
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci. , 16 November 2023
Sec. Zoological Medicine
Volume 10 - 2023 | https://doi.org/10.3389/fvets.2023.1161093
This article is part of the Research Topic Wildlife Parasitology: Emerging Diseases and Neglected Parasites View all 20 articles
 Ezequiel Hidalgo-Hermoso1*
Ezequiel Hidalgo-Hermoso1* Paulina Sepúlveda-García2,3
Paulina Sepúlveda-García2,3 Javier Cabello4
Javier Cabello4 Sebastian Celis5
Sebastian Celis5 Carola Valencia6
Carola Valencia6 Carolina Ortiz5
Carolina Ortiz5 Ignacio Kemec5
Ignacio Kemec5 Dario Moreira-Arce7,8
Dario Moreira-Arce7,8 Miguel Orsola9
Miguel Orsola9 Nivia Canales10
Nivia Canales10 Antonio Garnham11
Antonio Garnham11 Frank Vera6
Frank Vera6 Ananda Muller12,13
Ananda Muller12,13Introduction: Recent evidence shows a high diversity of infectious agents in wildlife that represent a threat to human, domestic, and wild animal health. In Chile, wild populations of the most common cervid species, pudu (Pudu puda), have been reported as hosts for novel pathogens such as Mycoplasma ovis-like and a novel ecotype of Anaplasma phagocytophilum. A better understanding of the epidemiology of this group and other intracellular bacteria that might have cervids as hosts would enlighten their population relevance. This study aimed to determine the occurrence and genetic diversity of Bartonella spp., hemotropic mycoplasmas, and Coxiella burnetii in pudus from Chile.
Methods: The DNA was extracted from the blood samples of 69 wild free-ranging and 30 captive pudus from Chile. A combination of real-time (nouG gene for Bartonella and IS1111 element for C. burnetii) and conventional PCR (16S rRNA for hemotropic Mycoplasma spp. and rpoB, gltA, and ITS for Bartonella spp.) was used for pathogen screening and molecular characterization.
Results: DNA of Bartonella spp. was detected in 10.1% [95% CI (5.2–18.2%)] samples, hemotropic Mycoplasma spp. in 1.7% [95% CI (0.08–10.1%)], and C. burnetii in 1.0% [95% CI (0.05–6.3%)] samples. Two sequenced samples were identified as Mycoplasma ovis-like, and one free-ranging pudu was positive for C. burnetii. While one captive and two free-ranging pudus were positive for Bartonella henselae, one wild pudu was co-positive for B. henselae and Bartonella sp., similar to Bartonellae identified in ruminants.
Discussion: To the best of our knowledge, this is the first report of B. henselae in wild ungulate species, and C. burnetii and Bartonella spp. in wild ungulate species in South America. Further research will be necessary to evaluate the potential role of pudu as reservoirs of infection and identify the sources for disease transmission among humans and wild and domestic animals.
The recognition of the role of wildlife as reservoirs of pathogens that threaten the health of humans and/or livestock species has increased in the past several decades; accordingly, the relevance of infectious agents in the wildlife conservation field has also amplified (1, 2). As expected, there are differences between regions of the world. In South America, for example, there is a lack of scientific publications on infectious diseases in wildlife when compared with more developed countries (3–6).
Bartonella spp., Coxiella burnetii, and hemotropic Mycoplasma spp. are intracellular bacteria that infect a wide range of animals (7–9) and humans. Hemotropic mycoplasmas (hemoplasmas) are obligate epi-erythrocytic, cell wall-deficient bacteria that usually generate hemolytic anemia in numerous animal species. Routes of transmission are not fully elucidated, but aggressive interactions and possibly fleas and ticks might be involved. The pathogenic potential of hemotropic mycoplasmas, as a cause of human disease, has not been clearly defined; the public health implications derived from these emerging zoonotic pathogens are underestimated (10). Bartonella is composed of gram-negative fastidious, facultative intracellular microorganisms transmitted by fleas and other vectors that provoke a long-lasting bacteremia in the mammal host. The zoonotic potential of these bacteria is well described, and the term bartonellosis has been implemented to refer to human diseases (11). Coxiella burnetii is a zoonotic, strictly intracellular gram-negative bacterium that infects a wide range of animals. In its sylvatic cycle, it can be transmitted by ticks. In humans, it is considered the causal agent of query fever (Q-fever), and the Centers for Disease Control and Prevention (CDC) has classified this microorganism as a potential bioterrorism agent (12).
There is increased evidence that wildlife species are also susceptible (13–15) and have the potential to be zoonotic (7, 16, 17). In Chile, several domestic and wildlife species have been identified as potential hosts for several hemoplasmas (18–22) and Bartonella spp. (23–28). Information on Coxiella burnetii is much more limited, with only one report with molecular evidence in bats from Chile (26). Despite being commonly reported in domestic and wild ruminants in Europe and North America (13, 29, 30), there are no studies for the detection of Bartonella spp. and C. burnetii in these taxa in Chile, and only until recently has it been possible to identify hemoplasmas in domestic camelids, llamas (Lama glama), and alpacas (Vicugna pacos) (31). Finally, the native pudu (Pudu puda) has been identified as the potential host species of several hemoplasmas (10), including Mycoplasma ovis-like, in the Chilean template forest.
Pudu is the most common cervid in Argentina and Chile and is considered threatened in both countries (32, 33), as shown in CITES Appendix I. In Chile, pudus inhabit temperate forests heavily affected by anthropic factors such as deforestation, housing construction, free-ranging dogs, and livestock (34). Additionally, a high diversity of infectious agents that could be a threat to their health status has recently been identified (10, 35–38). Recently, pudus were identified as potential reservoir hosts for the bovine viral diarrhea virus, which is a cause of major disease in cattle (39). This study aimed to determine the occurrence and genetic diversity of Bartonella spp., hemotropic mycoplasmas, and C. burnetii in free-ranging and captive pudus from Chile.
Blood samples from frozen banks in rescue centers and zoos/breeding centers were used. The frozen bank samples were opportunistically collected from 69 free-ranging pudus between 2016 and 2022 on admission day from two wildlife rehabilitation centers in the template forest ecosystem of southern Chile in Los Lagos District, one (USS: Cerefas, Universidad San Sebastian) located in the continental area and the other (Ch. S: Chiloe Silvestre NGO) in Chiloe island. Additionally, blood samples from 30 captive pudus were collected between 2017 and 2021 during preventive medicine procedures in two facilities, one located in the Mediterranean ecosystem of Central Chile in the Metropolitan District and the other in Los Lagos District, and do not have contact between centers. Blood samples were obtained by venipuncture of the jugular vein using an evacuated tube system (Vacutainer, Beckon, Dickson and Company, Franklin Lakes, New Jersey, USA) and stored at −20°C within 6 h of collection. For extensive sampling details, refer to the study mentioned in the reference (10).
The 99 frozen EDTA-blood samples were thawed at room temperature and vortexed at the UACh Veterinary Clinical Pathology Laboratory, Valdivia, Chile. DNA extraction from 200 μl of blood was performed using an E.Z.N.Z. Tissue DNA Kit (E.Z.N.A. Omega BioTek®, Norcross, GA, U.S.A.), according to the manufacturer's instructions, to obtain a concentration between 20 and 50 ng/μl of purified DNA. Concentration and purity of DNA were measured (NanoDrop ND-1000, Thermo Scientific, Waltham, MA, U.S.A.). The 260/280 nm absorbance ratio (OD260/OD280) provided an estimate of sample purity, accepting a ratio of 1.8 ± 0.2 as pure. DNA was stored at −20°C before performing PCR assays.
DNA samples were subjected to qPCR targeting the irbp gene (interphotoreceptor retinoid-binding protein) using the primers IRBP-CF_FWD (5′-TCCAACACCACCACTGAGATCTGGAC-3′) and IRBP-CF-REV (5′-GTGAGGAAGAAATCGGACTGGCC-3′), with the aim to check DNA template integrity and discard the presence of PCR inhibitors, as previously described (40). All cPCRs were performed with nuclease-free water as a negative control in a T100TM Thermal Cycler (Bio-Rad).
To detect and quantify Bartonella spp., the DNA of all irbp cPCR-positive samples were subsequently subjected to an initial screening by quantitative real-time PCR (qPCR) targeting the nuoG gene of Bartonella spp. (83 bp), using primers (F-Bart [5′-CAATCTTCTTTTGCTTCACC-3′] and R-Bart [5′- TCAGGGCTTTATGTGAATAC-3′], hydrolysis probe (TexasRed-5′- TTYGTCATTTGAACACG-3′[BHQ2a-Q]3′) as previously described (41). qPCR amplifications were conducted in Hard-Shell PCR plates (Bio-Rad©, CA, USA) using Thermal Cycler CFX96 Touch Real Time (Bio-Rad, CA, USA). Amplification efficiency (E) was calculated from the standard curve slope in each run using the following formula: (E = 10–1/slope). Copy numbers were estimated using 10-fold serial dilutions of gBlock® (Integrated DNA Technologies, Coralville, IA, U.S.A.), encoding the nuoG B. henselae sequence (insert containing 83 bp). Bartonella henselae genomic DNA from a cat tested in a previous study was used as a positive control (42). All PCR runs were performed with nuclease-free water (Promega®, Madison, WI, USA) as a negative control. Replicates showing a Cq difference higher than 0.5 were retested.
