- 1Guangdong Provincial Key Laboratory of Livestock Disease Prevention, Institute of Animal Health Guangdong Academy of Agricultural Sciences, Scientific Observation and Experiment Station of Veterinary Drugs and Diagnostic Techniques of Guangdong Province, Guangzhou, Guangdong, China
- 2Maoming Branch Center of Guangdong Laboratory for Lingnan Modern Agricultural Science and Technology, Maoming, China
Currently, porcine coronaviruses are prevalent in pigs, and due to the outbreak of COVID-19, porcine coronaviruses have become a research hotspot. porcine epidemic diarrhea virus (PEDV), Transmissible Gastroenteritis Virus (TGEV), and Porcine Deltacoronavirus (PDCoV) mentioned in this study mainly cause diarrhea in pigs. These viruses cause significant economic losses and pose a potential public health threat. In this study, specific primers and probes were designed according to the M gene of PEDV, the S gene of TGEV, and the M gene of PDCoV, respectively, and TaqMan probe-based multiplex real-time quantitative reverse transcription-polymerase chain reaction (qRT-PCR) was developed for the simultaneous detection of PEDV, TGEV, and PDCoV. This method has high sensitivity and specificity, and the detection limit of each virus can reach 2.95 × 100 copies/μl. An assay of 160 clinical samples from pigs with diarrhea showed that the positive rates of PEDV, TGEV, and PDCoV were 38.13, 1.88, and 5.00%; the coinfection rates of PEDV+TGEV, PEDV+PDCoV, TGEV+PDCoV, PEDV+TGEV+PDCoV were 1.25, 1.25, 0, 0.63%, respectively. The positive coincidence rates of the multiplex qRT-PCR and single-reaction qRT-PCR were 100%. This method is of great significance for clinical monitoring of the porcine enteric diarrhea virus and helps reduce the loss of the breeding industry and control the spread of the disease.
Introduction
Coronaviruses belonging to the family Coronaviridae, the order Nidovirales, are single-stranded, positive-sense RNA viruses with the largest genome among known RNA viruses (1–3). Within swine enteric viruses, coronaviruses are the most devastating pathogens responsible for acute diarrhea, vomiting, dehydration, and high mortality in neonatal and suckling piglets (4–6).
According to the genetic and antigenic characteristics, all coronaviruses were divided into four genera: Alphacoronaviruses, Betacoronaviruses, Gammacoronaviruses, and Deltacoronaviruses (7). Novel coronavirus whole genome sequencing analysis showed that alpha-coronaviruses and beta-coronaviruses infected mammals; gamma-coronaviruses and delta-coronaviruses mainly infected birds, but some viruses could also infect mammals (6).
Coronaviruses, including PEDV, TGEV, and PDCoV, can cause diarrhea in piglets. PEDV and TGEV are alphacoronaviruses, while PDCoV is delta coronavirus.
PEDV was first isolated from porcine intestinal contents in the United Kingdom in 1978. Since then, PEDV has spread worldwide and isolated in many countries, including the USA, the UK, Argentina, Russia, and China, resulting in heavy economic losses to the porcine industry (8–10).
TGEV was first reported in 1946 in the United States, followed by outbreaks in many countries in the Americas, Asia, and Europe. TGEV has the greatest impact on piglets, especially those under 2 weeks of age, who are most susceptible to infection. Piglets often excrete feces containing undigested curds, with mortality rates often reaching 100% (11–13).
PDCoV was identified in pigs in Hong Kong in late 2012 (14). To date, PDCoV has been detected in at least 20 states in the United States, as well as in Canada, South Korea, China, Thailand, Lao People's Democratic Republic, Vietnam, and Mexico, posing a significant threat to the global swine industry (11, 15–19). In addition, PDCoV has been detected in poultry and humans, reflecting the potential for cross-species transmission (20, 21).
The clinical symptoms of the intestinal diseases caused by these swine coronaviruses are highly similar, and it can be challenging to distinguish them. It is important to note that coronaviruses tend to interspecies transmission, as exemplified by SARS-CoV-2 and PDCoV (22–27). By monitoring the epidemiology of coronaviruses in pigs, the potential for cross-species transmission of these viruses and the trend of cross-regional transmission of viruses can be well studied.
So far, PEDV, TGEV, and PDCoV have caused huge economic losses to the pig industry worldwide. In addition, some viruses' cross-species transmission ability may threaten public health. Therefore, developing a simple, rapid, accurate, and high-throughput detection method is necessary to distinguish porcine enteric coronaviruses. A multiplex qRT-PCR detection method based on TaqMan probes was established to detect three viruses, PEDV, TGEV, and PDCoV. This method will improve the virus's detection efficiency and accuracy while reducing the detection cost.
Experimental section
Primers and probes
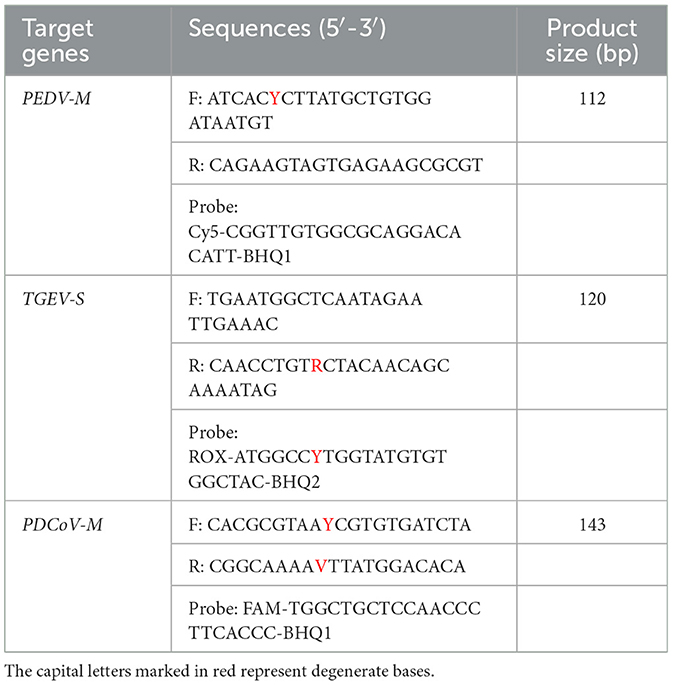
To ensure the detection performance of the primers used in the multiplex qRT-PCR method, all available PEDV, TGEV, and PDCoV sequences were obtained and analyzed from GenBank. The primers and probes of PEDV, TGEV, and PDCoV were designed according to the M gene of PEDV, the S gene of TGEV, and the M gene of PDCoV. Three primer pairs and probes were designed using Oligo 6 (Version 6.44) software (Table 1). TaqMan probes for PEDV, TGEV, and PDCoV were labeled at the 5′-end with the reporter molecule: X-Rhodamine maleimide (ROX), pentamethylene cyanine (CY5) and 5(6)-carboxyfluorescein (FAM), respectively. The 3′-end of TaqMan probes were labeled with the quenchers: 8-Bromo-7-hydroxyquinoline 1 (BHQ1), 8-Bromo-7-hydroxyquinoline 2 (BHQ2), 8-Bromo-7-hydroxyquinoline 2 (BHQ2). Primers and probes were synthesized by Sangon Biotechnology (Shanghai) Co., Ltd. Primers were also used to construct plasmid standards.
