
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci., 11 May 2023
Sec. Veterinary Infectious Diseases
Volume 10 - 2023 | https://doi.org/10.3389/fvets.2023.1169323
This article is part of the Research TopicPathogenic and Symbiotic Bacteria in Ruminants: Antimicrobial Resistance and Microbial HomeostasisView all 10 articles
Anaplasmosis is a severe tickborne disease of ruminants caused by Anaplasma marginale. A. marginale is distributed worldwide and attacks erythrocytes, resulting in an increased body temperature, anemia, jaundice, abortion, and, in some cases, death. Animals infected with this pathogen become lifelong carriers. In this study, we aimed to detect and characterize A. marginale isolated from cattle, buffalo, and camel populations using novel molecular techniques in southern Egypt. In total, 250 samples (from 100 cattle, 75 water buffaloes, and 75 camels) were analyzed by PCR for the presence of Anaplasmataceae, specifically A. marginale. The animals varied in breed, age, and gender, with most showing no signs of severe disease. By species, A. marginale was found in 61 out of 100 (61%) cattle, 9 out of 75 (12%) buffaloes, and only 5 out of 75 (6.66%) camels. All A. marginale-positive samples were examined for the heat-shock protein groEL gene and, additionally, for major surface protein 4 (msp4) and major surface protein 5 (msp5) genes to enhance specificity. Phylogenetic analysis of A. marginale targeted three genes (groEL, msp4, and msp5). This study provides the first report on using three genes for A. marginale detection in Camelus dromedarius in southern Egypt and generated new phylogenetic data for A. marginale infections in camels. A. marginale infection is endemic in different animal species in southern Egypt. Screening herds for A. marginale is recommended even when the signs of anaplasmosis are absent.
Tickborne diseases are a serious challenge to global health. In Egypt alone, they pose a significant threat to animal health, in particular to local exotic and crossbred cattle and buffalo, thus potentially undermining the livelihoods of their owners (1). Anaplasma species are the most common tickborne pathogens in cattle and are endemic across six continents with a high incidence in tropical and subtropical areas of the world (2). The disease they cause is termed anaplasmosis, which is particularly common in ruminants (3). Among Anaplasma species (Rickettsiales: Anaplasmataceae), Anaplasma marginale (A. marginale) may be the most dangerous.
Anaplasmosis causes progressive hemolytic anemia and significant economic losses in tropical and subtropical areas (4). Ticks are known carriers of A. marginale, and approximately 20 tick species have been implicated as vectors of anaplasmosis (5). Bovine anaplasmosis is an economically devastating disease that results in losses to the dairy and beef industries through reduced milk production, weight loss, abortion, jaundice, and sometimes death (6, 7). The disease is mainly spread to cattle by ixodid ticks, but other routes of infection include fly bites and blood-stained objects, such as needles, ear tags, and castration equipment. Placental transmission may feature in the disease's epidemiology in specific areas (8). Fever, anemia, weakness, enlarged lymph nodes, abortion, reduced milk production, and jaundice are signs of anaplasmosis in cattle, and the disease can be fatal in severe cases (9). Cattle recovering from acute infection remain carriers for the rest of their lives and can act as sources of infection for previously naïve livestock populations, triggering endemic infection or epizootics (10).
The camel is a multipurpose animal playing crucial roles in the transport and provision of milk and meat in arid and semi-arid regions of the world. Although camels are hardy animals and can withstand the harsh conditions of dry areas due to their unique, adaptive physiology, their health can be adversely affected by a range of specific diseases (11, 12), including those transmitted by bloodborne parasites. Such diseases can cause anemia, emaciation, and even death in severe cases when camels are infected (13). Camel anaplasmosis has been reported as a subclinical disease in dromedary camels of Tunisia, India, and Saudi Arabia (14).
Anaplasma species are longevous microorganisms, potentially surviving in hosts for months or years, and the consequences of this phenomenon include increased transmission and the occurrence of new anaplasmosis outbreaks (15). Control measures include frequent surveillance, prompt treatment, and eradication of arthropod vectors, and their feasibility depends on several variables, including geographic location, husbandry practices, and implementation costs (encompassing items such as the vaccine or antibiotic treatment programs) (16). Variability in vectoring capacity and limited understanding of the tick's immune response (particularly with regard to arthropod–microbe interactions for bacteria) have impeded control efforts (17). Although current knowledge is limited, vaccines against ticks are being developed (18). A. marginale infections in cattle and buffalo have previously been recorded in different parts of Egypt (19–23). In this study, we report on A. marginale in three governorates in the southern part of Egypt, which we targeted because of the lack of research on A. marginale and its host species in this part of the country. Specifically, we applied molecular techniques to detect and characterize A. marginale infecting cattle, buffaloes, and camels.
This study focused on anaplasmosis infection in local breeds of cattle, buffaloes, and camels of various ages (from 1 to 3 years) and both genders. It was conducted from April 2021 to January 2022, in three southern governorates in Egypt: Sohag, Qena, and Red Sea governates (Figure 1).