All positive Bartonella spp. nuoG-qPCR positive samples were subjected to cPCR amplification of a fragment of three loci [gltA (43), rpoB (44), and ITS (45)] by cPCR with the aim to molecularly characterize Bartonella spp. cPCR amplification reactions were performed in a T100 Bio-Rad thermocycler (Bio-Rad©, Hercules, CA, U.S.A.), and the details of the amplification conditions are presented in Table 1. Bartonella henselae genomic DNA from a cat tested in a previous study was used as a positive control (42).
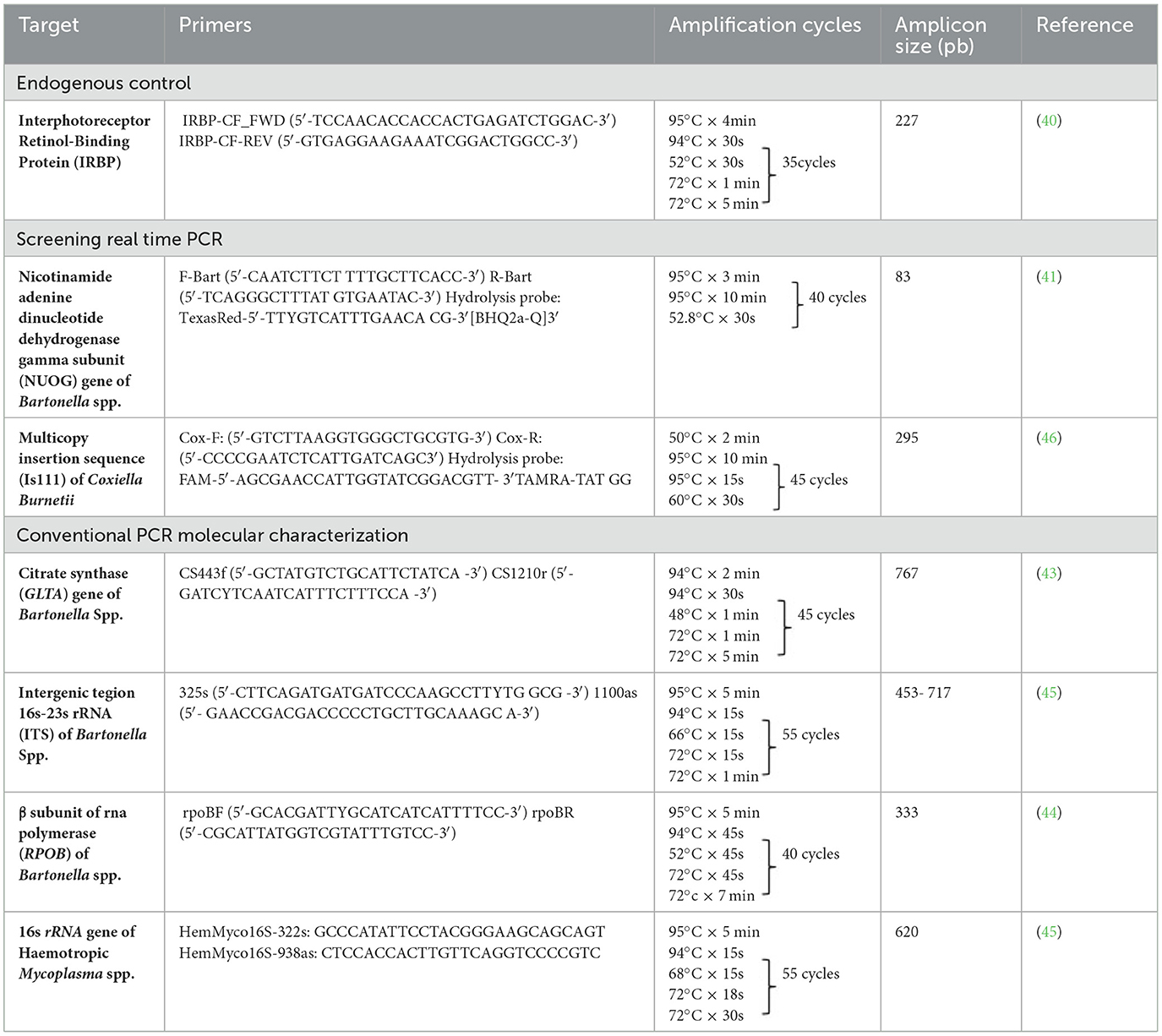
Table 1. Summary information of the conventional and Real time PCR primer sets, amplification conditions and their amplicon sizes used in the present study.
The screening real-time qPCR targeted a 295-bp fragment of the multicopy insertion element IS1111 and is used for sensitive detection of C. burnetii in biological samples (46) (Table 1). Primers Cox-F (GTC TTA AGG TGG GCT GCG TG) and Cox-R (CCC CGA ATC TCA TTG ATC AGC) and hydrolysis probe Cox-TM (FAM-AGC GAA CCA TTG GTA TCG GAC GTT–TAMRA–TAT GG) were used. Standard curves were constructed using 10-fold serial dilutions (2.0 × 107 to 2.0 × 100) of a gBlock® (Integrated DNA Technologies, Coralville, IA, USA), encoding a 295-bp fragment of the IS1111 element of C. burnetii (Integrated DNA Technologies, Coralville, IA, USA). Amplification efficiency (E) was calculated from the standard curve slope in each run using the following formula (E = 10–1/slope). Coxiella burnetii genomic DNA from a cow was used as a positive control. All PCR runs were performed with nuclease-free water (Thermo Scientific©, Waltham, MA, USA) as a negative control. Replicates showing a Cq difference higher than 0.5 were retested.
All positive samples in the irbp cPCR were subjected to a cPCR protocol targeting the 16S rRNA hemotropic Mycoplasma spp. gene (620 bp), using HemMycop16S-322s and HemMycop16S-938as primers (Table 1), according to a previously described protocol (7). All cPCR runs were performed with nuclease-free water (Thermo Scientific©) as a negative control, and a cat sample known to be infected by M. haemofelis was used as a positive control. This protocol was used for screening and later sequencing for molecular characterization of detected hemoplasmas.
Conventional PCR products were separated by 1.5% agarose gel electrophoresis (LE Agarose Seakem®, Lonza) and stained with SYBR© safe DNA gel stain (Thermo Scientific©). The DNA products with the expected size were purified and sequenced.
Only positive samples presenting strong band intensity (Bartonella spp. and hemotropic Mycoplasma spp.) were purified by enzymatic reaction using ExoSAP- ITTM PCR Product Cleanup Reagent (Thermo Scientific©, Carlsbad, CA, U.S.A.), following the manufacturer's instructions. Purified DNA was sent to MACROGEN (Seoul, Korea) for sequencing by the Sanger method in an automatic sequencer (A.B.I Prism 310 genetic analyzer; Applied Biosystem ©/PerkinElmer) for species identification. Forward and reverse sequences were analyzed in Geneious 7.1 (https://www.geneious.com), to obtain consensus sequences. Identity percentages were obtained using BLASTn (47).
Before constructing the phylogenetic inference, sequences belonging to different samples, but representing the same bacterial species, were aligned with Geneious 7.1 (https://www.geneious.com) using the MAFFT alignment method (48) and subsequently analyzed for detection of polymorphism and haplotype identification using DnaSP v5 software (49).
The sequences of the present study were aligned with other sequences from the database (GenBank) through the MAFFT program (Multiple Alignment by Fast Fourier Transform) (48) incorporated in Geneious 7.1 software (https://www.geneious.com). Then, multiple alignments were analyzed using BMGE (Block Mapping and Gathering with Entropy) software to remove ambiguously aligned regions (50).
For the phylogenetic analysis, the best evolutionary model was selected according to the Bayesian Information Criterion (BIC) for each one of the codon positions (partition) for the encoded genes (gltA and rpoB) (51). Thus, the best evolutionary models for Bartonella spp. gltA were K3P+G4 (partition 1), TIM3+F+G4 (partition 2), and TNe+G4 (partition 3). For Bartonella spp. rpoB, the best models were TPM3u+F+G4 (partition1), TNe+G4 (partition 2), and TIM3e+G4 (partition 3). For the non-coding genes (ITS), the best evolutionary model was selected according to the Akaike information criterion (AIC) (52). The best model for Bartonella spp. ITS was TPM2u+F+G4. For ITS, the best evolutionary model selection was assessed using Model Finder (53). Finally, all trees were inferred with a bootstrapping of 1,000 by the maximum likelihood (ML) method with IQ-TREE (54). To enroot the trees, the outgroups were the following for the Bartonella spp. trees: Ochrobactrum anthropii (gltA, ITS, rpoB), Brucella abortus (gltA, rpoB), and Brucella melitensis (ITS). Mycoplasma pneumoniae was used as an outgroup for the construction of the 16S rRNA hemoplasma tree.
All DNA samples (median and standard deviation (SD) of DNA concentration = 31.5 ± 56.2 ng/uL; mean and SD 260/280 ratio = 1.3 ± 0.35) were positive for the irbp gene.
Molecular occurrence of Bartonella spp. DNA in pudu detected by qPCR (mean and SD of reactions' efficiency = 100 ± 5.04%; r2 = 0.99 ± 0.005; slope = −3.32 ± 0.11; Y-intercept = 39.26 ± 1.09) was 10.1% (10/99) [95% CI (5.2–18.2%)]. Only three samples had consistent Cq, and the quantification of Bartonella spp. was 18.5 ± 14.02 nuoG-copies/μl (mean ± standard deviation, SD).
Representative sequences of Bartonella spp. gltA, ITS, and rpoB genes were deposited in GenBank (55) under the accession numbers OQ162290, OQ137267, and OQ162291. Within sequences that represented the same haplotype, only one representative sequence (with a higher size) was deposited in GenBank and used for phylogenetic analysis.