Virus strains and field samples
RNA extraction and reverse transcription treat intestinal tissue or stool samples with 3 to 5 times PBS, vortex to mix, and collect supernatant after centrifugation at 12,000 × g for 15 min at 4°C. Nucleic acids were extracted using the E.Z.N.A.® Total RNA Kit (Omega Bio-Tek, China) following the manufacturer's instructions. Reverse transcription was performed using the HiScript III RT SuperMix for qPCR (+gDNAwiper) kit (Nanjing Vazyme Biotechnology Co., Ltd.).
Construction of plasmid standards
The target fragments of PEDV, TGEV, and PDCoV were amplified by PCR using PrimeScript™ High-Fidelity RT-PCR Kit [Bao Biomedical Technology (Beijing) Co., Ltd]. The PCR fragment was then cloned into a pUC57 vector [Takara Biomedical Technology (Beijing) Co., Ltd.] by TA colony and confirmed by DNA sequencing, p-PEDV-TGEV-PDCoV. The plasmid copy number was calculated, diluting the plasmids from 2.95 × 107 to 2.95 × 100 copies/μl. Single-reaction qRT-PCR was performed for each virus using the 10-fold diluted plasmids to generate standard curves.
Reaction conditions of the single-plex qRT-PCR
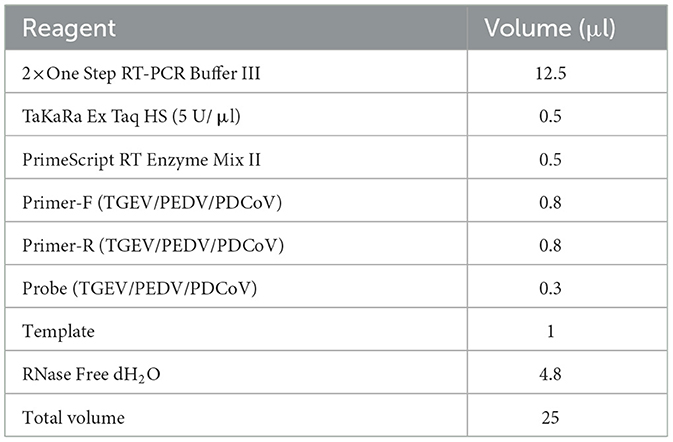
As shown in Table 2, the total volume of the single-reaction qRT-PCR reaction was 25 μl. Amplification was carried out on Gentier 96R (Xi'an Tianlong Science and Technology) using the following program: 95°C 10 s; 40 cycles of 95°C 5 s, 60°C 30 s. The fluorescence signal was automatically collected at the end of each cycle, 35°C 30 s.
Optimization of the multiplex qRT-PCR
The multiplex reaction system was optimized using different volumes of primers (10 μM) and probes (10 μM). In the optimization stage, the number of primers added to the system was 0.4, 0.6, 0.8, 1.0, and 1.2 μl, respectively. The number of probes added to the system was 0.1, 0.2, 0.3, 0.4, 0.5, and 0.6 μl, respectively. The recombinant plasmid (2.95 × 107 copies/μl) was used as the standard plasmid. Finally, each primer and probe concentration's fluorescence intensity and cycle threshold (Ct) value were compared. The same instrument and qRT-PCR program were used as described above. To obtain the best amplification efficiency, the annealing temperature was optimized. Twelve annealing temperature gradients were set up from 44 to 64°C, and each annealing temperature's fluorescence intensity and Ct values were compared.
Sensitivity of the multiplex qRT-PCR assay
