- 1College of Life Science, Guizhou University, Guiyang, China
- 2Key Laboratory of Animal Genetics, Breeding and Reproduction in the Plateau Mountainous Region, Ministry of Education, Guizhou University, Guiyang, China
- 3Key Laboratory of Animal Genetics, Breeding and Reproduction, Guiyang, China
- 4College of Animal Science, Guizhou University, Guiyang, China
Circular RNAs (circRNAs) play an important role in regulating the mammalian reproductive system, especially testicular development and spermatogenesis. However, their functions in testicular development and spermatogenesis in the Qianbei Ma goat, the Guizhou endemic breed are still unclear. In this study, tissue sectioning and circRNAs transcriptome analysis were conducted to compare the changes of morphology and circular RNAs gene expression profile at four different developmental stages (0Y, 0-month-old; 6Y, 6-month-old; 12Y, 12-month-old; 18Y, 18-month-old). The results showed that the circumferences and area of the seminiferous tubule gradually increased with age, and the lumen of the seminiferous tubule in the testis differentiated significantly. 12,784 circRNAs were detected from testicular tissues at four different developmental stages by RNA sequencing, and 8,140 DEcircRNAs (differentially expressed circRNAs) were found in 0Y vs. 6Y, 6Y vs. 12Y, 12Y vs. 18Y and 0Y vs. 18Y, 0Y vs. 12Y, 6Y vs. 18Y Functional enrichment analysis of the source genes showed that they were mainly enriched in testicular development and spermatogenesis. In addition, the miRNAs and mRNAs associated with DECircRNAs in 6 control groups were predicted by bioinformatics, and 81 highly expressed DECircRNAs and their associated miRNAs and mRNAs were selected to construct the ceRNA network. Through functional enrichment analysis of the target genes of circRNAs in the network, some candidate circRNAs related to testicular development and spermatogenesis were obtained. Such as circRNA_07172, circRNA_04859, circRNA_07832, circRNA_00032 and circRNA_07510. These results will help to reveal the mechanism of circRNAs in testicular development and spermatogenesis, and also provide some guidance for goat reproduction.
1. Introduction
Reproductive traits are important economic traits in goats, and improving the fecundity of breeding rams is an important guarantee to improve the efficiency of goat breeding (1). As an important organ for the maintenance of male reproductive ability, the main biological function of testis is to produce sperm, and testicular development is a necessary process for sperm production. It is the guarantee for the establishment and maintenance of the male reproductive system (2), while, the contribution of ram testes to reproduction is as high as 50% (3). During testicular development, the genome of testicular cells is actively transcribed into mRNA and many noncoding RNAs(ncRNAs) composed of miRNAs, circRNAs, and lncRNA. circRNAs can act as a competitive endogenous RNA (ceRNA) to regulate gene expression by regulating the activity of miRNA (4), thereby participating in the biological processes of testicular development and spermatogenesis (5, 6).
CircRNAs are a prominent new class of closed-loop RNA molecules found in eukaryotes, that are produced by the alternate shearing of precursor mRNAs (7), and are involved in the regulation of mammalian development (8), growth (9), reproduction (10), and immunity (11) in animals. For example, circPPARA can act as an endogenous sponge to adsorb mir-429 and mir-200b, thereby promoting intramuscular adipogenesis in pigs (12), and reduced expression of circZNF609 significantly inhibits myoblast proliferation in mice in vivo (13). Holdt et al. (14) discovered that circ-ANRIL could bind to ribosome biogenesis factor 1 (PES1) to prevent ribosome formation, thereby promoting cell apoptosis and inhibiting macrophage proliferation. Liu et al. (15) identified 388 differentially expressed (DE) circRNAs in pituitary tissues of high-yield and low-yield black goats and found that circ_0014542, circ_0031209 and circ_0019448 played important roles in goat reproduction. Zhang et al. (16) detected that circRNAs in goat endometrial epithelial cells could influence the reproductive performance of goats by regulating the embryo implantation process. The above studies suggest that circRNAs play an important role in the regulation of reproduction. In goats, the research on the reproductive performance of circRNA has mainly focused on the ovary, pituitary and uterus of ewes, while its effects and mechanism testicular development and spermatogenesis of rams have not been fully clarified.
Qianbei Ma goats are a unique goat variety in the mountainous area of the Guizhou Plateau. It is a large size, strong physique, compact structure, in meat, wool and dual-purpose goat. It has many advantages, such as high fertility, strong disease resistance, strong adaptability and stable genetic performance. It is an ideal animal for studying testicular development in highland meat goats (17). Based on the importance of the testis to the reproduction of goats, the testis of Qianbei Ma goats at four developmental stages (0Y, 6Y, 12Y, and 18Y) were selected for histomorphology and transcriptomic sequencing (NA-SEQ) analysis to explore the testis developmental rules. Through DEcircRNAs analysis, circRNAs regulation network construction s, and functional enrichment analysis the key circRNAs related to testis development and spermatogenesis at different developmental stages were identified. This study aims to reveal the underlying mechanism of circRNA regulation of testicular development and spermatogenesis, and provide theoretical basis and data support for improving the reproductive performance of rams.
2. Materials and methods
2.1. Collection of animal samples headings
Twenty healthy Qianbei Ma goats were randomly selected from the same flock at Fuxing Herding Co Ltd., Xishui County, Guizhou Province, China, and divided into four age groups (n = 5 for each age group): 0Y, 0-month-old (weight: 1.66 kg ± 0.5 kg); 6Y, 6-month-old (Weight: 30.30 kg ± 1.0 kg); 12Y, 12-month-old (Weight: 43.32 kg ± 1.5 kg); 18Y, 18-month-old (Weight: 47.53 kg ± 2.0 kg). After anesthesia, the left testicular tissue was castrated and collected with sterile scissors forceps immediately, and trimmed to approximately 5 mm testicular tissue fixed in 4% paraformaldehyde solution for H&E staining, with three biological replicates at each age. Testicular tissue samples were collected and washed with phosphate buffered salt solution within 20 min. Then all samples were snap frozen in liquid nitrogen and subsequently transferred to-80°C for storage for further studies.
2.2. Histomorphological analysis
Hematoxylin-eosin (H&E) staining: paraffin sections were stained with hematoxylin using conventional methods, restained with eosin, and sealed with neutral resin before being placed under a biological microscope for observation and photographed. The images were scanned and saved using CaseViewer 2.4 software, followed by physicochemical index analysis using Image-ProPlus 6.0 software to measure the circumference and area of the seminiferous tubules and count the number of supporting and leydig cells per unit area, using millimeter as the standard unit.
2.3. RNA extraction and library construction
Total RNA was isolated from testis samples using TRIzol RNA (Thermo Fisher Scientific, China) kit under aseptic conditions.Ribosomal RNA was removed from total RNA in strict accordance with the instructions for the EvoM-MLV RT Kit with gDNA Clean for qPCR (Hunan Acres Biological Company, China). RNase R was applied to remove linear RNA from total RNA (Servicebio Technology Co., Ltd., China), and the RNA was broken into short fragments by adding the breaking reagent. Using the RNA as the template, one-strand cDNA was synthesized by using random primers of six bases, and then two-strand cDNA was synthesized by the reaction system of two-strand synthesis. In the process of cDNA synthesis, DUTP was substituted for DTTP, and then different linkers were ligated. One strand containing dUTP was digested by the UNG enzyme, and only one strand of cDNA connecting different linkers was retained one strand of cDNA was purified by using the kit, and then the end was repaired. A tail was added, and the sequencing linker was connected. Then, the fragment size was selected, and PCR amplification was carried out. The library was screened by an Agilent 2,100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA) and sequenced by an Illumina sequencer.
2.4. circRNA identification and differential circRNA recognition
The raw sequencing data were processed to obtain high-quality pure sequences as follows: (a) reads containing adapters were filtered by fastp (version 0.23.0); (b) sequences containing more than 10% N; (c) all sequences containing baseA were removed; and (d) low-quality sequences (q ≤ 20). TopHat2 was used to map the clean reads to a reference genome from the National Center for Biotechnology Information (NCBI)1 (18), and the resulting comparisons were employed for the identification of circRNAs using Find_circ software (19). The expression of circRNAs was quantified by reads per million mapped (RPM) as a normalization method, and the DEseq2R package (1.10.1) was employed to analyze the differential expression of circRNAs. p value <0.05 and circRNAs with a |fold change| > 2.0 were regarded as significantly differentially expressed.
2.5. Go, and KEGG enrichment analysis
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of differentially expressed circRNA host genes were used for the note. GOseq software was applied for GO function analysis. Kobas software2 was applied to test the statistical enrichment of differential gene expression in the KEGG pathway. GO terms and KEGG pathways were considered significantly when at p < 0.05.
2.6. ceRNA network construction
miRanda3 and circBank4 analysis software was used to predict circRNA-targeted relationships with microRNAs. After the intersection of the prediction results of each analysis software, the CircrNA-targeted miRNA set was obtained. TargetScan5 and miRDB6 were used to predict the targeting relationship between miRNA and mRNA. After the prediction results were intersected, the set of miRNA-targeted mRNAs was obtained. Further integrating the relationship pairs among circRNA, miRNA and mRNA, to build the ceRNA regulatory network related to testicular development, their interaction network was visualized in Cytoscape software (v3.7.1).