Bartonella spp. DNA was successfully amplified by cPCR in 60% (6/10) of qPCR-positive samples, and six sequences were obtained [4 rpoB (samples: #6235, #5144, #8184, and 902020), 1 gltA (sample #6235), 1 ITS (sample #6235)] from four pudus [one captive (902,020) and three free-ranging (6,235, 5,144, and 8,184)]. The rpoB sequences were 100% similar to each other and showed 98.2% similarity with B. henselae from cats in Brazil (MN107418), 99.7% identity with B. henselae from a cat from Paraguay (MW514669), and 100% identity with B. henselae from Urva auropunctata from St. Kitts (MW728257). The gltA sequence showed 95.05% identity with uncultured Bartonella sp. from a cattle tail louse from Israel (KJ522487), and the ITS sequence showed 93.1% identity with Bartonella sp. from deer ked (DQ485307). As such, wild pudu #6235 was co-positive to B. henselae and Bartonella sp., similar to Bartonellae identified in ruminants.
The rpoB sequences of the present study were allocated in the same taxa, sharing a clade with B. henselae Houston 1 (AF171070), B. henselae from a cat from Paraguay (MW514660), and B. henselae from Urva auropunctata from St. Kitts and Nevis (MW728257) (Figure 1). The rpoB diversity analyses are represented on Table 2.
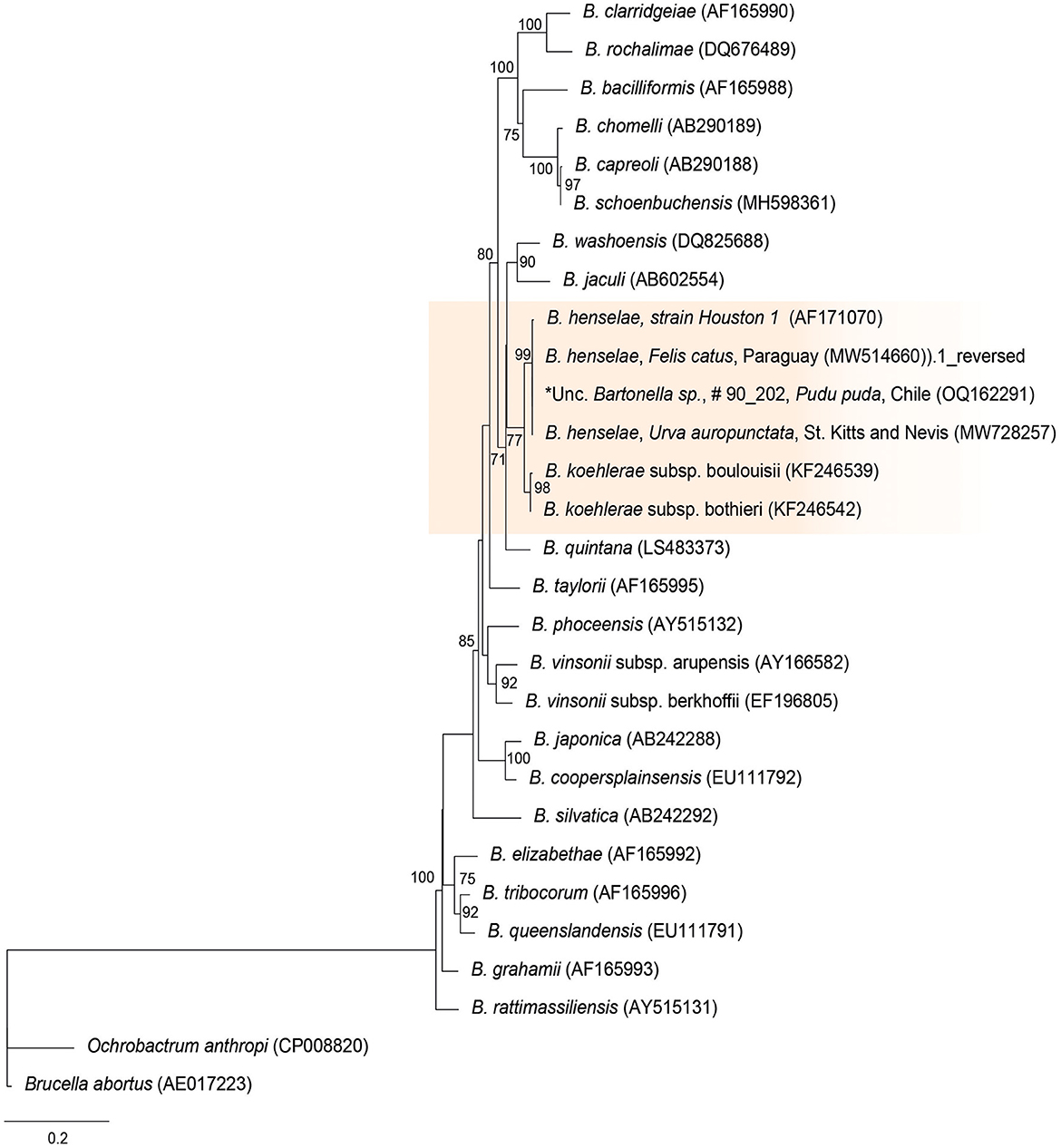
Figure 1. Maximum likelihood phylogenies for a subset of Bartonella spp. inferred using an alignment (1,152 bp) of the gene encoding the β subunit of RNA polymerase (rpoB). Calculated substitution model was TPM3u+F+G4 (partition 1), TNe+G4 (partition 2), and TIM3e+G4 (partition 3). Best models were chosen using the Bayesian information criterion (BIC).

Table 2. Polymorphism and genetic diversity of rpoB Bartonella species sequences identified in pudu from Chile.
The gltA phylogenetic reconstruction evidenced that the sequence of the present study was allocated to the same clade with Bartonella sp. from a Cervus from Japan (CP019781), Bartonella sp. from a cattle tail louse from Israel (KJ522487), and B. capreoli, B. schoenbuchensis, and B. chomeli (Figure 2).
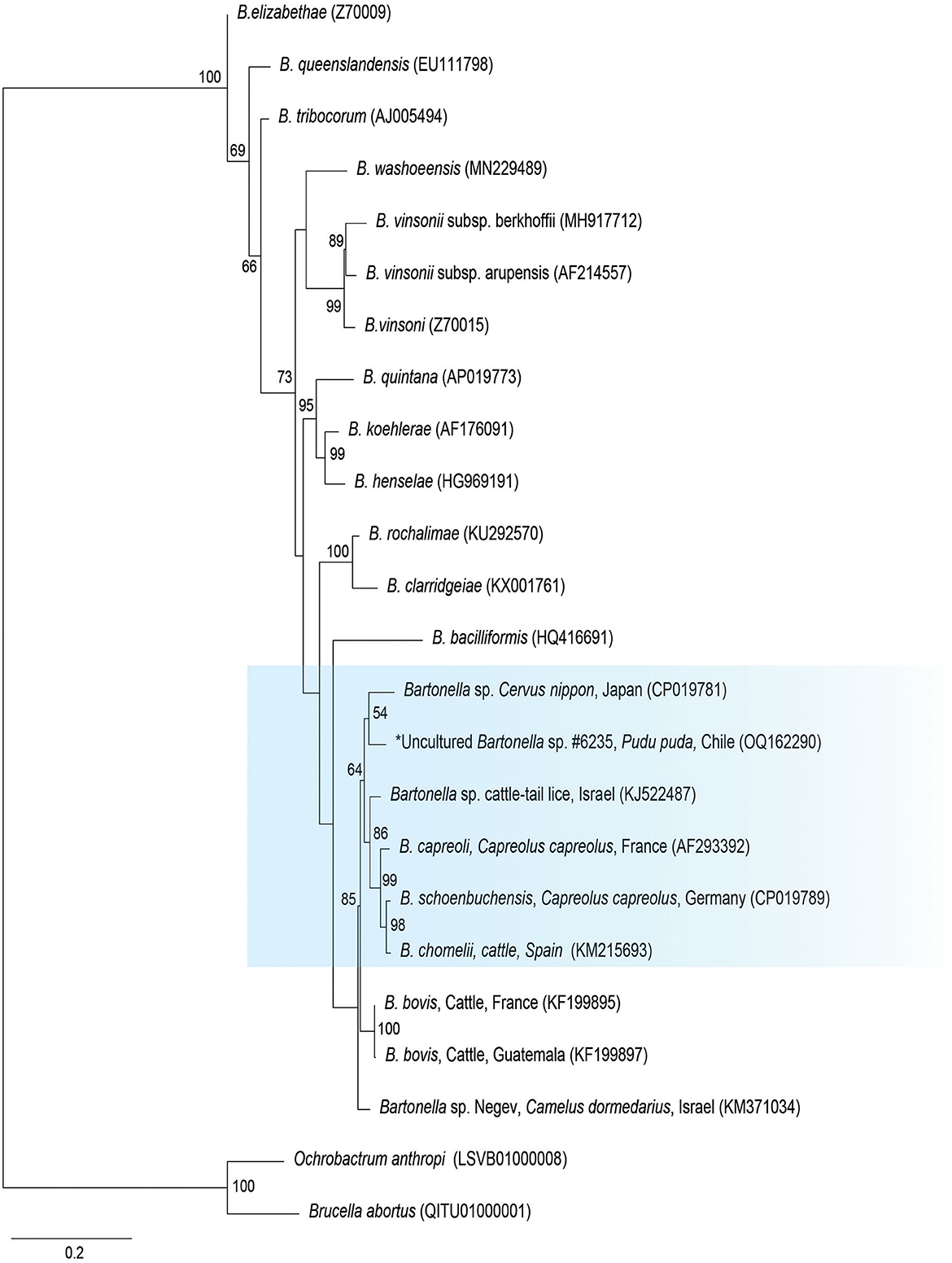
Figure 2. Maximum likelihood phylogenies for a subset of Bartonella spp. inferred using an alignment (1,290 bp) of the gene encoding citrate synthase (gltA). Calculated substitution model was K3P+G4 (partition 1), TIM3+F+G4 (partition 2), and TNe+G4 (partition 3). Best models were chosen using the Bayesian information criterion (BIC).