The standard plasmids were serially diluted 10-fold from 2.95 × 107 to 0.295 × 100 copies/μl (final concentration) for the standard plasmid and used as templates to evaluate the sensitivity of the developed assay, and the reaction does three repetitions at a time.
Specificity of the multiplex qRT-PCR assay
To verify the specificity of the assay, positive samples for PEDV, TGEV, PDCoV, SADS-CoV, porcine rotavirus (PoRV), porcine pseudorabies virus (PRV), porcine circovirus 2 (PCV-2), porcine reproductive and respiratory syndrome (PRRSV) were used for detection by the developed multiplex qRT-PCR. All the samples were previously stored in our laboratory.
Repeatability of the multiplex qRT-PCR assay
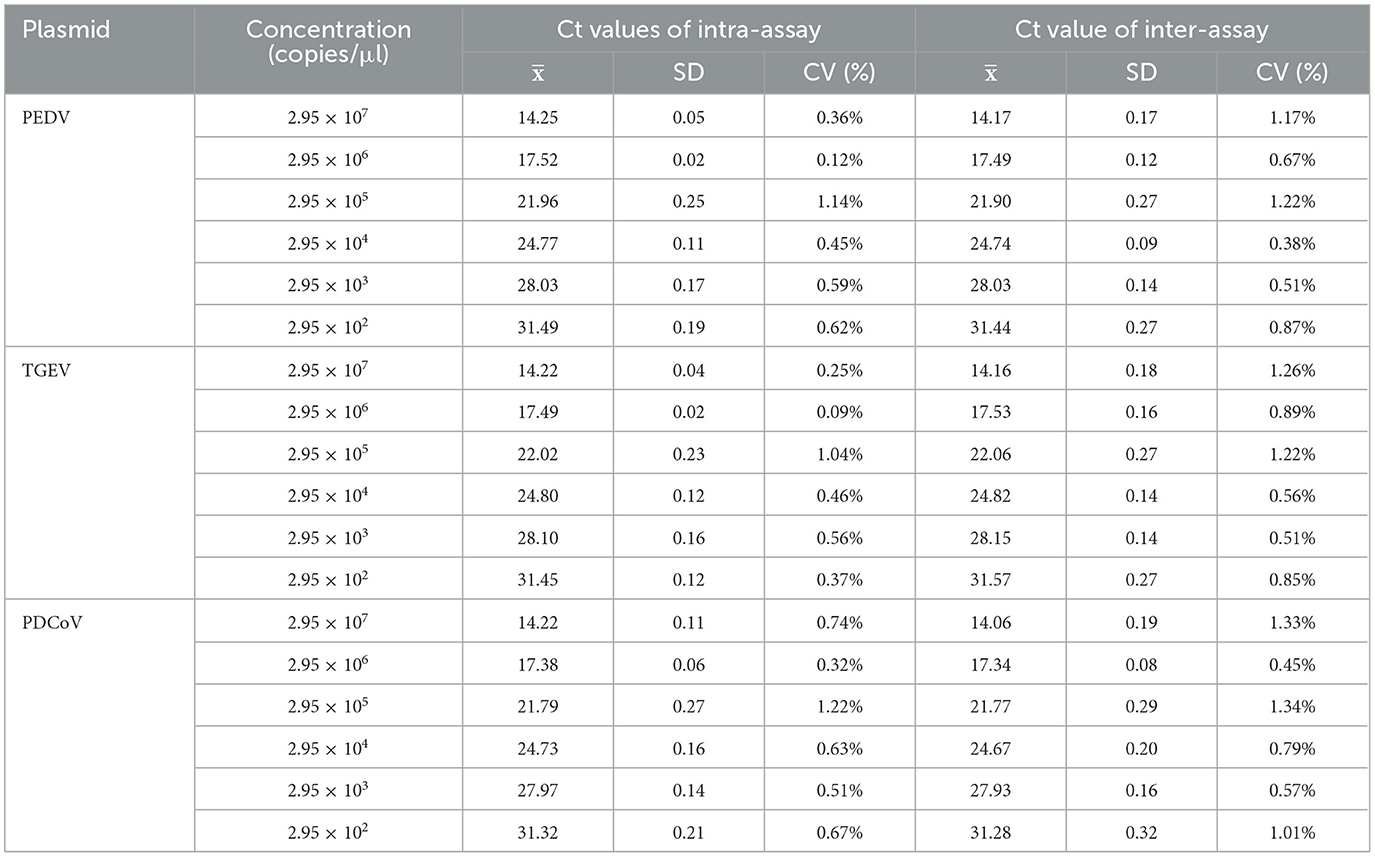
The assay was repeated three times with a 7-days interval, using 10-fold dilutions of the standard plasmid of each pathogen ranging from 2.95 × 107 to 2.95 × 102 copies/μl with three replicates per reaction. The coefficient of variation (CV) of the Ct values of the samples at each concentration in the three experiments was calculated to estimate repeatability.
Clinical sample detection
To test the clinical application effect of the developed method, a total of 160 clinical samples collected from pigs with symptoms of diarrhea, including 70 small intestinal tissues and 90 anal swabs, were used for detection. At the same time, a single-plex qRT-PCR was used to verify the results, and the results were compared and analyzed.
Results
Establish a standard curve
The standard plasmids, ranging from 2.95 × 107 to 2.95 × 102 copies/μl, were used to create standard curves. The standard curves showed an acceptable amplification efficiency and correlation coefficient: PEDV, R2 = 0.9983; TGEV, R2 = 0.9986 and PDCoV, R2 = 0.9978, and these results showed that the designed primers and probes were effective (Figure 1).
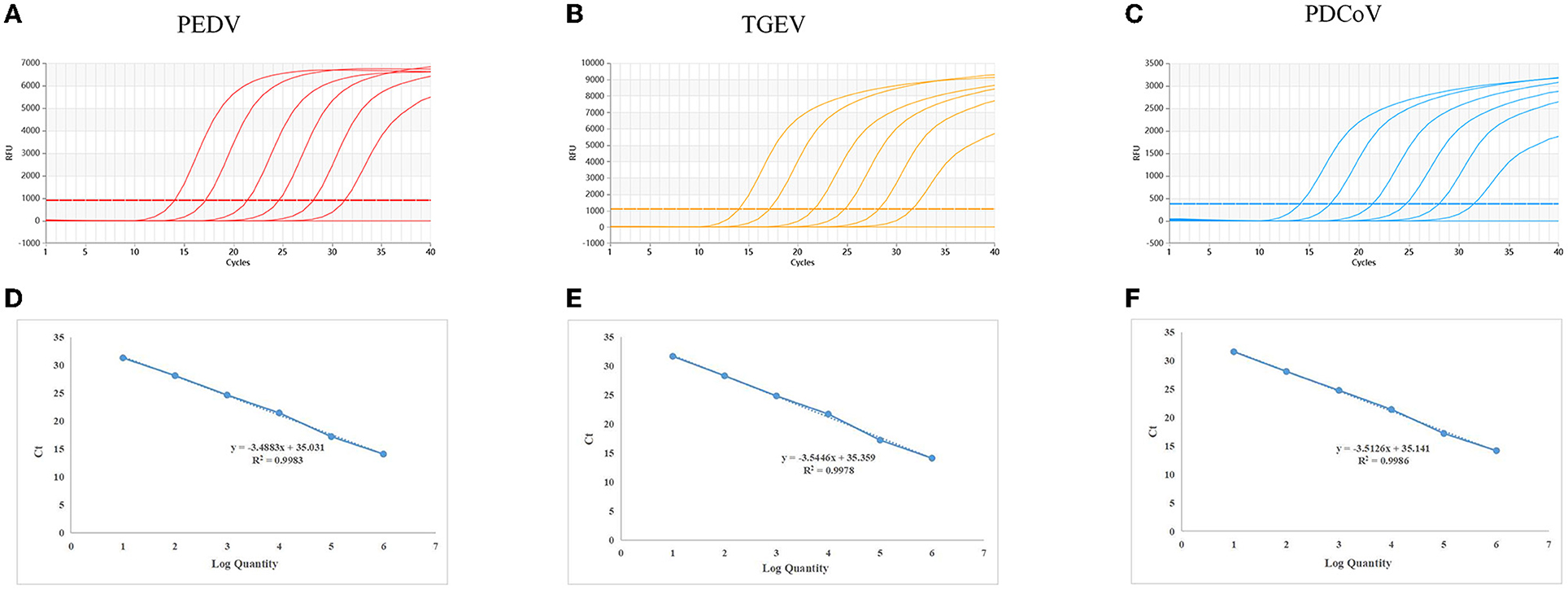
Figure 1. Dynamic curves and standard curves of the multiplex qRT-PCR: The dynamic curves were generated using the recombinant standard plasmids p-PEDV-TGEV-PDCoV for PEDV, TGEV, and PDCoV. The standard curves (D–F) were generated from the dynamic curves. In (A–C), the plasmid concentrations of curves 1 to 6 ranged from 2.95 × 107 to 2.95 × 102 copies/μl; 7: Negative control.
Optimization of the multiplex reaction system
The standard plasmids p-PEDV-TGEV-PDCoV carrying the target fragments were used as templates to optimize the reaction conditions of the multiplex qRT-PCR. The optimal annealing temperature and the concentrations of primers and probes were acquired based on orthogonal experiments.