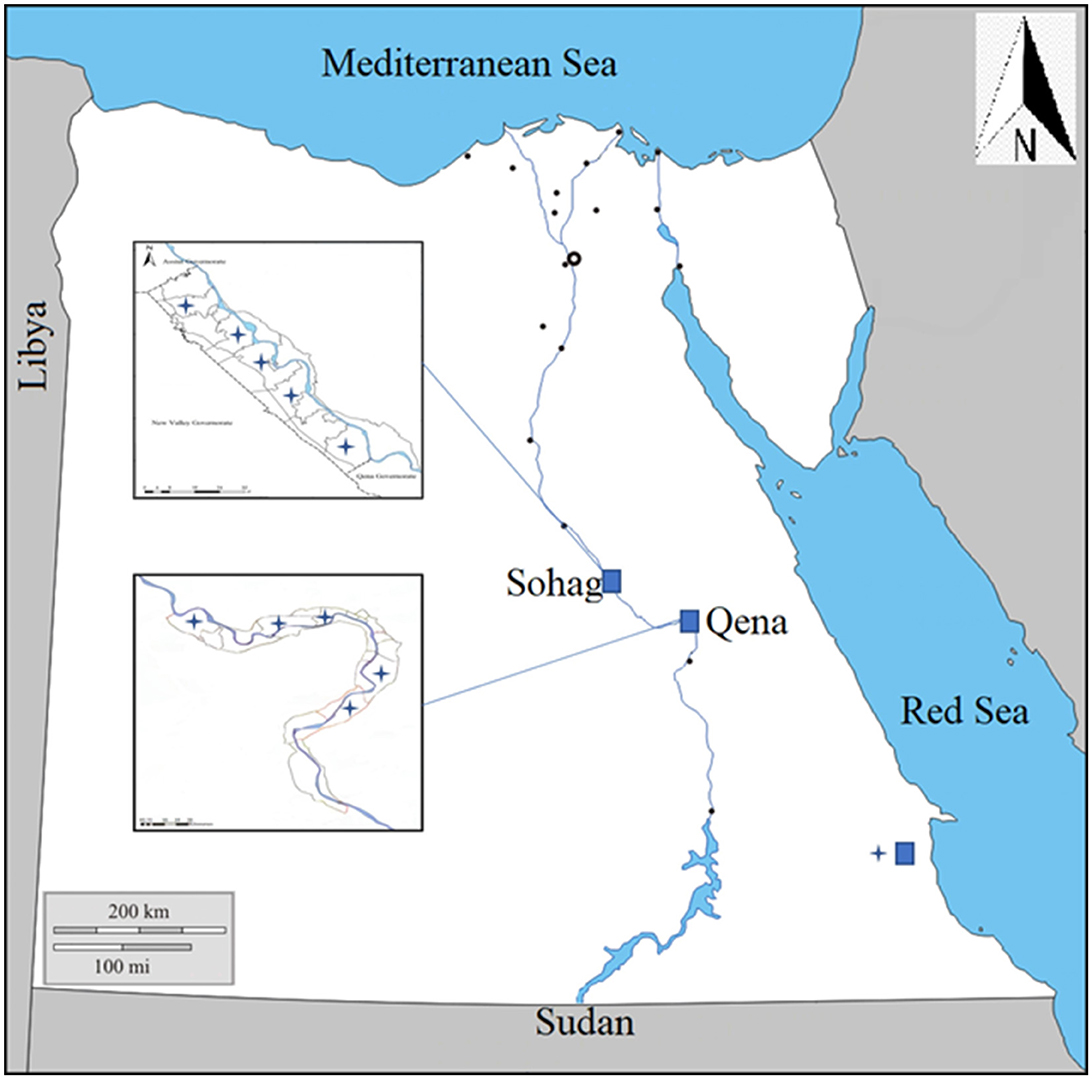
Figure 1. Map showing the southern part of Egypt where blood samples were collected from animals in three governorates, Sohag, Qena, and Red Sea.
Animals underwent clinical examinations before blood sample collection. The examination involved identifying age and gender and evaluating body mass index, body temperature, heart rate, respiratory rate, and visible mucous membranes. Some cattle were presented with pale mucous membranes, and respiratory disorders were noted in a small number of animals. All animals were infested with ticks, although buffaloes and camels showed no visible specifically associated clinical manifestations.
Samples were collected from animals selected at random. Small flock breeding mainly determines the species of animal raised by farmers in southern Egypt, which imposes some limitations on sample collection in this region. Accordingly, the number of samples in this study was set so as to provide a clear picture of the epidemiology of the relevant diseases in local animal populations. Whole blood samples were collected from the jugular vein of each animal with clean, sterile vacutainer tubes containing heparin for DNA extraction, as a target for PCR amplification. Samples were kept at −20°C until use.
All primers used in this study are listed in Table 1, and the PCR conditions are shown in Table 2. The amplification of bovine β-actin for DNA extract was confirmed by amplifying the bovine and camel β-actin–encoding genes (housekeeping genes) to ensure that the genomic DNA had been extracted from all samples (24, 25). Negative controls were samples containing nuclease-free water. Electrophoresis of PCR products was performed with 1.5% agarose gel in 1 × Tris–acetate–EDTA (TAE) buffer using a Mupid electrophoresis device (Mupid Co., Ltd., Tokyo, Japan), and bands were visualized through a gel documentation system UV device, WUV-M20 (ATTO Co., Ltd., Tokyo, Japan), after being stained with 5 g/ml ethidium bromide in 1 × TAE.
For this study, 250 samples (from 100 cattle, 75 water buffaloes, and 75 camels) were analyzed by PCR for the presence of Anaplasmataceae, specifically A. marginale. The animals varied in breed, age, and gender, with most showing no signs of severe disease when samples were collected using commercial extraction kits (Wizard® Genomic DNA Purification Kit, Promega, Madison, WI, USA). DNA was then extracted from whole blood samples. A. marginale was detected by screening using nested PCR amplification of the heat-shock protein groEL gene using the relevant primers (26). Selected A. marginale-positive samples were also subjected to conventional PCR targeting the msp4 and msp5 genes (26, 27). The PCR was performed with a total volume of 10 μl, using Tks Gflex DNA Polymerase (TaKaRa), 10 pmol each of the forward and reverse primers, and nuclease-free water. A template (1 μl DNA) was used. The PCR conditions are shown in Table 2. A negative control containing nuclease-free water was added to each PCR. The electrophoresis of the PCR products was performed using 1.5% gel and 1 × TAE buffer. The observation was made using a gel documentation system UV device, WUV-M20 (Atto Co., Ltd.), after the gel was stained with 5 μg/ml ethidium bromide in 1 × TAE.
The selected A. marginale groEl, msp4, and msp5 genes were subjected to PCR or 50 μl mixtures for sequence analysis. The amplicons were purified using a NucleoSpin Gel and PCR Clean-up kit (Macherey-Nagel, Leicestershire, Duren, Germany), following the manufacturer's protocol. Sequence readings were compared to sequences of reported isolates from a gene bank. A maximum-likelihood phylogenetic tree was constructed using MEGAX software (28), with bootstrap values estimated using 1,000 replicates based on Kimura's two-parameter substitution model (29).
