- Biosecurity Affairs Division, Development and Innovation Sector, Abu Dhabi Agriculture and Food Safety Authority (ADAFSA), Abu Dhabi, United Arab Emirates
Background: The study of coronaviruses has grown significantly in recent years.
Middle East respiratory syndrome coronavirus (MERS-CoV) replicates in various cell types, and quick development has been made of assays for its growth and quantification. However, only a few viral isolates are now available for investigation with full characterization. The current study aimed to isolate MERS-CoV from nasal swabs of dromedary camels and molecularly analyze the virus in order to detect strain-specific mutations and ascertain lineage classification.
Methods: We isolated the virus in Vero cells and adapted it for in vitro cultivation. The isolates were subjected to complete genome sequencing using next-generation sequencing followed by phylogenetic, mutation, and recombination analysis of the sequences.
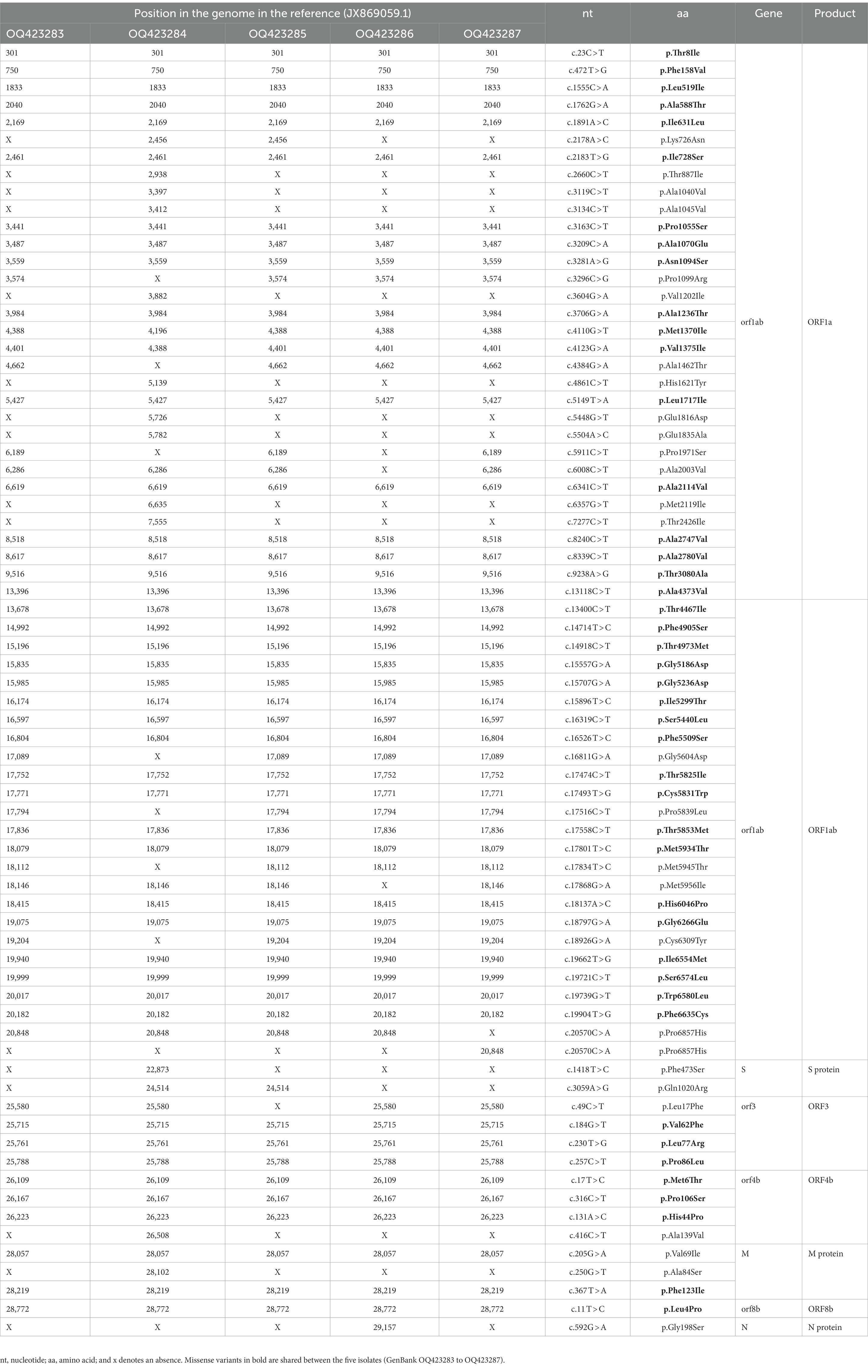
Results: A total of five viral isolates were obtained in Vero cells and adapted to in vitro cultures. Phylogenetic analysis classified all the isolates within clade B3. Four isolates clustered close to the MERS-CoV isolate camel/KFU-HKU-I/2017 (GenBank ID: MN758606.1) with nucleotide identity 99.90–99.91%. The later isolate clustered close to the MERS-CoV isolate Al-Hasa-SA2407/2016 (GenBank ID: MN654975.1) with a sequence identity of 99.86%. Furthermore, the isolates contained several amino acids substitutions in ORF1a (32), ORF1ab (25), S (2), ORF3 (4), ORF4b (4), M (3), ORF8b (1), and the N protein (1). The analysis further identified a recombination event in one of the reported sequences (OQ423284/MERS-CoV/dromedary/UAE-Al Ain/13/2016).
Conclusion: Data presented in this study indicated the need for continuous identification and characterization of MERS-CoV to monitor virus circulation in the region, which is necessary to develop effective control measures. The mutations described in this investigation might not accurately represent the virus’s natural evolution as artificial mutations may develop during cell culture passage. The isolated MERS-CoV strains would be helpful in new live attenuated vaccine development and efficacy studies.
1. Introduction
Middle East respiratory syndrome (MERS) is a viral respiratory infection of humans and dromedary camels that is caused by a coronavirus called the Middle East Respiratory Syndrome Coronavirus (MERS-CoV) (1). During the last two decades, the world witnessed the emergence of three novel coronaviruses: severe acute respiratory syndrome coronavirus 1 (SARS CoV-1) in 2002–2003, Middle East respiratory syndrome coronavirus (MERS-CoV) in 2012, and the ongoing pandemic caused by SARS-CoV-2 in 2019, resulting in a very significant impact on both humans and animals as well as the global economy. Coronaviruses can have a devastating impact on the health of humans and animals. Coronaviruses are important RNA viruses, and members of the subfamily Orthocoronavirinae are classified into four genera: Alphacoronavirus, Betacoronavirus, Gammacoronavirus, and Deltacoronavirus (2, 3).
