- 1Fisheries College, Key Laboratory of Healthy Mariculture for the East China Sea, Ministry of Agriculture, Jimei University, Xiamen, Fujian, China
- 2Fourth Institute of Oceanography, Key Laboratory of Tropical Marine Ecosystem and Bioresource, Ministry of Natural Resources, Beihai, China
- 3Institute of Electromagnetics and Acoustics, School of Electronic Science and Engineering, Xiamen University, Xiamen, China
Psychrophilic Aeromonas salmonicida could not grow above 25°C and therefore thought unable to infect mammals and humans. In our previous study, a mesophilic A. salmonicida SRW-OG1 was isolated from Epinephelus coioides with furunculosis. Through the analysis of preliminary RNA-seq, it was found that the Zn2+ uptake related genes znuA, znuB and znuC might be involved in the virulence regulation of A. salmonicida SRW-OG1. Therefore, the purpose of this study was to explore the effect of znuABC silencing on the virulence regulation of A. salmonicida SRW-OG1. The results showed that the growth of the znuA-RNAi, znuB-RNAi, and znuC-RNAi strains was severely restricted under the Fe2+ starvation, but surprisingly there was no significant difference under the Zn2+ restriction. In the absence of Zn2+ and Fe2+, the expression level of znuABC was significantly increased. The motility, biofilm formation, adhesion and hemolysis of the znuA-RNAi, znuB-RNAi, and znuC-RNAi strains were significantly reduced. We also detected the expression of znuABC under different growth periods, temperatures, pH, as well as Cu2+ and Pb2+ stresses. The results showed that znuABC was significantly up-regulated in the logarithmic phase and the decline phase of A. salmonicida. Interestingly, the trend of expression levels of the znuABC at 18, 28, and 37°C was reversed to another Zn2+ uptake related gene zupT. Taken together, these indicated that the znuABC was necessary for A. salmonicida SRW-OG1 pathogenicity and environmental adaptability, and was cross regulated by iron starvation, but it was not irreplaceable for A. salmonicida SRW-OG1 Zn2+ uptake in the host.
1. Introduction
Aeromonas salmonicida is a facultative anaerobic Gram-negative bacterium, widely existing in aquatic environments and soils (1), which can cause furunculosis in a variety of marine and freshwater fish, especially salmon (Salmonidae) and carp (Cyprinidae), as well as rainbow trout (Onco-rhynchus mykiss), turbot (Scophthalmus maximus L.), carp (Cyprinus carpio), goldfish (Carassius auratus) and so on (2–5). Since A. salmonicida has been proven unable to grow above 25°C, and most of them are isolated from cold water teleost fish (6–8), researchers believe that A. salmonicida cannot infect mammals and humans for quite a long time (9, 10). However, we previously isolated a mesophilic pathogenic A. salmonicida strain from orange-spotted grouper (Epinephelus coioides) with furunculosis and named it as A. salmonicida SRW-OG1 (11). Meanwhile, there have been some reported cases of human infection with mesophilic A. salmonicida. Based on the whole genome comparison between mesophilic A. salmonicida C947 and AJ83 isolated from human patients in Spain (12) and A. salmonicida SRW-OG1, we found that the genomes of C947, AJ83 and SRW-OG1 were highly similar and shared the vast majority of pathogen-host interaction factors (13), which indicated the possible pathogenicity of A. salmonicida SRW-OG1 to mammals and human.
Among all the essential trace elements in living organisms, iron (Fe) is the first abundant transition metal element, followed by zinc (Zn) (14–16). Zn is present in the vast majority of proteins. In prokaryote organisms, Zn-binding proteins account for about 4–8% of the total protein (17–19). It has been found that at least two zinc transport systems existing in bacterial pathogens (20). The Znu-like system, which is homologous to the znuABC system of the Zn2+ transport system of Escherichia coli found in a variety of gram-positive and gram-negative bacteria (21), is currently the most commonly used (22). znuABC is a high-affinity Zn2+ transporter composed of the soluble periplasmic binding protein ZnuA (Formerly YcdH), the high-permeability transmembrane protein ZnuB (Formerly YceA) and the ATPase ZnuC (Formerly YcdI), and is regulated by Zur (zinc uptake system) (23). znuABC belongs to the ABC transporter, which is one of the largest transporter gene families that transport a variety of substrates across the membrane by utilizing ATP energy (24, 25). These transporters have two transmembrane domains (TMDs) and two nucleotide binding domains (NBDs). TMD binds to the transported substrate, and NBD provides the energy needed for transport by binding and hydrolyzing two (and sometimes only one) ATP molecules (26). In Gram-positive and Gram-negative bacteria, Zn2+ input through the plasma membrane is primarily facilitated by the ABC family transporter znuABC (14, 27). Zinc transporters protein znuABC have been shown to be useful for a variety of pathogens to overcome nutritional immunity and calmodulin-mediated zinc chelation (28), such as Pseudomonas aeruginosa (29) and Salmonella typhimurium (30). Studies have shown that the deletion of the znuA of Salmonella enterica serovar Typhimurium does not limit the growth of pathogens in the culture medium, but greatly reduces the reproductive capacity in the Zn2+-limited culture medium (28). Compared with wild-type strain, the biofilm formation ability, movement ability and bacterial competition ability of the ΔznuABC mutant strains of Chromobacterium violaceum are weakened to a certain degree (31). Yersinia ruckeri ΔznuABC mutant strains does not slow down under zinc restriction but is significantly weaker than wild-type strains in bacterial competition (32). According to our previous RNA-seq analysis, znuABC might be key virulence genes during A. salmonicida SRW-OG1 infection at high temperature. But so far, there are no researches studies about the virulence effect of znuABC on A. salmonicida and whether it is related in the regulation of anti-nutritional immunity.
Therefore, we hypothesized that znuABC might play important roles in the anti-nutritional immunity and environmental adaptability, thus affecting the pathogenicity of mesophilic A. salmonicida SRW-OG1. The aim of this study was to investigate: (1) the relationship between the znuABC and A. salmonicida SRW-OG1 Zn2+ acquisition, (2) the sensitivity of the A. salmonicida SRW-OG1 znuABC to environmental stresses, and (3) the effects of znuABC on the virulence regulation of A. salmonicida SRW-OG1.
2. Materials and methods
2.1. Bacterial strain and growth conditions
A. salmonicida SRW-OG1 is a mesophilic pathogenic strain we previously isolated from E. coioides with furunculosis (11). The strain was regularly grown in Luria-Bertani (LB) medium at 28°C, 220 rpm (33). E. coli DH5α was purchased from TransGen Biotech (Beijing, China), and grown in LB medium at 37°C, 220 rpm. DH5α pCM 130 was preserved in our laboratory at −80°C.
2.2. Knockdown of A. salmonicida SRW-OG1 znuABC genes
Knocked-down strains of A. salmonicida SRW-OG1 znuA, znuB and znuC were constructed using RNAi, according to the method described by Bittel et al. (34)and Girard et al. (35, 36) with slight modifications. The sequence of A. salmonicida SRW-OG1 znuA, znuB and znuC genes were introduced into Invitrogen Block-iTTM RNAi Designer (https://rnaidesigner.thermofisher.com/rnaiexpress/design.do) to design shRNA online (Supplementary Table 1). T4 DNA ligase was used to ligate the shRNA with the linear pCM130, and the recombinant pCM130 vectors were first transformed into E. coli DH5α by heat shock according to the description of Huang et al. (37), and then extracted and transformed into the A. salmonicida SRW-OG1 by electroporation. The A. salmonicida SRW-OG1 was then cultured in LB with 100 μg/mL tetracycline (TET) at 28°C overnight. Positive colonies were verified by gene sequencing and the expression level of znuA, znuB and znuC was detected by qRT-PCR (38).
2.3. Growth ability detection in Zn2+ restriction environment
A. salmonicida SRW-OG1 wild-type strain and znuA, znuB, znuC-RNAi strains were cultured overnight at 28 °C, adjusted to OD600nm = 0.2, then diluted 1,000 times in gradient, and then 10 μL of bacterial dilution was mixed with 190 μL of Zn2+ chelating Luria-Bertani (LB) medium [adding 2 μM Tetrakis-(2-pyridylmethyl)-ethylenediamine (TPEN)] in each well of 96-well plate, and then incubated at 28°C. The concentration of bacterial solution was measured every hour for a total of 36 h and the values of OD600nm was recorded. From the OD600nm value, growth curves were plotted to compare the growth trends of wild-type, znuA, znuB and znuC-RNAi strains of A. salmonicida SRW-OG1 (39). Six independent biological replicates were performed for each time point. Data were presented as mean ± standard deviation (SD).
2.4. Growth ability detection in Fe2+ restriction environment
The wild-type and znuA, znuB, znuC-RNAi strains of A. salmonicida SRW-OG1 were incubated at 28°C at 220 rpm to OD600nm = 0.2. Then, the bacterial suspension was diluted 1,000 times, and 10 μL of bacterial dilution was mixed with 190 μL of Fe2+ chelating Luria-Bertani (LB) medium (adding 1 μmol/L 2,2-bipyridine) in each well of 96-well plate, and then incubated at 28°C. OD600nm was then recorded hourly for a total of 36 h to compare changes in growth capacity of wild-type and the knocked-down strains (40). Six independent biological replicates were performed for each time point. Data were presented as mean ± standard deviation (SD).