A total of 250 blood samples from cattle, buffaloes, and camels were collected from three governorates in southern Egypt. All 250 samples (100%) were confirmed to contain DNA, as they exhibited bands at the expected 227 bp for cattle and buffalo. The expected 438 bp for camels with the β-actin gene demonstrated that DNA had been successfully extracted from all samples.
All samples were then subjected to nested PCR to detect the presence of the A. marginale groEL gene. The prevalence of A. marginale was 75 out of 250 (30%) samples. By species, 61 out of 100 (61%) cattle were A. marginale positive, while 9 out of 75 (12%) buffaloes and only 5 out of 75 (6.66%) camels were A. marginale positive (Table 3). All samples positive for the A. marginale groEL gene were further examined for two additional genes (msp4 and msp5) to provide an enhanced degree of specificity for the identification of A. marginale.
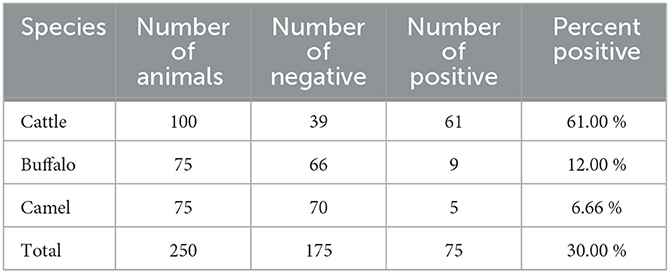
Table 3. Detection of Anaplasma marginale in cattle, buffaloes, and camels from southern Egypt based on PCR detection in blood samples.
Furthermore, a higher prevalence of A. marginale infection was found in Qena than in Sohag and Red Sea governates. We found no sex difference in any species in this study, based on the relative prevalence of A. marginale in males and females. Further investigations of risk factors should encompass univariate and multivariate analyses targeting animal and farm levels. Even so, we found a high prevalence (36% infection rate) in young animals (1 year old or less) relative to the adult animals. The breeding system also appears to be associated with the risk of A. marginale infection. Individually, bred animals had a lower infection rate than intensively bred animals (25 vs. 33.3%; Table 4).
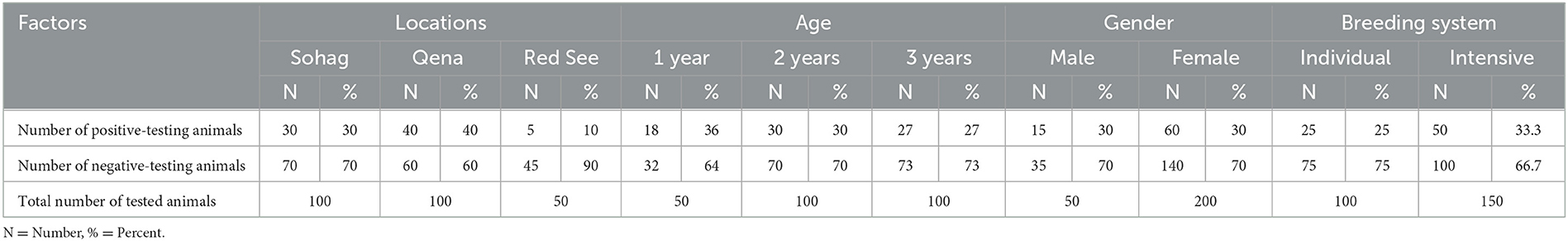
Table 4. Anaplasma marginale infection in cattle, buffaloes, and camels in three governorates in southern Egypt.
The A. marginale heat-shock protein groEL gene and major surface proteins msp4 and msp5 genes were sequenced for phylogenetic analysis and genotyping in cattle, buffaloes, and camels from three different governates in southern Egypt. All sequences were also submitted to a gene bank, and the following accession numbers can be used to access them: for the groEL gene (cattle: OP081155.1, OP081156.1, and OP081157.1; buffalo: OP081158.1 and OP081159.1; camel: OP081160.1 and OP081161.1); msp4 gene (cattle: OP142721.1 and OP142722.1; camel: OP142723.1 and OP142724.1; buffalo: OP142725.1 and OP142726.1); and msp5 gene (cattle: OP142716.1 and OP142717.1; buffalo: OP142718.1 and OP142719.1; camel: OP142720.1). Phylogenetic analysis established the relationships for A. marginale with the sequences identified for this study, and various isolates from other countries or other geographic locations in Egypt (Figures 2–4).

Figure 2. Phylogenetic relationships based on the sequences of the heat-shock protein (groEL) gene from A. marginale using the maximum-likelihood method and Kimura's two-parameter model, with branch lengths measured in the number of substitutions per site. The tree is depicted to scale. The percentage of trees in which the connected taxa clustered together is displayed next to the branches. Red, green, and blue circles represent A. marginale obtained in the present study.

Figure 3. Phylogenetic relations of A. marginale using the maximum-likelihood method and Kimura's two-parameter model based on major surface protein 4 (msp4) gene sequences. The percentage of trees in which the connected taxa clustered together is displayed next to the branches. Branch lengths are expressed in terms of the number of substitutions per site, and the tree is drawn to scale. Red, green, and blue circles represent A. marginale obtained in this study.