From April 2012 to July 2022, a total of 2,591 laboratory-confirmed cases of Middle East respiratory syndrome (MERS) were reported globally, with 894 associated deaths at a case-fatality ratio (CFR) of 34.5% (4). Via direct or indirect contact, humans can become infected with the MERS-CoV from dromedary camels (Camelus dromedarius), the virus’s natural host and zoonotic source. MERS-CoV has been identified in dromedaries in several countries in the Middle East, Africa, and South Asia. Some dromedary camels have shown minor upper respiratory symptoms related to MERS-CoV. While MERS-CoV has little effect on animal health, human infections adversely affect public health. MERS is an emerging disease with severe implications for human public health. MERS-CoV phylogeny currently comprises three major clades, provisionally named clades A, B, and C. The clade B strains currently infect humans and dromedary camels in the Arabian Peninsula, in contrast to clades A and C, which contain extinct strains and strains that are not circulating in this region. Clade B is divided into six phylogenetic lineages. In dromedary camels, the recombination between lineages 3 and 4 led to the emergence of the circulating recombinant lineage 5 around the year 2014 (5–8). More than 41,000 nasal swabs taken from camels in the United Arab Emirates (UAE) between 2013 and 2021 were tested for MERS-CoV by quantitative reverse transcription polymerase chain reaction (RT-qPCR) at the veterinary laboratories of Abu Dhabi Agriculture and Food Safety Authority (ADAFSA). Additionally, we conducted numerous research on various aspects of MERS-CoV infection, including the evaluation of diagnostic tests, epidemiology, and investigation of infection events in humans linked to sick camels, identification of the diversity of the virus in dromedary camels, zoonotic origin and transmission of the virus, and risk factors for the virus seropositivity among animal market and slaughterhouse workers (8–12).
This study aimed to isolate MERS-CoV from nasal swabs of dromedary camels, and propagate and molecularly characterize the isolated virus. The newly isolated virus serves as a good starting point for developing diagnostic tests and inactivated vaccines.
2. Materials and methods
2.1. Study animals
Dromedary camels were sampled at an open-air animal market in Al Ain, United Arab Emirates for MERS-CoV between September 2016 and November 2017 to investigate risk factors and genetic diversity of the virus. A nasal swab was taken from each of the 372 sampled camels together with information on age, sex, and animal origin. In order to minimize risk when handling MERS-CoV-suspected camel specimens and ensure correct specimen management, prompt communication between ADAFSA collecting veterinarians and laboratory staff was set up.
2.2. Sample collection and processing
Nasal samples were obtained by using a Dacron swab kit containing Puritan UniTranz-RT 1 mL Universal Transport Solution (Puritan, Brescia, Italy). All samples were barcoded and then transported to the third-level biological safety laboratory (BSL3) of ADAFSA in Abu Dhabi within 8 h using cool boxes maintained between 4–8°C. Each sample was mixed by pulse-vortexing for 15 s, the solution was transferred to a sterile 5 mL cryovial, and the swab kit was autoclaved and discarded. Samples were centrifuged at 1500 rpm for 10 min at 4°C, and the supernatants were filtered with a 0.45 μ filter (Millipore, Billerica, MA, United States).
2.3. Nucleic acid extraction and RT-qPCR
A volume of 400 μL from each nasal swab was used for the nucleic acid extraction with the EZ1 Virus Mini Kit v2.0 (48) kit (Qiagen), and the final elution was done in 60 μL. Screening for MERS-CoV was performed using the Coronavirus MERS-CoV RT-PCR (ModularDx Kit Coronavirus SA1 (EMC) upstream E-gene and Light Mix Modular MERS-CoV Orf1a), targeting the upstream region of the envelope gene (upE) and open reading frame 1a (ORF1a) (13, 14). The RT-qPCR was performed on LightCycler 2.0 using Roche’s LightCycler RNA Virus Master Chemistry (Basel, Switzerland).
2.4. Virus isolation
All the nasal swab samples were tested for MERS-CoV by RT-qPCR. Only samples with Cp (cycle threshold) values below 22 were subsequently used for virus isolation. A total of 10 nasal swabs positive by PCR were inoculated into Vero E6 cells (African green monkey kidney) obtained initially from American Type Culture Collection (ATCC, Rockville, MD, United States) specimen CCL-81™. Cells were maintained in Dulbecco’s modified Eagle’s medium with 4.5 g/L D-glucose, and l-glutamine (DMEM, Gibco, Thermo Fisher Scientific, Waltham, MA, United States) supplemented with 10% heat-inactivated fetal bovine serum (Gibco, United States) and 5 mL of Gibco Antibiotic-Antimycotic solution (containing 10,000 units/mL of penicillin, 10,000 μg/mL of streptomycin, and 25 μg/mL of Amphotericin B).
We inoculated Vero E6 cells seeded at 80–90% confluence in 6-well cell culture plates (Corning Inc., United States) with 100 μL of the samples and incubated the cells at 37°C in a 5% carbon dioxide atmosphere. A control well inoculated with sterile phosphate-buffered saline (PBS) was included on each plate. Plates were centrifuged at 1000 rpm (18 × g for 60 min) using a small benchtop centrifuge in a sealed biocontainment bucket (SL 8R Small Benchtop Centrifuge, Thermo Scientific, Germany). This system works because the low-speed centrifugation enhances viral adsorption to the susceptible cells. It is thought that the minor trauma to the cell surface produced because of low-speed centrifugation mechanical force enhances the viral entry into the cells, reducing the total time taken for the virus to produce infection of cells (15).
The inoculum was removed, cells were gently washed three times with DMEM, and a fresh medium was added. The infected cells were monitored daily for 7 days to check for cytopathic effects (CPE) using an inverted microscope, and the medium was changed on alternate days. When 70–90% CPE was observed, the supernatant and cells were harvested by freezing and thawing three times, and confirmation of virus isolation was done by RT-qPCR as described above. For those inoculated samples that showed no CPE, a blind passage was made using the freeze–thaw method. If no CPEs were observed until three successive blind passages, the sample was considered negative. Subsequent passages were performed in T-25 flasks.
2.5. Whole genome sequencing
2.5.1. Nucleic acid extraction and sequence-independent, single-primer-amplification
The cell culture extracts (400 μL) of MERS-CoV positive samples were used for the nucleic acid extraction with the EZ1 Virus Mini Kit v2.0 (48) kit (Qiagen). The final elution was done in 60 μL.
Random amplified cDNA was prepared for each sample using an approach that was previously described in another study (16). For reverse transcription, 4 μL of RNA and 1 μL of primer A (5´-GTTTCCCACTGGAGGATA-N9-3′, 40 pmol/μL) were mixed and incubated for 5 min at 65°C followed by cooling to room temperature. To synthesize the first-strand, the volumes of 2 μL SuperScript IV first-strand buffer, 1 μL of 12.5 mM dinucleoside triphosphates (dNTPs), 0.5 μL of 0.1 M dithiothreitol (DTT), 1 μL H2O, and 0.5 μL SuperScript IV (Thermo Fisher) were mixed and incubated for 10 min at 42°C. The following substances were mixed to synthesize the second strand: 1 μL Sequenase buffer, 0.85 μL H2O, and 0.15 μL Sequenase (Affymetrix) and incubated at 37°C for 8 min, followed by the addition of 0.45 μL Sequenase dilution buffer and 0.15 μL Sequenase. The mixture was incubated for 8 min at 37°C. Amplification of cDNA was performed in triplicate using 5 μL of the reaction mixture as input to a 50 μL of LA Taq (Sigma) reaction mixture, according to the manufacturer’s instructions, using 1 μL primer B (5´-GTTTCCCACTGGAGGATA-3′). The amplification conditions consisted of 98°C for 30 s, and 30 cycles of 94°C for 15 s, 50°C for 20 s, and 68°C for 2 min followed by 68°C for 10 min. The amplified cDNA was purified using AMPure XP beads (Beckman Coulter, Brea, CA) and quantified with a Qubit high-sensitivity double-stranded DNA (dsDNA) kit (Thermo Fisher) following the manufacturer’s instructions.