The results show that the optimal volume of probe and primer for PEDV and TGEV are 0.3 and 0.8 μl, respectively. The optimal volume of probes and primers for PDCoV were 0.2 and 0.6 μl, respectively (Figure 2). The results of annealing temperature optimization showed that 50.5°C was the optimal annealing temperature and the reaction had the best amplification efficiency (Figure 3).
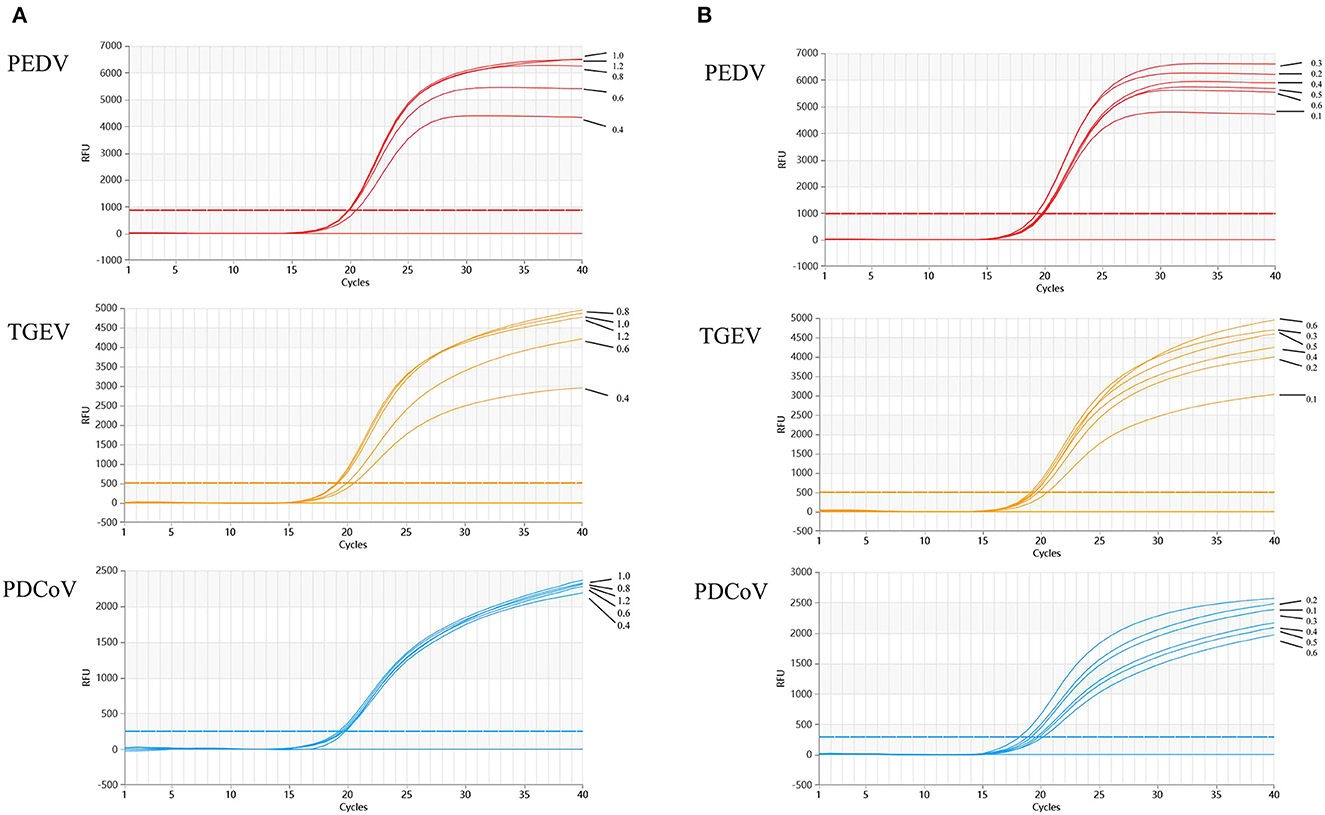
Figure 2. Optimization of multiplex qRT-PCR system. (A) shows the results of primers optimization in the reaction system, which showed that the optimal addition amount of upstream and downstream primers for TGEV and PEDV are 0.8 μl; the optimal addition amount of upstream and downstream primers for PDCoV is 0.6 μl. Optimization of multiplex qRT-PCR system. (B) Multiplex qRT-PCR probe volume optimizations. Probes optimization in the reaction system showed that the optimal addition number of probes for TGEV and PEDV is 0.3 μl; the optimal amount of probes for PDCoV is 0.2 μl.
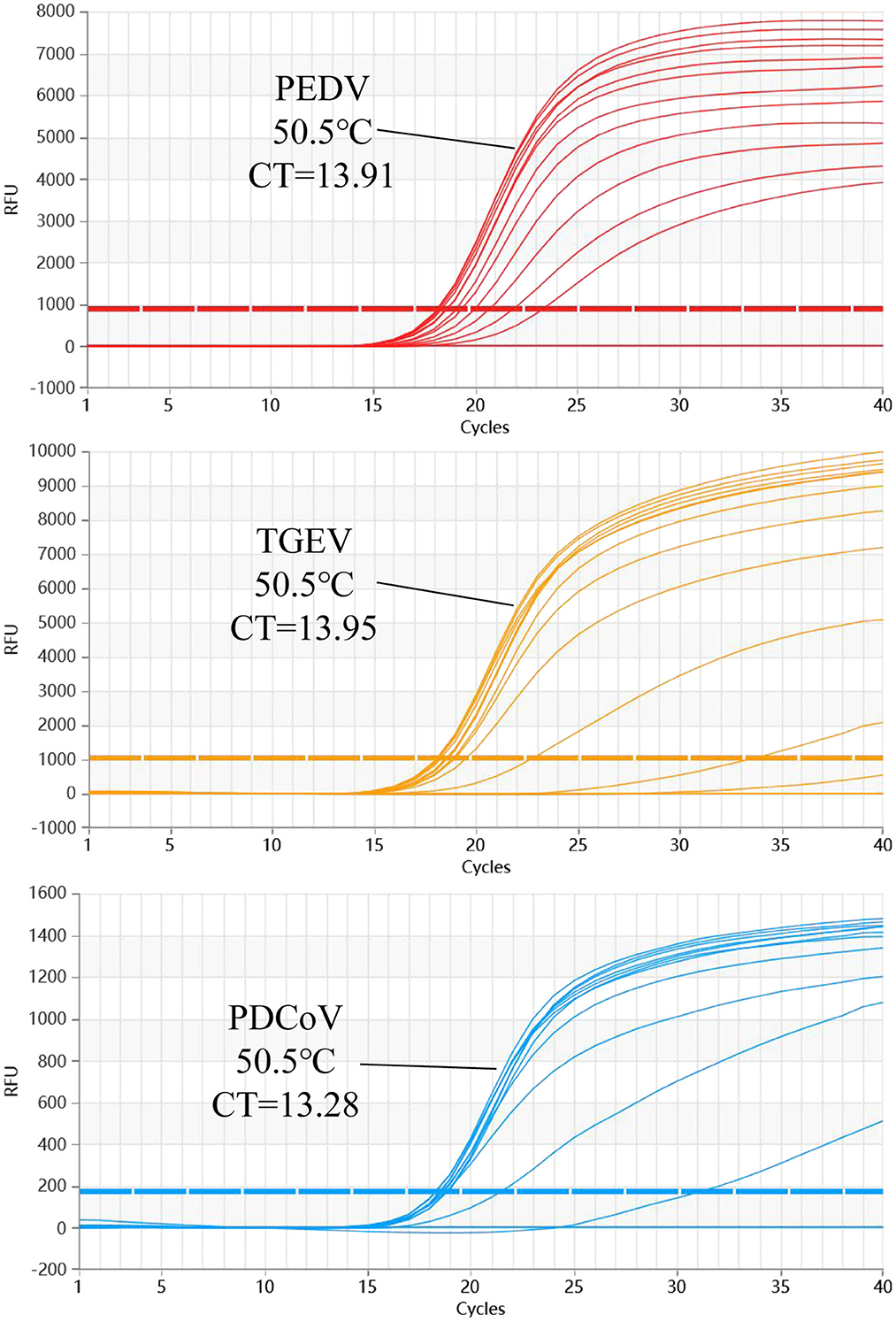
Figure 3. The multiplex qRT-PCR amplification curves of different Tm values. The annealing temperature optimization results of multiplex qRT-PCR. The results show that the amplification efficiency of this reaction is the highest when Tm = 50.5°C.