Finally, the ITS sequence was closely positioned to Bartonella sp. sequence from a deer-ked (DQ485307), B. schoenbuchensis (CP019789, HG77197), B. chomeli (KM215718), B. melophagi (JF834886), and B. bovis (KF218234, KR733201) (Figure 3).
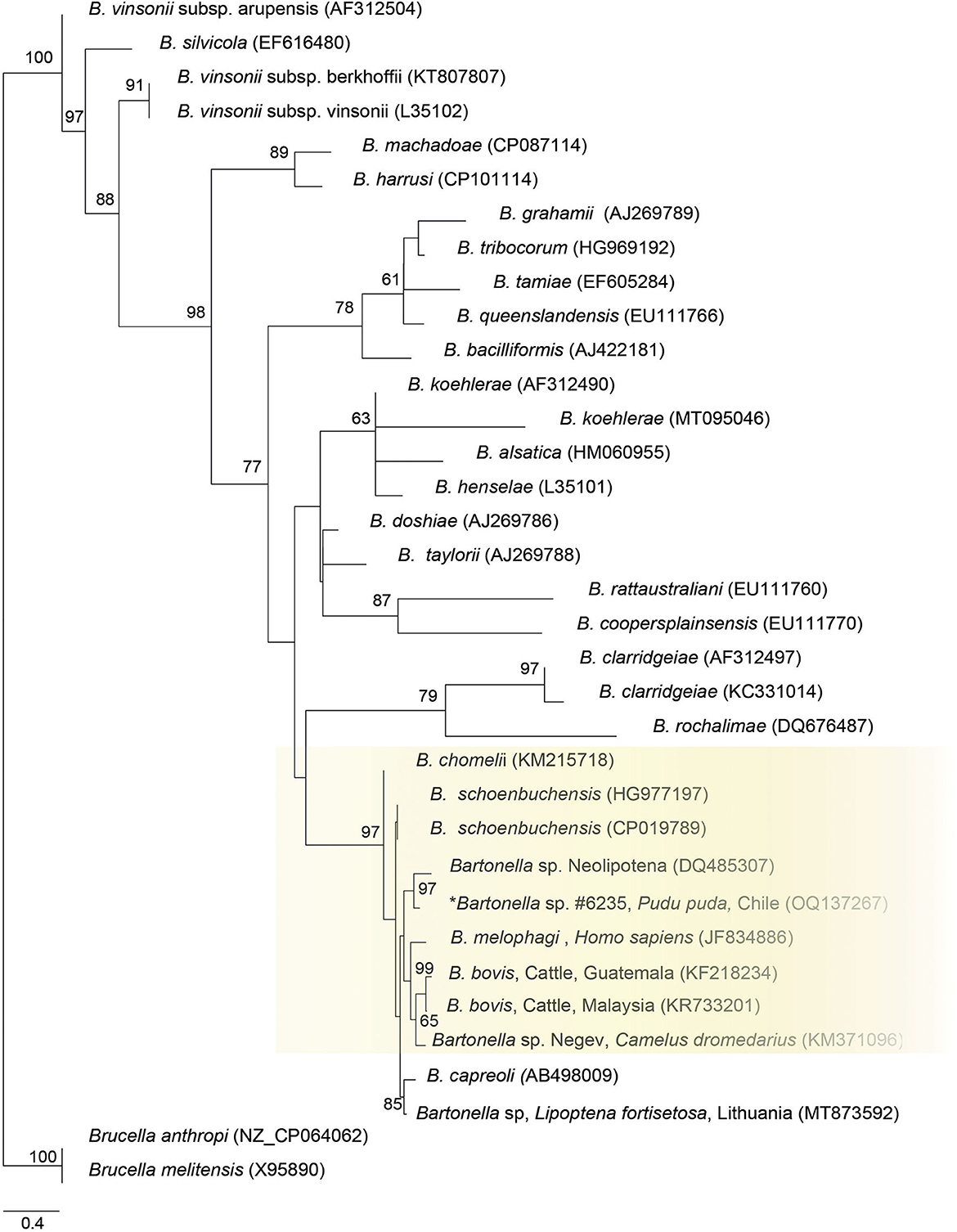
Figure 3. Maximum likelihood phylogenies for a subset of Bartonella spp. inferred using an alignment (522 bp) of the internal transcribed spacer (ITS). Calculated substitution model was TPM2u+F+G4. Best models were chosen using the Akaike information criterion (AIC).
Molecular occurrence of C. burnetii DNA in pudu detected by qPCR (mean and SD of reactions' efficiency = 100.6 ± 5.08%; r2 = 1.0 ± 0.005; slope = −3.31 ± 0.12; Y-intercept = 37.38 ± 0.88) was 1.0% (1/99) [95% CI (0.05–6.3%)].
Molecular occurrence of hemotropic Mycoplasma spp. in pudu by cPCR was 1.7% (1/60) [95% CI (0.08–10.1%)]. The sequence of the 16S rRNA fragment showed 100% BLASTn identity with Mycoplasma ovis-like amplified previously from Chilean pudu (MW532816) (Figure 4).
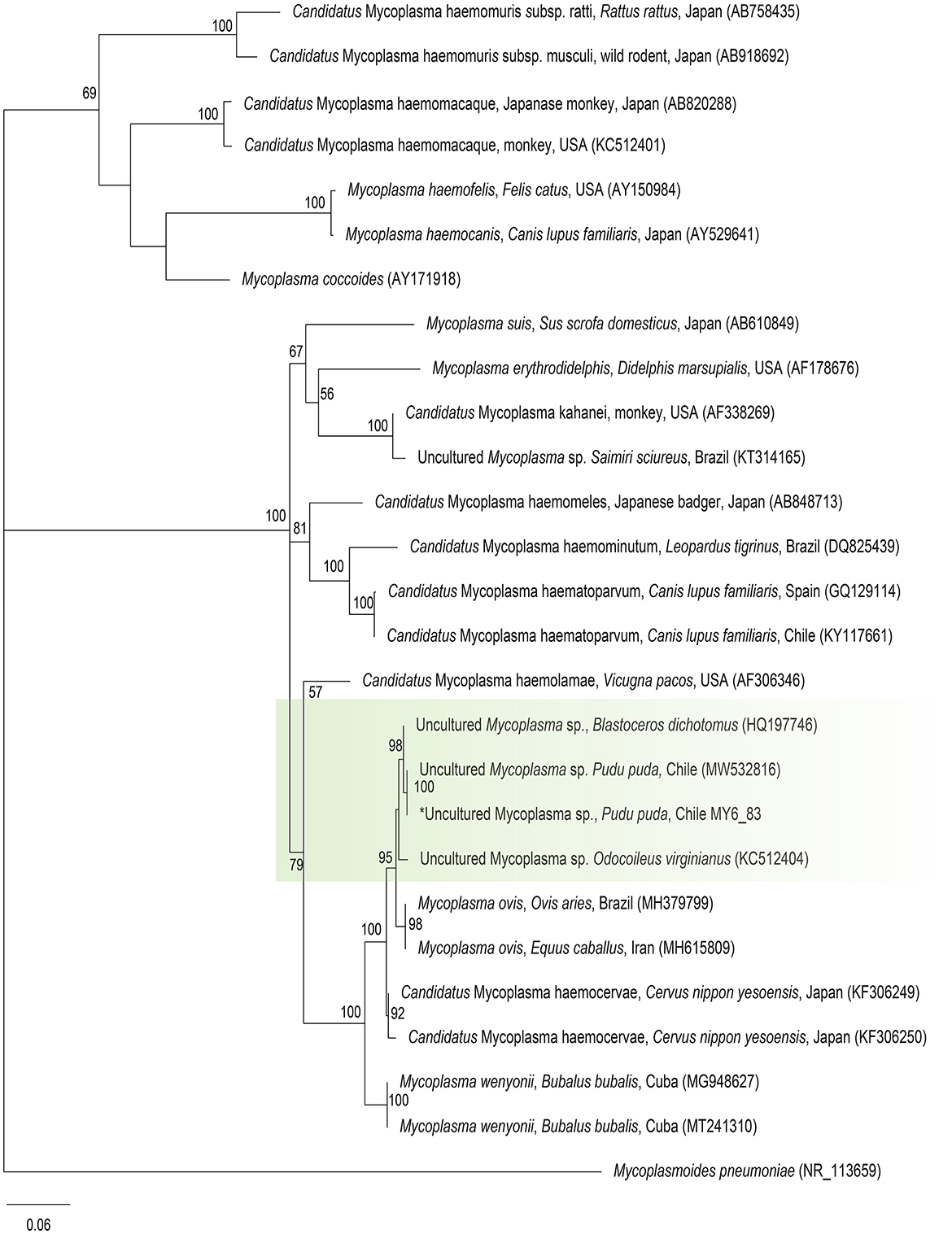
Figure 4. Maximum likelihood phylogenies for a subset of Mycoplasma spp. inferred using an alignment (620 bp) of the 16S rRNA gene. Calculated substitution model was GTR+F+G4. Best models were chosen using the Akaike information criterion (AIC).