2.5. Biofilm forming ability
Biofilm assays for A. salmonicida SRW-OG1 was carried out according to Huang et al. (41, 42) with slight modifications. First, the znuA, znuB, znuC-RNAi and wild-type strains of A. salmonicida SRW-OG1 were cultured under the same conditions (28°C, 220 rpm) overnight, and the OD600nm was adjusted to 0.2. The mixture of 100 μL of Luria-Bertani (LB) medium and 100 μL of bacterial suspension was incubated in the dark at 28°C for 24 h, washed 3 times with phosphate buffered saline (PBS, pH = 7.4, without Calcium and Magnesium), stained with 200 μL of 0.1 % crystal violet for 15 min, then rinsed with PBS, and air-dried at 25°C. The biofilm was dissolved with 200 μL of ethanoic acid (33 %) and quantified with OD590nm. Six independent biological replicates were performed for each strain.
2.6. Semi-solid agar plate movement ability
The semi-solid agar method was used to determine the motility of A. salmonicida SRW-OG1 (39, 43). Wild-type and znuA, znuB, znuC-RNAi strains of A. salmonicida SRW-OG1 were grown under the same conditions (28°C, 220 rpm) overnight, and adjusted to OD600nm of 0.3. 1 μL of bacterial suspension was dropped vertically on the center of semi-solid agar plate [Luria-Bertani (LB) + 0.3 % agar]. After incubation at 25°C for 30 min, transfer it to 28°C for 20 h. Colony diameters were measured instrumentally, while three independent biological replicates were performed for each strain.
2.7. Adhesion ability in vitro
The determination of adhesion ability of wild-type and znuA, znuB, znuC-RNAi strains of A. salmonicida SRW-OG1 were carried out according to the methods of Huang et al. (37, 44) with slight modifications. Briefly, 30 μL of sterile healthy E. coioides epidermal mucus was spread evenly on the glass slides, incubated overnight at 25°C in the dark, subsequently fixed with 4% methanol at 25°C for 30 min. The bacterial suspension was adjusted to OD600nm of 0.4. After that, 200 μL of bacterial suspension was taken to cover the area with mucus evenly, and incubated under dark and damp at 28°C for 2 h, then rinsed with phosphate buffered saline for 3 times, air-dried and fixed again with 4% methanol at 25°C for 30 min. Lastly, the slid glasses were stained with 0.1% crystal violet for 3 min, and observed with optical microscope and imaged with digital camera (magnification, × 1000) (45). The number of bacteria was counted from the image using IPwin software (11).
2.8. Hemolytic ability
Changes in hemolytic capacity of A. salmonicida SRW-OG1 wild-type, znuA, znuB, and znuC-RNAi strains were compared according to the methods described by Porto-Fett et al. (46, 47). A. salmonicida SRW-OG1 were cultured at 28°C, 220 rpm to the logarithmic growth period. A 6 mm round hole was punctured into the Columbia blood agar plate with an oxford cup, and then 100 μL of bacterial suspension was transferred into the center of the hole (48). Afterwards, the blood agar plates were cultured at 37°C for 12 h with the front side up. Finally, whether there was a hemolytic circle was observed and the diameter of the hemolytic circle was measured (49). Three independent biological replicates were performed for each strain.
2.9. The expression levels of znuABC under different stresses environment
In order to validate the effect of temperature on the expression of znuABC, the bacterial solution was adjusted to OD600nm = 0.3–0.4, and the expression level of znuABC was detected by qRT-PCR after being cultured at 18, 28, and 37°C for 2–4 h.
In order to validate the effect of pH on the expression of znuABC, the bacterial solution was adjusted to OD600nm = 0.3–0.4, then added 1mol/L HCl or NaOH to LB medium and adjusted the medium as pH = 4, 5, 6, 7, or 8. After this, they were incubated at 28°C for 2–4 h, and the expression level of znuABC was detected by qRT-PCR.
In order to validate the effect of Cu2+ and Pb2+ on the expression of znuABC, the bacterial solution was adjusted to OD600nm = 0.3–0.4, and added 30 mmol/L of Cu2+ or Pb2+ to LB medium and they were incubated at 28°C for 2–4 h. Finally, the expression level of znuABC was detected by qRT-PCR.
2.10. Total RNA extraction and reverse transcription
Total RNA was extracted using TRIzol reagent (Invitrogen, USA) following the manufacturer's instructions (11, 50). Reverse transcription was performed from 2.0 mg of total RNA using A Reverse Mu-MLV cDNA Synthesis Kit (TransGen Biotech Co., Ltd., Beijing, China) following the manufacturer's instructions (11, 51).
2.11. Quantitative real-time polymerase chain reaction (qRT-PCR)
The qRT-PCR was performed using a QuantStudio 6 Flex real-time PCR system (Life Technologies Inc., Carlsbad, CA, USA) with the Power Green qPCR Mix (Dongsheng Biotech Co., Guangzhou, China) (52). The total volume of the reaction was 10 μL, which included 0.25 μL forward primer (10 μM), 0.25 μL reverse primer (10 μM), 0.5 μL template, and 9.0 μL 2×Power Green qPCR Mix. The thermal cycler conditions were 95°C for 2 min, followed by 40 cycles of 95°C for 20 s, 58°C for 20 s, and 72°C for 20 s. All qRT-PCR experiments were performed in triplicate using independent samples. Supplementary Table 2 showed all the primers (designed by Primer 5 software, synthesized by Shanghai Sangon Biotechnology) used in the experiment. The relative expression levels of the znuA, znuB and znuC genes were calculated using the 2−ΔΔCt calculation and the gene expression levels were normalized to 16S rRNA and housekeeping gene gyrB (51, 53).
2.12. Statistical analysis
Data were presented as mean ± standard deviation (SD) (n = 3), and analyzed by SPSS 24.0 software (IBM, Armonk, NY, USA). Differences were compared by t-test, one-way ANOVA followed by the Dunnett's test. P < 0.05 was considered statistically significant (45).
2.13. Ethics statement
All animal experiments were carried out strictly under the recommendations in the “Guide for the Care and Use of Laboratory Animals” set by the National Institutes of Health. The animal protocols were approved by the Animal Ethics Committee of Jimei University (Acceptance NO JMULAC201159).
3. Result
3.1. Construction of znuA, znuB, and znuC-RNAi strains of A. salmonicida SRW-OG1
In the present study, three znuA, znuB and znuC knocked-down strains were successfully constructed. After the verification by qRT-RCR, the best silencing efficiency was reduced by 97.09, 90.30, and 96.16% (Figure 1A), respectively and was chosen for further research. To verify whether the silencing of znuA, znuB, and znuC genes affect the function of the low-affinity Zinc-transport system ZupT, the expression levels of zupT was determined by qRT-PCR in A. salmonicida SRW-OG1 wild-type, znuA, znuB, and znuC-RNAi strains. The results showed that (Figure 1B) the expression levels of zupT in znuA-RNAi, znuB-RNAi and znuC-RNAi were significantly up-regulated by 32.31 times, 16.08 times and 24.33 times, respectively, which indicated that the Zinc-transporter znuABC might interact with low-affinity Zinc-transport system ZupT. The increase of zupT expression might be a compensation mechanism for znuABC silencing.
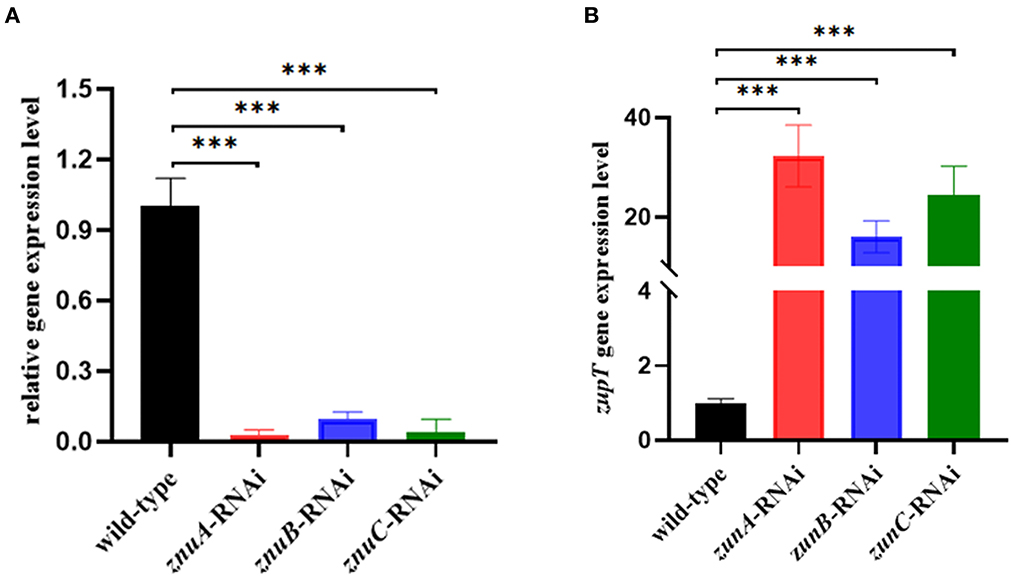
Figure 1. Construction of A. salmonicida SRW-OG1 znuA, znuB, and znuC silencing strains and expression of zupT in wild-type and the silencing strains. (A) was qRT-RCR validation of silencing efficiency of znuA-RNAi, znuB-RNAi, and znuC-RNAi. (B) was the expression levels of zupT in wild-type, znuA, znuB, znuC-RNAi strains of A. salmonicida SRW-OG1; Data were presented as mean ± standard deviation (SD) (n = 3). ***P < 0.001.