Sensitivity of the multiplex qRT-PCR assay
The sensitivity results showed that the detection limit of the developed multiplex qRT-PCR assay was 2.95 × 100 copies/μl of the standard plasmid (Figure 4).
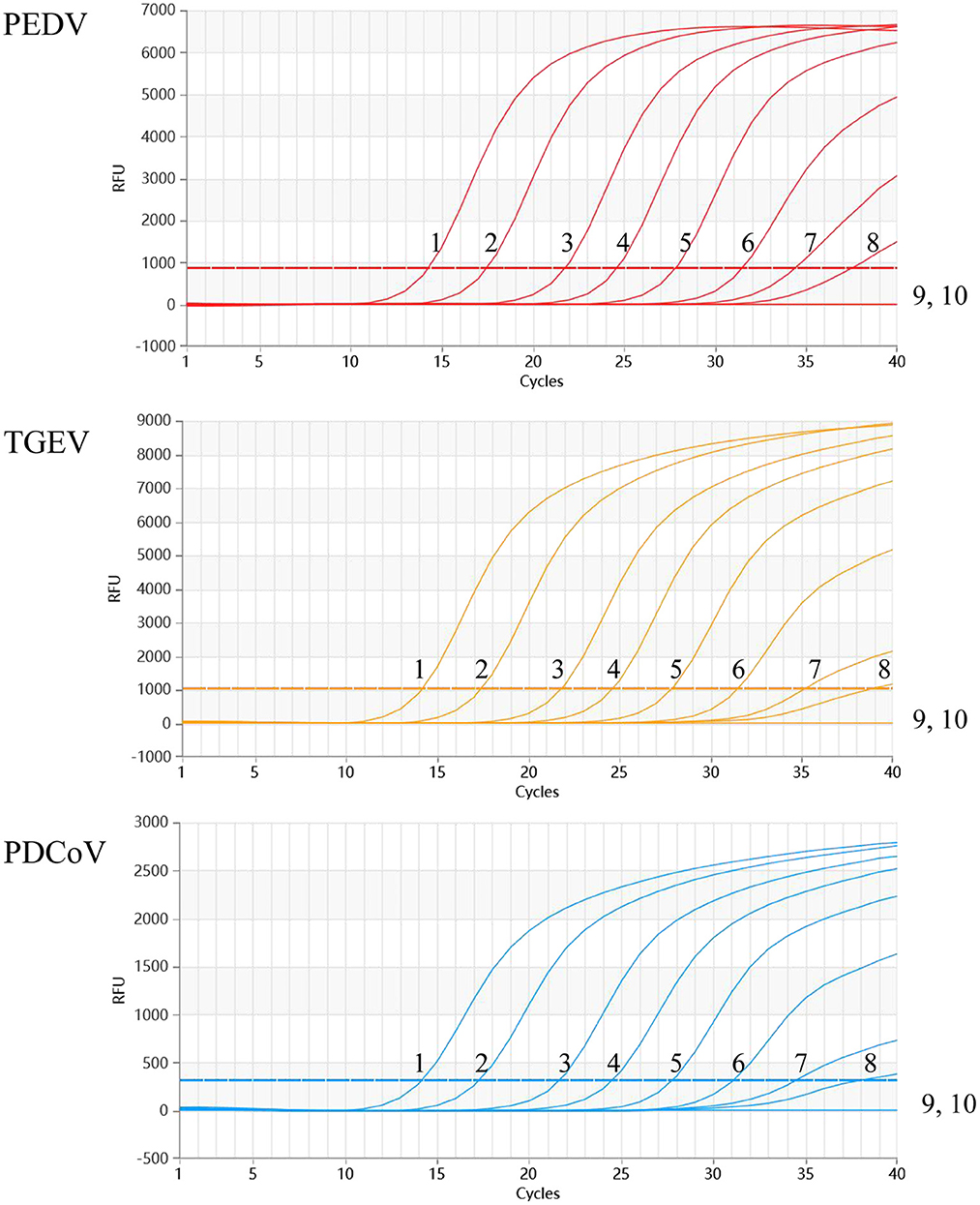
Figure 4. Sensitivity of the multiplex qRT-PCR assay. The dynamic curves were generated by using the recombinant standard plasmid p-PEDV-TGEV-PDCoV. 1-9: 2.95 × 107-0.219 × 100 copies/μl (final concentration); 10: Negative control.
Specificity of the multiplex qRT-PCR assay
As shown in Figure 5, the positive samples for PEDV, TGEV, and PDCoV could be detected, while no amplification curve was observed with other viruses of pigs used in the assay.