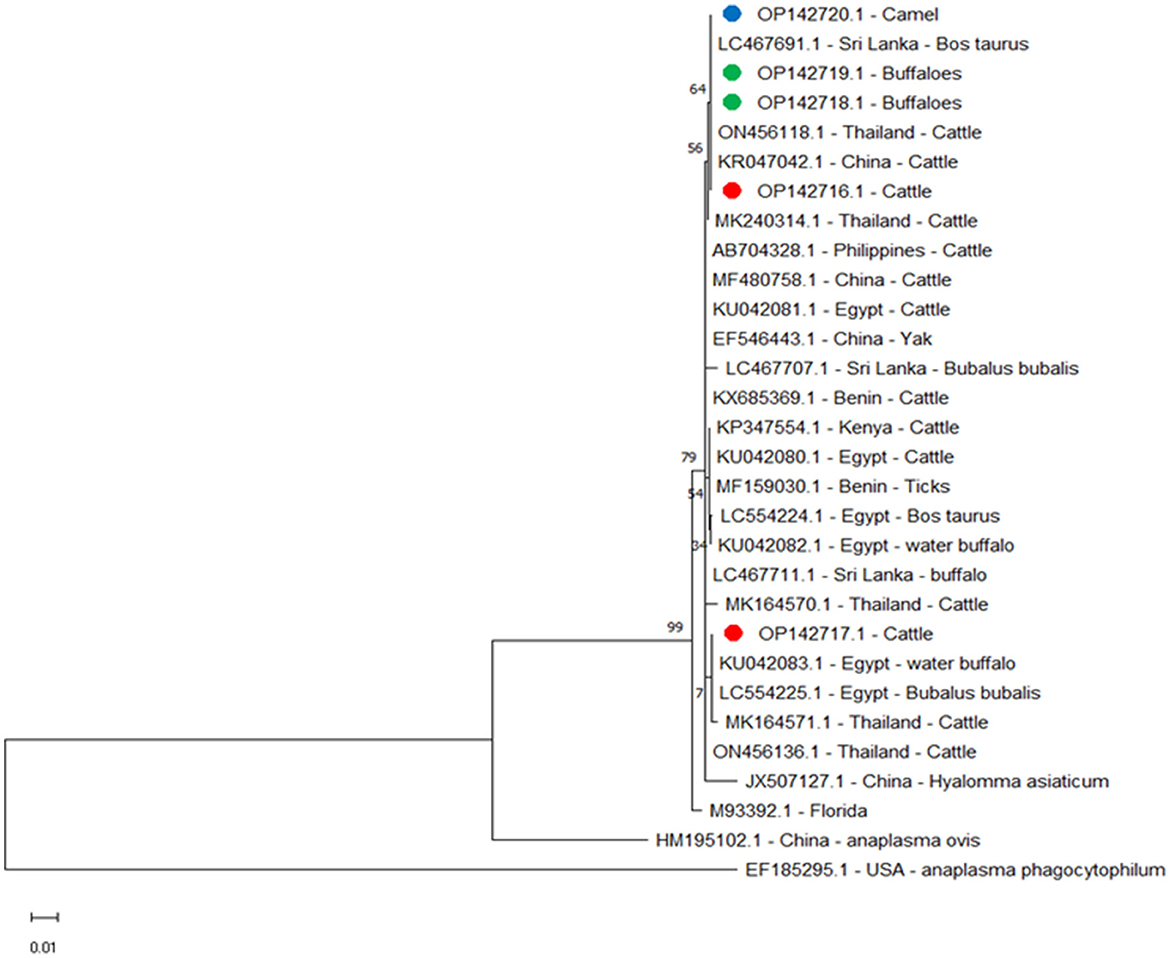
Figure 4. Phylogenetic relationships of A. marginale using the maximum-likelihood method and Kimura's two-parameter model based on major surface protein 5 (msp5) gene sequences. The percentage of trees in which the associated taxa clustered together is shown next to the branches. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. Red, green, and blue circles represent A. marginale obtained in this study.
A phylogenetic tree was constructed to compare the groEL gene for cattle, buffaloes, and camels with amplicons separated from other reported isolates. We found the following alignment identities: 100% to the Philippines, JQ839014.1 and LC461539.1; Malawi, LC664078.1; China, KX987398.1; Uganda, KY523021.1; 99.79% to Japan, FJ226455.1; and 98.07% to Mozambique, KR492655.1 (Figure 2).
For msp4 amplicons from this study for buffaloes, OP142725.1 and OP142726.1 were identified by the following alignment identities: 100% with amplicons isolated in Western Cuba MK809382.1 and Cuba MK809389.1; 99.88% with Western Cuba MK809387.1; 99.63% with Thailand MH939155.1; 99.38% with USA AY010253.1; 99.25% with Italy EU436159.1; 99.13% with USA AY127072.1; and 99% with Zimbabwe AY666010.1, India KX989521.1, and Hungary HM063432.1. However, for cattle and camel msp4 amplicons from this study, cattle OP142721.1 and OP142722.1 and camel OP142723.1 and OP142724.1 did not show 100% identity with any amplicon data in the gene bank. They showed 99.75% identity with Zimbabwe AY666010.1 and Hungary HM063432.1; 99.63% identity with India KX989521.1 and Sudan KU497715.1; 99.50% with USA AY010253.1; 99.38% with USA AY127072.1; 99.25% with Italy EU436159.1; 99.13% with Thailand MH939155.1; and 99.00% with Western Cuba MK809382.1 and Cuba MK809389.1. The phylogenetic tree for the msp4 gene showed that the amplicons from this study for cattle and camel were clustered in a single branch and closely related to a separate branch for other reported isolates from cattle in Sri Lanka and China (Figure 3).
We compared msp5 amplicons from this study for cattle OP142716.1, buffalo OP142718.1 and OP142719.1, and camel OP142720.1 with amplicons from other reported isolates and found alignment identities of 100% with Sri Lanka LC467691.1 and China KR047042.1; 99.61% with Egypt LC554225.1 and KU042081.1, Philippines AB704328.1, and Benin KX685369.1; 99.59% with Thailand MK240314.1; 99.42% with Egypt LC554224.1 and KU042080.1 and Kenya KP347554.1; 99.40% with Thailand MK164571.1; and 97.86% with the USA M93392.1 (Figure 4).
In this study, we addressed a paucity of complete data on Anaplasma species in southern Egypt distributed among cattle, buffaloes, and camel populations. A. marginale infection may be more common than previously believed, possibly due to misdiagnosis and undetected carrier animals. We suggest that more detailed information on the distribution of anaplasmosis in southern Egypt is urgently required.