2.7. RT–qPCR validation
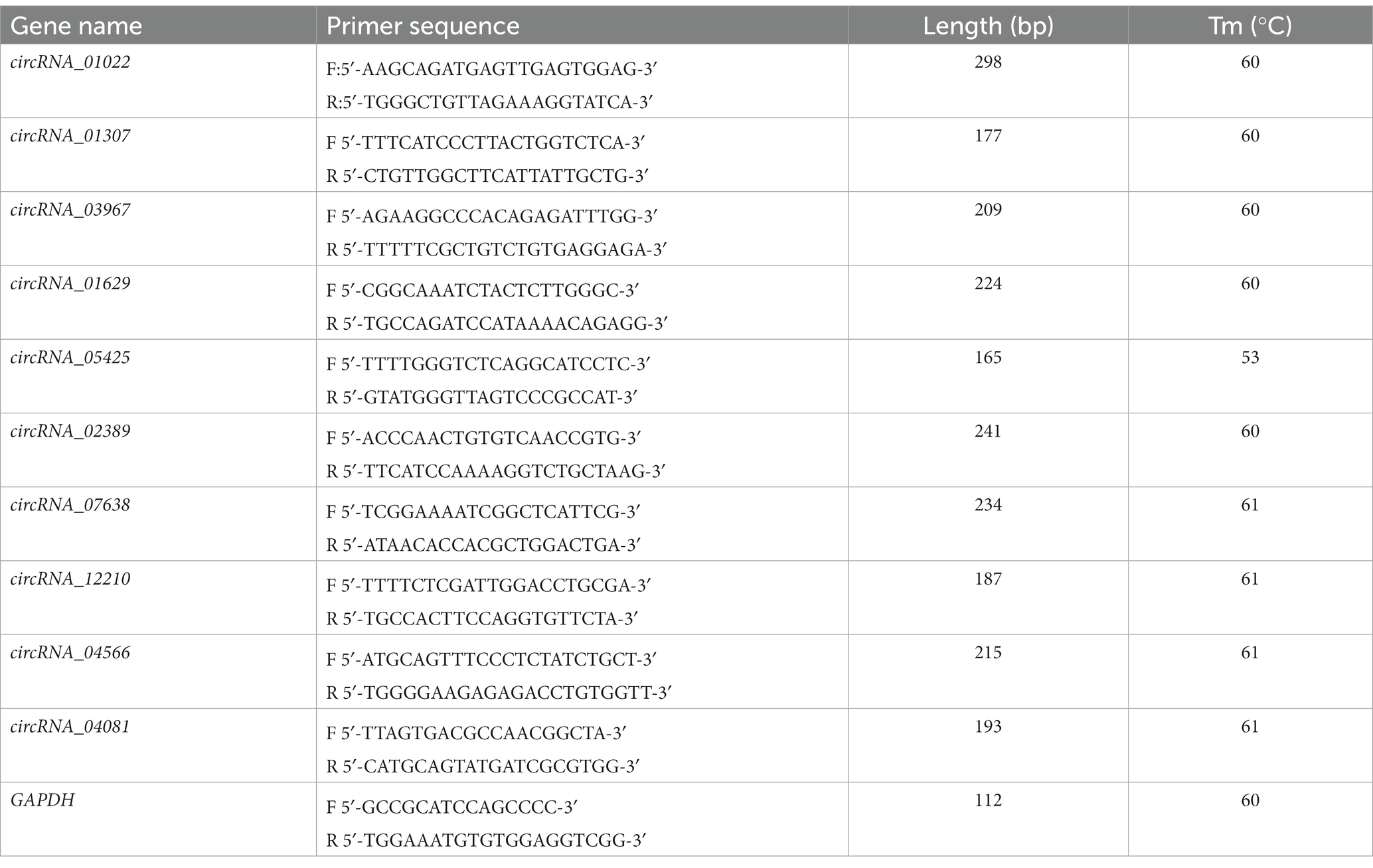
Ten DEcircRNAs were randomly selected for RT qPCR sequencing verification. The FirstChoice™ RLM-RACE kit (Thermo Fisher Scientific, China) was used synthesize total RNA into complementary DNA (cDNA). Primer information used in RT qPCR is listed in Table 1. CFX 96 Real Time PCR system (Bio-Rad, Foster City, CA, USA) was employed to detect the relative gene expression.
The reaction system was 20 μL: 2× RealStar Green Fast Mixture 10 μL, Primer Sense (10 pmol/μL) 1 μL, Primer Anti-sense (10 pmol/μL) 1 μL, cDNA (ng/μL) 2 μL, ddH2 O 6 μL. The reaction conditions were: 95°C for 2 min, 95°C for 15 s, annealing temperature (see Table 1 for details) for 30 s, and 72°C for 30 s. Solubility curve analysis was performed after 40 cycles, with a rate of 0.5°C rise every 5 s from 65°C to 95°C. Three biological replicates were set up. The specificity of the PCRs was determined by a single peak on the melting curve. Gene expression levels and normalized by GAPDH, and relative gene expression levels were calculated using the 2−ΔΔCt method. PCR products were confirmed by 1.5% agarose gel electrophoresis, and Sanger sequencing was then performed with PCR products to verify circRNA cyclization loci.
2.8. Statistics
SPSS 21.0 software (SPSS, Inc., Chicago, IL, USA) was used for statistical analysis (20). All values are expressed as the mean ± SEM. The difference between the two groups was expressed with *p < 0.05 and **p < 0.01 means that each experiment should be repeated at least 3 times.
3. Results
3.1. Histomorphological analysis of the testes at different ages in Qianbei Ma goats
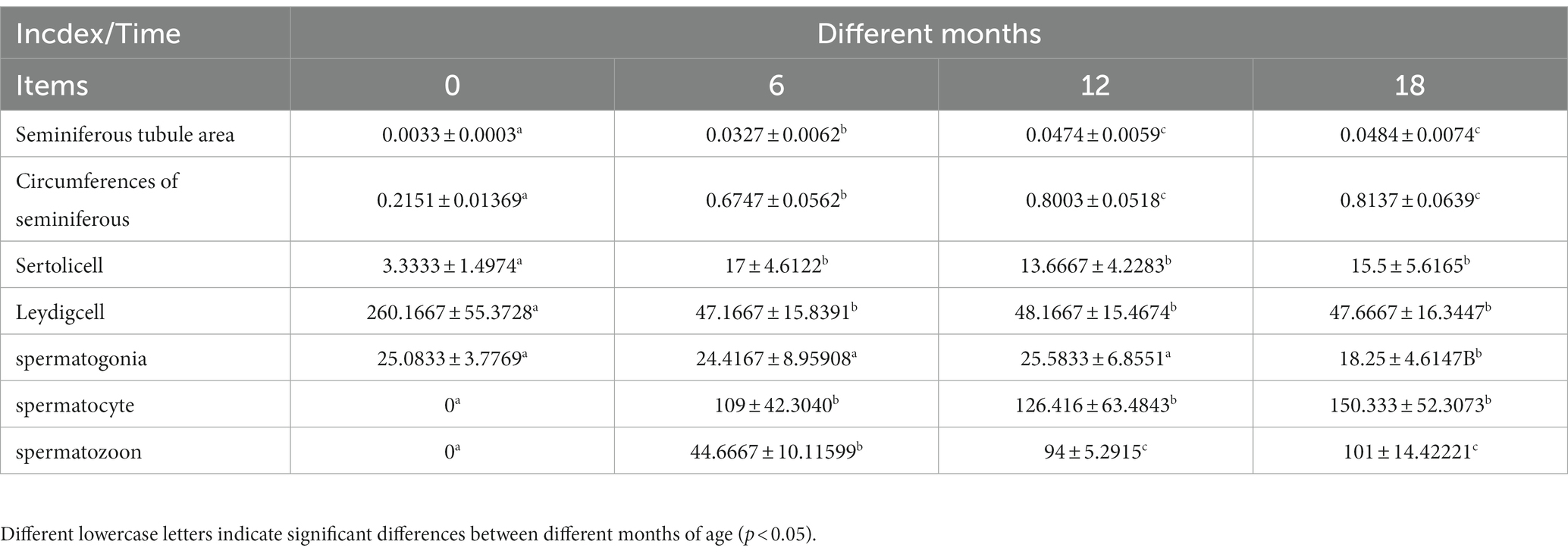
The testis tissue is composed of seminiferous tubules and testis stroma (21). As showed in Figure 1, only Sertoli cells and spermatogonia appear at 0 months in goats, and spermatozoa appear in the seminiferous tubule lumen at 6 months, indicating the onset of puberty in Qianbei Ma goats. Compared with 0-month-old goats, the lumen of convoluted seminiferous tubules in the testis differentiated significantly at the age of 6, 12, and 18 months, and the volume of Sertoli cells increased significantly. Compared with 6 months goats, the number of layers of spermatogenic cells and the number of sperm in the lumen of the testis increased significantly in 12-month-old and 18-month-old goats. Statistical analysis of seminiferous tubules and related cells in testis also showed that, with increasing age, the circumference and area of seminiferous tubules gradually increased, and the number of spermatocytes, Sertoli cells and sperm in testis gradually increased, while the number of leydig cells gradually decreased. However, the number of Sertoli cells and Leydig cells reached the limit in 6 months goats, and the number of spermatogonia was almost the same from 12 months to 18 months, and decreased at 18 months (see Table 2).
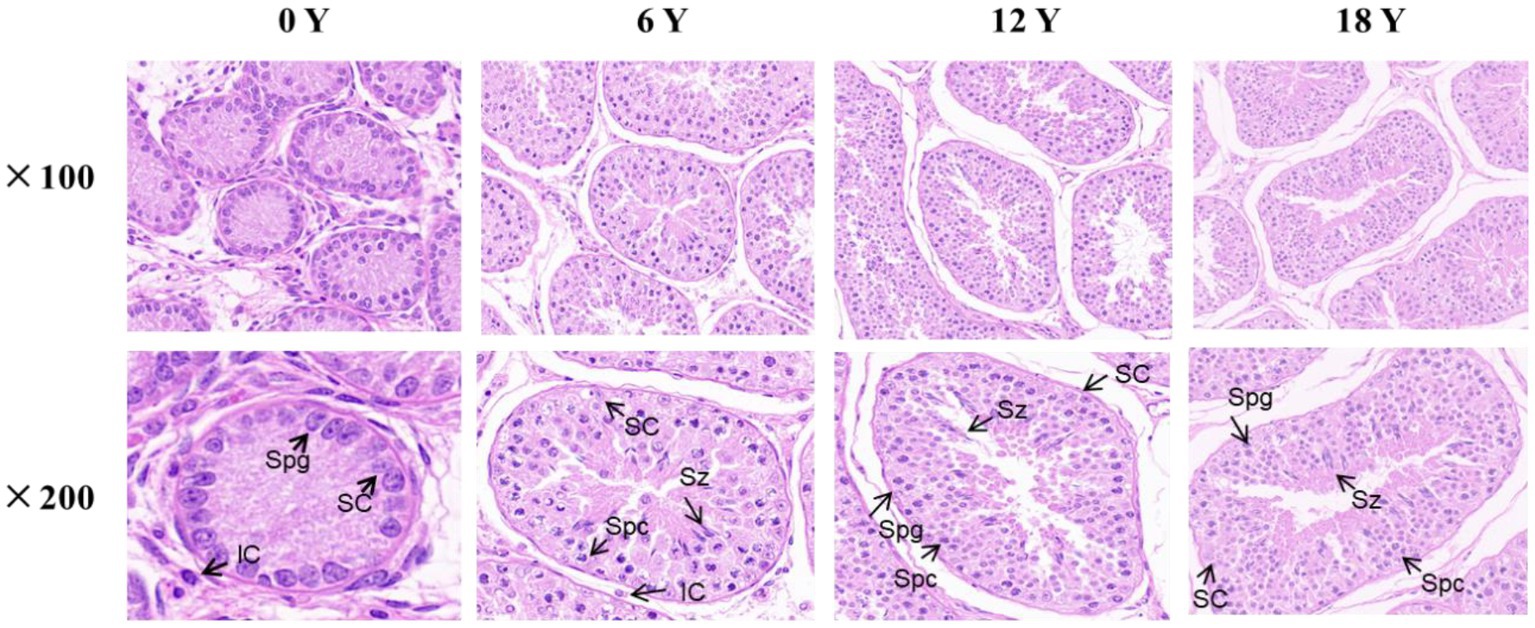
Figure 1. HE staining of testicular tissue from rams at different months of age. SC: supporting cells. Spg: spermatogonia. Spc: spermatocytes. LC: Leydig cells. Sz: spermatozoa.