This is the first study to document the presence of DNA of B. henselae in a wild ungulate species and Bartonella spp. and C. burnetii in wild ungulate species in South America. The circulation of Mycoplasma ovis-like in free-ranging pudu in Chile is also confirmed (10). The presence of these intracellular bacteria in free-living pudu could suggest an increase in the interaction between domestic species and their ectoparasites and these native species in their natural habitats. Unlike studies in wildlife in other regions (56–58), no co-infection with the three evaluated pathogens was found in pudus. However, one pudu was possibly co-infected with more than one Bartonella species, since B. henselae and Bartonella sp., similar to Bartonellae identified in ruminants, were detected. The co-occurrence of different Bartonella species in the bloodstream of reservoir animals such as pudus was earlier described in cats and rodents (59–61), and it illustrates the outstanding tolerance of these hosts to harbor mixed Bartonella infections. This could be mediated by an arthropod vector via multiplication and interaction of different Bartonella genetic variants in their digestive tract, with subsequent simultaneous transmission to the mammal host (62, 63). Culture and further molecular characterization of the isolates (64) should be attempted with these samples in future to confirm the co-positivity with multiple species of Bartonella.
Coxiella burnetii is an important bacterial zoonotic pathogen that can cause Q fever in humans. The bacterium has the potential to cause large-scale outbreaks due to its low infectious dose, environmental resistance, and ability to spread airborne through aerosolization of the pathogen, and is a potential biological threat classified as a “Select Agent” in the USA. Coxiella burnetii has a worldwide geographical distribution, apart from Antarctica and New Zealand, and has a wide and diverse host range. The pathogen primarily affects sheep, goats, and cattle, which are considered their primary reservoirs and sources for human outbreaks (65). The livestock species can be infected with C. burnetii and appear healthy, and people often become exposed by breathing in dust contaminated with animal feces, urine, and birth products. Wild ungulate species have been reported commonly exposed to C. burnetii infection in Europe and North America (66), including eight cervid species, but this report in pudu represents the first in deer from the Southern Hemisphere (65). In the Basque region in Spain, the prevalence has been categorized as stable throughout time. Therefore, the roe deer (Capreolus capreolus) plays a role in the sylvatic cycle of Q fever (67). In South America, there is no evidence of C. burnetii DNA in blood samples of wild boar (Sus scrofa), marsh deer (Blastocerus dichotomus), brown brocket deer (Mazama gouazoubira), small red brocket deer (Mazama bororo), red brocket deer (Mazama americana), and pampas deer (Ozotocerus bezoarticus) (12, 68). A recent study (12) found that 5.32% of the sampled deer was seropositive for C. burnetii by an indirect immunofluorescence assay (IFA) for IgG antibodies (anti-phase I); to date, it is the only evidence of exposure to this pathogen in deer in the region.
In Chile, DNA findings of C. burnetii have been reported in samples of animal origin only in bats and bulk tank milk from cows (26, 69). The last human Q fever outbreak in Chile was declared in 2017 in the Los Lagos District, the same region where molecular evidence was found in pudu in our study (70). This district is a part of the southern macrozone where seropositivity for humans (6%) was significantly higher than in other regions of the country (70). It is likely that the source of infection for pudu is of anthropogenic origin (livestock), or from exotic deer species, red deer, and/or fallow deer, which have been reported in the area (71) and have been commonly reported infected by C. burnetii in Europe (60, 65), or from rodent species previously found to be a source of livestock coxiellosis (72). Other serological or molecular studies in dogs and Darwin fox (Lycalopex fulvipes) in the southern macrozone found no evidence of C. burnetii infection (18, 73, 74). The finding of only one pudu being positive for the bacterium and the low prevalence of C. burnetii in Chile make serological and molecular screening necessary for a much larger number of pudu samples from the Los Lagos region, to evaluate their potential role as a host of infection for transmission to animals and humans. Additionally, the reports of infectious abortions in captive pudus in Chile (39) added to the evidence that C. burnetii has been involved in reproductive loss in captive exotic ungulates, mainly in bovid species (65), making it necessary to include in the differential diagnosis of possible causes of abortion in pudu. Moreover, future studies should attempt to molecularly characterize via sequencing the C. bunetii found in pudus from Chile.
In this study, B. henselae, an emerging zoonotic pathogen that causes scratch disease in humans and whose transmission mainly involves domestic cats as the main reservoir and cat fleas (Ctenocephalides felis) as the main vector (75), is described for the first time in wild ungulate species. Otherwise, to a lesser extent, it is reported in other mammals, bovines (76, 77) and rodents among them (78–81), suggesting that they have a permissive cycle in nature, being detected in several ecological niches (hosts and vectors) (77). Thus, this finding could indicate the circulation of B. henselae in an infected vector, favoring B. henselae transmission among domestic and wild mammals. Nonetheless, further epidemiological and genotyping studies are necessary to confirm this hypothesis. The DNA of Bartonella bacteria has been widely described in cervid species from Europe (30, 58, 82–84), North America (29, 85–87), and Asia (88, 89), usually with a higher prevalence (between 4.9 and 77.7%) than reported in our study. In South America, there are reports of Bartonella spp. in vector species of wild ungulates (68, 90) but not in their blood samples (68). In Chile, during the last decade, there have been reports of the presence of Bartonella spp. in cats, dogs, minks, and bats (23–26, 28, 91, 92). Bartonella henselae in pudu was similar to B. henselae reported in small Indian mongooses in the Caribbean (93) and cats from Brazil (94) and Paraguay (27). Bartonella sp., related to those reported infecting ruminants, such as B. schoenbuchensis [CP019789, HG977197 (95)], B. chomeli (KM215718) (96), B. melophagi (JF834886) (97), B. bovis, and B. capreoli, was also detected in a pudu in this study. More screening will be necessary to confirm the role of pudu in the epidemiology of this infectious agent and its impact on animal health.
Hemoplasma bacteria have been extensively studied in wild and domestic carnivores in Chile during the last decade. Darwin foxes (Lycalopex fulvipes) present a high prevalence of M. haemocanis causing enzootic and asymptomatic infections (18, 19) that could be a source of infection for pudu since both share the same habitat within the Los Lagos region. Hemotropic Mycoplasma spp. have been recently reported in llamas (12.8%) and alpacas (6.3%) (Candidatus Mycoplasma haemolamae) (31) and Mycoplasma ovis-like in free-living pudu in southern Chile (14%) (10). Molecular screening of hemotropic Mycoplasma spp. in sheep, livestock, and native (huemul) and exotic ungulates (wild boars, red deer) in the Los Lagos District is recommended to understand the epidemiology of these infectious agents and the possible role of pudu as a host. It is also recommended to evaluate the pathogenicity of Mycoplasma ovis-like in pudu.
For the first time in pudu, the finding of B. henselae and C. burnetii, both zoonotic pathogens, could be relevant to public health. Both B. henselae and C. burnetii are pathogens related to occupational diseases, with evidence of health risks for those working with infected species through occupational exposure in rehabilitation centers, breeding centers, and zoological parks (98, 99), representing an important factor to consider in medical and management practices with this animal species.
This study expands the knowledge of bacteria with zoonotic potential carried by pudu. Mycoplasma ovis-like was confirmed in pudus, while Bartonella spp., Bartonella henselae, and C. burnetii were described for the first time in South American ungulates. The results of this study suggest an anthropic impact on wildlife species with domestic species interacting epidemiologically with pudus in their natural habitats. Further research will be necessary to evaluate the potential role of pudu as a host and reservoir of infection, and identify the sources for disease transmission among humans and wild and domestic animals.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/nuccore; OQ162290, OQ137267, OQ162291.
Ethical approval was not required for the study involving animals in accordance with the local legislation and institutional requirements because we only use samples from Frozen Banks of the rehabilitation centers and zoos. We don't have involved in any managements animals procedure.
EH-H: conceptualization, supervision, funding acquisition, investigation, resources, data curation, writing—original draft preparation, and writing—reviewing and editing. PS-G: methodology, investigation, resources, data curation, writing—original draft preparation, and writing—reviewing and editing. JC, CV, FV, and IK: resources and data curation. SC and CO: resources. DM-A: writing—original draft preparation, and writing—reviewing and editing. MO and NC: methodology and investigation. AG: investigation and writing—reviewing and editing. AM: conceptualization, supervision, funding acquisition, methodology, investigation, writing—original draft preparation, and writing—reviewing and editing. All authors contributed to the article and approved the submitted version.
This study was supported by Fundacion Buin Zoo, Chiloe Silvestre, and ANID/CONICYT FONDECYT Regular 1191462 and 1231261. DM-A thanks to Grant ANID/BASAL FB210006.