2.5.2. MiSeq™ library preparation and sequencing
Amplified Round B cDNA from all samples were purified using AMPure XP beads (Beckman Coulter), and 2 ng was used as input into the Nextera XT kit (Illumina) previously described (17). After 13 cycles of amplification, Illumina library concentration and average fragment size were determined using the Agilent 4,200 TapeStation System. Sequencing was performed on an Illumina MiSeq using a V3 MiSeq™ (600 cycles) Reagent kit (Illumina, San Diego, CA, United States).
2.5.3. Long read sequencing
The cDNA of the sample with GenBank ID: OQ423284 (Lab ID: VL_13) and the sample with GenBank ID: OQ423287 (Lab ID: VL_765) were further subjected to long-read sequencing. The library was prepared using Ligation Sequencing Kit (SQK-LSK109), and samples were barcoded with Native Barcoding Expansion 1–12 (EXP-NBD104). Finally, 30 ng of the prepared library was sequenced on a Nanopore MinION device using the R9.4.1 flow cell.
2.5.4. Bioinformatics analysis
The Illumina sequences were first evaluated for their quality using FastQC software,1 and the low-quality reads were trimmed with trimmomatic software2 using the following settings: TRAILING:28 SLIDINGWINDOW:4:15 MINLEN:35 to remove the reads with quality below 15 or length less than 35 bp. The adaptors were removed from long reads using the Porechop tool.3
De novo assembly of short reads was performed with the shovill pipeline4 with a command: shovill --outdir out --R1 path/to/read1.fasta --R2 path/to/read2.fastq. The accuracy of the assembly was evaluated with the Quast tool5 with a command: quast -o assembly/quast path/to/scaffolds.fasta. The hybrid assembly for samples (GenBank IDs: OQ423284 and OQ423287) was accomplished with Spades.6 The consensus sequence of the final assembly was first analyzed by BLAST services, followed by phylogenetic analysis. The variant calling against the MERS-CoV reference Saudi isolate JX869059 (EMC/2012) was performed with the snippy tool with an option --mincov 20 (minimum coverage of 20). The nucleotide genome sequences obtained in this study were deposited in GenBank under the accession numbers OQ423283 to OQ423287.
2.5.5. Phylogenetic analysis of MERS-CoV
The BLAST tool available in the NCBI was used for each strain to determine the most closely related sequences of MERS-CoV, and the corresponding sequences, along with the newly isolated sequences, were then used to construct a phylogenetic tree. The final alignment contained 118 sequences from a nearly complete genome (~30,000 nt in length), including human cases of animal origin. The multiple sequence alignment was performed with the ClustalW tool, and the tree was constructed with the Maximum Likelihood method (ML) and Kimura 2-parameter model (18) with 1,000 Bootstrap confidence using MEGA X software (19).
2.5.6. Recombination analysis using RDP5
The Recombination Detection Program (RDP5) v.5.3 (20) was used to detect the possible recombination events using the default settings of algorithms GENECONV, BOOTSCAN, MaxChi, Chimaera, and 3Seq implemented in the RDP5 program. The sequences from this study were aligned with the reference sequences of MERS-CoV representing different clades (~30.000 bp were used for phylogenetic analyses), and the recombination events, likely parental isolates of recombinants, and recombination breakpoints were analyzed.
3. Results
3.1. Virus isolation
After 3 days, we observed virus-induced cytopathic effects (CPE) in three samples, and on the first blind passage, two more isolates were obtained. Table 1 provides information on the five virus isolates and the dromedary camels used for sample collection. Cells infected with all five isolates showed focal rounding and swelling of cells from the fourth day of inoculation, followed by detachment, plaque, and syncytium formation (Figure 1). The presence of MERS-CoV was confirmed by RT-qPCR.
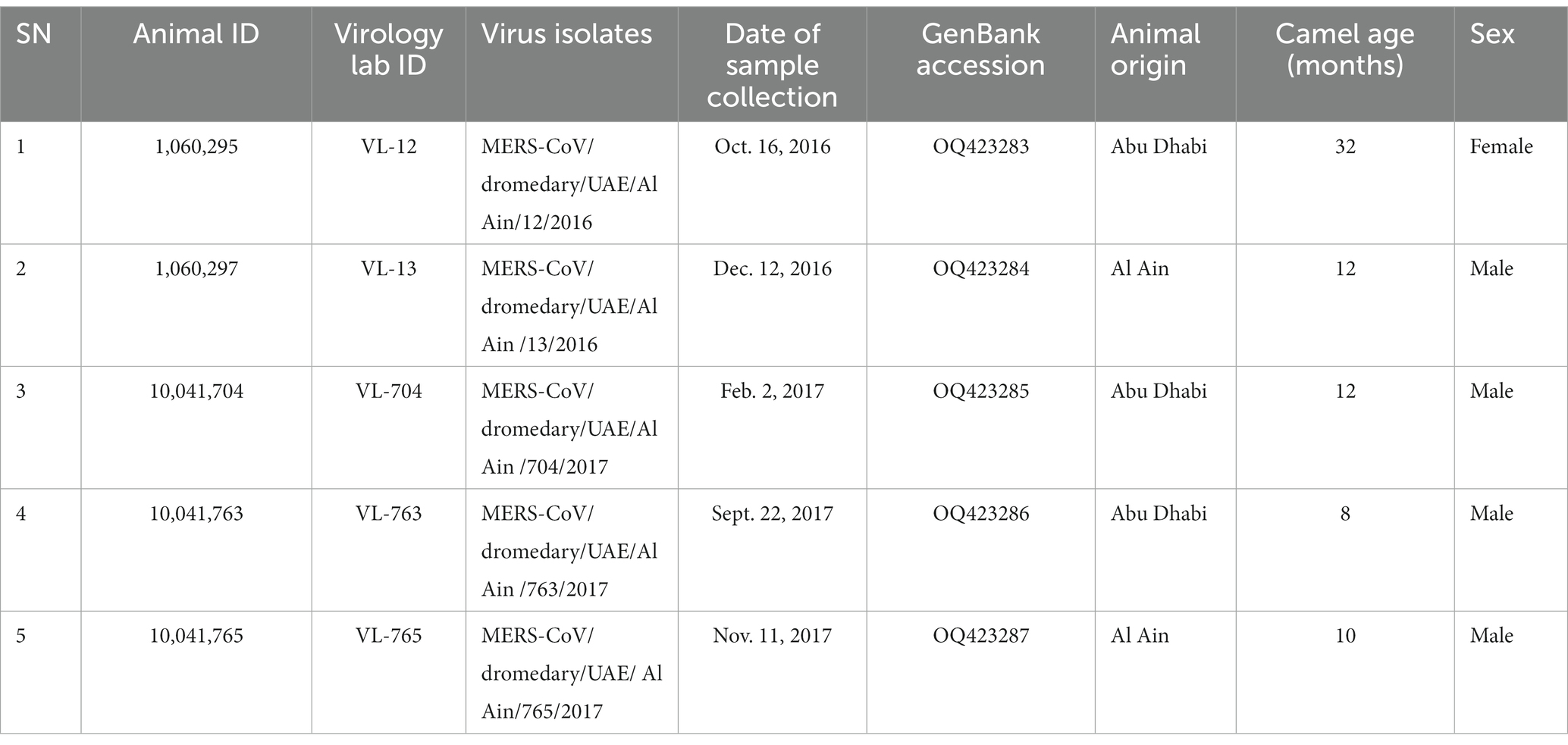
Table 1. Details on the five virus isolates and the dromedary camels from which samples were collected.