3.2. The effect of znuA, znuB, and znuC gene on the growth of A. salmonicida SRW-OG1 under Zn2+ and Fe2+ restriction environment
We determined the growth trend of the znuA, znuB, and znuC of A. salmonicida under Zn2+ and Fe2+ restriction conditions and the expression levels of znuA, znuB, and znuC under these limited conditions. In the Zn2+ restriction environment, the growth trend of znuA-RNAi, znuB-RNAi and znuC-RNAi were not significantly different than that of the wild-type in the whole growth period (Figure 2A). In the Fe2+ restriction environment, the growth trend of znuA-RNAi, znuB-RNAi and znuC-RNAi were significantly weaker than that of the wild-type from logarithmic growth period to decay period (Figure 2B). Among them, under the condition of Fe2+ limitation, the growth trends of the three strains declined to different extents, with the reduction of znuA-RNAi by 10%, znuB-RNAi by 40% and znuC-RNAi by 50 %, which might be related to the different functions of the three genes in the transportation process. The expression levels of znuA, znuB, and znuC in Zn2+ and Fe2+ limited environment by qRT-PCR (Figure 3). The results revealed that the expression levels of znuA, znuB and znuC were significantly up-regulated by 6.013, 2.42, and 1.91 times under Zn2+ restriction and 55.70, 2.09, and 2.16 times under Fe2+ restriction.
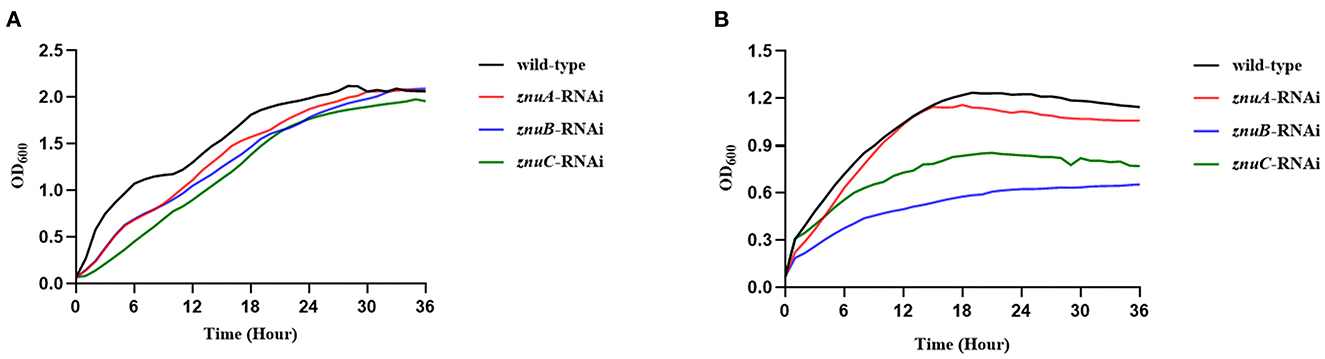
Figure 2. Determination of growth ability in metal ion limited environment. (A) was the growth curve under the condition of Zn2+ restriction environment. (B) was the growth curve under the condition of Fe2+ restriction environment.
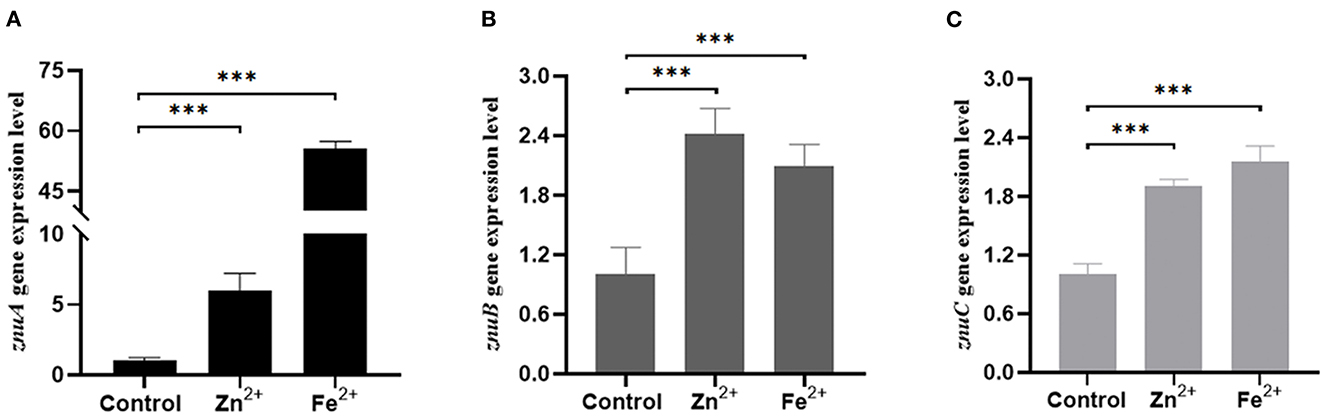
Figure 3. Determination of the expression levels of znuA, znuB, and znuC in Zn2+ and Fe2+ limitation. (A) was the expression level of znuA in Zn2+ and Fe2+ restriction. (B) was the expression level of znuB in Zn2+ and Fe2+ restriction. (C) was the expression level of znuC in Zn2+ and Fe2+ restriction. Data were presented as mean ± standard deviation (SD) (n = 3). ***P < 0.001.
3.3. The effect of znuA, znuB, and znuC on the biofilm forming ability of A. salmonicida SRW-OG1
The capability of biofilm formation was determined by crystal violet staining. Our results showed that the biofilm forming of znuA-RNAi, znuB-RNAi and znuC-RNAi were significantly weaker than that of the wild-type after culturing for 24 h under the same conditions, and the biofilm formation ability decreased by 2.18, 4.30, and 4.93 times, respectively (Figure 4).
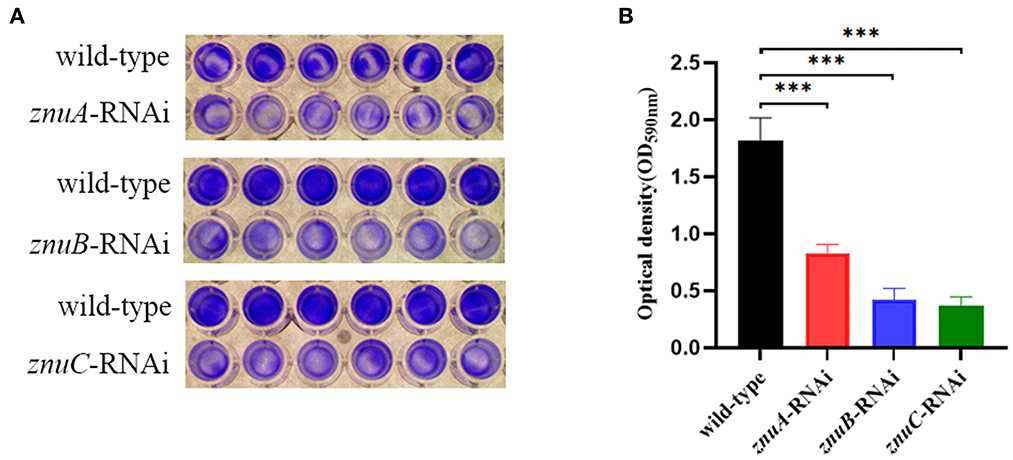
Figure 4. Biofilm forming ability assay of wild-type, znuA, znuB, and znuC-RNAi strains of A. salmonicida SRW-OG1. (A) was the typical figure of the biofilm formation of wild-type, znuA, znuB and znuC-RNAi strains of A. salmonicida SRW-OG1; (B) was the biofilm formation histogram of wild-type, znuA, znuB, and znuC-RNAi strains of A. salmonicida SRW-OG1. Data were presented as mean ± standard deviation (SD) (n = 3). ***P < 0.001.
3.4. The effect of znuA, znuB, and znuC on motility of A. salmonicida SRW-OG1
The motility assay showed (Figure 5) that the single colony diameters of wild-type, znuA, znuB and znuC-RNAi strains of A. salmonicida SRW-OG1 placed on semi-solid agar plates for 18 h were about 6.826 ± 0.21 mm, 3.271 ± 0.12 mm, 3.006 ± 0.15 mm and 3.594 ± 0.36 mm, respectively. Statistical analysis revealed a significant 2.09, 2.27, and 2.05 times reduction, respectively in motility of znuA-RNAi, znuB-RNAi and znuC-RNAi compared with wild-type strain. Therefore, it seemed that znuA, znuB, and znuC were positively related to the motility of A. salmonicida SRW-OG1.