The authors would like to thank all staff from Universidad San Sebastian Wildlife Rescue Center and Chiloe Silvestre for their support in sample collection.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Rhyan JC, Spraker TR. Emergence of diseases from wildlife reservoirs. Vet Pathol. (2010) 47:34–9. doi: 10.1177/0300985809354466
2. Barroso P, Acevedo P, Vicente J. The importance of long-term studies on wildlife diseases and their interfaces with humans and domestic animals: a review. Transbound Emerg Dis. (2021) 68:1895–909. doi: 10.1111/tbed.13916
3. Han BA, Kramer AM, Drake JM. Global patterns of zoonotic disease in mammals. Trends Parasitol. (2016) 32:565–77. doi: 10.1016/j.pt.2016.04.007
4. Sanchez-Vazquez MJ, Hidalgo-Hermoso E, Cacho-Zanette L, de Campos-Binder L, Rivera A, MolinaFlores B, et al. Characteristics and perspectives of disease at the wildlife-livestock interface in Central and South America. In:Vicente J, Vercauteren KC, Gortazar C, , editors Diseases at the wildlife-livestock interface: Research and perspectives in a changing world, Wildlife Research Monographs 3. Cham, Switzerland: Springer (2021). p. 271–304. doi: 10.1007/978-3-030-65365-1_9
5. Tompkins DM, Carver S, Jones ME, Krkošek M, Skerratt LF. Emerging infectious diseases of wildlife: a critical perspective. Trends Parasitol. (2015) 31:149–59. doi: 10.1016/j.pt.2015.01.007
6. Wiethoelter AK, Beltrán-Alcrudo D, Kock R, Mor SM. Global trends in infectious diseases at the wildlife-livestock interface. Proc Natl Acad Sci U S A. (2015) 112:9662–7. doi: 10.1073/pnas.1422741112
7. Maggi RG, Compton SM, Trull CL, Mascarelli PE, Mozayeni BR, Breitschwerdt EB. Infection with hemotropic Mycoplasma species in patients with or without extensive arthropod or animal contact. J Clin Microbiol. (2013) 51:3237–41. doi: 10.1128/JCM.01125-13
8. Cheslock MA, Embers ME. Human Bartonellosis: an underappreciated public health problem? Trop Med Infect Dis. (2019) 4:69. doi: 10.3390/tropicalmed4020069
9. Eldin C, Mélenotte C, Mediannikov O, Ghigo E, Million M, Edouard S, et al. From Q Fever to Coxiella burnetii infection: a paradigm change. Clin Microbiol Rev. (2017) 30:115–90. doi: 10.1128/CMR.00045-16
10. Hidalgo-Hermoso E, Cabello J, Novoa-Lozano I, Celis S, Ortiz C, Kemec I, et al. Molecular detection and characterization of hemoplasmas in the pudu (Pudu puda), a native cervid from chile. J Wildl Dis. (2022) 58:8–14. doi: 10.7589/JWD-D-21-00057
11. Shamshiri Z, Goudarztalejerdi A, Zolhavarieh SM, Greco G, Sazmand A, Chomel BB. Molecular detection and identification of Bartonella species in cats from Hamedan and Kermanshah, Western Iran. Comp Immunol Microbiol Infect Dis. (2022) 89:101879. doi: 10.1016/j.cimid.2022.101879
12. Zanatto DCdS, Duarte JMB, Labruna MB, Tasso JB, Calchi AC, Machado RZ, et al. Evidence of exposure to Coxiella burnetii in neotropical free-living cervids in South America. Acta Trop. (2019) 197:105037. doi: 10.1016/j.actatropica.2019.05.028
13. Yon L, Duff JP, Ågren EO, Erdélyi K, Ferroglio E, Godfroid J, et al. Recent changes in infectious diseases in European wildlife. J Wildl Dis. (2019) 55:3–43. doi: 10.7589/2017-07-172
14. Westmoreland LS, Stoskopf MK, Maggi RG. Detection and prevalence of four different hemotropic Mycoplasma spp. in Eastern North Carolina American black bears (Ursus americanus). Comp Immunol Microbiol Infect Dis. (2017) 50:106–9. doi: 10.1016/j.cimid.2016.12.002
15. Kosoy M, Goodrich I. Comparative ecology of Bartonella and Brucella infections in wild carnivores. Front Vet Sci. (2019) 5:322. doi: 10.3389/fvets.2018.00322
16. González-Barrio D, Hagen F, Tilburg JJ, Ruiz-Fons F. Coxiella burnetii genotypes in Iberian wildlife. Microb Ecol. (2016) 72:890–7. doi: 10.1007/s00248-016-0786-9
17. Billeter SA. A review of bartonella infections in california-implications for public and veterinary health. J Med Entomol. (2022) 59:1154–1163. doi: 10.1093/jme/tjac056
18. Cabello J, Altet L, Napolitano C, Sastre N, Hidalgo E, Dávila JA, et al. Survey of infectious agents in the endangered Darwin's fox (Lycalopex fulvipes): high prevalence and diversity of hemotrophic mycoplasmas. Vet Microbiol. (2013) 167:448–54. doi: 10.1016/j.vetmic.2013.09.034
19. Di Cataldo S, Hidalgo-Hermoso E, Sacristán I, Cevidanes A, Napolitano C, Hernández CV, et al. Hemoplasmas are endemic and cause asymptomatic infection in the endangered darwin's fox (Lycalopex fulvipes). Appl Environ Microbiol. (2020) 86:e00779–20. doi: 10.1128/AEM.00779-20
20. Di Cataldo S, Cevidanes A, Ulloa-Contreras C, Sacristán I, Peñaloza-Madrid D, Vianna J, et al. Widespread infection with hemotropic mycoplasmas in free-ranging dogs and wild foxes across six bioclimatic regions of Chile. Microorganisms. (2021) 9:919. doi: 10.3390/microorganisms9050919
21. Millán J, Cevidanes A, Sacristán I, Alvarado-Rybak M, Sepúlveda G, Ramos-Mella CA, et al. Detection and characterization of hemotropic mycoplasmas in bats in Chile. J Wildl Dis. (2019) 55:977–81. doi: 10.7589/2018-12-290
22. Sacristán I, Acuña F, Aguilar E, García S, López MJ, Cevidanes A, et al. Assessing cross-species transmission of hemoplasmas at the wild-domestic felid interface in Chile using genetic and landscape variables analysis. Sci Rep. (2019) 9:16816. doi: 10.1038/s41598-019-53184-4
23. Müller A, Walker R, Bittencourt P, Machado RZ, Benevenute JL, DO Amaral RB, et al. Prevalence, hematological findings and genetic diversity of Bartonella spp. in domestic cats from Valdivia, Southern Chile. Parasitology. (2017) 144:773–82. doi: 10.1017/S003118201600247X
24. Müller A, Soto F, Sepúlveda M, Bittencourt P, Benevenute JL, Ikeda P, et al. Bartonella vinsonii subsp. berkhoffii and B henselae in dogs. Epidemiol Infect. (2018) 146:1202–4. doi: 10.1017/S0950268818001127
25. Müller A, Gutiérrez R, Seguel M, Monti G, Otth C, Bittencourt P, et al. Molecular survey of Bartonella spp. in rodents and fleas from Chile. Acta Trop. (2020) 212:105672. doi: 10.1016/j.actatropica.2020.105672
26. Müller A, Sepúlveda P, Di Cataldo S, Cevidanes A, Lisón F, Millán J. Molecular investigation of zoonotic intracellular bacteria in Chilean bats. Comp Immunol Microbiol Infect Dis. (2020) 73:101541. doi: 10.1016/j.cimid.2020.101541
27. Sepúlveda-García P, Pérez-Macchi S, Gonçalves LR, do Amaral RB, Bittencourt P, André MR, et al. Molecular survey and genetic diversity of Bartonella spp in domestic cats from Paraguay. Infect Genet Evol. (2022) 97:105181. doi: 10.1016/j.meegid.2021.105181
28. Weinborn-Astudillo RM, Pau N, Tobar BZ, Jaffe DA, Boulouis HJ, Sepúlveda P, et al. Bartonella infection in stray dogs from central and southern Chile (Linares and Puerto Montt). Vector Borne Zoonotic Dis. (2020) 20:187–92. doi: 10.1089/vbz.2019.2505
29. Izenour K, Zikeli S, Kalalah A, Ditchkoff SS, Starkey LA, Wang C, et al. Diverse Bartonella spp. detected in white-tailed deer (Odocoileus virginianus) and associated keds (Lipoptena mazamae) in the Southeastern USA. J Wildl Dis. (2020) 56:505–11. doi: 10.7589/2019-08-196
30. Wijburg SR, Fonville M, de Bruin A, van Rijn PA, Montizaan MGE, van den Broek J, et al. Prevalence and predictors of vector-borne pathogens in Dutch roe deer. Parasit Vectors. (2022) 15:76. doi: 10.1186/s13071-022-05195-w
31. Ramos P, Sepulveda-Garcia P, Alabí A, Romero A, Pinto T, Rojas A, et al. Molecular survey and genetic characterization of 'Candidatus Mycoplasma haemolamae' in llamas (Lama glama) and alpacas (Vicugna pacos) from Southern Chile. Acta Trop. (2021) 222:106046. doi: 10.1016/j.actatropica.2021.106046
32. Biblioteca del Congreso Nacional de Chile. (2007) Supreme decrete no. 151 of the Ministerio Secretaria General de la Presidencia de Chile. [First Species Classification Process, MMA]. Spanish. Available online at: https://www.bcn.cl/leychile/navegar?idNorma=259402 (accessed December 2020).
33. MAD 2021 (Ministerio de Ambiente y Desarrollo Sustentable). Resolución 316/2021. Categorización de los Mamíferos de Argentina según su riesgo de extinción. (2021).
34. Silva-Rodriguez E, Pastore H, Jimenez J. Pudu puda. In: The International Union for Conservation of Nature red list of threatened species. (2016). Available online at: http://dx.doi.org/10.2305/IUCN.UK.2016-1.RLTS.T18848A22164089 (accessed June 2019).