Figure 1. Isolation of Middle East respiratory syndrome coronavirus (MERS-CoV) from nasal swabs of dromedary camels. Vero E6 cells were incubated at 37°C and observed daily for cytopathic effect (CPE) under a light microscope. (A) Normal uninfected cells. (B) Early CPE of focal cell rounding at day three post-inoculation (PI) (arrow). (C) Extensive CPE and plaque formation (arrows) observed on day 5 PI. (D) Multinucleated giant cell formation (syncytial formation) (arrows). Original magnification ×10.
3.2. Phylogenetic analysis
A phylogenetic analysis using nearly entire genome sequences (30.000 bp) of 118 MERS-CoV sequences, including our isolates, was conducted to determine which group and lineage our isolates belonged to. The result showed that all five isolates clustered within clade B3 with variable relatedness to the previous UAE isolates. The samples with GenBank IDs OQ423283, OQ423285, OQ423286, and OQ423287 formed a separate cluster and were close to the MERS-CoV isolate camel/KFU-HKU-I/2017 (GenBank ID: MN758606.1), as shown in (Figure 2). The sample GenBank ID: OQ423284 was clustered close to the MERS-CoV isolate Al-Hasa-SA2407/2016 (GenBank ID: MN654975.1), as shown in Figure 2.
Figure 2. Phylogenetic relationship of MERS-CoV genomes. The tree was constructed based on near-complete MERS-CoV genomes (~30,000 nt in length, n = 118), including those of five newly sequenced isolates from the UAE. The tree was constructed with the Maximum Likelihood method and the Kimura 2-parameter model (18) using MEGAX (19). Alignments were performed with ClustalW (21) impeded in MEGAX. Bootstrapping was performed with 1,000 replicates and the value is indicated between 0.7–1 (70–100%). Clade A and clade B are labeled as (A1–A2) and (B1–B6), respectively, while clade C is not shown as it mainly circulates in the African region (22). The sequences obtained in this study are grouped with clade B3 and are marked with the red color. Branch lengths reflect the number of nucleotide substitutions per site.
The MERS-CoV isolates shared 99.69–99.99% nucleotide sequence intragroup identity, 99.73–99.91% identity with the MERS-CoV isolate camel/KFU-HKU-I/2017 (sequence ID: MN758606.1), 99.74–99.86% with the MERS CoV isolate Al-Hasa-SA2407/2016 (GenBank ID: MN654975.1), and 99.42–99.47% identity with the Human betacoronavirus 2c EMC/2012 (GenBank ID: JX869059.2) (Table 2).
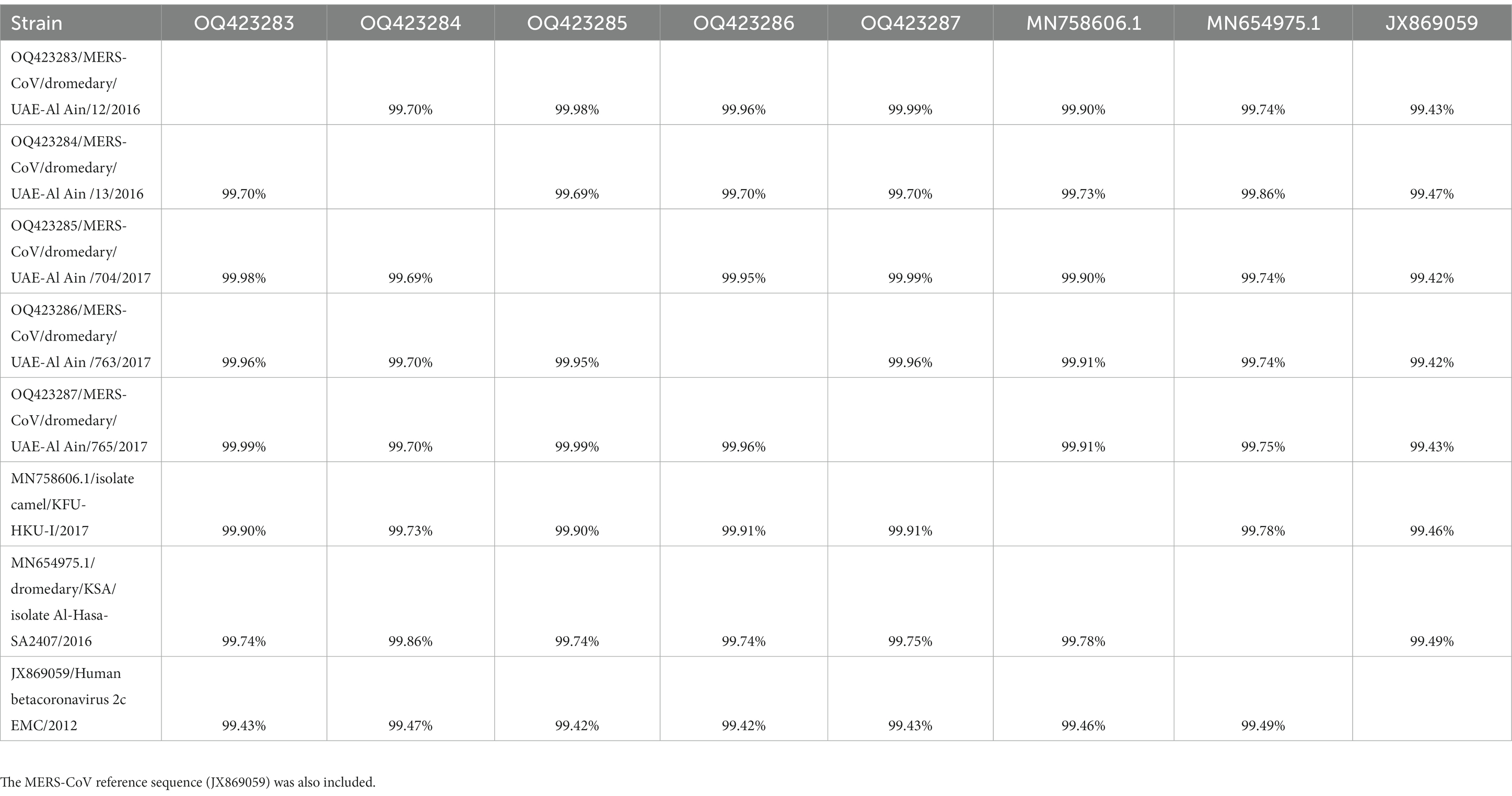
Table 2. The percent identity matrix created by ClustalW for the MERS-CoV isolates along with close sequence obtained after BLAST analysis.
3.3. Mutational analysis
The single nucleotide polymorphism (SNP) was called based on the original Saudi MERS-CoV reference isolate JXJX869059, where several mutations were reported. Some are unique for these isolates.