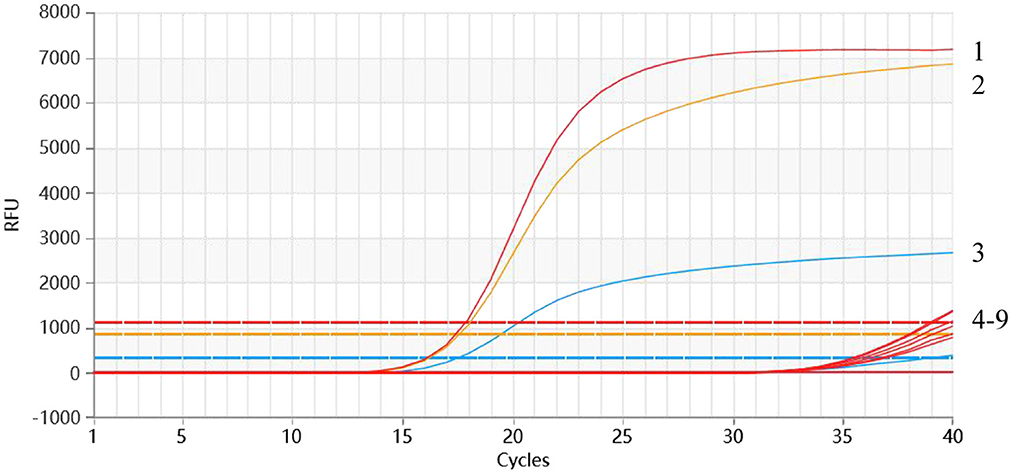
Figure 5. Specificity analysis of the multiplex qRT-PCR. 1-3: p-PEDV-TGEV-PDCoV (2.95 × 106 copies/μl); 4: PRRSV; 5: SADS; 6: PRV; 7: PoRV; 8: PCV2; 9: Negative control.
Repeatability of the multiplex qRT-PCR assay
The repeatability experiment was carried out with the serially diluted standard plasmids (2.95 × 107 to 2.95 × 102 copies/μl) as templates, and the Ct value of the experiment was calculated. As shown in Table 3, the coefficients of variation (CVs) of the intra- and inter-assay ranged from 0.12 to 1.22% and 0.38–1.34%, respectively, <2%. The experimental results showed that the developed multiplex method was stable.
Clinical sample detection
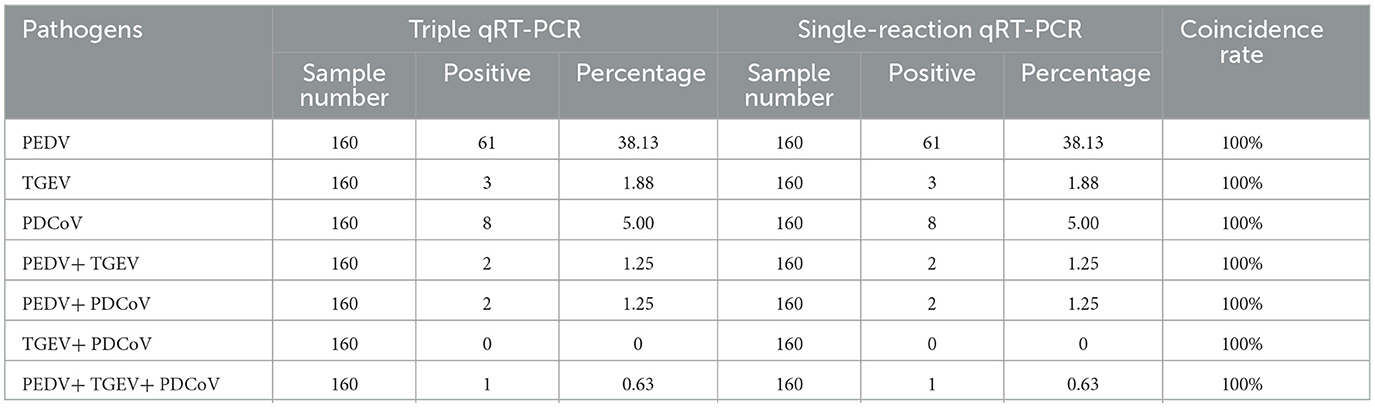
A total of 160 clinical samples, including 70 intestinal tissue samples and 90 anal swab samples from pig farms where diarrhea occurred in southern China, were tested by the developed multiplex qRT-PCR. The results of clinical sample tests showed that the positive rates of PEDV, TGEV, and PDCoV were 38.13% (61/160), 1.88% (3/160), and 5.00% (8/160), respectively. Additionally, the results also showed that 2 (1.25%) samples were coinfected by PEDV and PDCoV; 2 (1.25%) samples were coinfected by PEDV and TGEV; no coinfection of TGEV and PDCoV, and 1 (0.63%) sample was coinfected with PEDV, TGEV, and PDCoV. To verify the accuracy of the developed method, the clinical samples were detected by single-plex qRT-PCR, and the results of single-plex and multiplex qRT-PCR were compared and analyzed. The coincidence rate of the two methods was 100%, which confirmed that the results of the developed method were accurate and reliable (Table 4).
Discussion
In recent years, the viral diarrhea of piglets has still seriously threatened the development of the pig industry, causing major economic losses to global pig farmers. Clinically, PEDV, TGEV, and PDCoV are piglets' main pathogens causing viral diarrhea (28, 29). The epidemiological survey of pig diarrhea viruses showed that PEDV, TGEV, and PDCoV exhibited a trend of hybrid infection, including PEDV and PDCoV hybrid infections, PEDV and TGEV hybrid infections, and PEDV, TGEV, and PDCoV mixed infections (12, 30). The cause of the intestinal disease of piglets with hybrid infection of these viruses is becoming increasingly complicated. The clinical symptoms caused by PEDV, TGEV, and PDCoV exhibited high similarities, leading to the difficulty of determining the pathogens through clinical symptoms. Studies have shown that hybrid infections of multiple viruses may accelerate the common evolution of single and hybrid viruses or the reorganization of multiple viruses into new viruses, as reported by TGEV and PEDV reorganized strains in 2016 (31–34). Reorganization may produce intestinal virus strains or new viruses, which may cause potential outbreaks or popularity of pig viral diarrhea. Due to the global prevalence of SARS-CoV-2, coronavirus has attracted the great attention of scientists. It is worth noting that the newly discovered pork intestine coronavirus PDCoV has been detected in the infection cases of other species (including humans), indicating the great species crossing potential of PDCoV and a great threat to human public health (20).
Real-time quantitative fluorescent PCR is more sensitive, faster, and more accurate for detecting viral pathogens (35, 36). The multiplex RT-qPCR can detect and differentiate more than one pathogen in a single assay, which is particularly suitable for detecting mixed infection of multiple pathogens. In this study, a real-time multiplex PCR based on three pairs of specific primers and probes was developed for the differential detection of PEDV, TGEV, and PDCoV in one reaction. The developed multiplex qRT-PCR could specifically detect PEDV, TGEV, and PDCoV with the LOD of 2.95 × 100 copies/μl for each pathogen. In multiplex qRT-PCR systems, the primers and probes must have good specificity. Because multiplex groups of oligonucleotides in the system will increase the possibility of non-specific amplification, it is necessary for probes and primers with excellent specificity.