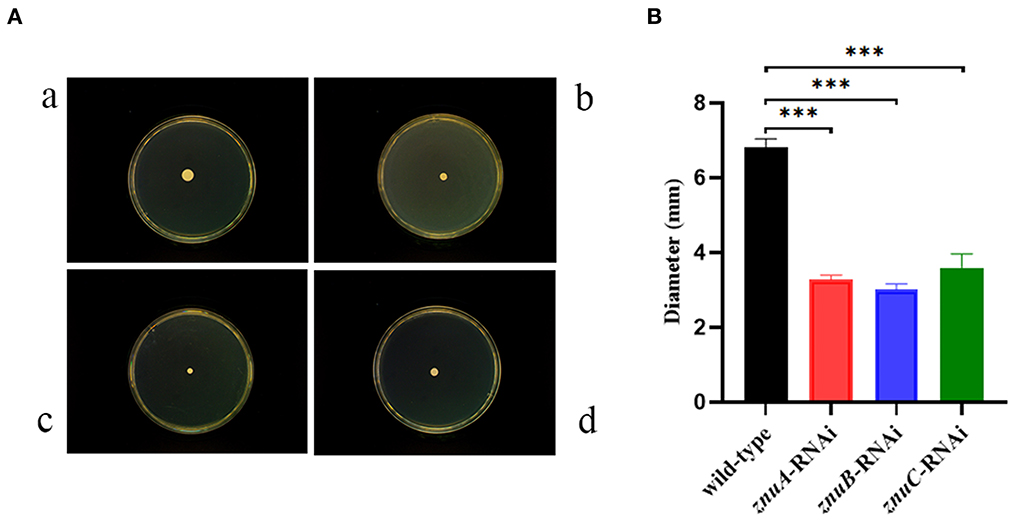
Figure 5. Single colony motility assay of wild-type and znuA-RNAi, znuB-RNAi, znuC-RNAi of A. salmonicida SRW-OG1. (A) was the typical observation of plate movement of the wild-type (a), znuA-RNAi (b), znuB-RNAi (c) and znuC-RNAi (d) strains; (B) was the colony diameter histogram. Data were presented as mean ± standard deviation (SD) (n = 3). *** P < 0.001.
3.5. The effect of znuA, znuB, and znuC on hemolytic ability of A. salmonicida SRW-OG1
According to our recently published study, the A. salmonicida SRW-OG1 contains the aerolysin and hemolysin (5). In the present study, the hemolysis activity of A. salmonicida SRW-OG1 wild-type, znuA-RNAi, znuB-RNAi and znuC-RNAi strains were compared. As compared with wild-type strain, znuA-RNAi, znuB-RNAi and znuC-RNAi strains have no obvious hemolysis circle, which indicated that the hemolytic capacity of the znuA-RNAi, znuB-RNAi and znuC-RNAi were significantly lower than that of the wild-type (Figure 6A).
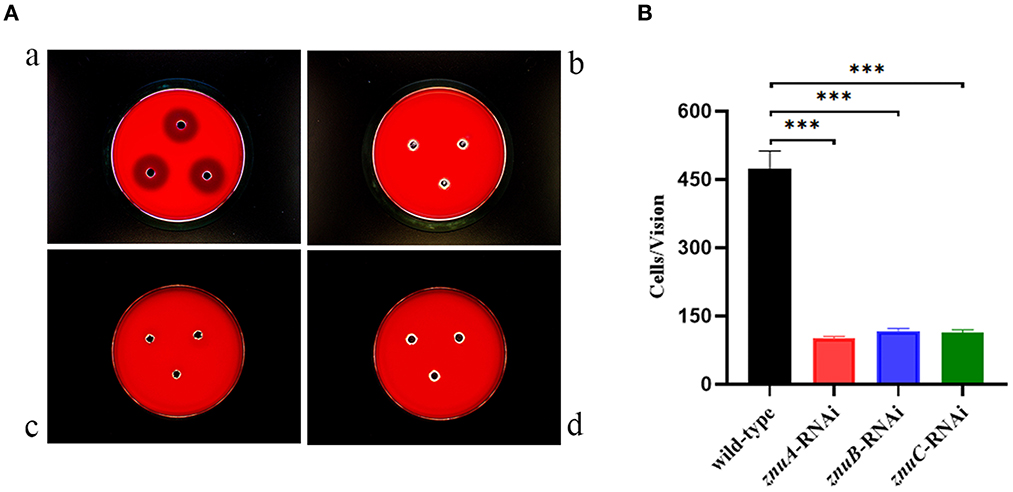
Figure 6. Determination of hemolytic ability and adhesion ability in vitro of wild-type, znuA-RNAi, znuB-RNAi and znuC-RNAi strains of A. salmonicida SRW-OG1. (A) was the typical observation of the hemolytic assay of wild-type (a), znuA-RNAi (b), znuB-RNAi (c) and znuC-RNAi (d) of A. salmonicida SRW-OG1 under the optical microscope; (B) is the histogram of the number of wild-type, znuA-RNAi, znuB-RNAi and znuC-RNAi strains of A. salmonicida SRW-OG1 in the field of optical microscope during in vitro adhesion test. Data were presented as mean ± standard deviation (SD) (n = 3). *** P < 0.001.
3.6. The effect of znuA, znuB, and znuC on adhesion ability of A. salmonicida SRW-OG1
As compared with wild-type strain, the adhesion ability of the znuA-RNAi, znuB-RNAi and znuC-RNAi strains in vitro decreased by 4.65, 4.06, and 4.16 times, respectively (Figure 6B), which showed that the number of wild-type adhered to the mucus was significantly more than that of znuA-RNAi, znuB-RNAi and znuC-RNAi, and indicated that the adhesion ability in vitro of znuA-RNAi, znuB-RNAi and znuC-RNAi were significantly lower than that of wild-type.
3.7. The expression levels of znuA, znuB, and znuC at different growth stages of A. salmonicida SRW-OG1
We determined the expression level of znuA, znuB and znuC in different growth periods (Figure 7). In the logarithmic phase (14 h), compared with the growth adaptation phase (3 h), the expression levels of znuA, znuB and znuC were increased significantly by 5.73, 5.39, and 3.08 times, respectively. At the growth stability stage (26 h), the expression of znuB was significantly increased by 2.62 times, while the differences between znuA and znuC were not statistically significant. During the growth recession (33 h), the expression levels of znuA, znuB, and znuC were significantly increased by 7.17, 5.93, and 11.92 times, respectively.
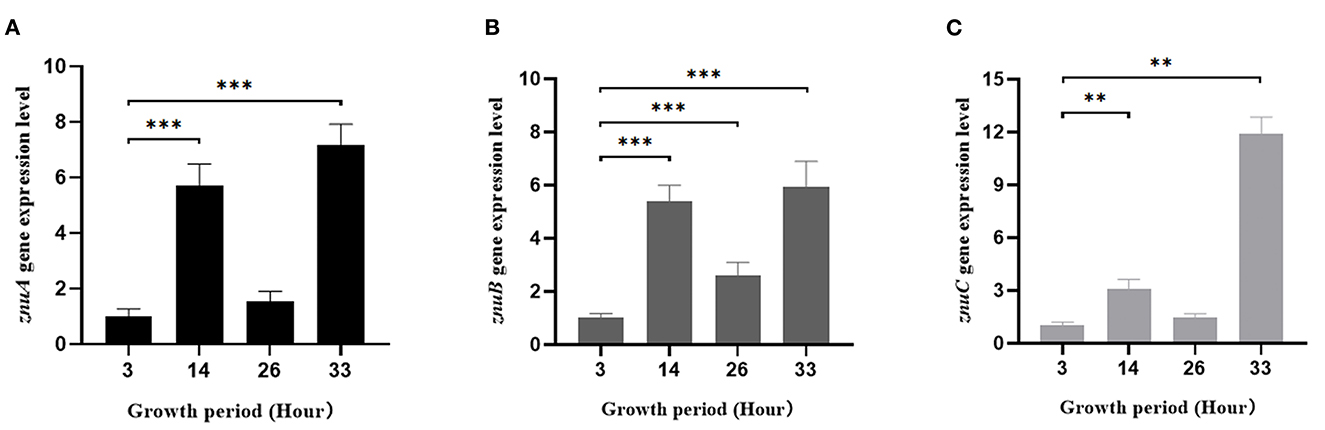
Figure 7. Determination of the expression levels of znuA, znuB, and znuC in different growth stages. The expression levels of znuA (A), znuB (B) and znuC (C) in different growth period. Data were presented as mean ± standard deviation (SD) (n = 3). **P < 0.01, ***P < 0.001.
3.8. Determination of znuA, znuB, and znuC expression levels under different stress environments
According to our previous RNA-seq analysis, compared with the expression levels of znuA, znuB, and znuC at 18°C, the expression levels at 28°C were down-regulated by 3.85, 3.13 and 2.08 times but there was no significant difference at 37°C. Verified by qRT-PCR, the znuA, znuB and znuC gene expression levels were consistent with the RNA-seq (Figure 8): compared with the expression levels of znuA, znuB, and znuC genes at 18°C, the expression level at 28°C was down-regulated by 6.76, 2.81, and 2.34 times but there was no significant difference at 37°C.