3.3.1. Missense variants
In coding region ORF1a, there were 32 missense variants, of which 18/32 were shared between the five isolates, 9/32 were found only in OQ423284, 1/32 was found in samples OQ423284 and OQ423285, 1/32 was found in samples OQ423283, OQ423285 and OQ423287, 2/32 were found in samples OQ423283, OQ423285, OQ423286 and OQ423287, and 1/32 was found in samples OQ423283, OQ423284, OQ423285, and OQ423287. In ORF1ab, there were 25 missense variants, of which 18/25 were shared between the five isolates, 4/25 were found in samples OQ423283, OQ423285, OQ423286 and OQ423287, 1/25 was found in samples OQ423283, OQ423284, OQ423285, and OQ423287, and 1/25 was found in samples OQ423283, OQ423284, OQ423285, and OQ423286. In the spike, there were two missense variants: one variant (p.Phe473Ser) was found in OQ423283 only, while the other (p.Gln1020Arg) was found in samples OQ423283, OQ423284, and OQ423285. In ORF3, there were four missense variants, and three variants (p.Val62Phe, p.Leu77Arg, and p.Pro86Leu) were shared with five samples. Another variant (p.Leu17Phe) was found in samples OQ423283, OQ423284, OQ423286, and OQ423287. In ORF4b, there were four missense variants, of which three variants (p.Met6Thr, p.Pro106Ser, and p.His44Pro) were shared with five samples. Another variant (p.Ala139Val) was found in OQ423283 only. In M, there were three missense variants, of which two variants (p.Val69Ile and p.Phe123Ile) were found in all samples. Another variant (p.Ala84Ser) was found in OQ423284 only. In ORF8b, there was a single missense variant (p.Leu4Pro) detected in all samples. In N, there was a single missense variant (p.Gly198Ser) found in OQ423286 only. The summary of the missense mutations is shown in Table 3 and more details are provided in Supplementary Table S1.
3.3.2. Synonymous variants
In ORF1a, there were 46 synonymous variants, of which 25 were shared between the five isolates. In ORF1ab, there was only one synonymous variant shared between the isolates. There were 17 synonymous variants in S, of which 11 were found in all isolates. In ORF3, there were two synonymous variants. In ORF4a, there were two synonymous variants with one shared between the isolates. In ORF4b, there were four synonymous variants, with only one shared between the isolates. In ORF5, there were five synonymous variants, of which four were shared between the isolates. In M, one synonymous variant was shared between the isolates. In N, there were four synonymous variants. The summary of synonymous variants across the viral genome is provided in Supplementary Table S2.
3.4. Analysis of recombination
The recombination analysis indicated that OQ423284/MERS-CoV/dromedary/ UAE-Al Ain/13/2016 is a recombinant strain (Figure 3) with close similarity with B3.MN654975.1/Camelus dromedarius/KSA/isolate Al-Hasa-SA2407/2016 as a major parent (99.9%) and B3.MN655008.1/Camelus dromedarius/KSA/isolate Al-Hasa-SA2696/2017 as a minor parent (99.9%), both of which are nearly identical. The recombination event was detected with six algorithms (RDP, GENECONV, BootScan, MaxiChi, Chimaera, and 3Seq). The beginning and end of the recombination event detected by each algorithm, along with the significant p-values, are shown in Table 4.
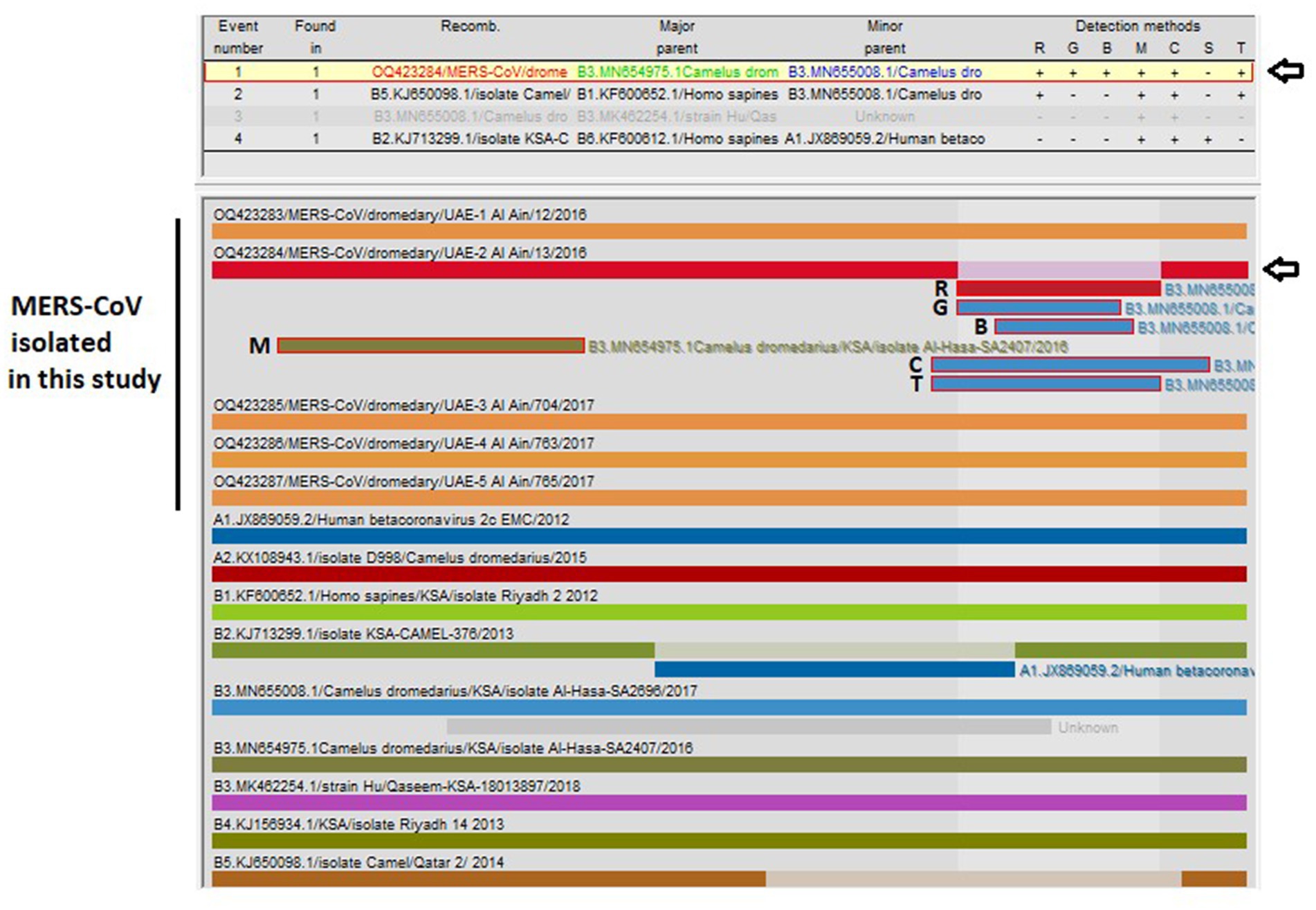
Figure 3. Analysis of possible recombination events in a nearly complete MERS-CoV genome (~30,000 bp). A single recombination event was detected in OQ423284/MERS-CoV/dromedary/UAE-2/Al Ain/13/2016 (indicated by a black arrow). The event detected by the six programs impeded in RDP5 program v.5.3 are indicated (R, RDP; G, GENECONV; B, BootScan; M, MaxiChi; C, Chimaera; T, 3Seq). Detailed information for recombination was also provided in Table 4. Each segment is indicated by a different color bar.