The developed multiplex assay was also used to detect clinical diarrhea samples of piglets from southern China. Among the 160 samples, 61 samples were positive for PEDV, 3 samples for TGEV, and 8 samples for PDCoV, indicating that PEDV was still the major pathogen of piglet diarrhea, while the sporadic infections of PDCoV should also be highly concerned, as the increased detection rate of PDCoV in recent clinical samples. Furthermore, coinfection of PEDV, TGEV, and PDCoV had existed in certain pig herds, confirmed by the detection results that coinfection rates of PEDV/TGEV, PEDV/PDCoV, PEDV/TGEV/PDCoV were 1.25, 1.25, and 0.63%, respectively. Based on the results mentioned above, the developed multiplex real-time PCR could be a useful tool for rapid differentiation of PEDV, TGEV, and PDCoV in clinical samples from piglets with diarrhea and warning the infection of PDCoV.
Conclusion
In conclusion, an excellent detection method should accurately reflect the pathogen's epidemiological data and help researchers monitor and prevent the virus more effectively. Although multiplex qRT-PCR has the advantages of saving time and capital costs, due to the high requirements for primers and probes, it still needs to be improved to develop effective multiplex qRT-PCR, often accompanied by many challenges. The development method can quickly and accurately detect three kinds of porcine diarrhea viruses, PEDV, TGEV, and PDCoV, simultaneously, effectively saving time and cost. The prevalence of different diarrhea viruses in piglets will increase the complexity of diarrhea and make it difficult to prevent and control. As a potential zoonotic pathogen, the prevalence of PDCoV in piglets should also attract continuous attention. Above all, more attention should be given to the molecular prevalence of PEDV, TGEV, and PDCoV, guiding precise prevention and control more effectively in the field.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding authors.
Author contributions
YL, J-WN, and XZ performed the experiments and drafted the manuscript. J-WN, P-PC, and D-XY prepared materials for the experiments. YL and J-WN participated in the experiments. K-LZ, H-CG, C-LL, and S-LZ contributed to the data analysis. J-FZ, ML, and S-LZ conceived the study. All authors read and approved the final manuscript.
Funding
We are supported by the grants from Guangzhou Science and Technology Bureau (202206010192), Natural Science Foundation of China (No. U22A20520), Guangdong Academy of Agricultural Sciences (XTXM202202, R2020PY-JC001, and 202122TD), Guangdong Provincial Department of Science and Technology (2021B1212050021), Department of Agriculture and Rural Affairs of Guangdong Province (2022KJ114), and the open competition program of top ten critical priorities of Agricultural Science and Technology Innovation for the 14th Five-Year Plan of Guangdong Province (No. 2022SDZG02).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Brian DA, Baric RS. Coronavirus Genome Structure and Replication. Berlin, Heidelberg: Springer Berlin Heidelberg (2005). p. 1–30.
2. Kroneman A, Cornelissen LAHM, Horzinek MC, de Groot RJ, Egberink HF. Identification and characterization of a porcine Torovirus. J Virol. (1998) 72:3507–11. doi: 10.1128/JVI.72.5.3507-3511.1998
3. Liao ME, Xie Y, Shi M, Cui J. Over two decades of research on the marine RNA virosphere. iMeta. (2022). doi: 10.1002/imt2.59
4. Huang X, Chen J, Yao G, Guo Q, Wang J, Liu G, et al. TaqMan-probe-based multiplex real-time RT-qPCR for simultaneous detection of porcine enteric coronaviruses. Appl Microbiol Biotechnol. (2019) 103:4943–52. doi: 10.1007/s00253-019-09835-7
5. Li G, Wu M, Li J, Cai W, Xie Y, Si G, et al. Rapid detection of porcine deltacoronavirus and porcine epidemic diarrhea virus using the duplex recombinase polymerase amplification method. J Virol Methods. (2021) 292:114096. doi: 10.1016/j.jviromet.2021.114096
6. Woo PCY, Huang Y, Lau SKP, Yuen K. Coronavirus genomics and bioinformatics analysis. Viruses. (2010) 2:1804–20. doi: 10.3390/v2081803
7. Wang Q, Vlasova AN, Kenney SP, Saif LJ. Emerging and re-emerging coronaviruses in pigs. Curr Opin Virol. (2019) 34:39–49. doi: 10.1016/j.coviro.2018.12.001
8. Ma L, Lian K, Zhu M, Tang Y, Zhang M. Visual detection of porcine epidemic diarrhea virus by recombinase polymerase amplification combined with lateral flow dipstrip. BMC Vet Res. (2022) 18:140. doi: 10.1186/s12917-022-03232-5
9. Pensaert MB, de Bouck P. A new coronavirus-like particle associated with diarrhea in swine. Arch Virol. (1978) 58:243–47. doi: 10.1007/BF01317606
10. Yang Y, Yu J, Huang Y. Swine enteric alphacoronavirus (swine acute diarrhea syndrome coronavirus): An update three years after its discovery. Virus Res. (2020) 285:198024. doi: 10.1016/j.virusres.2020.198024
11. Hu X, Li N, Tian Z, Yin X, Qu L, Qu J. Molecular characterization and phylogenetic analysis of transmissible gastroenteritis virus HX strain isolated from China. Bmc Vet Res. (2015) 11:72. doi: 10.1186/s12917-015-0387-8
12. Liu Q, Wang H. porcine enteric coronaviruses: an updated overview of the pathogenesis, prevalence, and diagnosis. Vet Res Commun. (2021) 45:75–86. doi: 10.1007/s11259-021-09808-0
13. Xia L, Yang Y, Wang J, Jing Y, Yang Q. Impact of TGEV infection on the pig small intestine. Virol J. (2018) 15:102. doi: 10.1186/s12985-018-1012-9
14. Woo PCY, Lau SKP, Lam CSF, Lau CCY, Tsang AKL, Lau JHN, et al. Discovery of seven novel mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J Virol. (2012) 86:3995–4008. doi: 10.1128/JVI.06540-11
15. Liu C, Zhang X, Zhang Z, Chen R, Zhang Z, Xue Q. Complete genome characterization of novel Chinese porcine deltacoronavirus strain SD. Genome Announcements. (2017) 5:17. doi: 10.1128/genomeA.00930-17