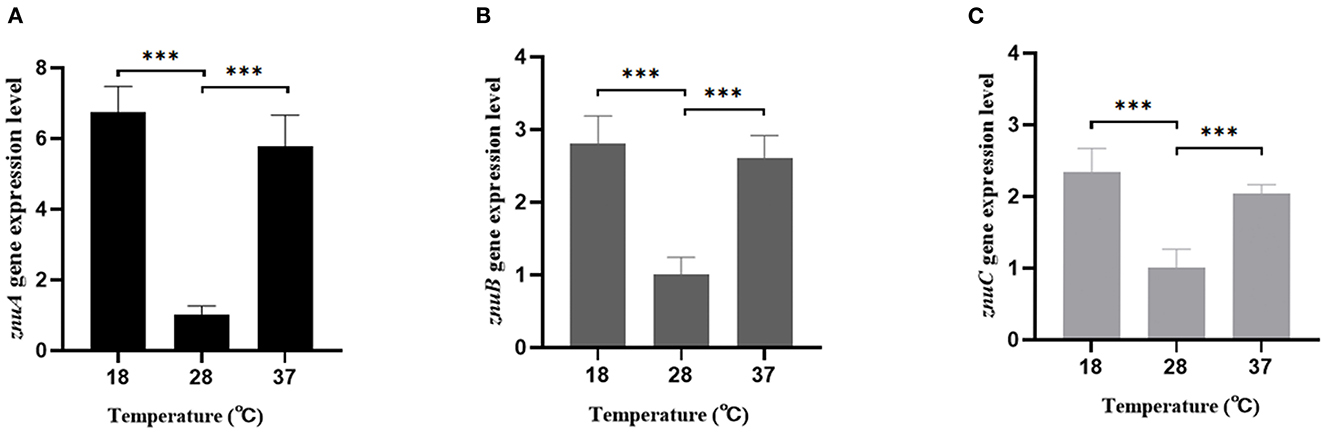
Figure 8. The expression levels of znuA, znuB, and znuC under different temperature. (A) was the expression levels of znuA at 18, 28, and 37°C; (B) was the expression levels of znuB at 18, 28, and 37°C; (C) was the expression levels of znuC at 18, 28, and 37°C. Data were presented as mean ± standard deviation (SD) (n = 3). ***P < 0.001.
In comparison with the pH = 7 group, the expression level of znuA was significantly up-regulated by 2.05 and 3.92 times at pH = 4 and 8, respectively, and it was significantly down-regulated by 3.23 times at pH = 5, although there was a slight increase of znuA expression at pH = 6 (Figure 9A). The expression level of znuB was significantly down-regulated by 13.07, 5.06, 11.17, and 6.89 times at pH = 4, 5, 6, and 8, respectively (Figure 9B). The expression level of znuC was significantly down-regulated by 1.57, 4.16, 1.89, and 2.97 times at pH = 4, 5, 6, and 8, respectively (Figure 9C). The expression of znuA decreased by 2.52 times under Cu2+ stress, while under Pb2+ stress, there was no significant difference (Figure 10A). The expression of znuB decreased by 1.87 and 1.77 times under Cu2+ and Pb2+ stress, respectively (Figure 10B). The expression of znuC decreased by 3.53 and 2.15 times under Cu2+ and Pb2+ stress, respectively (Figure 10C).
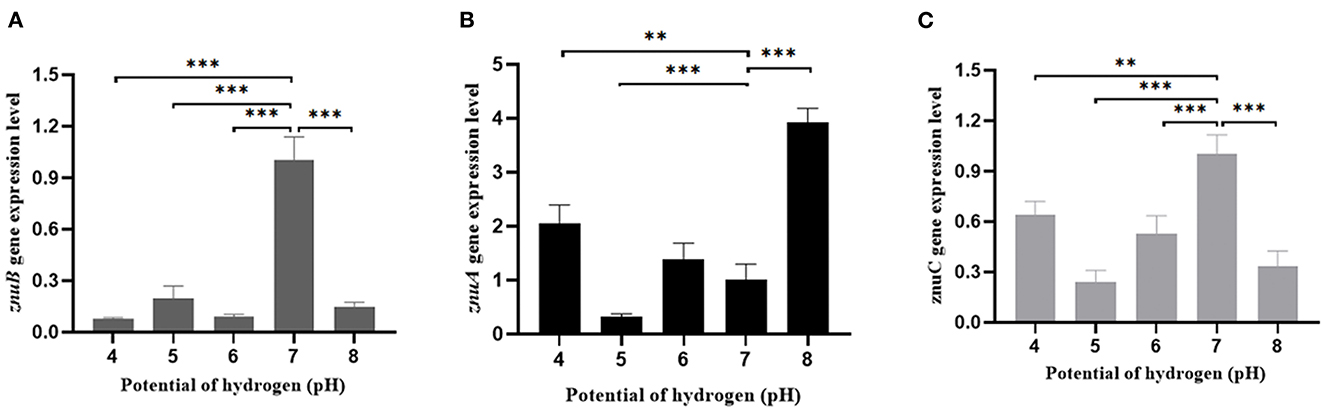
Figure 9. The expression levels of znuA, znuB and znuC gene under different potential of hydrogen. (A) was the expression levels of znuA gene at pH = 4, 5, 6, 7 and 8; (B) was the expression levels of znuB gene at pH = 4, 5, 6, 7 and 8; (C) was the expression levels of znuC gene at pH = 4, 5, 6, 7, and 8. Data were presented as mean ± standard deviation (SD) (n = 3). **P < 0.01, ***P < 0.001.
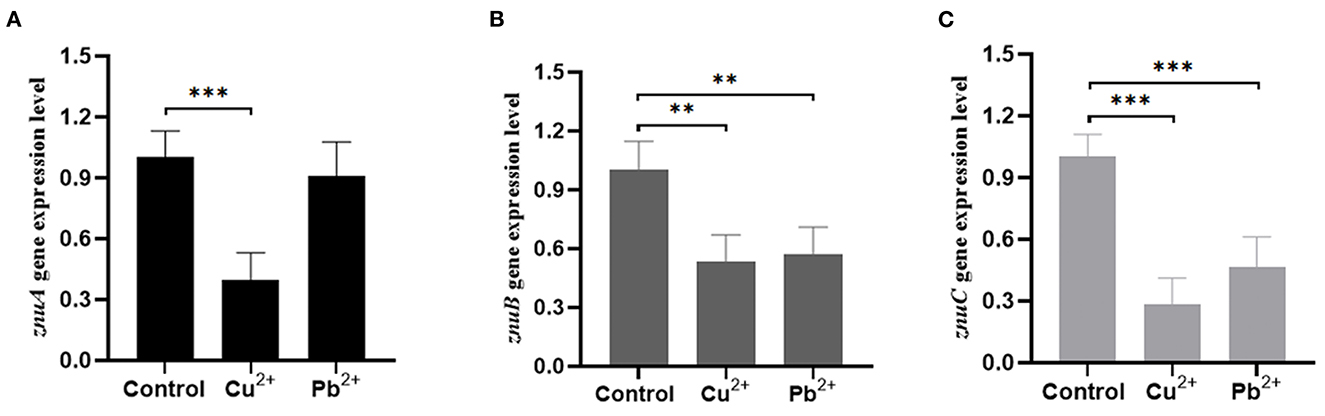
Figure 10. The expression levels of znuA, znuB and znuC gene under different heavy metal ion stresses. (A) was the expression levels of znuA under Cu2+ and Pb2+ stresses; (B) was the expression levels of znuB under Cu2+ and Pb2+ stresses; (C) was the expression levels of znuC under Cu2+ and Pb2+ stresses. Data were presented as mean ± standard deviation (SD) (n = 3). **P < 0.01, ***P < 0.001.
4. Discussion
Divalent transition metal ions such as Fe, Mn, Zn, Cd, etc., are necessary for all living organisms. During the infection, pathogens must compete with the host for micronutrients in order to survive and proliferate. However, the host utilizes the micronutrient requirement of pathogen to evolve a complex mechanism to battle with them, which known as nutritional immunity. At present, most of the research on nutritional immunity focuses on mammals, but there is little research on nutritional immunity in fish, and research on zinc nutritional immunity in fish is almost non-existent. Although zinc is essential for organisms, excessive amounts of zinc can be harmful to cells (54). At physiological concentrations, zinc may have a protective effect against free radical formation (55, 56), however, excess zinc has been shown to induce cytotoxicity by competing for protein binding with other metals (57), and exposure to high concentrations of zinc has been hypothesized to cause protein degeneration and dysfunction (58). There are mainly two Zinc transporters for zinc-uptake in most gram-negative bacteria, one is the znuABC, a high-affinity Zinc transport system, and the other is the low-affinity ZIP family Zn transporter ZupT (59). znuABC is composed of soluble periplasmic binding protein ZnuA, high-permeability transmembrane protein ZnuB and ATP enzyme ZnuC, and is regulated by Zur (zinc uptake system) (23). Zur is a member of the Fur-like family of transcriptional regulators with the ability to sense and respond to minute changes in intracellular zinc concentration (60). Zur has a structurally stable zinc binding site (C site) and 1 or 2 binding sites (M and D sites) that regulate DNA binding (61–63). Therefore, there are two situations: under the condition of sufficient zinc, excessive Zn2+ interact with M and / or D sites, causing conformational changes, and binding regulatory factors to znuA promoter, thus inhibiting its transcription, and in the case of Zn2+ deficiency, the M and / or D sites are not occupied, which leads to the instability of Zur, and the regulator cannot inhibit the transcription of znuA, thus allowing Zn2+ to be absorbed (64, 65).
Studies have shown that the growth of Salmonella enterica serovar Typhimurium without zupT is not restricted under normal conditions (66), but there is a growth defect under zinc restriction, and that S. Typhimurium without znuABC is not restricted under zinc restriction (67), which is approximately the same as our results, and that the growth of A. salmonicida SRW-OG1 znuA-RNAi, znuB-RNAi and znuC-RNAi silent strains in Zn2+ chelating medium is not restricted. However, the growth of A. salmonicida SRW-OG1 was significantly inhibited in the Fe2+ chelating medium. Therefore, we speculated that the uptake of zinc by the znuABC system was limited under the Zn2+ limitation condition, but another Zn transport system ZupT played a role to compensate for the znuABC. Studies have shown that excessive Zn2+ interferes with the steady state of Fe2+ in E. coli, and excessive Zn2+ instantaneously up-regulates the Fe2+ absorption genes and down-regulates the Fe2+ storage genes, thereby increasing the cellular iron quota (68). According to Huang et al., the znuA, znuB and znuC of Pseudomonas plecoglossicida were transcribed into an mRNA molecule and negatively regulated by the Fur binding box located upstream of znuC, and the regulation of Fur on ZnuCBA indicates that there is cross-talk between the acquisition of Zn2+ and Fe2+ and the nutritional immunity of the host, and it also proves that the separation of iron in the host may occur before the separation of zinc (41). Under the condition of Zn2+ and Fe2+ restriction, the znuA, znuB and znuC of A. salmonicida SRW-OG1 were significantly up-regulated, which indicated an interaction between Fur and znuABC during uptake of zinc and iron by A. salmonicida, but whether the transporter znuABC of A. salmonicida SRW-OG1 was regulated by Fur needed further research and verification.