Table 4. Summary of the recombination event identified by the Recombination Detection Program (RDP5) v.5.3.
4. Discussion
MERS-CoV replicates in a wide range of cell types (23, 24) and in vitro, and assays for MERS-CoV growth and quantification have been rapidly developed. However, few virus isolates are currently available with full characterization for research work (25–29). Besides, the high mortality rate of ~34% and the continuous introduction of the virus from dromedary camels to humans urge for the establishment of research infrastructure to aid in research and development. Hence, in the current study, attempts were made to isolate the MERS-CoV from nasal swabs of dromedary camels. In the present study, five viral isolates were obtained from nasal swabs of dromedary camels in the UAE and then adapted to in vitro cultures and further molecularly characterized to identify strain-specific variations and to determine lineage classification.
The phylogenetic analysis performed in this study indicated that all five isolates belonged to clade B3 of MERS-CoV and were clustered close to the MERS-CoV isolated from camels in Saudi Arabia (KSA) between the years 2016 to 2017. One sample (GenBank ID OQ423284) clustered close to the MERS-CoV detected in the Al Hasa region (GenBank ID: MN654975.1), which was reported in 2016, while other isolates clustered close to the MERS-CoV isolate camel/KFU-HKU-I/2017 (GenBank: MN758606.1), which was reported in 2017.
The clustering of MERS-CoV sequences identified in this study was closely related to camel isolates, which may suggest camel-to-camel transmission and supports camels as an animal reservoir for MERS-CoV. Of note, the sequences of the current isolates were found to cluster distantly in the phylogenetic tree far from the previously known MERS-CoV isolated from camels in the UAE, which were located in lineage 5 and lineage 7 within clade B (8). Therefore, this study may provide insight into the risk of trades and livestock markets in the spread of MERS-CoV between camels.
In many studies, a high frequency of mutations and recombinant events were observed in MERS-CoV camel isolates (30) as well as human isolates, and this is necessary for the virus fitness and adaptation in the host.
In our study, where the virus was sequenced from cell culture-adapted isolates, several mutations were observed. For instance, 32 amino acid substitutions in the ORF1a, 25 amino acid substitutions in the ORF1ab, and a single substitution (p.Gly198Ser) in the N region were reported in samples with GenBank IDs: OQ423285 and OQ423287. The latter mutation was also previously reported in human isolates in KSA (31). Generally, amino acid substitutions in the nsp proteins, as well as the N protein, were found to reduce interaction and subsequent virus replication and progeny production (32).
Moreover, two missense amino acid substitutions in the S protein (p.Phe473Ser in sample 2 and p.Gln1020Arg in samples 2 and 3) were reported. It is known that the p.Phe473Ser is located in the receptor-binding domain (RBD) of the spike gene, and exclusively detected in camel strains in Jeddah and Riyadh (22, 33). The mutation p.Gln1020Arg (Q1020R) is located in the heptad repeat region (HR1) of the spike protein. Other mutations detected in our isolates such as Q1020R, R1020Q, or Q1020H were also reported in camel strains in Egypt, Nigeria, the UAE, and some KSA human strains in Jeddah (34–36); these were found to be associated with major MERS-CoV outbreaks in these countries (22). Therefore, changes in the coronavirus spike should be carefully monitored for their effects on receptor binding, virus transmission (37), or even interspecies jump and spillover to humans (31).
This study also identified three mutations in ORF4b in all isolates and one mutation in samples with GenBank IDs OQ423284 and OQ423287 only. Amino acid substitutions in the ORF4b protein were found to alter the host’s immune response to disease and viral pathogenesis (35, 38).
In addition, one mutation in ORF3 (p.Pro86Leu) observed in all current isolates was also reported previously in MERS-CoV strains in camels from the UAE (39). The biological impact of such amino acid changes in ORF3 (4), M (3), ORF8b (1), and among MERS-CoV strains needs to be fully examined.
Analysis of recombination indicated one of the reported sequences (OQ423284/MERS-CoV/dromedary/UAE-Al Ain/13/2016) is a recombinant strain containing sequence from Al Hasa strains (GenBank ID: MN654975.1 and GenBank ID: MN655008.1) within lineage 3 (Clade B3) with high similarity (99.9%). This is not surprising as recombination frequency due to the exchange of functional motifs or even entire genes is quite high (40) within closely related coronaviruses and between different coronaviruses (3, 41, 42). Recombination events generally affect the evolution and transmission of CoVs, including MERS-CoV, so positive selection sites in the spike protein of MERS-CoV in camels may have enabled virus spillover to humans (6, 26, 43). Therefore, the existence of a recombinant virus in our isolates implies the necessity of continuous surveillance of MERS-CoV infections and variants of concern in camels as well as humans, coupled with efforts toward the development of a MERS vaccine.
Artificial mutations could probably develop during cell culture passage even though the five MERS-CoV isolates analyzed in the current study were from early Vero cell passages and were still regarded as wild-type viruses. The fact that cell culture adaptation causes numerous artificial mutations when SARS-CoV-2 is isolated and multiplied in Vero-related cells is well documented (44). The recovery of MERS-CoV from synthesized RNA via the Baric approach in Vero cells is known to easily cause the T1015N artificial mutation in the S protein (45). Additionally, the repetitive passage of MERS-CoV on BHK cells that express DPP4 results in tailored mutations in the spike protein, even if Vero cells are not involved (46). Accordingly, the mutations described in this investigation might not accurately represent the virus’s natural evolution. It is recommended that directly sequencing the wild-type virus from the positive samples using the amplicon method (36) or the SISPA method (16) is the next step to validate mutations reported in this study.
5. Conclusion
In conclusion, MERS-CoV genomic sequences determined in this study are similar to those of viruses detected in camels in Saudi Arabia during the period 2016–2018. Phylogenetic analysis of MERS-CoV isolates indicated the virus lineage 3 clade B viruses continue to be dominant among camels in the UAE. Sequence analysis identified several mutations in different virus proteins as well as a recombination event, raising concerns regarding new viral outbreaks and disease severity. The results of this study support the necessity for the complete genome sequencing of all new cases of MERS-CoV to monitor virus circulation in the region and to develop effective control measures. Although the mutations detected in this study may not reflect the true natural evolution of the virus, as artificial mutations may occur during cell culture passage, the isolated MERS-CoV would be useful in new vaccine development and efficacy studies, pathogenicity, and antiviral research.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Ethics statement
The current study involving swab samples from dromedary camels was reviewed and approved by the ADAFSA Ethics Committee for Monitoring the use of Animals in Scientific Research (ADAFSA-EA-08-2016) and the use of these samples for virus isolation was permitted without additional ethical approval. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
AK and HI: conceptualization and data analysis. AK, HI, and SM: methodology, data curation, and writing-original draft preparation. HA and ZA-H: sample preparation and testing. AK, HI, and AS: writing-review and editing. AS and SM: supervision and funding acquisition. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by Abu Dhabi Agriculture and Food Safety Authority (ADAFSA), Abu Dhabi, UAE.