16. Madapong A, Saeng-chuto K, Lorsirigool A, Temeeyasen G, Srijangwad A, Tripipat T, et al. Complete genome sequence of porcine deltacoronavirus isolated in Thailand in 2015. Genome Announcements. (2016) 4:16. doi: 10.1128/genomeA.00408-16
17. Pérez Rivera C, Ramírez Mendoza H, Mendoza Elvira S, Segura Velázquez R, Sánchez Betancourt JI. First report and phylogenetic analysis of porcine deltacoronavirus in Mexico. Transbound Emerg Dis. (2019) 66:1436–41. doi: 10.1111/tbed.13193
18. Wang L, Byrum B, Zhang Y. Detection and genetic characterization of deltacoronavirus in Pigs, Ohio, USA, 2014. Emerg Infect Dis. (2014) 20:1227–30. doi: 10.3201/eid2007.140296
19. Woo PCY, Lau SKP, Yip CCY, Huang Y, Tsoi H, Chan K, et al. Comparative analysis of 22 coronavirus HKU1 genomes reveals a novel genotype and evidence of natural recombination in coronavirus HKU1. J Virol. (2006) 80:7136–45. doi: 10.1128/JVI.00509-06
20. Boley PA, Alhamo MA, Lossie G, Yadav KK, Vasquez-Lee M, Saif LJ, et al. porcine delta coronavirus infection and transmission in poultry, United States1. Emerg Infect Dis. (2020) 26:255–65. doi: 10.3201/eid2602.190346
21. Lednicky JA, Tagliamonte MS, White SK, Elbadry MA, Alam MM, Stephenson CJ, et al. Emergence of porcine delta-coronavirus pathogenic infections among children in Haiti through independent zoonoses and convergent evolution. medRxiv. (2021). doi: 10.1101/2021.03.19.21253391
22. Gralinski LE, Menachery VD. Return of the coronavirus: 2019-nCoV. Viruses. (2020) 12:135. doi: 10.3390/v12020135
23. Li G, Zhai SL, Zhou X, Chen TB, Niu JW, Xie YS, et al. Phylogeography and evolutionary dynamics analysis of porcine delta-coronavirus with host expansion to humans. Transbound Emerg Dis. (2022) 69:14503. doi: 10.1111/tbed.14503
24. Pekar JE, Magee A, Parker E, Moshiri N, Izhikevich K, Havens JL, et al. The molecular epidemiology of multiple zoonotic origins of SARS-CoV-2. Science. (2022) 377:960–66. doi: 10.1126/science.abp8337
25. Sun J, He W, Wang L, Lai A, Ji X, Zhai X, et al. COVID-19: epidemiology, evolution, and cross-disciplinary perspectives. Trends Mol Med. (2020) 26:483–95. doi: 10.1016/j.molmed.2020.02.008
26. Zhai SL Li CL, Sun MF, Zhang JF, Zheng C, Liao M. Natural infections of SARS-CoV-2 increased in animals: how should humans interact with animals? J Med Virol. (2022) 94:3503–05. doi: 10.1002/jmv.27772
27. Zhai SL, Sun MF, Zhang JF, Zheng C, Liao M. Spillover infection of common animal coronaviruses to humans. Lancet Microbe. (2022) 3:e808. doi: 10.1016/S2666-5247(22)00198-7
28. Bevins SN, Lutman M, Pedersen K, Barrett N, Gidlewski T, Deliberto TJ, et al. Spillover of Swine Coronaviruses, United States. Emerg Infect Dis. (2018) 24:1390–92. doi: 10.3201/eid2407.172077
29. Zhao Y, Qu H, Hu J, Fu J, Chen R, Li C, et al. Characterization and pathogenicity of the porcine delta coronavirus isolated in Southwest China. Viruses. (2019) 11:1174. doi: 10.3390/v11111074
30. Si G, Niu J, Zhou X, Xie Y, Chen Z, Li G, et al. Use of dual priming oligonucleotide system-based multiplex RT-PCR assay to detect five diarrhea viruses in pig herds in South China. AMB Express. (2021) 11:99. doi: 10.1186/s13568-021-01255-z
31. Akimkin V, Beer M, Blome S, Hanke D, Höper D, Jenckel M, et al. New chimeric porcine coronavirus in swine feces, Germany, 2012. Emerg Infect Dis. (2016) 22:1314–15. doi: 10.3201/eid2207.160179
32. Belsham GJ, Rasmussen TB, Normann P, Vaclavek P, Strandbygaard B, Bøtner A. Characterization of a novel chimeric swine enteric coronavirus from diseased pigs in central eastern Europe in 2016. Transbound Emerg Dis. (2016) 63:595–601. doi: 10.1111/tbed.12579
33. Ding G, Fu Y, Li B, Chen J, Wang J, Yin B, et al. Development of a multiplex RT-PCR for the detection of major diarrhoeal viruses in pig herds in China. Transbound Emerg Dis. (2020) 67:678–85. doi: 10.1111/tbed.13385
34. Zhao P, Bai J, Jiang P, Tang T, Li Y, Tan C, et al. Development of a multiplex TaqMan probe-based real-time PCR for discrimination of variant and classical porcine epidemic diarrhea virus. J Virol Methods. (2014) 206:150–55. doi: 10.1016/j.jviromet.2014.06.006
35. Boniotti MB, Papetti A, Lavazza A, Alborali G, Sozzi E, Chiapponi C, et al. porcine epidemic diarrhea virus and discovery of a recombinant swine enteric coronavirus, Italy. Emerg Infect Dis. (2016) 22:83–7. doi: 10.3201/eid2201.150544
Keywords: porcine epidemic diarrhea virus, Transmissible Gastroenteritis Virus, Porcine Deltacoronavirus, porcine coronaviruses, multiplex real-time qRT-PCR
Citation: Li Y, Niu J-W, Zhou X, Chu P-P, Zhang K-L, Gou H-C, Yang D-X, Zhang J-F, Li C-L, Liao M and Zhai S-L (2023) Development of a multiplex qRT-PCR assay for the detection of porcine epidemic diarrhea virus, porcine transmissible gastroenteritis virus and porcine Deltacoronavirus. Front. Vet. Sci. 10:1158585. doi: 10.3389/fvets.2023.1158585
Received: 04 February 2023; Accepted: 27 February 2023;
Published: 16 March 2023.
Edited by:
Xiangdong Li, Yangzhou University, ChinaReviewed by:
Lang Gong, South China Agricultural University, ChinaBin Li, Jiangsu Academy of Agricultural Sciences (JAAS), China
Copyright © 2023 Li, Niu, Zhou, Chu, Zhang, Gou, Yang, Zhang, Li, Liao and Zhai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chun-Ling Li, bGNsY2xhcmVAMTYzLmNvbQ==; Ming Liao, bWxpYW9Ac2NhdS5lZHUuY24=; Shao-Lun Zhai, emhhaXNoYW9sdW5AMTYzLmNvbQ==
†These authors have contributed equally to this work
 Yan Li1†
Yan Li1† Jia-Wei Niu
Jia-Wei Niu Xia Zhou
Xia Zhou Kun-Li Zhang
Kun-Li Zhang Hong-Chao Gou
Hong-Chao Gou Jian-Feng Zhang
Jian-Feng Zhang Ming Liao
Ming Liao Shao-Lun Zhai
Shao-Lun Zhai