depletion of butyrate-producing Clostridia from the gut microbiota drives an aerobic luminal expansion of Salmonella. Cell Host Microbe. (2016) 19:443–54. doi: 10.1016/j.chom.2016.03.004
39. Vaishnava S, Yamamoto M, Severson KM, Ruhn KA Yu X, Koren O, et al. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science. (2011) 334:255–8. doi: 10.1126/science.1209791
40. Fournout S, Dozois CM, Yerle M, Pinton P, Fairbrother JM, Oswald E, et al. Cloning, chromosomal location, and tissue expression of the gene for pig interleukin-18. Immunogenetics. (2000) 51:358–65. doi: 10.1007/s002510050630
41. Müller AA, Dolowschiak T, Sellin ME, Felmy B, Verbree C, Gadient S, et al. An NK cell perforin response elicited via IL-18 controls mucosal inflammation kinetics during Salmonella gut infection. PLoS Pathog. (2016) 12:e1005723. doi: 10.1371/journal.ppat.1005723
42. Nowarski R, Jackson R, Gagliani N, de Zoete MR, Palm NW, Bailis W, et al. Epithelial IL-18 equilibrium controls barrier function in colitis. Cell. (2015) 163:1444–56. doi: 10.1016/j.cell.2015.10.072
43. Bellido-Carreras N, Argüello H, Zaldívar-López S, Jiménez-Marín Á, Martins RP, Arce C, et al. Salmonella Typhimurium infection along the porcine gastrointestinal tract and associated lymphoid tissues. Vet Pathol. (2019) 56:681–90. doi: 10.1177/0300985819843682
44. Collado-Romero M, Aguilar C, Arce C, Lucena C, Codrea MC, Morera L, et al. Quantitative proteomics and bioinformatic analysis provide new insight into the dynamic response of porcine intestine to Salmonella Typhimurium. Front Cell Infect Microbiol. (2015) 5:64. doi: 10.3389/fcimb.2015.00064
Keywords: diarrhea, microbiome, raw potato starch, resistant starch, Salmonella Typhimurium, weaned pig, cytokine, transcriptome
Citation: Yi S-W, Lee HG, Kim E, Jung Y-H, Bok E-Y, Cho A, Do YJ, Hur T-Y and Oh S-I (2023) Raw potato starch diet supplement in weaned pigs could reduce Salmonella Typhimurium infection by altering microbiome composition and improving immune status. Front. Vet. Sci. 10:1183400. doi: 10.3389/fvets.2023.1183400
Received: 10 March 2023; Accepted: 02 May 2023;
Published: 23 May 2023.
Edited by:
Balamuralikrishnan Balasubramanian, Sejong University, Republic of KoreaReviewed by:
Muhammad Akbar Shahid, Bahauddin Zakariya University, PakistanCopyright © 2023 Yi, Lee, Kim, Jung, Bok, Cho, Do, Hur and Oh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sang-Ik Oh, c2lvaEBqYm51LmFjLmty
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.