Acknowledgments
The authors thank veterinarians of ADAFSA and Al Ain municipality for sample collection.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2023.1182165/full#supplementary-material
Footnotes
1. ^https://github.com/s-andrews/FastQC
2. ^https://github.com/usadellab/Trimmomatic
3. ^https://github.com/rrwick/Porechop
4. ^https://github.com/tseemann/shovill
References
1. World Organization for Animal Health (WOAH) (2022). Case definition for reporting to WOAH. Available at: https://www.woah.org/en/disease/middle-east-respiratory-syndrome-mers/ (Accessed September 25, 2022).
2. V’kovski, P, Kratzel, A, Steiner, S, Stalder, H, and Thiel, V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat Rev Microbiol. (2021) 19:155–70. doi: 10.1038/s41579-020-00468-6
3. Woo, PC, Huang, Y, Lau, SK, and Yuen, K-Y. Coronavirus genomics and bioinformatics analysis. Viruses. (2010) 2:1804–20. doi: 10.3390/v2081803
4. World Health Organization (WHO) (2022). MERS situation update, July 2022. Available at: https://www.emro.who.int/health-topics/mers-cov/mers-outbreaks.html (Accessed September 25, 2022).
5. Assiri, AM, Midgley, CM, Abedi, GR, Bin Saeed, A, Almasri, MM, Lu, X, et al. Epidemiology of a novel recombinant Middle East respiratory syndrome coronavirus in humans in Saudi Arabia. J Infect Dis. (2016) 214:712–21. doi: 10.1093/infdis/jiw236
6. Chu, DK, Hui, KP, Perera, RA, Miguel, E, Niemeyer, D, Zhao, J, et al. MERS coronaviruses from camels in Africa exhibit region-dependent genetic diversity. Proc Natl Acad Sci. (2018) 115:3144–9. doi: 10.1073/pnas.1718769115
7. Schroeder, S, Mache, C, Kleine-Weber, H, Corman, VM, Muth, D, Richter, A, et al. Functional comparison of MERS-coronavirus lineages reveals increased replicative fitness of the recombinant lineage 5. Nat Commun. (2021) 12:5324. doi: 10.1038/s41467-021-25519-1
8. Yusof, MF, Queen, K, Eltahir, YM, Paden, CR, Al Hammadi, ZMAH, Tao, Y, et al. Diversity of Middle East respiratory syndrome coronaviruses in 109 dromedary camels based on full-genome sequencing, Abu Dhabi, United Arab Emirates. Emerg Microbes Infect. (2017) 6:1–10. doi: 10.1038/emi.2017.89
9. Khudhair, A, Killerby, ME, Al Mulla, M, Abou Elkheir, K, Ternanni, W, Bandar, Z, et al. Risk factors for MERS-CoV seropositivity among animal market and slaughterhouse workers, Abu Dhabi, United Arab Emirates, 2014–2017. Emerg Infect Dis. (2019) 25:927–35. doi: 10.3201/eid2505.181728
10. Muhairi, SA, Hosani, FA, Eltahir, YM, Mulla, MA, Yusof, MF, Serhan, WS, et al. Epidemiological investigation of Middle East respiratory syndrome coronavirus in dromedary camel farms linked with human infection in Abu Dhabi emirate, United Arab Emirates. Virus Genes. (2016) 52:848–54. doi: 10.1007/s11262-016-1367-1
11. Paden, C, Yusof, M, Al Hammadi, Z, Queen, K, Tao, Y, Eltahir, Y, et al. Zoonotic origin and transmission of Middle East respiratory syndrome coronavirus in the UAE. Zoonoses Public Health. (2018) 65:322–33. doi: 10.1111/zph.12435
12. Song, D, Ha, G, Serhan, W, Eltahir, Y, Yusof, M, Hashem, F, et al. Development and validation of a rapid immunochromatographic assay for detection of Middle East respiratory syndrome coronavirus antigen in dromedary camels. J Clin Microbiol. (2015) 53:1178–82. doi: 10.1128/JCM.03096-14
13. Corman, V, Eckerle, I, Bleicker, T, Zaki, A, Landt, O, Eschbach-Bludau, M, et al. Detection of a novel human coronavirus by real-time reverse-transcription polymerase chain reaction. Eur Secur. (2012) 17:20285. doi: 10.2807/ese.17.39.20285-en
14. Corman, V, Müller, MA, Costabel, U, Timm, J, Binger, T, Meyer, B, et al. Assays for laboratory confirmation of novel human coronavirus (hCoV-EMC) infections. Eur Secur. (2012) 17:20334. doi: 10.2807/ese.17.49.20334-en
15. Engler, HD, and Preuss, J. Laboratory diagnosis of respiratory virus infections in 24 hours by utilizing shell vial cultures. J Clin Microbiol. (1997) 35:2165–7. doi: 10.1128/jcm.35.8.2165-2167.1997
16. Alirezaie, B, Bahador, D, Mohammadi, A, and Foroughi, A. Sequence-independent single-primer-amplification (SISPA) as a screening technique for detecting unexpected RNA viral adventitious agents in cell cultures. Arch Biotechnol Biomed. (2021) 5:008–12. doi: 10.29328/journal.abb.1001022
17. Katusiime, MG, Halvas, EK, Wright, I, Joseph, K, Bale, MJ, Kirby-McCullough, B, et al. Intact HIV proviruses persist in children seven to nine years after initiation of antiretroviral therapy in the first year of life. J Virol. (2020) 94:e01519–9. doi: 10.1128/JVI.01519-19
18. Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. (1980) 16:111–20. doi: 10.1007/BF01731581
19. Kumar, S, Stecher, G, Li, M, Knyaz, C, and Tamura, K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. (2018) 35:1547–9. doi: 10.1093/molbev/msy096
20. Martin, DP, Varsani, A, Roumagnac, P, Botha, G, Maslamoney, S, Schwab, T, et al. RDP5: a computer program for analyzing recombination in, and removing signals of recombination from, nucleotide sequence datasets. Virus Evol. (2021) 7:veaa087. doi: 10.1093/ve/veaa087
21. Larkin, MA, Blackshields, G, Brown, NP, Chenna, R, McGettigan, PA, McWilliam, H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. (2007) 23:2947–8. doi: 10.1093/bioinformatics/btm404
22. Mostafa, A, Kandeil, A, Shehata, M, El Shesheny, R, Samy, AM, Kayali, G, et al. Middle east respiratory syndrome coronavirus (mers-cov): state of the science. Microorganisms. (2020) 8:991. doi: 10.3390/microorganisms8070991
23. Chan, JF-W, Chan, K-H, Choi, GK-Y, To KK-W, Tse, H, Cai, J-P, et al. Differential cell line susceptibility to the emerging novel human betacoronavirus 2c EMC/2012: implications for disease pathogenesis and clinical manifestation. J Infect Dis. (2013) 207:1743–52. doi: 10.1093/infdis/jit123
24. Müller, MA, Raj, VS, Muth, D, Meyer, B, Kallies, S, Smits, SL, et al. Human coronavirus EMC does not require the SARS-coronavirus receptor and maintains broad replicative capability in mammalian cell lines. MBio. (2012) 3:e00515–2. doi: 10.1128/mBio.00515-12