3.2. Transcriptome sequencing of the circRNA in testis of the Qianbei Ma goats
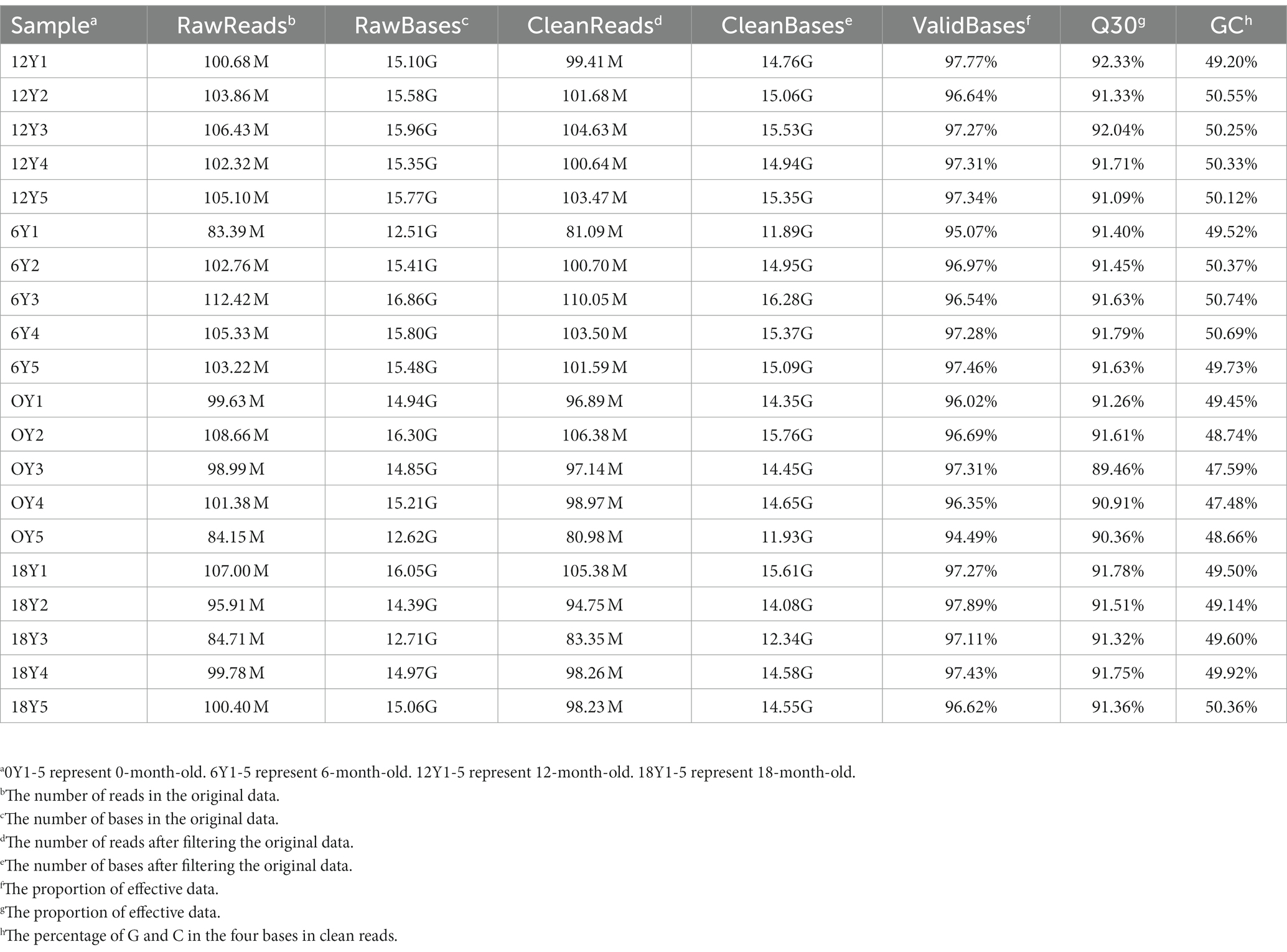
After removing low-quality reads, 19,706,582 clean reads were obtained (480, 343, 016 from the 0Y group; 496, 921, 424 from the 6Y group; 509, 827, 848 from the 12Y group; 479, 974, 294 from the 18Y group) (Table 3). In the 0Y, 6Y, 12Y and 18Y samples, 98.34, 97.97, 97.42 and 98.4% of the reads were located in the reference genome, and the base rates of Q30 were 91.7, 91.58, 90.72 and 91.54%, respectively. A total of 12,874 circRNAs were identified and designated by numbering from circRNA_00001 to circRNA_12,874. Based on their position in the goat genome, 83% of circRNAs were found to be sense overlapping, while the other 17% were intergenic, exon, antisense and intronic (Figure 2A). The majority of circRNAs were distributed within 2000 nt in length, but the most circRNAs clustered at more than 2000 nt. (Figure 2B). These circRNAs were distributed on 29 autosomes, with higher distribution on chromosomes 1, 3 and 10 (Figure 2C).
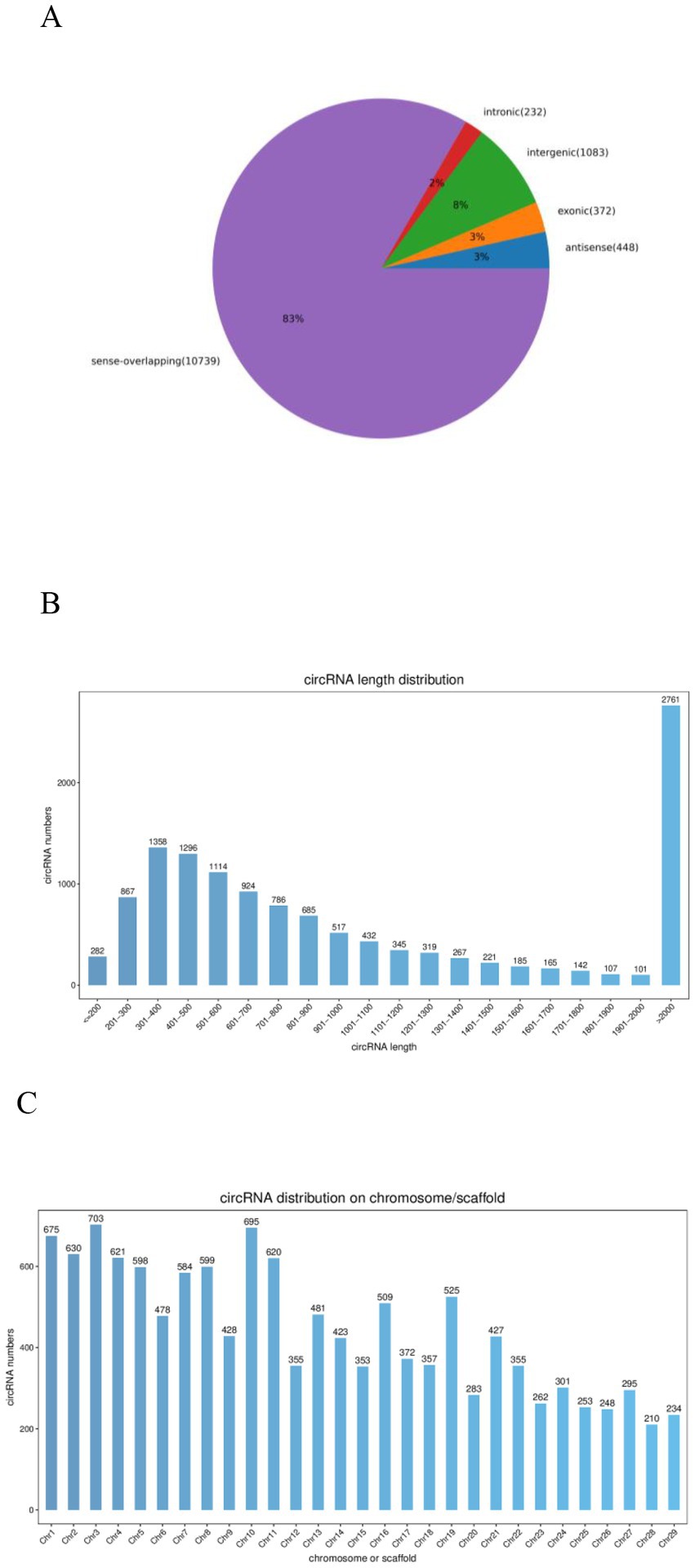
Figure 2. Qianbei Ma Goat testicular circRNAs. (A) CircRNAs from distinct genomic sites. (B) Length distributions of the known circRNAs. (C) Chromosome distribution map of circRNAs.
3.3. Differential expression analysis of circRNAs in the testis of Qianbei Ma goats
In the circRNA differential expression analysis, we found 8,140 differentially expressed circrnas in the six comparison groups. (Figure 3A). Specifically, 0Y vs. 6Y had 4,026 differentially expressed circRNAs (up: 2596, down: 1430), 6Y vs. 12Y had 4,030 (up: 2152, down: 1878), 12Y vs. 18Y had 3,226 (up: 1645, down: 1581), 0Y vs. 18Y had 3,760 (up: 2359, down: 1401), 0Y vs. 12Y had 3,878 (up: 2465, down: 1413), and 6Y vs. 18Y had 3,721 (up: 1839 down: 1882), of which 4 circRNAs were coexpressed in four age groups (Figure 3B). Ten DEcircRNAs were then randomly selected to validate the sequencing results by RT–qPCR, and the expression trends of the detected circRNAs were consistent with the RNA-seq sequencing results (Figure 4A). The PCR products were subjected to Sanger sequencing, verifying that the selected circRNA had the correct cyclization linker site (Figure 4B). These results indicated that the circRNA sequencing data were reliable, and could be used for further analysis.