Anaplasmosis is a tickborne rickettsial disease that can adversely affect livestock health and performance worldwide (30). Anaplasmosis reportedly incurs an average cost of $793 per head of cattle, 54% of which can be attributed to death, followed by 15% attributable to treatment, 14% to weight loss, 8% to chronic disease, and 9% to abortion (31). Previous studies in Egypt have shown a wide distribution of A. marginale in cattle and water buffalo; however, the data are still incomplete for camels. In Egypt, A. marginale is the second-most common tickborne disease in cattle (21.2%); the infection rate in buffaloes is 37.5%, and dromedaries have reportedly been infected with Babesia (11.0%), Theileria (71.8%), and Anaplasma species, as well as C. burnetti (20.8%) and Rickettsia spp. (31.9%) (32). For anaplasmosis in Egypt, the highest proportions of seropositive animals have been reported in Gharbia (100%), Suez (83.3%), and Port Said (33.3%), whereas the lowest proportions have been recorded in Sohag (4.7%) and Aswan (5.2%) (23). This study was performed to determine the presence of A. marginale in cattle, buffaloes, and camels in southern Egypt. The discovery of high prevalence rates of Piroplasma and Anaplasmataceae among animals that appeared to be in good health—when considered together with the recent rise in international animal trading—suggests the possibility of new genotypes of infections emerging and re-emerging in Egypt following a spread of pathogens from surrounding endemic countries (33, 34). Buffalo from southern Egypt show lower infection rates than cattle from similar areas, and these results may indicate a natural resistance against A. marginale in Egyptian buffalo. Previous studies have also demonstrated that water buffalo may show reduced infectivity and cellular replication for this pathogen, resist natural tick infestation, and have a reduced potential for transmitting tickborne diseases (35). The immune system can protect buffalo against high rickettsia levels and related diseases in their acute phase (36). Furthermore, we found that camels had a lower infection rate than cattle and buffaloes, with only five out of 75 camels (6.66%) positive for A. marginale. At least four anaplasma species (A. marginale, A. platys, A. phagocytophilum, and Candidatus A. camelii) have been identified as infecting Camelus dromedarius. However, infection with A. marginale in camel was detected primarily by conventional blood testing with stained blood smears, whereas other Anaplasma species in camel were either identified serologically or molecularly (37–40), using Anaplasmataceae 16S rRNA-based amplification, sequencing, and phylogenetic tree construction for A. phagocytophilum, A. platys, A. ovis, and Candidatus A. camelii (41). In only one study, in the Riyadh region of Saudi Arabia, have Arabian camels been shown to have Anaplasma species, by amplifying the particular msp5 gene; the pathogen species was determined to be A. marginale (42). To the best of our knowledge, this study presents the first report on using molecular methods and phylogenetic analysis to identify A. marginale in dromedary camels in southern Egypt.
Based on these epidemiological results and the genetic variation of A. marginale detected in loci different from previous studies (43, 44), we conclude that the prevalence and epidemiological characteristics of A. marginale infection are closely related to its geographic location.
Major surface protein genes are under selective pressure from the host immune system and play a significant role in the interaction of Anaplasma species with host cells (45–47). All Anaplasma species studied thus far have orthologs of the immunodominant outer membrane protein msp4 (46). Both prokaryotes and eukaryotes have a highly conserved housekeeping gene called groEL. Ehrlichia, Rickettsia, and Anaplasma species are all members of the Rickettsiales bacterial family, and this gene has recently been used in phylogenetic analyses of these species (47, 48). Six membrane surface proteins of the initial bodies of this organism (carriers of epitopes B and T) have been characterized. Major surface proteins have been named and identified as 1a, 1b, 2, 3, 4, and 5 (49); these proteins were recognized by neutralizing antibodies, and they have a strong intermolecular relationship in the membranes of the initial bodies, performing essential functions (36). Genes encoding these proteins have been studied, showing their protein products to have a variable polymorphism. They can be represented in the genome by a single copy gene or by forming part of multigenic families (50). All Anaplasma species and all examined isolates of A. marginale have shown a single copy of the msp5 gene present in their genomes. This gene is highly conserved, and it is a strong candidate for diagnosing bovine anaplasmosis, since the msp5 protein it produces has low structural complexity, is similarly conserved, and elicits high antibody titers (51).
Whenever infected cattle are moved, a new genotype of A. marginale is imported to their new location, and this genotype may then spread to susceptible cattle either mechanically or biologically. Few genotypic variations are detected in A. marginale isolates from places like Australia where cattle movements are rare (52). Dogs were shown to be carriers of ticks that disseminated A. marginale infection to cattle herds, and it is thus likely that physical contact between animals could result in tick transmission from one host to another, spreading tickborne diseases between them (53). The close contact between cattle and buffalo, particularly in the individual breeding system, could be a factor in the transmission of A. marginale between the different animal species. On the other hand, there is minimal interaction between camels and other animals; however, camels can be transported by the same vehicles as used to transport cattle and buffaloes, and such vehicles may become a path of infection and contribute to the spread of A. marginale in animal populations in southern Egypt and elsewhere.
The prevalence of A. marginale is known to vary according to environmental conditions, sample site, vector species, host breed, and breeding system (54). According to our research, intensive breeding systems had a higher infection rate than individual breeding systems (33.3 vs. 25%). This may be because there is more animal contact in intensive breeding systems than in individual breeding systems, making it easier for ticks to spread from one animal to another. Management practices differ from farm to farm in the southern area, where most farms house small numbers of co-reared animals because most farmers implement a multidisciplinary system encompassing pastoral and arable farming. Accordingly, predicting direct effects on disease epidemiology is challenging due to limitations in potential study populations.
Phylogenetic analyses using the msp4 gene have been used to elucidate the biogeography and evolution of the Anaplasma species (46). Phylogenetic analysis based on msp4 for A. marginale showed that the amplicons from this study for cattle and camel cluster in a single branch and have a close relationship with separate branches of other reported animal amplicons. According to one report in 2022, A. marginale was detected in camels in southern Egypt using the msp5 gene; however, that report did not provide any data on phylogenetic analyses of A. marginale in camels (55).