Bacteria had specific resistance genes against heavy metal ions such as Ag+, AsO3−, Cd2+, Co2+, Cu2+, Hg2+, Ni2+, and Pb2+, common membrane cation pumps were Cd2+-resistance ATPase of Gram-positive bacteria (CadA) and other bacteria, animal and plant P-type ATPases (54). Cd2+, Zn2+, Co2+ and Ni2+ are pumped from Gram-bacteria by three polypeptide membrane complex that is NOT an ATPase but functions as a divalent cation/2H+ antiporter. The complex consists of an inner membrane protein (CzcA), an outer membrane protein (CzcC) and a protein associated with both membranes (CzcB) (69). The Zur regulon of E. coli contains four operons: znuABC, zinT, ykgMO and pliG (70, 71). Many bacteria such as Paracoccus denitrificans (72), Mycobacterium tuberculosis (73) and Bacillus subtilis (74) contain znuABC or Znu-like members of the ABC superfamily. The metal-resistant betaproteobacterium Cupriavidus metallidurans had multiple metal resistance systems, including the CzcCBA transmembrane efflux system, which although it does not contain znuABC has the ZupT of the ZIP family that maintains zinc homeostasis (75). According to our results, znuA, znuB, and znuC were all significantly up-regulated to varying degrees under heavy metal ion stresses, which suggested that znuA, znuB, and znuC might be potential toxic ion resistance genes.
A. salmonicida SRW-OG1, isolated from warm water marine fish E. coioides, could normally grow and infect at high temperatures (28–37°C). A. salmonicida was first isolated from human blood in 2014, before it was determined to infect a variety of freshwater fish but was not pathogenic to mammals and humans because it could not grow at 37°C (76). Cases of human infection by A. salmonicida have been reported, such as the first case of endocarditis caused by A. salmonicida infection (77). The isolation of C947 strains from patients with acute gastroenteritis and the isolation of AJ83 strains from patients with post-traumatic cellulitis of the foot (12). Based on pathogen-host interaction factor analysis by Qi et al. (13) against A. salmonicida SRW-OG1, C947, and AJ83 strains and transcriptome data analysis by Chen et al. (5) on A. salmonicida SRW-OG1 at different temperatures, A. salmonicida SRW-OG1 has a great potential to infect mammals and humans. Notably, the remarkably high expression of znuA, znuB, and znuC genes at 18 and 37°C also indicated that they might be related to the pathogenesis under different temperatures. Considering that SRW-OG1 had more than one zinc uptake and transport system, we speculated that the emergence of this U-type expression trend might be an adaptive mechanism for different hosts/environments formed by A. salmonicida in the long-term evolution process. At the same time, it also indicates that A. salmonicida SRW-OG1 can infect marine and freshwater fish at low temperature (18 and 28°C) and mammals or human at high temperature (37°C).
The biofilm formation ability, adhesion ability, hemolytic activity and motility of A. salmonicida SRW-OG1 are the basis for judging the virulence. According to our results, the biofilm formation ability, motility, adhesion and hemolysis ability of znuA-RNAi, znuB-RNAi and znuC-RNAi strains were weakened to different degrees, and these abilities were the basic conditions for bacteria to infect the host or cells. These indicated that znuA, znuB, and znuC genes were involved in the pathogenic process of A. salmonicida SRW-OG1 and played a key role in its survival in adversity.
5. Conclusion
Our results showed that znuA, znuB, and znuC were closely related to the biofilm formation, motility, adhesion and hemolytic activity of A. salmonicida SRW-OG1. Meanwhile, the expression of znuA, znuB, and znuC were significantly affected by the environmental factors such as temperature, pH and heavy metals. Under the Zn2+ and Fe2+ limitation, the uptake of Zn2+ by znuABC was limited, but ZupT could compensate for it, and the high expression of znuA, znuB, and znuC might indicate a cross-talk of Fur and znuABC. Therefore, znuA, znuB, and znuC appears to be vital for the general fitness of A. salmonicida SRW-OG1, so as to indirectly affect its efficiency of infecting the host.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Ethics statement
All animal experiments were carried out strictly under the recommendations in the Guide for the Care and Use of Laboratory Animals set by the National Institutes of Health. The animal protocols were approved by the Animal Ethics Committee of Jimei University (Acceptance NO JMULAC201159).
Author contributions
YQ and YZ conceived the experiments. JW, LX, and YQ conducted the experiments. JW and YZ wrote the manuscript. All authors assisted in the collection and interpretation of data.
Funding
This research was funded by the Project of Subsidy Funds for Marine Economic Development in Fujian Province under grant FJHJF-L-2022-20, the major program of Science and Technology Planning of Xiamen under contract No. 3502Z20211004, and the Natural Science Foundation of Fujian Province (Nos. 2019J01695 and 2021J01823).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2023.1172123/full#supplementary-material
References
1. Jin S, Fu S, Li R, Dang H, Gao D, Ye S, et al. Identification and histopathological and pathogenicity analysis of Aeromonas salmonicida salmonicida from goldfish (Carassius auratus) in North China. Aquacult Fisheries. (2019) 5:36–41. doi: 10.1016/j.aaf.2019.04.004
2. Connors E, Soto-Dávila M, Hossain A, Vasquez I, Gnanagobal H, Santander J. Identification and validation of reliable Aeromonas salmonicida subspecies salmonicida reference genes for differential gene expression analyses. Infect Genet Evol. (2019) 73:314–21. doi: 10.1016/j.meegid.2019.05.011
3. Coscelli GA, Bermúdez R, Losada AP, Santos Y. Quiroga MI. Vaccination against Aeromonas salmonicida in turbot (Scophthalmus maximus L): Study of the efficacy, morphological changes and antigen distribution. Aquaculture. (2015) 445:22–32. doi: 10.1016/j.aquaculture.2015.04.011
4. Nakayama K, Yamashita R, Kitamura S I. Use of common carp (Cyprinus carpio) and Aeromonas salmonicida for detection of immunomodulatory effects of chemicals on fish. Mar Pollut Bull. (2017) 124:710–3. doi: 10.1016/j.marpolbul.2016.12.060
5. Chen Y, Wang J, Cai H, Lin M, Zhang Y, Huang L. Enhanced hemolytic activity of mesophilic Aeromonas salmonicida SRW-OG1 Is brought about by elevated temperatures. Microorganisms. (2022) 10:2033. doi: 10.3390/microorganisms10102033
6. Austin B, Austin DA. Aeromonadaceae Representative (Aeromonas salmonicida). Berlin: Springer International Publishing. (2016), p. 215–321. doi: 10.1007/978-3-319-32674-0_5
7. Vincent AT, Bernatchez A, Frey J, Charette SJ, A. Mesophilic Aeromonas salmonicida strain isolated from an unsuspected host, the migratory bird pied avocet. Microorganisms. (2019) 7:592. doi: 10.3390/microorganisms7120592
8. Mendoza-Barberá E, Merino S. Tomás J. Surface Glucan Structures in Aeromonas spp. Marine Drugs. (2021) 19:649. doi: 10.3390/md19110649
9. Vincent A T, Hosseini N, Charette S J. The Aeromonas salmonicida plasmidome: a model of modular evolution and genetic diversity. Ann N Y Acad Sci. (2021) 1488:16–32. doi: 10.1111/nyas.14503
10. Jun JW, Kang JW, Giri SS, Kim SW, Kim SG, Kwon J, et al. Preventive effect of starch hydrogel-based oral vaccine against Aeromonas salmonicida infection in rainbow trout (Oncorhynchus mykiss). Aquaculture. (2022) 555:738202. doi: 10.1016/j.aquaculture.2022.738202
11. Huang L, Qi W, Zuo Y, Alias SA, Xu W. The immune response of a warm water fish orange-spotted grouper (Epinephelus coioides) infected with a typical cold water bacterial pathogen Aeromonas salmonicida is AhR dependent. Develop Comparat Immunol. (2020) 113:103779. doi: 10.1016/j.dci.2020.103779