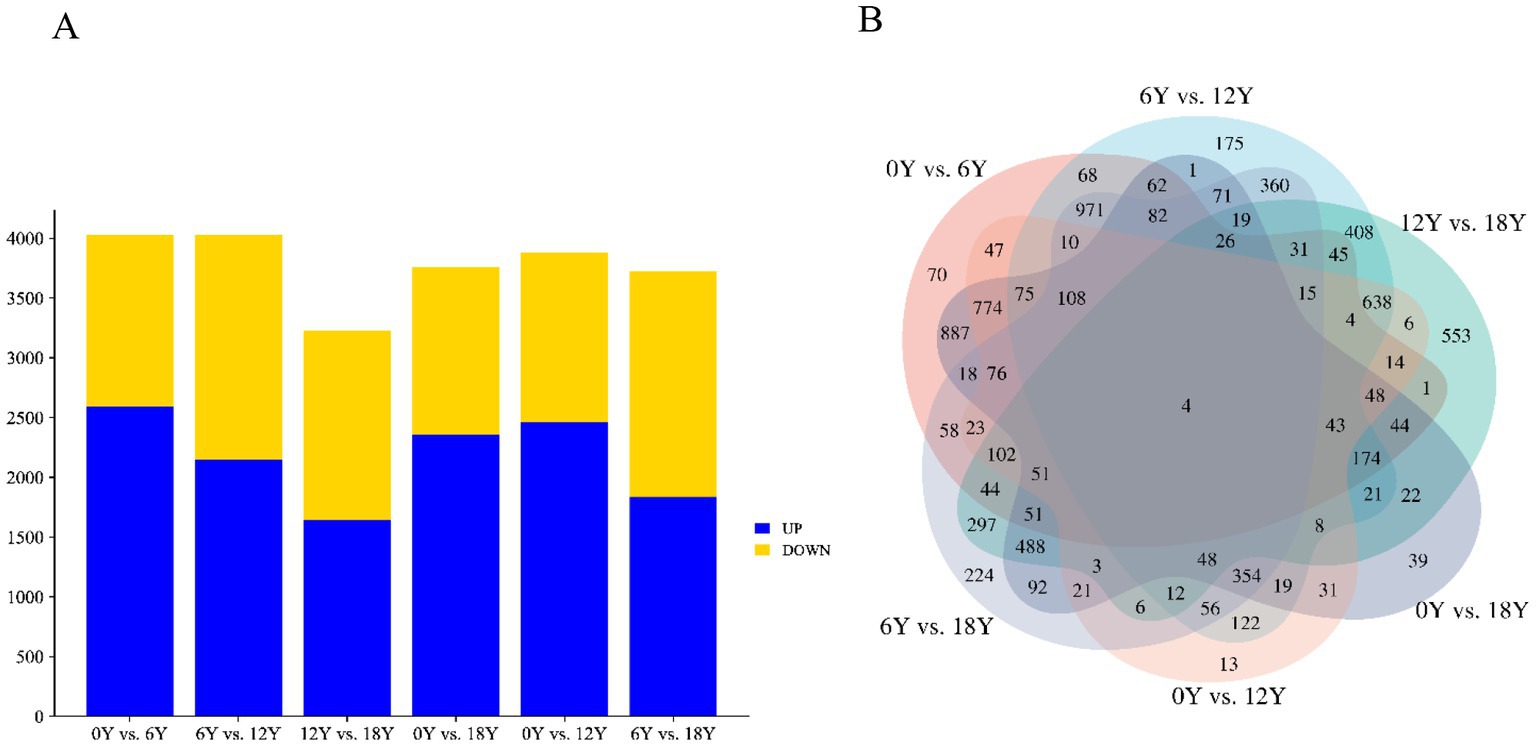
Figure 3. Summary of differential expression analysis of annotated circRNAs. (A) Number of DEcircRNAs in different control groups. (B) Wayne diagram of DEcircRNAs in different control groups.
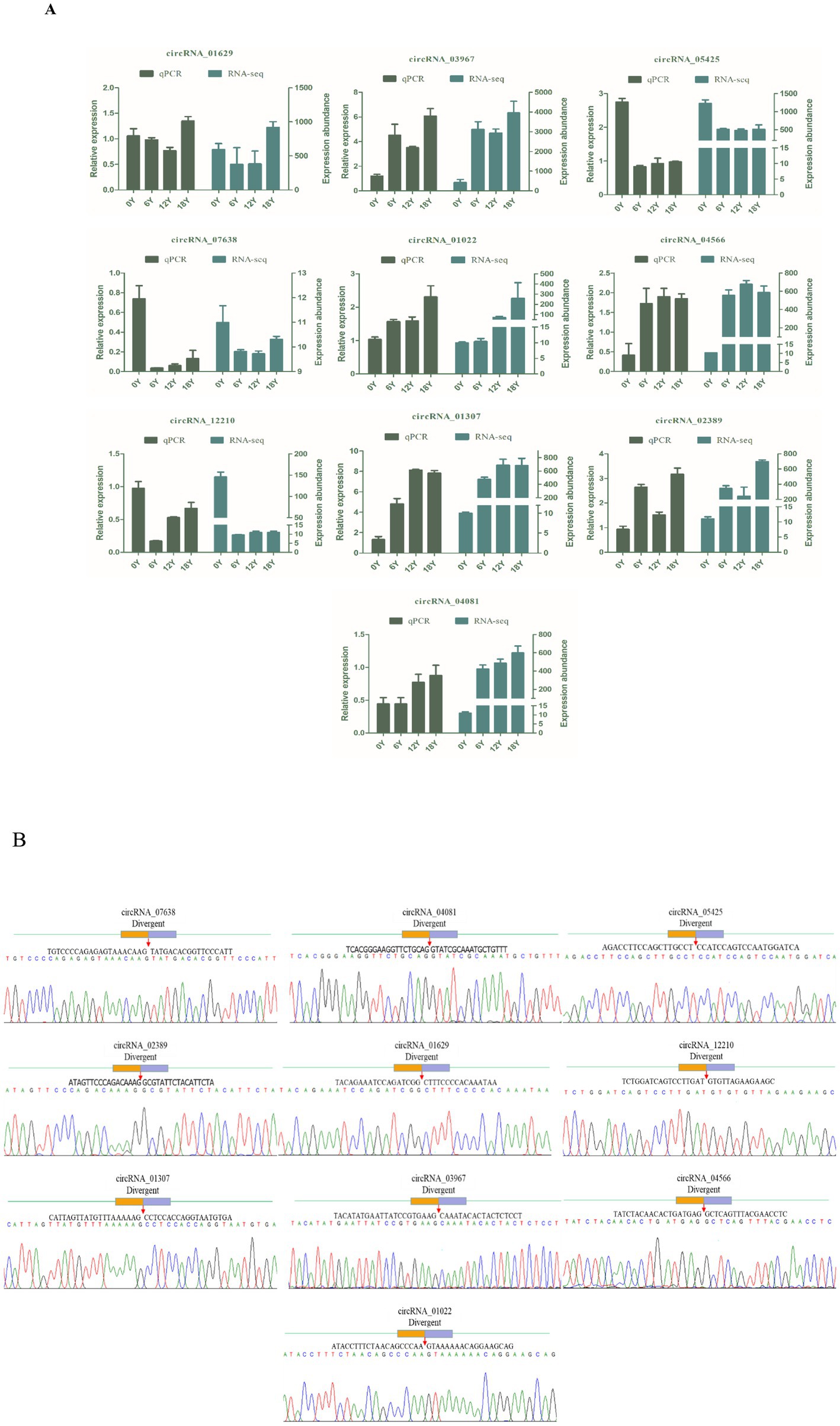
Figure 4. circRNA RT–qPCR validation and Sanger sequencing. (A) Comparison of qPCR and RNA-seq results for 10 circRNAs. (B) Circular junctions were confirmed by Sanger sequencing.
3.4. Functional enrichment and annotation analysis of host genes of DEcircRNAs
Analysis of the origin of the DEcircRNAs we obtained showed that in the 0Y vs. 6Y comparison, 3,729 of the 4,026 identified were from 2,221 source genes, with the remaining 297 circRNAs from intergenic regions. In the 6Y vs. 12Y group, 3,725 of the 4,030 DEcircRNAs identified were from 2,168 source genes and the remaining 305 circRNAs were from intergenic. In the 12Y vs. 18Y group, 2,963 of the 3,226 DEcircRNAs identified were from 1852 source genes and the remaining 263 circRNAs were from intergenic regions. In the 0Y vs. 18Y group, 3,760 of the 3,760 of the 3,497 DEcircRNAs identified were from 2040 source genes, with the remaining 263 circRNAs from intergenic regions. In the 0Y vs. 12Y group, 3601of the 3,878 DEcircRNAs identified were from 1885 source genes and the remaining 277 circRNAs were from intergenic regions. In the 6Y vs. 18Y group, 3428of the 3,721 DEcircRNAs identified were from 2034 source genes and the remaining 293 circRNAs were from intergenic regions.
The results of GO enrichment analysis showed that 0Y vs. 6Y, 0Y vs. 12Y and 0Y vs. 18Y were mainly involved in cilia assembly (GO:0060271), gonad development (GO:0008406), protein transport (GO:0015031) and regulation of cell morphogenesis (GO:0022604). In terms of cellular components, they mainly included centrosome (GO: 0005813), cytoplasm (GO: 0005737) and Golgi apparatus (GO: 0005794). In terms of molecular function, the main focus was on ATP binding (GO: 0005524), and microtubule binding (GO: 0008017) (Figures 5A,D,E). The source genes of the DEcircRNAs 6Y vs. 12Y, 6Y vs. 18Y and 12Y vs. 18Y were dominated by spermatogenesis (GO: 0007283), protein phosphorylation (GO: 0006468), cilia assembly (GO: 0060271), spermatocyte development (GO: 0007286), which was very similar to 0Y vs. 6Y in terms of cellular composition and molecular function (Figures 5B,C,F). The above terms such as GO:0008406, GO: 0007283 and GO: 0007286 are closely related to with gonad and germ cell development, indicating that the genes enriched in these cellular components may play an important role in testis development and spermatogenesis.