35. Moreno-Beas E, Abalos P, Hidalgo-Hermoso E. Seroprevalence of nine Leptospira interrogans serovars in wild carnivores, ungulates, and primates from a zoo population in a metropolitan region of Chile. J Zoo Wildl Med. (2015) 46:774–8. doi: 10.1638/2014-0139.1
36. Hidalgo-Hermoso E, Celis S, Cabello J, Kemec I, Ortiz C, Lagos R, et al. Molecular survey of selected viruses in Pudus (Pudu puda) in Chile revealing first identification of caprine herpesvirus-2 (CpHV-2) in South American ungulates. Vet Q. (2023) 43:1–7. doi: 10.1080/01652176.2022.2149879
37. Santodomingo A, Robbiano S, Thomas R, Parragué-Migone C, Cabello-Stom J, Vera-Otarola F, et al. search for piroplasmids and spirochetes in threatened pudu (Pudu puda) and associated ticks from Southern Chile unveils a novel Babesia sp. and a variant of Borrelia chilensis. Transbound Emerg Dis. (2022) 69:3737–48. doi: 10.1111/tbed.14743
38. Santodomingo A, Thomas R, Robbiano S, Uribe JE, Parragué-Migone C, Cabello-Stom J, et al. Wild deer (Pudu puda) from Chile harbor a novel ecotype of Anaplasma phagocytophilum. Parasit Vectors. (2023) 16:38. doi: 10.1186/s13071-023-05657-9
39. Salgado R, Hidalgo-Hermoso E, Pizarro-Lucero J. Detection of persistent pestivirus infection in pudú (Pudu puda) in a captive population of artiodactyls in Chile. BMC Vet Res. (2018) 14:37. doi: 10.1186/s12917-018-1363-x
40. Ferreira EC, Gontijo CM, Cruz I, Melo MN, Silva AM. Alternative PCR protocol using a single primer set for assessing DNA quality in several tissues from a large variety of mammalian species living in areas endemic for leishmaniasis. Mem Inst Oswaldo Cruz. (2010) 105:895–8. doi: 10.1590/S0074-02762010000700009
41. André MR, Dumler JS, Herrera HM, Gonçalves LR, de Sousa KC, Scorpio DG. Assessment of a quantitative 5′ nuclease real-time polymerase chain reaction using the nicotinamide adenine dinucleotide dehydrogenase gamma subunit (nuoG) for Bartonella species in domiciled and stray cats in Brazil. J Feline Med Surg. (2016) 18:783–90. doi: 10.1177/1098612X15593787
42. Sepúlveda-García P, Alabi A, Álvarez K, Rojas L, Mella A, Gonçalves LR, et al. Bartonella spp. in households with cats: Risk factors for infection in cats and human exposure. One Health. (2023) 16:100545. doi: 10.1016/j.onehlt.2023.100545
43. Billeter SA, Gundi VAKB, Rood MP, Kosoy MY. Molecular detection and identification of Bartonella species in Xenopsylla cheopis fleas (Siphonaptera: Pulicidae) collected from Rattus norvegicus rats in Los Angeles, California. Appl Environ Microbiol. (2011) 77:7850–2. doi: 10.1128/AEM.06012-11
44. Paziewska A, Harris PD, Zwolinska L, Bajer A, Sinski E. Recombination within and between species of the alpha proteobacterium Bartonella infecting rodents. Microb Ecol. (2011) 61:134–45. doi: 10.1007/s00248-010-9735-1
45. Maggi RG, Breitschwerdt E. Potential limitations of the 16S-23S rRNA intergenic region for molecular detection of Bartonella species. J Clin Microbiol. (2005) 43:1171–6. doi: 10.1128/JCM.43.3.1171-1176.2005
46. Klee SR, Tyczka J, Ellerbrok H, Franz T, Linke S, Baljer G, et al. Highly sensitive real-time PCR for specific detection and quantification of Coxiella burnetii. BMC Microbiol. (2006) 6:1–8. doi: 10.1186/1471-2180-6-2
47. Altschul S. Basic local alignment search tool. J Mol Biol. (1990) 215:403–10. doi: 10.1016/S0022-2836(05)80360-2
48. Katoh K, Misawa K, Kuma K, Miyata T. MAFFT a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. (2002) 30:3059–66. doi: 10.1093/nar/gkf436
49. Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. (2009) 25:1451–2. doi: 10.1093/bioinformatics/btp187
50. Criscuolo A, Gribaldo S, BMGE. (Block Mapping and Gathering with Entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol Biol. (2010) 10:210. doi: 10.1186/1471-2148-10-210
51. Schwarz G. Estimating the dimension of a model. Ann Stat. (1978) 6:461–464. doi: 10.1214/aos/1176344136
52. Posada D, Buckley TR. Model selection and model averaging in phylogenetics: Advantages of akaike information criterion and bayesian approaches over likelihood ratio tests. Syst Biol. (2004) 53:793–808. doi: 10.1080/10635150490522304
53. Kalyaanamoorthy S, Minh BQ, Wong TKF, Von Haeseler A, Jermiin LS. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat Methods. (2017) 14:587–589. doi: 10.1038/nmeth.4285
54. Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ, IQ-TREE. A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. (2015) 32:268–74. doi: 10.1093/molbev/msu300
55. Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank: update. Nucleic Acids Res. (2004) 32:D23–26. doi: 10.1093/nar/gkh045
56. Gakuya F, Akoko J, Wambua L, Nyamota R, Ronoh B, Lekolool I, et al. Evidence of co-exposure with Brucella spp, Coxiella burnetii, and Rift Valley fever virus among various species of wildlife in Kenya. PLoS Negl Trop Dis. (2022) 16:e0010596. doi: 10.1371/journal.pntd.0010596
57. Razanske I, Rosef O, Radzijevskaja J, Krikstolaitis R, Paulauskas A. Impact of tick-borne Anaplasma phagocytophilum infections in calves of moose (Alces alces) in southern Norway. Folia Parasitol. (2021) 68:23. doi: 10.14411/fp.2021.023
58. Welc-Faleciak R, Werszko J, Cydzik K, Bajer A, Michalik J, Behnke JM. Co-infection and genetic diversity of tick-borne pathogens in roe deer from Poland. Vector Borne Zoonotic Dis. (2013) 13:277–88. doi: 10.1089/vbz.2012.1136
59. Gurfield AN, Boulouis HJ, Chomel BB, Heller R, Kasten RW, Yamamoto K, et al. Coinfection with Bartonella clarridgeiae and Bartonella henselae and with different Bartonella henselae strains in domestic cats. J Clin Microbiol. (1997) 35:2120–3. doi: 10.1128/jcm.35.8.2120-2123.1997
60. Inoue K, Maruyama S, Kabeya H, Kawanami K, Yanai K, Jitchum S, et al. Prevalence of Bartonella infection in cats and dogs in a metropolitan area, Thailand. Epidemiol Infect. (2009) 137:1568–73. doi: 10.1017/S095026880900257X
61. Jian R, Ren Q, Xue J, Xie GC, Wang J, Chen GQ, et al. Genetic diversity of Bartonella infection in residential and field rodents in Hebei, China. Front Microbiol. (2022) 13:1039665. doi: 10.3389/fmicb.2022.1039665
62. Furquim MEC, do Amaral R, Dias CM, Gonçalves LR, Perles L, Lima CAP, et al. Genetic diversity and multilocus sequence typing analysis of Bartonella henselae in domestic cats from Southeastern Brazil. Acta Trop. (2021) 222:106037. doi: 10.1016/j.actatropica.2021.106037
63. Huwyler C, Heiniger N, Chomel BB, Kim M, Kasten RW, Koehler JE. Dynamics of co-infection with Bartonella henselae genotypes I and II in naturally infected cats: implications for feline vaccine development. Microb Ecol. (2017) 74:474–84. doi: 10.1007/s00248-017-0936-8
64. Gutiérrez R, Vayssier-Taussat M, Buffet JP, Harrus S. Guidelines for the isolation, molecular detection, and characterization of Bartonella species. Vector Borne Zoonotic Dis. (2017) 17:42–50. doi: 10.1089/vbz.2016.1956
65. Celina SS, Cerný J. Coxiella burnetii in ticks, livestock, pets and wildlife: a mini-review. Front Vet Sci. (2022) 9:1068129. doi: 10.3389/fvets.2022.1068129
66. González-Barrio D, Ruiz-Fons F. Coxiella burnetii in wild mammals: a systematic review. Transbound Emerg Dis. (2019) 66:662–71. doi: 10.1111/tbed.13085
67. Zendoia II, Cevidanes A, Hurtado A, Vázquez P, Barral M, Barandika JF, et al. Stable prevalence of Coxiella burnetii in wildlife after a decade of surveillance in northern Spain. Vet Microbiol. (2022) 268:109422. doi: 10.1016/j.vetmic.2022.109422
68. Santana MS, Hoppe EGL, Carraro PE, Calchi AC, de Oliveira LB, do Amaral RB, et al. Molecular detection of vector-borne agents in wild boars (Sus scrofa) and associated ticks from Brazil, with evidence of putative new genotypes of Ehrlichia, Anaplasma, and haemoplasmas. Transbound Emerg Dis. (2022) 69:e2808–31. doi: 10.1111/tbed.14632
69. Cornejo J, Araya P, Ibáñez D, Hormazabal JC, Retamal P, Fresno M, et al. Identification of Coxiella burnetii in tank raw cow milk: first findings from Chile. Vector Borne Zoonotic Dis. (2020) 20:228–30. doi: 10.1089/vbz.2019.2535