12. Vincent AT, Fernández-Bravo A, Sanchis M, Mayayo E, Figueras MJ, Charette SJ. Investigation of the virulence and genomics of Aeromonas salmonicida strains isolated from human patients - ScienceDirect. Infect Genet Evolut. (2019) 68:1–9. doi: 10.1016/j.meegid.2018.11.019
13. Zhong Y, Qi W, Xu W, Zhao L, Xiao B, Yan Q, et al. Insights into mesophilic virulence, antibiotic resistant and human pathogenicity: a genomics study on the Aeromonas salmonicida SRW-OG1 newly isolated from the Asian fish Epinephelus coioides. Aquaculture. (2021) 539:736630. doi: 10.1016/j.aquaculture.2021.736630
14. Murdoch CC, Skaar EP. Nutritional immunity: the battle for nutrient metals at the host–pathogen interface. Nat Rev Microbiol. (2022) 20:1–14. doi: 10.1038/s41579-022-00745-6
15. Wang C, Zhang R, Wei X, Lv M, Jiang Z. Metalloimmunology: the metal ion-controlled immunity. Adv Immunol. (2020) 145:187–241. doi: 10.1016/bs.ai.2019.11.007
16. Paterson JR, Beecroft MS, Mulla RS, Osman D, Reeder NL, Caserta JA, et al. Insights into the antibacterial mechanism of action of chelating agents by selective deprivation of iron, manganese, and zinc. Appl Environ Microbiol. (2022) 88:e1621–41. doi: 10.1128/AEM.01641-21
17. Herzog S, Brinkmann H, Vences M, Fleißner A. Evidence of repeated horizontal transfer of sterol C-5 desaturase encoding genes among dikarya fungi. Mol Phylogenet Evol. (2020) 150:106850. doi: 10.1016/j.ympev.2020.106850
18. Ueta M, Wada C, Wada A. YkgM and YkgO maintain translation by replacing their paralogs, zinc-binding ribosomal proteins L31 and L36, with identical activities. Genes to Cells. (2020) 25:562–81. doi: 10.1111/gtc.12796
19. Zhang Y, Zheng J. Bioinformatics of metalloproteins and metalloproteomes. Molecules. (2020) 25:3366. doi: 10.3390/molecules25153366
20. Raza SH, Khan R, Cheng G, Long F, Bing S, Easa AA, et al. RNA-Seq reveals the potential molecular mechanisms of bovine KLF6 gene in the regulation of adipogenesis. Int J Biol Macromol. (2022) 195:198–206. doi: 10.1016/j.ijbiomac.2021.11.202
21. Hantke K. Bacterial zinc uptake and regulators. Curr Opin Microbiol. (2005) 8:196–202. doi: 10.1016/j.mib.2005.02.001
22. Nuonming P, Khemthong S, Sukchawalit R, Mongkolsuk S. Identification of Zur boxes and determination of their roles in the differential regulation of the Zur regulon in Agrobacterium tumefaciens C58. Appl Microbiol Biotechnol. (2020) 104:2109–23. doi: 10.1007/s00253-020-10346-z
23. Lilay GH, Persson DP, Castro PH, Liao F, Alexander RD, Aarts MGM, et al. Arabidopsis bZIP19 and bZIP23 act as zinc sensors to control plant zinc status. Nature Plants. (2021) 7:137–43. doi: 10.1038/s41477-021-00856-7
24. Younus I, Kochkina S, Choi CC, Sun W, Ford RC. ATP-Binding Cassette Transporters: Snap-on Complexes?[M]//Macromolecular Protein Complexes IV. Berlin: Springer (2022), p. 35–82. doi: 10.1007/978-3-031-00793-4_2
25. Chaptal V, Zampieri V, Wiseman B, Orelle C, Martin J, Nguyen KA, et al. Substrate-bound and substrate-free outward-facing structures of a multidrug ABC exporter. Sci Adv. (2022) 8:g9215. doi: 10.1126/sciadv.abg9215
26. Furuta T. Structural dynamics of ABC transporters: molecular simulation studies. Biochem Soc Trans. (2021) 49:405–14. doi: 10.1042/BST20200710
27. Serrano FA, Yukl ET. Contributions of conformational flexibility to high-affinity zinc binding in the solute binding protein AztC. ACS omega. (2022) 7:3768–74. doi: 10.1021/acsomega.1c06639
28. Behnsen J, Zhi H, Aron AT, Subramanian V, Santus W, Lee MH, et al. Siderophore-mediated zinc acquisition enhances enterobacterial colonization of the inflamed gut. Nat Commun. (2021) 12:1–15. doi: 10.1038/s41467-021-27297-2
29. D'Orazio M, Mastropasqua MC, Cerasi M, Pacello F, Consalvo A, Chirullo B, et al. The capability of Pseudomonas aeruginosa to recruit zinc under conditions of limited metal availability is affected by inactivation of the znuABC transporter. Metallomics. (2015) 7:1023–35. doi: 10.1039/C5MT00017C
30. Thompson MW. Regulation of zinc-dependent enzymes by metal carrier proteins. Biometals. (2022) 35:1–27. doi: 10.1007/s10534-022-00373-w
31. Santos RERS. Júnior WPd, Harrison S, Skaar EP, Chazin WJ, Neto JFd. The zinc transporter znuABC is critical for he virulence of Chromobacterium violaceum and contributes to diverse zinc-dependent physiological processes. Infect Immun. (2021) 89:e311–21. doi: 10.1128/IAI.00311-21
32. Dahiya I, Stevenson R. The znuABC operon is important for Yersinia ruckeri infections of rainbow trout, Oncorhynchus mykiss (Walbaum). J Fish Dis. (2010) 33:331–40. doi: 10.1111/j.1365-2761.2009.01125.x
33. Huang L, Zhao L, Liu W, Xu X, Su Y, Qin Y, et al. Dual RNA-Seq Unveils Pseudomonas plecoglossicida htpG gene functions during host-pathogen interactions with Epinephelus coioides. Front Immunol. (2019) 10:984. doi: 10.3389/fimmu.2019.00984
34. Bittel M, Reichert P, Sarfati I, Dressel A, Leikam S, Uderhardt S, et al. Visualizing transfer of microbial biomolecules by outer membrane vesicles in microbe-host-communication in vivo. J Extracellular Vesicles. (2021) 10:e12159. doi: 10.1002/jev2.12159
35. Girard R, Tremblay S, Noll C, St-Jean S, Jones C, Gélinas Y, et al. The transcription factor hepatocyte nuclear factor 4A acts in the intestine to promote white adipose tissue energy storage. Nat Commun. (2022) 13:1–14. doi: 10.1038/s41467-021-27934-w
36. He L, Wang L, Zhao L, Zhuang Z, Wang X, Huang H, et al. Integration of RNA-seq and RNAi reveals the contribution of znuA gene to the pathogenicity of Pseudomonas plecoglossicida and to the immune response of Epinephelus coioides. J Fish Dis. (2021) 44:1831–41. doi: 10.1111/jfd.13502
37. Huang L, Hu J, Su Y, Qin Y, Kong W, Ma Y, et al. Identification and characterization of three Vibrio alginolyticus non-coding RNAs involved in adhesion, chemotaxis, and motility processes. Front Cell Infect Microbiol. (2015) 5:56. doi: 10.3389/fcimb.2015.00056
38. Qi W, Gao Q, Tian J, Wu B, Lin M, Qi S, et al. Immune responses and inorganic ion transport regulations of Epinephelus coioides in response to L321_RS13075 gene of Pseudomonas plecoglossicida. Fish Shellfish Immunol. (2022) 120:599–609. doi: 10.1016/j.fsi.2021.12.036
39. Zhang M, Yan Q, Mao L, Wang S, Huang L, Xu X, et al. KatG plays an important role in Aeromonas hydrophila survival in fish macrophages and escape for further infection. Gene. (2018) 672:156–64. doi: 10.1016/j.gene.2018.06.029
40. Huang L, Huang L, Yan Q, Qin Y, Ma Y, Lin M, et al. The TCA pathway is an important player in the regulatory network governing vibrio alginolyticus adhesion under adversity. Front Microbiol. (2016) 7:40. doi: 10.3389/fmicb.2016.00040
41. Huang L, Zuo Y, Qin Y, Zhao L, Lin M, Yan Q. The zinc nutritional immunity of Epinephelus coioides contributes to the importance of znuC during Pseudomonas plecoglossicida infection. Front Immunol. (2021) 12:1575. doi: 10.3389/fimmu.2021.678699
42. Yang D, Zhao L, Li Q, Huang L, Qin Y, Wang P, et al. flgC gene is involved in the virulence regulation of Pseudomonas plecoglossicida and affects the immune response of Epinephelus coioides. Fish Shellfish Immunol. (2023) 132:108512. doi: 10.1016/j.fsi.2022.108512
43. Cai H, Ma Y, Qin Y, Zhao L, Yan Q, Huang L. Vvrr2: a new Vibrio ncRNA involved in dynamic synthesis of multiple biofilm matrix exopolusaccharides, biofilm structuring and virulence. Aquaculture. (2023) 563:738925. doi: 10.1016/j.aquaculture.2022.738925
44. Yuan B, Zhao L-M, Zhuang Z-X, Wang X-R, Fu Q, Huang H-B, et al. Transcriptomic and metabolomic insights into the role of the flgK gene in the pathogenicity of Pseudomonas plecoglossicida to orange-spotted grouper (Epinephelus coioides). Zoological Res. (2022) 43:952–65. doi: 10.24272/j.issn.2095-8137.2022.216
45. He R, Wang J, Lin M, Tian J, Wu B, Tan X, et al. Effect of ferredoxin receptor FusA on the virulence mechanism of Pseudomonas plecoglossicida. Front Cell Infect Microbiol. (2022) 12:225. doi: 10.3389/fcimb.2022.808800