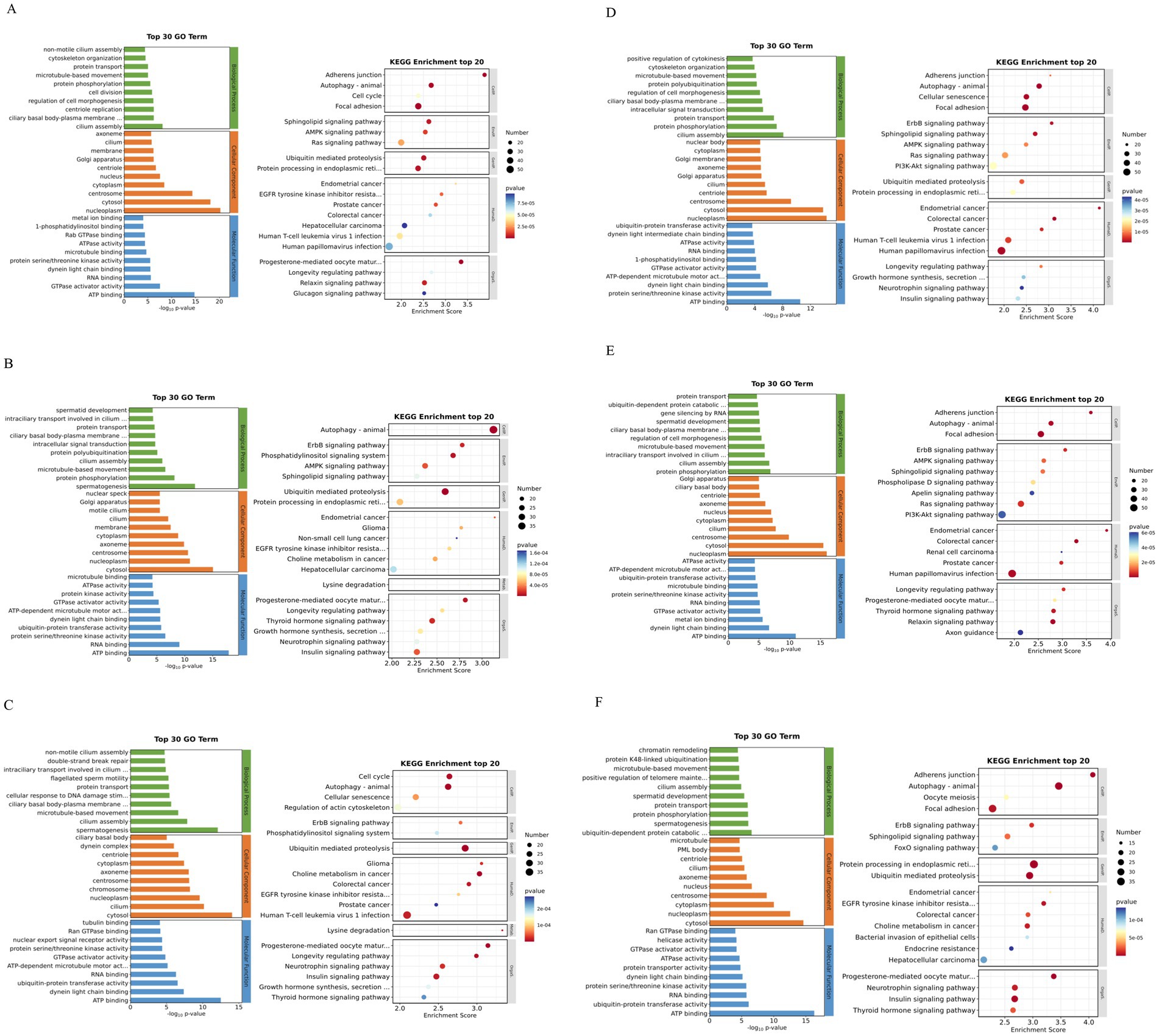
Figure 5. GO annotation and KEGG enrichment analysis of differential circRNA source genes in different control groups. (A) 0Y vs. 6Y, (B) 6Y vs. 12Y, (C) 12Y vs. 18Y, (D) 0Y vs. 18Y, (E) 0Y vs. 12Y, and (F) 6Y vs. 18Y; 0Y, 6Y, 12Y and 18Y refer to October, June, December and August, respectively.
KEGG enrichment results showed that the DEcircRNA-derived genes of 0Y vs. 6Y, 0Y vs. 12Y and 0Y vs. 18Y were mainly involved in Autophagy - animal (chx04140), Sphingolipid signaling pathway (chx04071), Longevity regulating pathway (chx04211), AMPK signaling pathway (chx04152), Ras signaling pathway (chx04014), and PI3K-Akt signaling pathway (chx04151) (Figures 5A,D,E); Host genes of DEcircRNA 6Y vs. 12Y, 6Y vs. 18Y and 12Y vs. 18Y were mainly involved in autophagy-animal (chx04140), ubiquitin mediated proteolysis (chx04120), phosphatidylinositol signaling system (chx04070), ErbB signaling pathway (chx04012), and thyroid hormone signaling pathway (chx04919) (Figures 5B,C,F). However, chx04152 (22), chx04014 (23) and chx04151 (24) are signaling pathways closely related to mammalian testicular development and spermatogenesis, suggesting that the differentially expressed genes in these pathways may be involved in the regulation of goat testicular development and spermatogenesis.
3.5. Bioinformatics analysis of ceRNA networks
ceRNAs are a regulatory mechanism that exists within organisms. Commonly circRNAs compete to bind miRNAs, inhibit their target mRNA translation or cause mRNA degradation, thereby regulating gene expression. Given the important of role circRNAs play in the ceRNA network, we bioinformatically predicted all DEcircRNAs in six comparison groups, yielding a total of 252 circRNA-miRNA relationship pairs and 3,722 miRNA-mRNA relationship pairs. We used 81 highly expressed DEcircRNAs, their corresponding target miRNAs and the mRNAs targeted by these miRNAs, to construct a regulatory network affecting testicular development (Figure 6A). In the ceRNA network, 66 candidate circRNAs, regulating 57 miRNA–mRNA relationship pairs, associated with spermatogenesis, they included 62 mRNAs and 22 miRNAs; 22 candidate were circRNAs were associated with germ cell development, regulating 10 miRNA–mRNA relationship pairs, including 9 mRNAs and 5 miRNAs; 43 were candidate circRNAs associated with the cell cycle, regulating 20 miRNA–mRNA relationship pairs, including 17 mRNAs and 11 miRNAs; and 36 candidate circRNAs associated with androgens, regulating 6 miRNA–mRNA relationship pairs, including 4 mRNAs and 6 miRNAs. Among them, sox8, KATNB1, STAG3, ZBTB7A and other mRNAs relevant to testicular development and spermatogenesis were identified. Finally, some important regulatory relationships linked to testicular development and spermatogenesis including were obtained through the ceRNA regulatory mechanism: circRNA_07172/ novel270_mature/sox8, circRNA_04859/novel270_mature/ sox8, circRNA_07832/chi-miR-433/KATNB1, circRNA_00032/novel24_mature/STAG3, circRNA_07510/chi-miR-133a-3p/ZBTB7A. We hypothesize that these circRNAs regulate genes associated with testicular development and spermatogenesis by competitive binding to their corresponding miRNAs and play important biological roles in the processes connected with testicular development and spermatogenesis.
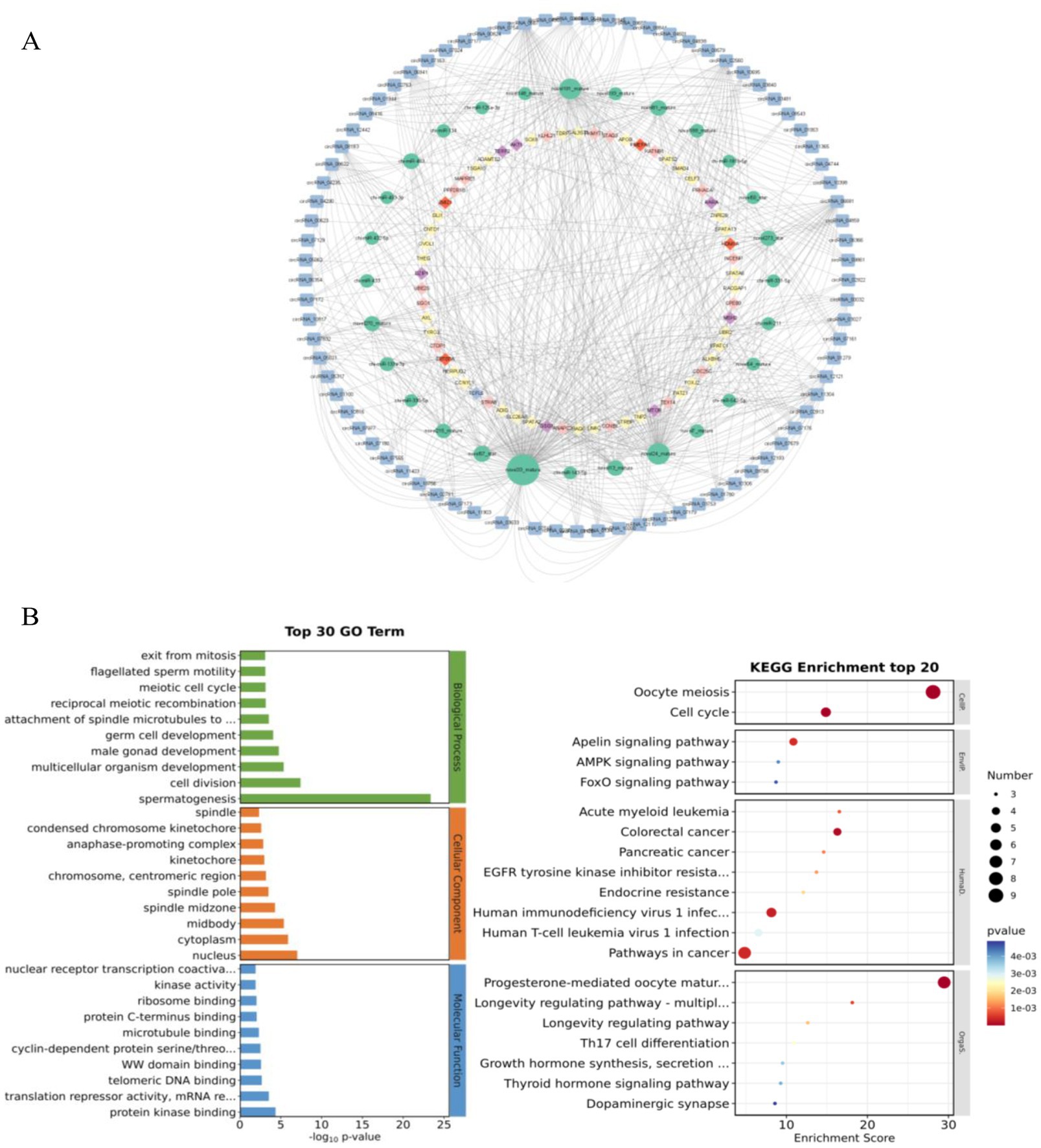
Figure 6. ceRNA network. (A) The ceRNA network related to testicular development and spermatogenesis in goats. (B) Enrichment analysis of genes in the ceRNA network by GO and KEGG.