The phylogenetic analyses based on groEL and msp5 genes produced very similar results to those on isolates from other locations, possibly due to the unregulated movement of live animals between locations in Egypt for slaughter and marketing. Such local circulation of pathogens should be considered even though the issue of globally circulating tick diseases has gained attention recently with the importation of live animals from other countries to Egypt.
The findings of this study indicate that A. marginale is highly prevalent in camels, cattle, and buffaloes in southern Egypt. The identity of the A. marginale was confirmed by amplifying the specific msp4 and msp5 genes in phylogenetic analysis, which provided new data for A. marginale in southern Egypt. According to obtained results, A. marginale infection is endemic in different animal species in southern Egypt. It is the first report using three genes for A. marginale in Camelus dromedarius in southern Egypt.
The datasets presented in this study can be found in online repositories. The names of the repository or repositories and accession number(s) can be found in the article or supplementary material.
The animal study was reviewed and approved by the Faculty of Veterinary Medicine, South Valley University (10/09.02.2021, 53/13.09.2022).
HM and TT: conceptualization, design, experiments, formal analysis, investigation, writing of the original draft, project administration, and funding acquisition. HM, AA, and TT: resources, shared materials, and writing – review and editing. All authors contributed to the article and approved the submitted version.
This work was supported by the JSPS KAKENHI (Grant numbers JP20KK0154 and JP22H02522), JSPS Bilateral Program (Grant number JPJSBP120206002), and the Heiwa Nakajima Foundation. HM received financial support from the Egyptian government, Ministry of Higher Education and Scientific Research (Faculty of Veterinary Medicine, South Valley University) in the form of a scholarship as a post-doctor.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. AL-Hosary A, Ahmed L, Ahmed J, Nijhof A, Clausen PH. Epidemiological study on tropical theileriosis (Theileria annulata infection) in the Egyptian Oases with special reference to the molecular characterization of Theileria spp. Ticks Tick-Borne Dis. (2018) 9:1489–93. doi: 10.1016/j.ttbdis.2018.07.008
2. Ruybal P, Moretta R, Perez A, Petrigh R, Zimmer P, Alcarez E, et al. Genetic diversity of Anaplasma marginale in Argentina. Vet Parasitol. (2009) 162:176–80. doi: 10.1016/j.vetpar.2009.02.006
3. Kocan KM, De La Fuente J, Blouin EF, Garcia-Garcia JC. Anaplasma marginale (Rickettsiales: Anaplasmataceae): recent advances in defining hostpathogenhost-pathogen of a tick-borne rickettsia. Parasitology. (2004) 129:285–300. doi: 10.1017/S0031182003004700
4. Hairgrove T, Schroeder ME, Budke CM, Rodgers S, Chung C, et al. Molecular and serological in-herd prevalence of Anaplasma marginale infection in Texas cattle. Prev Vet Med. (2015) 119:1–9. doi: 10.1016/j.prevetmed.2015.02.006
5. Ueti MW, Reagan JO, Knowles DP, Scoles GA, Shkap V, et al. Identification of midgut and salivary glands as specific and distinct barriers to efficient tick-borne transmission of Anaplasma marginale. Infect Immun. (2007) 75:2959–64. doi: 10.1128/IAI.00284-07
6. Hove P, Khumalo ZTH, Chaisi ME, Oosthuizen MC, Brayton KA, Collins NE. Detection and characterisation of Anaplasma marginale and A. centrale in South Africa Vet Sci. (2018) 5:26. doi: 10.3390/vetsci5010026
7. Okafor CC, Collins SL, Daniel JA, Harvey B, Sun X, et al. Factors associated with Seroprevalence of Anaplasma marginale in Kentucky cattle. Vet Parasitol Reg Stud Rep. (2018) 13:212–9. doi: 10.1016/j.vprsr.2018.07.003
8. Aubry P, Geale D, A. review of bovine anaplasmosis. Transbound Emerg Dis. (2011) 58:1–30. doi: 10.1111/j.1865-1682.2010.01173.x
9. Kocan KM. de la Fuente J, Blouin EF, Coetzee JF, Ewing SA. The natural history of Anaplasma marginale. Vet Parasitol. (2010) 167:95–107. doi: 10.1016/j.vetpar.2009.09.012
10. Kocan KM. de la Fuente J, Coburn LA. Insights into the development of Ixodes scapularis a resource for research on a medically important tick species. Parasit Vectors. (2015) 8:592. doi: 10.1186/s13071-015-1185-7
11. Kamani J, Usman TA, Onyeamaechi EG, Usman MA Kida SM, et al. Hemoparasites of camels (Camelus dromedarius) in Maiduguri, Nigeria. Anim Res Int. (2008) 5:838–9. doi: 10.4314/ari.v5i2.48726
12. Swelum AA, Ismael AB, Khalaf AF, Abouheif MA. Clinical and laboratory findings associated with naturally occurring babesiosis in dromedary camels. Bull Vet Inst Pulaway. (2014) 58:229–33. doi: 10.2478/bvip-2014-0034
13. Mahran OM. Some studies on blood parasites in camels (Camelus dromedarius) at Shalatin City, Red Sea Governorate. Assiut Vet Med J. (1980) 50:172–84. doi: 10.21608/avmj.2004.178964
14. Ben Said M, Belkahia H, Sayahi L, Aloui M, Jemli MH, Hadj Mohamed B, et al. First serological study of the prevalence of Anaplasma phagocytophilum in dromedary (Camelus dromedarius) in Tunisia. Bull Soc Pathol Exot. (2013) 107:1–6. doi: 10.1007/s13149-013-0323-8
15. Chung C, Wilson C, Bandaranayaka-Mudiyanselage CB, Kang E, Adams DS, et al. Improved diagnostic performance of commercial Anaplasma antibody competitive enzyme-linked immunosorbent assay using recombinant major surface protein 5–glutathione S-transferase fusion protein as antigen. J Vet Diagn Investig. (2013) 26:61–71. doi: 10.1177/1040638713511813
16. Kocan KM, Blouin EF, Barbet A. Anaplasmosis control. Past, present, and future. Ann NY Acad Sci. (2000) 916:501–9. doi: 10.1111/j.1749-6632.2000.tb05329.x