46. Porto-Fett ACS, Espuña E, Shane LE, Shoyer BA, McGeary L, Vinyard BT, et al. Viability of Shiga toxin–producing Escherichia coli, Salmonella spp, and Listeria monocytogenes during preparation and storage of fuet, a traditional dry-cured Spanish pork sausage. J Food Protect. (2022) 85:879–89. doi: 10.4315/JFP-21-356
47. Cai H, Yu J, Qiao Y, Ma Y, Zheng J, Lin M, et al. Effect of the Type VI secretion system secreted protein hcp on the virulence of Aeromonas salmonicida. Microorganisms. (2022) 10:2307. doi: 10.3390/microorganisms10122307
48. Gao Q, Ma X, Wang Z, Chen H, Luo Y, Wu B, et al. Seasonal variation, virulence gene and antibiotic resistance of Vibrio in a semi-enclosed bay with mariculture (Dongshan Bay, Southern China). Mar Pollut Bull. (2022) 184:114112. doi: 10.1016/j.marpolbul.2022.114112
49. Afzaal M, Saeed F, Ateeq H, Shah YA, Hussain M, Javed A, et al. Effect of cellulose–chitosan hybrid-based encapsulation on the viability and stability of probiotics under simulated gastric transit and in kefir. Biomimetics. (2022) 7:109. doi: 10.3390/biomimetics7030109
50. Xu W, Lin W, Wang Z, Gao Y, Luo Y, Grossart H-P, et al. Disentangling the abundance and structure of Vibrio communities in a semi-enclosed Bay with mariculture (Dongshan Bay, Southern China). Comput Struct Biotechnol J. (2021) 19:4381–93. doi: 10.1016/j.csbj.2021.07.040
51. Xin G, Wang F, Zhao L, Qin Y, Huang L, Yan Q, et al. Integration of RNA-seq and RNAi provides a novel insight into the effect of pvdE gene to the pathogenic of Pseudomonas plecoglossicida and on the immune responses of orange-spotted grouper (Epinephelus coioides). Aquaculture. (2020) 529:735695. doi: 10.1016/j.aquaculture.2020.735695
52. Huang L, Wang L, Lin X, Su Y, Qin Y, Kong W, et al. mcp, aer, cheB, and cheV contribute to the regulation of Vibrio alginolyticus (ND-01) adhesion under gradients of environmental factors. Microbiologyopen. (2017) 6:e517. doi: 10.1002/mbo3.517
53. Feng Y, Jonker MJ, Moustakas I, Brul S, Kuile BHT. Dynamics of mutations during development of resistance by Pseudomonas aeruginosa against five antibiotics. Antimicrob Agents Chemother. (2016) 60:4229–36. doi: 10.1128/AAC.00434-16
54. Pal A, Bhattacharjee S, Saha J, Sarkar M, Mandal P. Bacterial survival strategies and responses under heavy metal stress: a comprehensive overview. Crit Rev Microbiol. (2022) 48:327–55. doi: 10.1080/1040841X.2021.1970512
55. Yang J, Wang T, Lin G, Li M, Zhang Y, Mai K. The assessment of dietary organic zinc on zinc homeostasis, antioxidant capacity, immune response, glycolysis and intestinal microbiota in white shrimp (Litopenaeus vannamei Boone 1931). Antioxidants. (2022) 11:1492. doi: 10.3390/antiox11081492
56. Troxell B, Hassan H M. Transcriptional regulation by Ferric Uptake Regulator (Fur) in pathogenic bacteria. Front Cell Infect Microbiol. (2013) 3:59. doi: 10.3389/fcimb.2013.00059
57. Islam T, Albracht-Schulte K, Ramalingam L, Schlabritz-Lutsevich N, Park OH, Zabet-Moghaddam M, et al. Anti-inflammatory mechanisms of polyphenols in adipose tissue inflammation: Role of gut microbiota, intestinal barrier integrity and zinc homeostasis. J Nutr Biochem. (2022) 115:109242. doi: 10.1016/j.jnutbio.2022.109242
58. Gao R, Brokaw SE Li Z, Helfant LJ, Wu T, Malik M, et al. Exploring the mono-/bistability range of positively autoregulated signaling systems in the presence of competing transcription factor binding sites. PLoS Comput Biol. (2022) 18:e1010738. doi: 10.1371/journal.pcbi.1010738
59. Huynh U, Zastrow ML. Metallobiology of Lactobacillaceae in the gut microbiome. J Inorgan Biochem. (2022) 238:112023. doi: 10.1016/j.jinorgbio.2022.112023
61. Yang X, Wang Y, Liu G, Deng Z, Lin S, Zheng J. Structural basis of Streptomyces transcription activation by zinc uptake regulator. Nucleic Acids Res. (2022) 50:8363–76. doi: 10.1093/nar/gkac627
62. Sue A. Regulation and Distribution of Zinc in the Caenorhabditis elegans Reproductive System. Evanston: Northwestern University (2022).
63. Shin J-H, Jung HJ, An YJ, Cho Y-B, Cha S-S, Roe J-H. Graded expression of zinc-responsive genes through two regulatory zinc-binding sites in Zur. Proc Nat Acad Sci. (2011) 108:5045–50. doi: 10.1073/pnas.1017744108
64. Smith KF, Bibb LA, Schmitt MP, Oram DM. Regulation and activity of a zinc uptake regulator, Zur, in Corynebacterium diphtheriae. J Bacteriol. (2009) 191:1595–603. doi: 10.1128/JB.01392-08
65. Ellison ML, Parrish W, Danell AS, Pesci EC. The transcriptional regulator Np20 is the zinc uptake regulator in Pseudomonas aeruginosa. PLoS ONE. (2013) 8:e75389. doi: 10.1371/journal.pone.0075389
66. Cerasi M, Liu JZ, Ammendola S, Poe AJ, Petrarca P, Pesciaroli M, et al. The ZupT transporter plays an important role in zinc homeostasis and contributes to Salmonella enterica virulence. Metallomics. (2014) 6:845–53. doi: 10.1039/C3MT00352C
67. Moreau G B, Qin A, Mann B J. Zinc acquisition mechanisms differ between environmental and virulent Francisella Species. J Bacteriol. (2018) 200:e517–87. doi: 10.1128/JB.00587-17
68. Xu Z, Wang P, Wang H, Yu ZH, Au-Yeung HY, Hirayama T, et al. Zinc excess increases cellular demand for iron and decreases tolerance to copper in Escherichia coli. J Biol Chem. (2019) 294:16978–91. doi: 10.1074/jbc.RA119.010023
69. Ganguly T, Peterson AM, Burkholder M, Kajfasz JK, Abranches J, Lemos JA. ZccE is a Novel P-type ATPase that protects Streptococcus mutans against zinc intoxication. PLoS Pathog. (2022) 18:e1010477. doi: 10.1371/journal.ppat.1010477
70. Gilston BA, Wang S, Marcus MD, Canalizo-Hernández MA, Swindell EP, Xue Y, et al. Structural and mechanistic basis of zinc regulation across the E. coli Zur regulon PLoS biology. (2014) 12:e1001987. doi: 10.1371/journal.pbio.1001987
71. Ducret V, Gonzalez D, Perron K. Zinc homeostasis in Pseudomonas. Biometals. (2022). doi: 10.1007/s10534-022-00475-5. [Epub ahead of print].
72. Neupane DP, Jacquez B, Sundararajan A, Ramaraj T, Schilkey FD, Yukl ET, et al. Zinc-dependent transcriptional regulation in Paracoccus denitrificans. Front Microbiol. (2017) 8:569. doi: 10.3389/fmicb.2017.00569
73. Maciag A, Dainese E, Rodriguez GM, Milano A, Provvedi R, Pasca MR, et al. Global analysis of the Mycobacterium tuberculosis Zur (FurB) regulon. J Bacteriol. (2007) 189:730–40. doi: 10.1128/JB.01190-06
74. Bhatt K, Maheshwari D K. Insights into zinc-sensing metalloregulator ‘Zur'deciphering mechanism of zinc transportation in Bacillus spp. by modeling, simulation and molecular docking. J Biomol Struct Dyn. (2022) 40:764–79. doi: 10.1080/07391102.2020.1818625
75. Kirsten A, Herzberg M, Voigt A, Seravalli J, Grass G, Scherer J, et al. Contributions of five secondary metal uptake systems to metal homeostasis of Cupriavidus metallidurans CH34. J Bacteriol. (2011) 193:4652–63. doi: 10.1128/JB.05293-11
76. Tewari R, Dudeja M, Nandy S, Das AK. Isolation of Aeromonas salmonicida from Human Blood Sample: A Case Report. J Clin Diagn Res. (2014) 8:139–40. doi: 10.7860/JCDR/2014/6883.4032
Keywords: Aeromonas salmonicida, Epinephelus coioides, znuABC, virulence regulation, zinc
Citation: Wang J, Xiu L, Qiao Y and Zhang Y (2023) Virulence regulation of Zn2+ uptake system znuABC on mesophilic Aeromonas salmonicida SRW-OG1. Front. Vet. Sci. 10:1172123. doi: 10.3389/fvets.2023.1172123
Received: 23 February 2023; Accepted: 14 March 2023;
Published: 29 March 2023.
Edited by:
Michael Kogut, Agricultural Research Service (USDA), United StatesReviewed by:
Jiali Cai, Xiamen University, ChinaMingliang Chen, State Oceanic Administration, China
Dongxu Gao, Dalian Ocean University, China
Copyright © 2023 Wang, Xiu, Qiao and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Youyu Zhang, emhhbmd5b3V5dUB4bXUuZWR1LmNu
 Jiajia Wang1
Jiajia Wang1 Ying Qiao
Ying Qiao Youyu Zhang
Youyu Zhang