The enrichment analysis of relational genes in ceRNA network by GO entry and KEGG pathway showed that kindred genes in the ceRNA network mainly participated in the cell composition (GO: 0005634), cytoplasm (GO: 0005737), chromosome and centromeric region (GO: 0000775). In terms of cell composition, it mainly includes nucleus (GO: 0005634), cytoplasm (GO: 0005737), chromosome and centromeric region (GO: 0000775); Molecular functions mainly focus on protein kinase binding (GO: 0019901,), telomeric DNA binding (GO: 0042162) and microtubule binding (GO: 008017); In terms of spermatogenesis, it mainly includes nucleus (GO: 007283), cell division (GO: 0051301), germ cell development (GO: 007281,) and meiotic cell cycle (GO: 0051321) in biological process. Among them, sox8, KATNB1 and testicular development cognate genes were significantly enriched in GO: 007283 and GO: 0051301 (p<0.05) (Figure 6B). These results suggest that these GO functional genes may further regulate the development of goat testes.
KEGG showed that the related genes in the ceRNA network were mainly involved in meiosis (chx04114), cell cycle (chx04110), synthesis, secretion and function of growth hormone (chx04141), AMPK signal pathway (chx04935), and FoxO signal pathway (chx04068). This is consistent with GO entry of the DECircRNA source gene and enrichment analysis of the KEGG pathway (Figure 6B). Among them, chx04935 (25), chx04068 (26) and chx04141 (27) have been identified as regulatory pathways interrelated to testicular development and spermatogenesis. Akt3, the source gene of circRNA_03787 and circRNA_03789 in the chx04935 pathway, is involved in the regulation of SSCS self-renewal, proliferation and differentiation, and is essential for spermatogenesis in male animals (28, 29). In conclusion, based on the function of the source gene, circRNA may play an important role in goat testicular development and spermatogenesis.
4. Discussion
The testis is the most important reproductive organ in mammals and its main functions are sperm production and androgen secretion (30). As abundant noncoding RNAs in the eukaryotic transcriptome, circRNAs are important regulators of gene expression and miRNA function at the transcriptional, posttranscriptional and translational levels (31). They are important regulators of gene expression and miRNA function. In recent years, high-throughput RNA sequencing and bioinformatics analysis have revealed that circRNAs located in the cytoplasm can compete with mRNAs for the target binding sites of miRNAs, thereby regulating the expression of mRNAs. For example, 1,432 DEcircRNAs were identified in the spermatozoa of patients with weak spermatozoa, and circCIT, circUSP54, circTRMT2B and circTADA2A were identified as marker circRNAs for weak spermatozoa (32). RNA-Seq of Qinchuan cattle at different ages revealed that 2,225 circRNAs were upregulated and 2023 were downregulated in adult cattle testes, and these differentially expressed circRNAs may be involved in testicular development or spermatogenesis in Qinchuan cattle (33). In goats, it was demonstrated that circP13 significantly promoted differentiation and inhibited apoptosis of primary myogenic cells in goats (34), and circRNA8073 indirectly inhibited apoptosis of endometrial epithelial cells, which was beneficial to embryo implantation in goats (35).
In this study, Qianbei Ma goats were selected as the research object. H&E staining and RNA-seq were used to explore the development of the testes of Qianbei Ma goats at different age stages, and key candidate genes and signaling pathways affecting the growth and development of testes of Qianbeima goat were screened. We analyzed the testicular tissue sections of Qianbei Ma goats at different developmental stages and found that the circumference and area of the tubules of the curved sperm increased with age. Sertoli cells in testis increased and Leydig cells decreased, but the number of Sertoli cells and Leydig cells peaked at 6 months of age and maintained a dynamic balance, which was similar to other animal testicular histomorphology studies (36).
We used RNA-seq sequencing technology to further investigate the molecular mechanisms regulating testicular development and spermatogenesis in Qianbei Ma goatS. A total of 12,874 circRNAs were identified from testicular tissues at four different developmental periods. 8,140 differentially expressed circRNAs were detected in six comparison groups of the comparison group, with 4 circRNAs differentially expressed in all four comparison groups.
To explore the role of circRNAs in the testicular development, the source genes of DEcircRNAs were functionally analyzed and GO functional annotation revealed that the source genes of DEcircRNAs were mainly involved in biological processes such as development and reproduction. For example, circRNA_12624 in 0Y vs. 6Y, 0Y vs. 12Y and 0Y vs. 18Y was derived from the ATG5 gene. ATG5 is a cytoplasmic protein mainly expressed in the nucleus, which can be used as a key factor to induce autophagy, regulate cell growth cycle and spermatogenesis of male animals (37–39).The 10 DEcircRNAs in 6Y vs. 12Y, 6Y vs. 18Y and 12Y vs. 18Y were derived from four genes encoding genes associated with germ cell development and spermatogenesis [PTBP2 (40), PRMT5 (41), PLK4, SYCE1 (42)]. Of these, circRNA_04355, circRNA_04356 and circRNA_04357 were all derived from the PLK4 gene, which has been shown to play a role in the initiation of spermatogenesis (43). Some studies also pointed out that PLK4 mutation is likely to cause male pentaspermia and affect fertility (44). In addition, circRNA_06790 is produced from the CTNNB1 gene, which is a key factor in regulating germ cell growth and plays an important role in the stage of differentiation from spermatogonia to spermatocytes to ensure sound fertility in animals (45). In summary, the DEcircRNAs we identified may play important roles in a range of processes related to testicular development, including spermatogenesis, germ cell proliferation, androgen secretion and the cell cycle.
The KEGG analysis of the source genes of DE circRNA showed that in the comparison group of six comparison groups of 0Y vs. 6Y, 0Y vs. 18Y, 6Y vs. 12Y, 12Y vs. 18Y, 0Y vs. 12Y and 6Y vs. 18Y, the Ras, AMPK, Hippo, cAMP, TGF-BETA, PI3K-Akt, and Wnt signaling pathways were enriched in a series of signaling pathways associated with testicular development and spermatogenesis. For example, the Ras signaling pathway participates in the regulation of the CREB family member phosphorylation process, thus promoting the proliferation of spermatogonial stem cells (46); the AMPK/mTOR signaling pathway is involved in regulating autophagy and apoptosis of Sertoli cells. AMPK can also reduce the toxicity of ROS by enhancing the activity of antioxidant enzymes, thus ensuring spermatogenesis in male animals (47). Activation of the YAP1 protein in the Hippo signal pathway can effectively promote the proliferation of testicular germ cells and spermatogenesis (2). The PI3K/Akt signal transduction pathway can promote testosterone secretion through androgen receptor (AR) (48), and the Wnt signal pathway can increase the expression of StarD7, and improve the process of testosterone secretion in rat Leydig cells (49). Therefore, the DEcircRNA identified in this study is likely to participate in the biological process of regulating testicular development and spermatogenesis in Qianbei Ma goats.
The ceRNA (competing endogenous RNAs) hypothesis has revealed a new mechanism for endogenous interactions between Rnas. Relevant studies have been conducted in cattle (50), goats (33) and pigs (51). circBDP1 can regulate the proliferation and differentiation of bovine adipose cells through the mir-204/TRARG1 pathway (50). circTUT7 influences the development process of porcine embryonic muscle by regulating the expression of HMG20B in the ceRNA mechanism (51). To reveal the functions involved in DEcircRNAs in this study, 81 DEcircRNAs with high expression levels were selected from four aspects of germ cell development, spermatogenesis, androgen and cell cycle, and their corresponding target miRNAs and mRNAs with high scores were predicted. The ceRNA regulatory network affecting testicular development and spermatogenesis was constructed. sox8 may be regulated by circRNA_07172/novel81_mature and circRNA_04859/novel270_mature pairs in the network. novel81_mature and novel270_mature are novel miRNAs identified in this study, and there are no related studies. sox8 is a product of adult Sertoli cells. It is a regulatory component of the testicular barrier and a key factor in maintaining normal fertility in male animals (52). Its elimination not only leads to the disorder of the spermatogenic cycle, but also causes abnormal motility of sperm in the epididymis, which affects the reproductive ability of animals (53). STAG3, a protein encoding meiotic condensation complex, can regulate germ cell development and is a target gene of the circRNA_00032/novel24_mature regulatory axis in the network (54). Previous studies have found that STAG3 gene mutations can cause male infertility due to meiotic arrest, and STAG3 knockout can cause azoospermia in male mice (55, 56). Chi-miR-433 in the regulatory axis of circRNA_07832/chi-miR-433/KATNB1 has been confirmed to participate in animal growth and development processes such as goat hair follicle development and muscle differentiation, but its role in testicular development and spermatogenesis has rarely been studied (57, 58). In this study, the target gene KATNB1 regulated by circRNA_07832/chi-miR-433 is a key molecule involved in testicular development. It can regulate the growth, development and reproduction of mammals by regulating the cycle of fibroblasts and spermatocytes. (59). ZBTB7A is a transcription inhibitor belonging to the POK (POZ/BTB and Kruppel) protein family (60). Studies have shown that its family proteins ZBTB42 and ZBTB40 participate in biological processes such as animal germ cell development and sperm maturation (61, 62). In this study, ZBTB7A was regulated by the circRNA_07510/chi-miR-133a-3p relationship pair. Therefore, circRNA_07172, circRNA_04859, circRNA_00032, circRNA_07832, and circRNA_07510 are closely related to testicular development and spermatogenesis. They may inhibit the activity of miRNA and promote the expression of mRNA by binding to miRNA response elements (MREs), thereby affecting the biological process of testicular development and spermatogenesis, but the mechanism needs to be further studied.
After functional enrichment analysis of the relational genes in the ceRNA Network, GO entry analysis showed that the relevant genes were distributed to several major biological processes, including spermatogenesis, testicular development, cellular processes and reproduction. This was consistent with the enrichment entry of the source gene GO. The enrichment results of the KEGG pathway analysis showed that the corresponding genes were mainly enriched in the MAPK signaling pathway, cAMP signaling pathway, Wnt signaling pathway, Ras signaling pathway, and PI3K-Akt signaling pathways, which are related to testicular development and spermatogenesis. Studies have shown that the WNT signaling pathway can induce morphological changes and cell movement in mouse and human spermatogonia through its main member WNT3A triggering membrane extension (63). The cAMP signaling pathway can regulate the proliferation of sertoli cells through tight connections and adhesive connections, and Sertoli cells participate in the formation of the testicular barrier to ensure the stability of microenvironment in seminiferous tubules and promote spermatogenesis (47). Many studies have also demonstrated that the PI3K-Akt signaling pathway plays an important role in testicular development and spermatogenesis in animals (64). For example, Meroni S et al. detected that FSH positively regulates the proliferation of Sertoli cells by activating the PI3K-dependent pathway to increase the transcriptional activity of HIF 2 (hypoxia-inducible factor) and cyclin D1 expression, thus promoting spermatogenesis (65).Therefore, the above analysis indicates that the genes in the ceRNA network are inextricably linked to testicular development and spermatogenesis, and these circRNAs also play an important role in testicular development and spermatogenesis in Qianbei Ma goats, but their specific regulatory mechanisms need to be further investigated.