17. Kopácek P, Hajdusek O, Buresová V, Daffre S. Tick innate immunity. Adv Exp Med Biol. (2010) 708:137–62.
18. Merino O, Alberdi P, Pérez de. la Lastra JM, de la Fuente J. Tick vaccines and the control of tick-borne pathogens. Front Cell Infect Microbiol. (2013) 3:30. doi: 10.3389/fcimb.2013.00030
19. El-Naga T, Barghash S. Blood parasites in camels (Camelus dromedarius) in Northern West Coast of Egypt. J Bacteriol Parasitol. (2016) 7:1000258. doi: 10.1016/j.parepi.2016.07.002
20. AL-Hosary A, Răileanu C, Tauchmann O, Fischer S. Nijhof, AM, Silaghi, C Epidemiology and genotyping of Anaplasma marginale and co-infection with piroplasms and other Anaplasmataceae in cattle and buffaloes from Egypt. Parasit Vectors. (2020) 13:495. doi: 10.1186/s13071-020-04372-z
21. El-Dakhly KM, Arafa WM, Soliman S, Abdel-Fatah OR, Wahba AA, et al. Molecular detection, phylogenetic analysis, and genetic diversity of Theileria annulata, Babesia bigemina, and Anaplasma marginale in cattle in three districts of Egypt. Acta Parasitol. (2020) 65:620–7. doi: 10.2478/s11686-020-00189-z
22. Nasreldin N, Ewida RM, Hamdon H, Elnaker YF. Molecular diagnosis and biochemical studies of tick-borne diseases (anaplasmosis and babesiosis) in Aberdeen Angus Cattle in New Valley, Egypt. Vet World. (2020) 13:1884. doi: 10.14202/vetworld.2020.1884-1891
23. Parvizi O, El-Adawy H, Melzer F, Roesler U, Neubauer H, et al. Seroprevalence and molecular detection of bovine anaplasmosis in Egypt. Pathogens. (2020) 9:64. doi: 10.3390/pathogens9010064
24. Squillacioti C, De Luca A, Alì S, Ciarcia R, Germano G, Vittoria A, et al. Presence and distribution of urocortin and corticotrophin-releasing hormone receptors in the bovine thyroid gland. Anat Histol Embryol. (2014) 43:429–34. doi: 10.1111/ahe.12094
25. Elbers JP, Rogers MF, Perelman PL, Proskuryakova AA, Serdyukova NA, et al. Improving Illumina assemblies with Hi-C and long reads: An example with the North African dromedary. Mol Ecol Resour. (2019) 19:1015–26. doi: 10.1111/1755-0998.13020
26. Ybañez AP, Sivakumar T, Badgar B, Battur B, Altangerel K, Matsumoto K, et al. Specific molecular detection and characterization of Anaplasma marginale in Mongolian cattle. J Vet Med Sci. (2012) 75:399–406. doi: 10.1292/jvms.12-0361
27. de la Fuente J, Van Den Bussche RA, Kocan KM. Molecular phylogeny and biogeography of North American isolates of Anaplasma marginale (Rickettsiaceae: Ehrlichieae). Vet Parasitol. (2001) 97: 65–76. doi: 10.1016/S0304-4017(01)00378-8
28. Kumar S, Stecher G, Li M, Knyaz C, Tamura K, MEGA X. Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. (2018) 35:1547–9. doi: 10.1093/molbev/msy096
29. Kimura M, A. simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. (1980) 16:111–20. doi: 10.1007/BF01731581
30. Jongejan F, Uilenberg G. The global importance of ticks. Parasitology. (2004) 129:S3–14. doi: 10.1017/S0031182004005967
31. Railey AF, Marsh TL. Economic benefits of diagnostic testing in livestock: anaplasmosis in cattle. Front Vet Sci. (2021) 8:626420. doi: 10.3389/fvets.2021.626420
32. El-Alfy ES, Abbas I, Baghdadi HB, El-Sayed SAE, Ji S, Rizk MA, et al. Molecular epidemiology and species diversity of tick-borne pathogens of animals in Egypt: a systematic review and meta-Analysis. Pathogens. (2022) 11:912. doi: 10.3390/pathogens11080912
33. Bilgic HB, Karagenç T, Shiels B, Tait A, Eren H, Weir W. Evaluation of cytochrome b as a sensitive target for PCR based detection of T. annulata carrier animals. Vet Parasitol. (2010) 174:341–7. doi: 10.1016/j.vetpar.2010.08.025
34. Zeb J, Shams S, Din IU, Ayaz S, Khan A. Molecular epidemiology and associated risk factors of Anaplasma marginale and Theileria annulata in cattle from North-western Pakistan. Vet Parasitol. (2020) 279:109044. doi: 10.1016/j.vetpar.2020.109044
35. Obregon D, Corona BG, de la Fuente J, Cabezas-Cruz A, Goncalves LR, Matos CA, et al. Molecular evidence of the reservoir competence of water buffalo (Bubalus bubalis) for Anaplasma marginale in Cuba. Vet Parasitol Reg Stud Rep. (2018) 13:180–7. doi: 10.1016/j.vprsr.2018.06.007
36. Palmer GH, Rurangirwa FR, Kocan KM, Brown WC. Molecular basis for vaccine development against the ehrlichial pathogen Anaplasma marginale. Parasitol Today. (1999) 15:281–6. doi: 10.1016/S0169-4758(99)01469-6
37. Ismael AB, Swelum AA, Khalaf AF, Alowaimer AN. First evidence of natural anaplasmosis in Camelus dromedarius in Saudi Arabia. J Camel Pract Res. (2016) 23:95–100. doi: 10.5958/2277-8934.2016.00014.X
38. Ait Lbacha H, Zouagui Z, Alali S, Rhalem A, Petit E, et al. “Candidatus anaplasma camelii” in one-humped camels (Camelus dromedarius) in Morocco: a novel and emerging Anaplasma species. Infect Dis Poverty. (2017) 6:1. doi: 10.1186/s40249-016-0216-8