70. Tapia T, Olivares MF, Stenos J, Iglesias R, Díaz N, Vergara N, et al. National seroprevalence of Coxiella burnetii in Chile, 2016-2017. Pathogens. (2021) 10:531. doi: 10.3390/pathogens10050531
71. Flueck Werner T, Smith-Flueck Jo Anne M. A review of introduced cervids in Chile. Animal Prod Sci. (2012) 52:681–4. doi: 10.1071/AN11343
72. Reusken C, van der Plaats R, Opsteegh M, de Bruin A, Swart A. Coxiella burnetii (Q fever) in Rattus norvegicus and Rattus rattus at livestock farms and urban locations in the Netherlands; could Rattus spp. represent reservoirs for (re)introduction? Prev Vet Med. (2011) 101:124–30. doi: 10.1016/j.prevetmed.2011.05.003
73. Di Cataldo S, Cevidanes A, Ulloa-Contreras C, Hidalgo-Hermoso E, Gargano V, Cabello J, et al. serosurvey for spotted fever group Rickettsia and Coxiella burnetii antibodies in rural dogs and foxes, Chile. Comp Immunol Microbiol Infect Dis. (2022) 83:101769. doi: 10.1016/j.cimid.2022.101769
74. Hidalgo-Hermoso E, Cabello J, Verasay J, Moreira-Arce D, Hidalgo M, Abalos P, et al. Serosurvey for selected parasitic and bacterial pathogens in Darwin's fox (Lycalopex fulvipes): not only dog diseases are a threat. J Wildl Dis. (2022) 58:76–85. doi: 10.7589/JWD-D-21-00024
75. Breitschwerdt EB. Feline bartonellosis and cat scratch disease. Vet Immunol Immunopathol. (2008) 123:167–71. doi: 10.1016/j.vetimm.2008.01.025
76. Cherry NA, Maggi RG, Cannedy AL, Breitschwerdt EB. PCR detection of Bartonella bovis and Bartonella henselae in the blood of beef cattle. Vet Microbiol. (2009) 135:308–12. doi: 10.1016/j.vetmic.2008.09.063
77. Gutiérrez R, Cohen L, Morick D, Mumcuoglu KY, Harrus S, Gottlieb Y. Identification of different Bartonella species in the cattle tail louse (Haematopinus quadripertusus) and in cattle blood. Appl Environ Microbiol. (2014) 80:5477–83. doi: 10.1128/AEM.01409-14
78. Engbaek K, Lawson PA. Identification of Bartonella species in rodents, shrews and cats in Denmark: detection of two B. henselae variants, one in cats and the other in the long-tailed field mouse. Apmis. (2004) 112:336–41. doi: 10.1111/j.1600-0463.2004.apm1120603.x
79. Kim CM, Kim JY Yi YH, Lee MJ, Cho MR, Shah DH, Klein TA, et al. Detection of Bartonella species from ticks, mites and small mammals in Korea. J Vet Sci. (2005) 6:327–34. doi: 10.4142/jvs.2005.6.4.327
80. Matsumoto K, Cook JA, Goethert HK, Telford SR. Bartonella sp. infection of voles trapped from an interior Alaskan site where ticks are absent. J Wildlife Dis. (2010) 46:173–8. doi: 10.7589/0090-3558-46.1.173
81. Böge I, Pfeffer M, Htwe NM, Maw PP, Sarathchandra SR, Sluydts V, et al. First detection of Bartonella spp. in small mammals from rice storage and processing facilities in Myanmar and Sri Lanka. Microorganisms. (2021) 9:658. doi: 10.3390/microorganisms9030658
82. Razanske I, Rosef O, Radzijevskaja J, Klepeckiene K, Lipatova I, Paulauskas A. Infections with Bartonella spp. in free-ranging cervids and deer keds (Lipoptena cervi) in Norway. Comp Immunol Microbiol Infect Dis. (2018) 58:26–30. doi: 10.1016/j.cimid.2018.06.003
83. Sacristán C, das Neves CG, Suhel F, Sacristán I, Tengs T, Hamnes IS, Madslien K. Bartonella spp detection in ticks, Culicoides biting midges and wild cervids from Norway. Transbound Emerg Dis. (2021) 68:941–51. doi: 10.1111/tbed.13762
84. Pérez Vera C, Aaltonen K, Spillmann T, Vapalahti O, Sironen T. Geographic Distribution and molecular diversity of Bartonella spp. infections in moose (Alces alces) in Finland. J Wildl Dis. (2016) 52:209–16. doi: 10.7589/2015-05-131
85. Bai Y, Cross PC, Malania L, Kosoy M. Isolation of Bartonella capreoli from elk. Vet Microbiol. (2011) 148:329–32. doi: 10.1016/j.vetmic.2010.09.022
86. Chang CC, Chomel BB, Kasten RW, Heller RM, Kocan KM, Ueno H, et al. Bartonella spp. isolated from wild and domestic ruminants in North America. Emerg Infect Dis. (2000) 6:306–11. doi: 10.3201/eid0603.000313
87. Chitwood MC, Maggi RG, Kennedy-Stoskopf S, Toliver M, DePerno CS. Bartonella vinsonii subsp. berkhoffii in free-ranging white-tailed deer (Odocoileus virginianus). J Wildl Dis. (2013) 49:468–70. doi: 10.7589/2012-11-286
88. Pangjai D, Intachinda S, Maruyama S, Boonmar S, Kabeya H, Sato S, et al. Isolation and phylogenetic analysis of Bartonella species from Rusa deer (Rusa timorensis) in Thailand. Comp Immunol Microbiol Infect Dis. (2018) 56:58–62. doi: 10.1016/j.cimid.2017.12.005
89. Sato S, Kabeya H, Yamazaki M, Takeno S, Suzuki K, Kobayashi S, et al. Prevalence and genetic diversity of Bartonella species in sika deer (Cervus nippon) in Japan. Comp Immunol Microbiol Infect Dis. (2012) 35:575–81. doi: 10.1016/j.cimid.2012.07.001
90. Souza U, Dall'Agnol B, Michel T, Webster A, Klafke G, Martins JR, et al. Detection of Bartonella sp in deer louse flies (Lipoptena mazamae) on gray brocket deer (Mazama gouazoubira) in the neotropics. J Zoo Wildl Med. (2017) 8:532–5. doi: 10.1638/2016-0058R3.1
91. Sacristán I, Sieg M, Acuña F, Aguilar E, García S, López MJ, et al. Molecular and serological survey of carnivore pathogens in free-roaming domestic cats of rural communities in southern Chile. J Vet Med Sci. (2019) 81:1740–8. doi: 10.1292/jvms.19-0208
92. Sepúlveda-García P, Raffo E, Medina-Vogel G, Muñoz F, Muñoz P, Alabí A, et al. Molecular survey of Bartonella spp. and haemoplasmas in American minks (Neovison vison). Transbound Emerg Dis. (2021) 68:2094–110. doi: 10.1111/tbed.13857
93. Mau A, Calchi AC, Bittencourt P, Navarrete-Talloni MJ, Sauvé C, Conan A, et al. Molecular survey and genetic diversity of Bartonella spp. in small Indian mongooses (Urva auropunctata) and their fleas on Saint Kitts, West Indies. Microorganisms. (2021) 9:1350. doi: 10.3390/microorganisms9071350
94. Pedrassani D, Biolchi J, Gonçalves LR, Mendes NS, Zanatto DCS, Calchi AC, et al. Molecular detection of vector-borne agents in cats in Southern Brazil. Rev Bras Parasitol Vet. (2019) 28:632–43. doi: 10.1590/s1984-29612019077
95. Vayssier-Taussat M, Moutailler S, Féménia F, Raymond P, Croce O, La Scola B, et al. Identification of novel zoonotic activity of Bartonella spp, France. Emerg Infect Dis. (2016) 22:457–62. doi: 10.3201/eid2203.150269
96. Antequera-Gómez ML, Lozano-Almendral L, Barandika JF, González-Martín-Niño RM, Rodríguez-Moreno I, García-Pérez AL, et al. Bartonella chomelii is the most frequent species infecting cattle grazing in communal mountain pastures in Spain. Appl Environ Microbiol. (2015) 81:623–9. doi: 10.1128/AEM.03159-14
97. Maggi RG, Kosoy M, Mintzer M, Breitschwerdt EB. Isolation of Candidatus Bartonella melophagi from human blood. Emerg Infect Dis. (2009) 15:66–8. doi: 10.3201/eid1501.081080
98. Mathews KO, Toribio J-A, Norris JM, Phalen D, Wood N, Graves SR, et al. Coxiella burnetii seroprevalence and Q fever in Australian wildlife rehabilitators. One Health. (2021) 12:100197. doi: 10.1016/j.onehlt.2020.100197
Keywords: zoonotic diseases, wildlife host, endangered species, Coxiella burnetii, hemoplasmas
Citation: Hidalgo-Hermoso E, Sepúlveda-García P, Cabello J, Celis S, Valencia C, Ortiz C, Kemec I, Moreira-Arce D, Orsola M, Canales N, Garnham A, Vera F and Muller A (2023) Molecular survey and phylogenetic analysis of Bartonella sp., Coxiella sp., and hemoplamas in pudu (Pudu puda) from Chile: first report of Bartonella henselae in a wild ungulate species. Front. Vet. Sci. 10:1161093. doi: 10.3389/fvets.2023.1161093
Received: 07 February 2023; Accepted: 28 September 2023;
Published: 16 November 2023.
Edited by:
Georgiana Deak, University of Agricultural Sciences and Veterinary Medicine of Cluj-Napoca, RomaniaReviewed by:
Aleksandar Potkonjak, University of Novi Sad, SerbiaCopyright © 2023 Hidalgo-Hermoso, Sepúlveda-García, Cabello, Celis, Valencia, Ortiz, Kemec, Moreira-Arce, Orsola, Canales, Garnham, Vera and Muller. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ezequiel Hidalgo-Hermoso, ZXplcXVpZWxoaWRhbGdvdmV0QHlhaG9vLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.