5. Conclusion
In this study, we found that the circumference and area of seminiferous tubules and the number of sperm in the testis of Qianbei Ma goats increased with age. A total of 12,784 circRNAs were identified in the testis of Qianbei Ma goats, and 8,140 DEcircRNAs were obtained from six comparison groups(0Yvs6Y, 6Yvs12Y, 12Yvs18Y, 0Yvs18Y, 0Y vs. 12Y, 6Y vs. 18Y). The genes of origin of these DEcircRNAs are mainly involved in spermatogenesis and testicular development. circRNA_07172, circRNA_04859, circRNA_00032, circRNA_07832 and circRNA_07510 may play key roles in testicular development and spermatogenesis in goats. This study enriches the existing database of circRNAs in goat testis development and spermatogenesis.
For additional requirements for specific article types and further information please refer to “Article types” on every Frontiers journal page.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA917820.
Ethics statement
The studies involving animals have been reviewed and approved by the Animal Protection and Use Committee of Guizhou University, Guiyang, China (Approval number: EAE-GZU-2021-E021). The animal handling procedures were in line with the Chinese Animal Welfare Guidelines and were approved by the Animal Protection and Use Committee of Guizhou University, Guiyang, China (Approval number: EAE-GZU-2021-E021).
Author contributions
WT and XC conceived and designed the experiments. WT, KF, WG, and ZA conducted the experiments. WT and WG analyzed the data. WT wrote the paper. XC, WG, and ZA revised the paper. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by the National Natural Science Foundation of China (32260835), the Science and Technology Project of Guizhou Province [Qian Kehe Foundation—ZK (2021) General 151], and the Guizhou High-Level Innovative Talents Project [Qian Kehe Platform Talents (2022) 021–1].
Acknowledgments
We thank XC and QX for their help and support, and Guizhou University for providing the experimental platform.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
1. ^https://www.ncbi.nlm.nih.gov/genome/?term=Capra+hircus
2. ^http://kobas.cbi.pku.edu.cn/
References
1. Mueller, J, Getachew, T, Rekik, M, Rischkowsky, B, Abate, Z, Wondim, B, et al. Converting multi-trait breeding objectives into operative selection indexes to ensure genetic gains in low-input sheep and goat breeding programmes. Animal. (2021) 15:100198. doi: 10.1016/j.animal.2021.100198
2. Zhang, G-M, Zhang, T-T, An, S-Y, El-Samahy, MA, Yang, H, Wan, Y-J, et al. Expression of hippo signaling pathway components in Hu sheep male reproductive tract and spermatozoa. Theriogenology. (2019) 126:239–48. doi: 10.1016/j.theriogenology.2018.12.029
3. MacLaren, AP. Ram fertility in south-West Scotland. Br Vet J. (1988) 144:45–54. doi: 10.1016/0007-1935(88)90151-0
4. Zhong, Y, Du, Y, Yang, X, Mo, Y, Fan, C, Xiong, F, et al. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol Cancer. (2018) 17:79. doi: 10.1186/s12943-018-0827-8
5. Luo, H, Lv, W, Tong, Q, Jin, J, Xu, Z, and Zuo, B. Functional non-coding RNA during embryonic Myogenesis and postnatal muscle development and disease. Front Cell Dev Biol. (2021) 9:628339. doi: 10.3389/fcell.2021.628339
6. Santos, RM, Moreno, C, and Zhang, WC. Non-coding RNAs in lung tumor initiation and progression. Int J Mol Sci. (2020) 21:2774. doi: 10.3390/ijms21082774
7. Li, X, Yang, L, and Chen, LL. The biogenesis, functions, and challenges of circular RNAs. Mol Cell. (2018) 71:428–42. doi: 10.1016/j.molcel.2018.06.034
8. Zhao, XW, Huang, DW, Zhu, HL, Pan, XC, Wang, XX, Qi, YX, et al. Alterations of the circular RNA profile in the jejunum of neonatal calves in response to colostrum and milk feeding. J Dairy Sci. (2019) 102:7038–48. doi: 10.3168/jds.2018-15942
9. Huang, K, Chen, M, Zhong, D, Luo, X, Feng, T, Song, M, et al. Circular RNA profiling reveals an abundant circEch1 that promotes Myogenesis and differentiation of bovine skeletal muscle. J Agric Food Chem. (2021) 69:592–601. doi: 10.1021/acs.jafc.0c06400
10. Khan, IM, Liu, H, Zhuang, J, Khan, NM, Zhang, D, Chen, J, et al. Circular RNA expression and regulation profiling in testicular tissues of immature and mature Wandong cattle (Bos taurus). Front Genet. (2021) 12:685541s. doi: 10.3389/fgene.2021.685541
11. Xin, S, Lv, X, Wang, L, Zheng, WW, Xu, T, and Sun, Y. Circular RNA circRara promote the innate immune responses in miiuy croaker, Miichthys miiuy. Fish Shellfish Immunol. (2022) 128:557–64. doi: 10.1016/j.fsi.2022.08.026
12. Li, B, He, Y, Wu, W, Tan, X, Wang, Z, Irwin, DM, et al. Circular RNA profiling identifies novel circPPARA that promotes intramuscular fat deposition in pigs. J Agric Food Chem. (2022) 70:4123–37. doi: 10.1021/acs.jafc.1c07358
13. Legnini, I, Timoteo, GD, Rossi, F, Morlando, M, Briganti, F, Sthandier, O, et al. Circ-ZNF609 is a circular RNA that can be translated and functions in Myogenesis. Mol Cell. (2017) 66:22–37.e9. doi: 10.1016/j.molcel.2017.02.017
14. Holdt, LM, Stahringer, A, Sass, K, Pichler, G, Kulak, NA, Wilfert, W, et al. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat Commun. (2016) 7:12429. doi: 10.1038/ncomms12429
15. Liu, Y, Wang, P, Zhou, Z, He, X, Tao, L, Jiang, Y, et al. Expression profile analysis to identify circular RNA expression signatures in the prolificacy trait of Yunshang black goat pituitary in the estrus cycle. Front Genet. (2021) 12:801357. doi: 10.3389/fgene.2021.801357
16. Zhang, L, Liu, X, Ma, X, Liu, Y, Che, S, Cui, J, et al. Testin was regulated by circRNA3175-miR182 and inhibited endometrial epithelial cell apoptosis in pre-receptive endometrium of dairy goats. J Cell Physiol. (2018) 233:6965–74. doi: 10.1002/jcp.26614
17. Ao, Y, Chen, X, Zhou, Z, Luo, J, Wei, S, Zhang, Y, et al. TGFβ2 and PPP3CA gene expression in gonadal ax is tissue and regulation of ovarian granulosa cells in Qianbei Ma goat. J Anim Husbandry Vet Med. (2020) 11:2699–709. doi: 10.11843/j.issn.0366-6964.2020.11.009
18. Dong, Y, Xie, M, Jiang, Y, Xiao, N, Du, X, Zhang, W, et al. Sequencing and automated whole-genome optical mapping of the genome of a domestic goat (Capra hircus). Nat Biotechnol. (2013) 31:135–41. doi: 10.1038/nbt.2478
19. Memczak, S, Jens, M, Elefsinioti, A, Torti, F, Krueger, J, Rybak, A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. (2013) 495:333–8. doi: 10.1038/nature11928
20. Zhang, X, Yuan, Z, Li, F, and Yue, X. Integrating transcriptome and metabolome to identify key genes regulating important muscular flavour precursors in sheep. Animal. (2022) 16:100679. doi: 10.1016/j.animal.2022
21. Ishida, T, Tagatz, GE, and Okagaki, T. Gonadoblastoma: ultrastructural evidence for testicular origin. Case Rep. (2015) 37:1770–81. doi: 10.1002/1097-0142(197604)37
22. Salazar, G, Dong, L, Liao, C, Batkiewicz, L, Arbing, R, Chung, S, et al. Apoptosis in male germ cells in response to cyclin A1-deficiency and cell cycle arrest. Biochem Pharmacol. (2003) 66:1571–9. doi: 10.1016/s0006-2952(03)00513-6
23. Liu, ZJ, Liu, YH, Huang, SY, and Zang, ZJ. Insights into the regulation on proliferation and differentiation of stem Leydig cells.J. Stem Cell Rev. (2021) 17:1521–33. doi: 10.1007/s12015-021-10133-x
24. Kumar, S, Lee, HJ, Park, HS, and Lee, K. Testis-specific GTPase (TSG): an oligomeric protein. BMC Genomics. (2016) 17:1–10. doi: 10.1186/s12864-016-3145-9
25. Hga, B, Yo, A, Hy, A, Hcab, C, Hda, B, Hl, A, et al. Induction of autophagy via the ROS-dependent AMPK-mTOR pathway protects copper-induced spermatogenesis disorder. Redox Biol. (2021) 49:102227. doi: 10.1016/j.redox.2021.102227
26. Goertz, MJ, Wu, Z, Gallardo, TD, Hamra, FK, and Castrillon, DH. Foxo1 is required in mouse spermatogonial stem cells for their maintenance and the initiation of spermatogenesis. J Clin Invest. (2011) 121:3456–66. doi: 10.1172/JCI57984
27. Yuka, K, and Seiji, F. X-linked hypophosphatemia and FGF23-related Hypophosphatemic diseases: Prospect for new treatment. Endocr Rev. (2018) 39:274–91. doi: 10.1210/er.2017-00220
28. Sharma, M, and Braun, RE. Cyclical expression of GDNF is required for spermatogonial stem cell homeostasis. Development. (2018) 145:dev.151555. doi: 10.1242/dev.151555
29. Jia, H, Ma, T, Jia, S, and Ouyang, YJAPC. AKT3 and related molecules as potential biomarkers responsible for cryptorchidism and cryptorchidism-induced azoospermia. Transl Pediatr. (2021) 10:1805–17. doi: 10.21037/tp-21-31
30. La, Y, Ma, X, Bao, P, Chu, M, Yan, P, Guo, X, et al. Identification and characterization of Piwi-interacting RNAs for early testicular development in yak. Int J Mol Sci. (2022) 23:12320. doi: 10.3390/ijms232012320
31. Ebbesen, KK, Hansen, TB, and Kjems, J. Insights into circular RNA biology. RNA Biol. (2022) 14:1035–45. doi: 10.1080/15476286.2016.1271524
32. Manfrevola, F, Chioccarelli, T, Cobellis, G, Fasano, S, Ferraro, B, Sellitto, C, et al. CircRNA role and circRNA-dependent network (ceRNET) in Asthenozoospermia. Front Endocrinol (Lausanne). (2020) 11:395. doi: 10.3389/fendo.2020.00395
33. Gao, Y, Wu, M, Fan, Y, Li, S, Lai, Z, Huang, Y, et al. Identification and characterization of circular RNAs in Qinchuan cattle testis. R Soc Open Sci. (2018) 5:180413. doi: 10.1098/rsos.180413;180413
34. Zhang, Z, Fan, Y, Deng, K, Liang, Y, Zhang, G, Gao, X, et al. Circular RNA circUSP13 sponges miR-29c to promote differentiation and inhibit apoptosis of goat myoblasts by targeting IGF1. FASEB J. (2022) 36:e22097. doi: 10.1096/fj.202101317R
35. Zhang, L, Liu, X, Che, S, Cui, J, Ma, X, An, X, et al. Endometrial epithelial cell apoptosis is inhibited by a ciR8073-miR181a-Neurotensis pathway during embryo implantation. Mol Ther Nucleic Acids. (2019) 14:262–73. doi: 10.1016/j.omtn.2018.12.005
36. Tao, L T. (2021)Transcriptomics-based identification of functional genes and their expression regulation in Tibetan sheep testes. PHD. GanSu, China: GanSu Agricultal University.