39. Alshahrani M, Alanazi A, Alouffi A, Abdullah H, Allam A, et al. Molecular detection of Candidatus Anaplasma camelii in camels (Camelus dromedarius) from Asir Province, Saudi Arabia. Tropical Biomed. (2020) 37:587–98. doi: 10.47665/tb.37.3.587
40. Li Y, Yang J, Chen Z, Qin G, Li Y. Anaplasma infection of Bactrian camels (Camelus bactrianus) and ticks in Xinjiang, China. Parasit Vectors. (2015) 8:313. doi: 10.1186/s13071-015-0931-1
41. Azmat M, Ijaz M, Farooqi SH, Ghaffar A, Ali A, et al. Molecular epidemiology, associated risk factors, and phylogenetic analysis of anaplasmosis in camel. Microb Pathog. (2018) 123:377–84. doi: 10.1016/j.micpath.2018.07.034
42. Omer E, Elfehid M, Alwazan A, Alouffi AS, Alshammari FA, et al. Molecular detection of Anaplasma marginale in Arabian Camels (Camelus dromedarius) in Riyadh, Saudi Arabia. J Saudi Soc Agric Sci. (2022) 21:73–6. doi: 10.1016/j.jssas.2021.06.010
43. de la Fuente J, Atkinson MW, Naranjo V, Fernández de Mera IG, et al. Sequence analysis of the msp4 gene of Anaplasma ovis strains. Vet Microbiol. (2007) 119:375–81. doi: 10.1016/j.vetmic.2006.09.011
44. Palmer GH, Brown WC, Rurangirwa FR. Antigenic variation in the persistence and transmission of the ehrlichia Anaplasma marginale. Microbes Infect. (2000) 2:167–76. doi: 10.1016/S1286-4579(00)00271-9
45. Brayton KA, Palmer GH, Brown WC. Genomic and proteomic approaches to vaccine candidate identification for Anaplasma marginale. Expert Rev Vaccines. (2006) 5:95–101. doi: 10.1586/14760584.5.1.95
46. de la Fuente J, Lew A, Lutz H, Meli ML, Hofmann-Lehmann R, et al. Genetic diversity of Anaplasma species major surface proteins and implications for anaplasmosis serodiagnosis and vaccine development. Anim Health Res Rev. (2005) 6:75–89. doi: 10.1079/AHR2005104
47. Chisu V, Zobba R, Lecis R, Sotgiu F, Masala G, et al. GroEL typing and phylogeny of Anaplasma species in ticks from domestic and wild vertebrates. Ticks Tick Borne Dis. (2018) 9:31–6. doi: 10.1016/j.ttbdis.2017.10.012
48. Dumler JS, Barbet AF, Bekker CP, Dasch GA, Palmer GH, et al. Reorganization of genera in the families Rickettsiaceae and Anaplasmataceae in the order Rickettsiales: unification of some species of Ehrlichia with Anaplasma, Cowdria with Ehrlichia and Ehrlichia with Neorickettsia, descriptions of six new species combinations and designation of Ehrlichia equi and 'HGE agent' as subjective synonyms of Ehrlichia phagocytophila. Int J Syst Evol Microbiol. (2001) 2145–65. doi: 10.1099/00207713-51-6-2145
49. Corona B, Machado H, Rodríguez M, Martínez S. Characterization of recombinant MSP5 Anaplasma marginale Havana isolate. Braz J Microbiol. (2009) 40:972–9. doi: 10.1590/S1517-83822009000400032
50. Barbet AF, Blentlinger R, Yi J, Lundgren AM, Blouin EF, Kocan KM. Comparison of surface proteins of Anaplasma marginale grown in tick cell culture, tick salivary glands, and cattle. Infect Immun. (1999) 67:102–7. doi: 10.1128/IAI.67.1.102-107.1999
51. Knowles D, Torioni de. Echaide S, Palmer G, McGuire T, et al. Antibody against an Anaplasma marginale MSP5 epitope common to tick and erythrocyte stages identifies persistently infected cattle. J Clin Microbiol. (1996) 34:2225–30. doi: 10.1128/jcm.34.9.2225-2230.1996
52. de la Fuente J, Van Den Bussche RA, Prado TM, Kocan KM. Anaplasma marginale msp1alpha genotypes evolved under positive selection pressure but are not markers for geographic isolates. J Clin Microbiol. (2003) 41:1609–16. doi: 10.1128/JCM.41.4.1609-1616.2003
53. Ashraf S, Parveen A, Muhammad Awais M, Gillani Q, Aktas M, et al. A report on molecular detection and phylogenetic evaluation of Anaplasma marginale in ticks and blood samples collected from cattle in district Layyah in Punjab (Pakistan). Curr Microbiol. (2021) 78:274–81. doi: 10.1007/s00284-020-02256-0
54. Bursakov SA, Kovalchuk SN. Co-infection with tick-borne disease agents in cattle in Russia. Ticks Tick Borne Dis. (2019) 10:709–13. doi: 10.1016/j.ttbdis.2019.03.004
Keywords: Anaplasma marginale, cattle, buffalo, camel, Egypt, PCR, sequencing
Citation: Mahmoud HYAH, Ali AO and Tanaka T (2023) Molecular detection and characterization of Anaplasma marginale infecting cattle, buffalo, and camel populations in southern Egypt. Front. Vet. Sci. 10:1169323. doi: 10.3389/fvets.2023.1169323
Received: 19 February 2023; Accepted: 10 April 2023;
Published: 11 May 2023.
Edited by:
Yasser Mahmmod, Higher Colleges of Technology, United Arab EmiratesReviewed by:
Mohamed Nayel, University of Sadat City, EgyptCopyright © 2023 Mahmoud, Ali and Tanaka. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tetsuya Tanaka, azYxOTk0MzFAa2FkYWkuanA=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.