25. Lau, SK, Wernery, R, Wong, EY, Joseph, S, Tsang, AK, Patteril, NAG, et al. Polyphyletic origin of MERS coronaviruses and isolation of a novel clade a strain from dromedary camels in the United Arab Emirates. Emerg Microbes Infect. (2016) 5:1–9. doi: 10.1038/emi.2016.129
26. Ngere, I, Hunsperger, EA, Tong, S, Oyugi, J, Jaoko, W, Harcourt, JL, et al. Outbreak of Middle East respiratory syndrome coronavirus in camels and probable spillover infection to humans in Kenya. Viruses. (2022) 14:1743. doi: 10.3390/v14081743
27. Tamin, A, Queen, K, Paden, CR, Lu, X, Andres, E, Sakthivel, SK, et al. Isolation and growth characterization of novel full length and deletion mutant human MERS-CoV strains from clinical specimens collected during 2015. J Gen Virol. (2019) 100:1523–9. doi: 10.1099/jgv.0.001334
28. Teng, JLL, Wernery, U, Lee, HH, Fung, J, Joseph, S, Li, KSM, et al. Co-circulation of a novel dromedary camel parainfluenza virus 3 and Middle East respiratory syndrome coronavirus in a dromedary herd with respiratory tract infections. Front Microbiol. (2021) 12:739779. doi: 10.3389/fmicb.2021.739779
29. Zaki, AM, Van Boheemen, S, Bestebroer, TM, Osterhaus, AD, and Fouchier, RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. (2012) 367:1814–20. doi: 10.1056/NEJMoa1211721
30. Darriba, D, Taboada, GL, Doallo, R, and Posada, D. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. (2012) 9:772–2. doi: 10.1038/nmeth.2109
31. AlBalwi, MA, Khan, A, AlDrees, M, Udayaraja, G, Manie, B, Arabi, Y, et al. Evolving sequence mutations in the middle east respiratory syndrome coronavirus (MERS-CoV). J Infect Public Health. (2020) 13:1544–50. doi: 10.1016/j.jiph.2020.06.030
32. Hurst, KR, Koetzner, CA, and Masters, PS. Characterization of a critical interaction between the coronavirus nucleocapsid protein and nonstructural protein 3 of the viral replicase-transcriptase complex. J Virol. (2013) 87:9159–72. doi: 10.1128/JVI.01275-13
33. Fagbo, SF, Skakni, L, Chu, DK, Garbati, MA, Joseph, M, Peiris, M, et al. Molecular epidemiology of hospital outbreak of Middle East respiratory syndrome, Riyadh, Saudi Arabia, 2014. Emerg Infect Dis. (2015) 21:1981–8. doi: 10.3201/eid2111.150944
34. Cotten, M, Watson, SJ, Zumla, AI, Makhdoom, HQ, Palser, AL, Ong, SH, et al. Spread, circulation, and evolution of the Middle East respiratory syndrome coronavirus. MBio. (2014) 5:e01062–13. doi: 10.1128/mBio.01062-13
35. Forni, D, Filippi, G, Cagliani, R, De Gioia, L, Pozzoli, U, Al-Daghri, N, et al. The heptad repeat region is a major selection target in MERS-CoV and related coronaviruses. Sci Rep. (2015) 5:1–10. doi: 10.1038/srep14480
36. Cotten, M, Watson, SJ, Kellam, P, Al-Rabeeah, AA, Makhdoom, HQ, Assiri, A, et al. Transmission and evolution of the Middle East respiratory syndrome coronavirus in Saudi Arabia: a descriptive genomic study. Lancet. (2013) 382:1993–2002. doi: 10.1016/S0140-6736(13)61887-5
37. Kleine-Weber, H, Elzayat, MT, Wang, L, Graham, BS, Müller, MA, Drosten, C, et al. Mutations in the spike protein of Middle East respiratory syndrome coronavirus transmitted in Korea increase resistance to antibody-mediated neutralization. J Virol. (2019) 93:e01381–18. doi: 10.1128/JVI.01381-18
38. Yang, Y, Ye, F, Zhu, N, Wang, W, Deng, Y, Zhao, Z, et al. Middle East respiratory syndrome coronavirus ORF4b protein inhibits type I interferon production through both cytoplasmic and nuclear targets. Sci Rep. (2015) 5:1–13. doi: 10.1038/srep17554
39. Al Hammadi, ZM, Chu, DK, Eltahir, YM, Al Hosani, F, Al Mulla, M, Tarnini, W, et al. Asymptomatic MERS-CoV infection in humans possibly linked to infected dromedaries imported from Oman to United Arab Emirates, may 2015. Emerg Infect Dis. (2015) 21:2197–200. doi: 10.3201/eid2112.151132
40. Du, J, Yang, L, Ren, X, Zhang, J, Dong, J, Sun, L, et al. Genetic diversity of coronaviruses in Miniopterus fuliginosus bats. Sci China Life Sci. (2016) 59:604–14. doi: 10.1007/s11427-016-5039-0
41. Woo, PC, Lau, SK, Yip, CC, Huang, Y, Tsoi, H-W, Chan, K-H, et al. Comparative analysis of 22 coronavirus HKU1 genomes reveals a novel genotype and evidence of natural recombination in coronavirus HKU1. J Virol. (2006) 80:7136–45. doi: 10.1128/JVI.00509-06
42. Woo, PC, Lau, SK, and Yuen, K-y. Infectious diseases emerging from Chinese wet-markets: zoonotic origins of severe respiratory viral infections. Curr Opin Infect Dis. (2006) 19:401–7. doi: 10.1097/01.qco.0000244043.08264.fc
43. Su, S, Wong, G, Shi, W, Liu, J, Lai, AC, Zhou, J, et al. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. (2016) 24:490–502. doi: 10.1016/j.tim.2016.03.003
44. Sasaki, M, Uemura, K, Sato, A, Toba, S, Sanaki, T, Maenaka, K, et al. SARS-CoV-2 variants with mutations at the S1/S2 cleavage site are generated in vitro during propagation in TMPRSS2-deficient cells. PLoS Pathog. (2021) 17:e1009233. doi: 10.1371/journal.ppat.1009233
45. Scobey, T, Yount, BL, Sims, AC, Donaldson, EF, Agnihothram, SS, Menachery, VD, et al. Reverse genetics with a full-length infectious cDNA of the Middle East respiratory syndrome coronavirus. Proc Natl Acad Sci. (2013) 110:16157–62. doi: 10.1073/pnas.1311542110
Keywords: MERS-CoV, coronavirus, isolation, characterization, dromedary, camel, zoonotic disease
Citation: Khalafalla AI, Ishag HZA, Albalushi HIA, Al-Hammadi ZMA-H, Al Yammahi SMS, Shah AAM and Al Muhairi SSM (2023) Isolation and genetic characterization of MERS-CoV from dromedary camels in the United Arab Emirates. Front. Vet. Sci. 10:1182165. doi: 10.3389/fvets.2023.1182165
Edited by:
Thamir A. Alandijany, King Abdulaziz University, Saudi ArabiaReviewed by:
Mohamed Ahmed Ali, National Research Centre, EgyptAbdullah Al-Mubarak, King Faisal University, Saudi Arabia
Copyright © 2023 Khalafalla, Ishag, Albalushi, Al-Hammadi, Al Yammahi, Shah and Al Muhairi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Abdelmalik Ibrahim Khalafalla, abdelmalik.khalafalla@adafsa.gov.ae
†These authors have contributed equally to this work
 Abdelmalik Ibrahim Khalafalla
Abdelmalik Ibrahim Khalafalla