37. Li, H, Peng, X, Wang, Y, Cao, S, Xiong, L, Fan, J, et al. Atg5-mediated autophagy deficiency in proximal tubules promotes cell cycle G2/M arrest and renal fibrosis. Autophagy. (2016) 12:1472–86. doi: 10.1080/15548627.2016.1190071
38. Maskey, D, Yousefi, S, Schmid, I, Zlobec, I, Perren, A, Friis, R, et al. ATG5 is induced by DNA-damaging agents and promotes mitotic catastrophe independent of autophagy. Nat Commun. (2013) 4:2130. doi: 10.1038/ncomms3130
39. Huang, Q, Liu, Y, Zhang, S, Yap, YT, Li, W, Zhang, D, et al. Autophagy core protein ATG5 is required for elongating spermatid development, sperm individualization and normal fertility in male mice. Autophagy. (2021) 17:1753–67. doi: 10.1080/15548627
40. Feng, S, Li, J, Wen, H, Liu, K, Gui, Y, Wen, Y, et al. hnRNPH1 recruits PTBP2 and SRSF3 to modulate alternative splicing in germ cells.Nature. Communications. (2022) 13:3588. doi: 10.1038/s41467-022-31364-7
41. Chu, Z, Niu, B, Zhu, H, He, X, Bai, C, Li, G, et al. PRMT5 enhances generation of induced pluripotent stem cells from dairy goat embryonic fibroblasts via down-regulation of p53. Cell Prolif. (2015) 48:29–38. doi: 10.1111/cpr.12150
42. Ren, F, Xi, H, Qiao, P, Li, Y, Xian, M, Zhu, D, et al. Single-cell transcriptomics reveals male germ cells and Sertoli cells developmental patterns in dairy goats.Front cell. Dev Biol. (2022) 10:944325. doi: 10.3389/fcell.2022.944325
43. Harris, RM, Weiss, J, and Jameson, JL. Male Hypogonadism and germ cell loss caused by a mutation in polo-like kinase 4. Endocrinology. (2011) 152:3975–85. doi: 10.1210/en.2011-1106
44. Miyamoto, T, Bando, Y, Koh, E, Tsujimura, A, Miyagawa, Y, Iijima, M, et al. A PLK4 mutation causing azoospermia in a man with Sertoli cell-only syndrome. Andrology. (2016) 4:75–81. doi: 10.1111/andr.12113
45. Chassot, AA, Le Rolle, M, Jourden, M, Taketo, MM, Ghyselinck, NB, and Chaboissier, MC. Constitutive WNT/CTNNB1 activation triggers spermatogonial stem cell proliferation and germ cell depletion. Dev Biol. (2017) 426:17–27. doi: 10.1016/j.ydbio.2017.04.010
46. He, ZP, Jiang, JJ, Kokkinaki, M, Golestaneh, N, and Marie, CH. Gdnf Upregulates c-Fos transcription via the Ras/Erk1/2 pathway to promote mouse Spermatogonial stem cell proliferation. Stem Cells. (2008) 26:266–78. doi: 10.1634/stemcells.2007-0436
47. Zhang, X, Hu, ZT, Li, Y, Li, YX, Xian, M, Guo, SM, et al. Effect of Astragalus polysaccharides on the cryopreservation of goat semen. Theriogenology. (2022) 193:47–57. doi: 10.1016/j.theriogenology.2022.08.007
48. Li, D, Wang, Q, Shi, K, Lu, Y, Yu, D, Shi, X, et al. Testosterone promotes the proliferation of chicken embryonic myoblasts via androgen receptor mediated PI3K/Akt signaling pathway. Int J Mol Sci. (2020) 21:1152. doi: 10.3390/ijms21031152
49. Fu, H, Jing, J, Yi, N, Yu, S, Yun, S, Liang, Y, et al. Effect of StarD7 and Wnt/β-catenin signal pathway on the testosterone secretion stimulated by Annexin 5 in rat Leydig cells. J Peking Univ. Health Sci. (2012) 44:518–23.
50. Zhang, S, Jiang, E, Kang, Z, Bi, Y, Liu, H, Xu, H, et al. CircRNA profiling reveals an abundant circBDP1 that regulates bovine fat development by sponging miR-181b/miR-204 targeting Sirt1/TRARG1. J Agric Food Chem. (2022) 70:14312–28. doi: 10.1021/acs.jafc.2c05939
51. Hong, L, Gu, T, He, Y, Zhou, C, Hu, Q, Wang, X, et al. Genome-wide analysis of circular RNAs mediated ceRNA regulation in porcine embryonic muscle development. Front Cell Dev Biol. (2019) 7:289. doi: 10.3389/fcell.2019.00289
52. Kennedy, CL, Koopman, P, and Mishina, Y. Sox8 and Sertoli-cell function. Ann N Y Acad Sci. (2007) 1120:104–13. doi: 10.1196/annals.1411.007
53. Takada, S, Kennedy, CL, Scott, G, Harada, SI, Ray, MK, Dai, Q, et al. Sox8 is a critical regulator of adult Sertoli cell function and male fertility. Dev Biol. (2008) 316:359–70. doi: 10.1016/j.ydbio.2008.01.042
54. Zhang, LF, Jing, W, Li, XH, Liu, MF, Shi, HJ, DeLeon, M, et al. PHB regulates meiotic recombination via JAK2-mediated histone modifications in spermatogenesis. Nucleic Acids Res. (2020) 48:4780–96. doi: 10.1093/nar/gkaa203
55. Nam, Y, Kang, KM, Sung, SR, Park, JE, Shin, YJ, and Song, SH. The association of stromal antigen 3(STAG3) sequence variations with spermatogenic impairment in the male Korean population. Asian J Androl. (2020) 22:106–11. doi: 10.4103/aja.aja_28_19
56. Bijl, NVD, Rpke, A, Biswas, U, Wste, M, and Tüttelmann, F. Mutations in the stromal antigen 3 (STAG3) gene cause male infertility due to meiotic arrest. Hum Reprod. (2019) 34:2112–9. doi: 10.1093/humrep/dez204
57. Ye, J, Zhao, X, Xue, H, Zou, X, Liu, G, Deng, M, et al. RNA-Seq reveals miRNA and mRNA co-regulate muscle differentiation in fetal Leizhou goats. Front Vet Sci. (2022) 9:829769. doi: 10.3389/fvets.2022.829769
58. Ma, R, Shang, F, Rong, Y, Pan, J, Wang, M, Niu, S, et al. Expression profile of long non-coding RNA in inner Mongolian cashmere goat with putative roles in hair follicles development. Front Vet Sci. (2022) 9:995604–4. doi: 10.3389/fvets.2022.995604
59. Pleuger, C, Fietz, D, Hartmann, K, Weidner, W, Kliesch, S, O'Bryan, MK, et al. Expression of katanin p80 in human spermatogenesis. Fertil Steril. (2016) 106:1683–1690.e1. doi: 10.1016/j.fertnstert.2016.08.043
60. Cui, J, Yang, Y, Zhang, C, Hu, P, Kan, W, Bai, X, et al. FBI-1 functions as a novel AR co-repressor in prostate cancer cells. Cell Mol Life Sci. (2011) 68:1091–103. doi: 10.1007/s00018-010-0511-7
61. Wu, X, Yang, Y, Zhong, C, Guo, Y, Li, S, Lin, H, et al. Transcriptome profiling of laser-captured germ cells and functional characterization of zbtb40 during 17alpha-methyltestosterone-induced spermatogenesis in orange-spotted grouper (Epinephelus coioides). BMC Genomics. (2020) 21:73–13. doi: 10.1186/s12864-020-6477-4
62. Rocha, DR, Martins, JAM, van Tilburg, MF, Oliveira, RV, Moreno, FB, Monteiro-Moreira, ACO, et al. Effect of increased testicular temperature on seminal plasma proteome of the ram. Theriogenology. (2015) 84:1291–305. doi: 10.1016/j.theriogenology.2015.07.008
63. Golestaneh, N, Beauchamp, E, Fallen, S, Kokkinaki, M, Uren, A, and Dym, M. Wnt signaling promotes proliferation and stemness regulation of spermatogonial stem/progenitor cells. Reproduction. (2009) 138:151–62. doi: 10.1530/REP-08-0510
64. Yang, T, and Yang, W. The dynamics and regulation of microfilament during spermatogenesis. Gene. (2020) 744:144635. doi: 10.1016/j.gene.2020.144635
Keywords: testicular development, spermatogenesis, circRNA, Qianbei Ma goats, ceRNA
Citation: Tang W, Xu QH, Chen X, Guo W, Ao Z, Fu K, Ji T, Zou Y, Chen JJ and Zhang Y (2023) Transcriptome sequencing reveals the effects of circRNA on testicular development and spermatogenesis in Qianbei Ma goats. Front. Vet. Sci. 10:1167758. doi: 10.3389/fvets.2023.1167758
Edited by:
M. Sofia Ortega, University of Wisconsin-Madison, United StatesReviewed by:
Xiaoyun He, Chinese Academy of Agricultural Sciences, ChinaXiangpeng Yue, Lanzhou University, China
Copyright © 2023 Tang, Xu, Chen, Guo, Ao, Fu, Ji, Zou, Chen and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qiang Hou Xu, Z3pkeHhocUAxNjMuY29t; Xiang Chen, eGNoZW4yQGd6dS5lZHUuY24=
 Wen Tang
Wen Tang Qiang Hou Xu1,2,3,4*
Qiang Hou Xu1,2,3,4* Xiang Chen
Xiang Chen Wei Guo
Wei Guo