- Department of Biological Sciences, Graduate School of Science, The University of Tokyo, Tokyo, Japan
Mobile genetic elements (e.g., transposable elements and plasmids) and viruses display significant diversity with various life cycles, but how this diversity emerges remains obscure. We previously reported a novel and giant (180 kb long) mobile element, Teratorn, originally identified in the genome of medaka, Oryzias latipes. Teratorn is a composite DNA transposon created by a fusion of a piggyBac-like DNA transposon (piggyBac) and a novel herpesvirus of the Alloherpesviridae family. Genomic survey revealed that Teratorn-like herpesviruses are widely distributed among teleost genomes, the majority of which are also fused with piggyBac, suggesting that fusion with piggyBac is a trigger for the life-cycle shift of authentic herpesviruses to an intragenomic parasite. Thus, Teratorn-like herpesvirus provides a clear example of how novel mobile elements emerge, that is to say, the creation of diversity. In this review, we discuss the unique sequence and life-cycle characteristics of Teratorn, followed by the evolutionary process of piggyBac-herpesvirus fusion based on the distribution of Teratorn-like herpesviruses (relatives) among teleosts. Finally, we provide other examples of evolutionary associations between different classes of elements and propose that recombination could be a driving force generating novel mobile elements.
Introduction
Cellular organisms have been parasitized by selfish-replicating genetic entities in the history of life, which include transposable elements (TE) and viruses, as well as plasmids and self-splicing elements (1). Viruses and mobile genetic elements (MGEs) are very abundant biological entities on earth; viruses are estimated to outnumber 10 to 100-fold relative to cellular organisms (2), while TEs often occupy a large part of the host genome [e.g., ~45% in human (3) and ~ 85% in maize (4)]. TEs and viruses frequently cause harmful effects on host organisms, such as insertional mutation by TEs and pathogenesis by viruses. To counter selfish elements, cellular organisms have developed various defense systems, such as RNA interference, epigenetic silencing, and innate and adaptive immune systems (5–7). On the other hand, host organisms have sometimes co-opted selfish elements for their own functions, such as novel gene regulatory networks by many TEs (8–10), V(D)J recombination system in vertebrates from a Transib-like transposon (11), polydnaviruses in parasitoid wasps (12) and placenta in mammals from retroviruses (8, 13, 14). Together, MGEs and viruses have been key players in the evolution of cellular organisms. Despite their importance, however, we are far from full understanding of how MGEs and viruses themselves have evolved; that is, their origin, relationships among different classes of elements, and underlying mechanisms of the emergence of novel elements. This is attributed to the enormous sequence diversity caused by the rapid evolution rate and frequent gene gain/loss (15).
MGEs and viruses are often classified based on their genome constitution (either DNA or RNA of double-stranded (ds) or single-stranded (ss) nucleotides), replication manner (e.g., DNA polymerization, RNA-dependent RNA transcription, reverse transcription), and life cycle (with/without extracellular phase) (1, 16, 17). Eukaryotic TEs are categorized into two classes, according to whether or not they include RNA intermediates. Class I elements (retrotransposons) include long terminal repeat (LTR) and non-LTR elements, both of which transpose in a “copy-and-paste” manner mediated by RNA transcription and reverse transcription. Class II elements (DNA transposons) are further classified into two subclasses, depending on the type of transposition; i.e., in a “cut-and-paste” (e.g., P element, Tc1/mariner, hAT, piggyBac) or “copy-and-paste” manner (e.g., Helitron, Polintons/Mavericks) (16). Similarly, eukaryotic viruses are classified into the following groups according to their genomic entities and replication manner; dsDNA viruses (e.g., Poxviruses, Herpesviruses, Adenoviruses, and Polyomaviruses), ssDNA viruses (e.g., Geminiviruses and Circoviruses), dsRNA viruses (e.g., Reoviruses), positive-strand ssRNA viruses (e.g., Picornaviruses, Coronaviruses, and Tombusviruses), negative-strand ssRNA viruses (e.g., Orthomyxoviruses and Filoviruses), ssRNA-reverse transcriptase (RT) viruses (e.g., Retroviruses) and dsDNA-RT viruses (e.g., Pararetroviruses) (17). Given their enormous diversity, as well as the absence of genes conserved among all elements, it had long remained unclear the evolutionary relationships among different classes of elements, except for a few well-known cases (e.g., ssRNA-RT viruses and LTR retrotransposons) (14, 18). However, recent advances in whole-genome shotgun sequencing technologies and metagenomic analyses have enabled us to identify many novel MGEs and viruses, providing evolutionary insights into the mechanisms underlying the emergence of distinct classes of elements.
We previously identified a giant (~180 kb long) DNA transposon, named Teratorn, in the genome of a small teleost fish medaka (Oryzias latipes) (19), which was the biggest TE ever found at that time. Teratorn is unique in that it contains the transposition machinery of a piggyBac-like DNA transposon including active transposase, as well as the whole genome of an intact herpesvirus genome. A comprehensive genomic survey indicated that Teratorn-like elements are widely distributed among several teleost genomes, some of which were shown to be a fused form, suggesting the generality of piggyBac-herpesvirus fusion (20, 21). We proposed that fusion with the DNA transposon caused a shift in life cycle of the herpesvirus to an intragenomic life form, enabling successful intragenomic propagation in teleosts.
Here we review the characteristics of Teratorn and its relative elements. First, we introduce the identification of Teratorn in the medaka genome. We then describe the sequence characteristics of Teratorn and discuss its uniqueness such as its large size for a transposon and life cycle. Finally, with a likely evolutionary scenario of piggyBac-herpesvirus fusion, we propose that recombination among different classes of elements could be a driving force for generating novel mobile elements.
Identification of Teratorn from medaka spontaneous mutants
Medaka is a small egg-laying freshwater teleost fish that habitats in East Asia, including Japan, South Korea, Taiwan, and China. Medaka has long been utilized as an experimental vertebrate model organism along with zebrafish, because of transparent embryos, a short period of life cycle (2.5 to 3 months), high fecundity, easiness of genetic manipulation, and availability of the high-quality whole-genome sequencing data (22, 23). Furthermore, medaka has a strong advantage in the field of genetics in that in the long history of medaka research in Japan, various healthy inbred strains were established and various spontaneous mutants have been isolated [ref. Shimada and Takeda, 2010 (22)]. Importantly, active DNA transposons were isolated from medaka spontaneous mutants. For example, a series of genetic analyses of albino medaka mutants identified two active DNA transposons Tol1 and Tol2, inserted at the tyrosinase gene (the rate-limiting enzyme for melanin biosynthesis) (24–26). These two elements belong to the hAT superfamily DNA transposons, are 4.4 kb and 4.7 kb long respectively, and encode the transposase and terminal inverted repeats (TIRs), the conventional form of DNA transposon that transpose in a cut-and-paste manner. These two elements were the first active DNA transposons reported in vertebrates (24, 25), and are currently widely utilized as a vector for transgenesis such as Tol2-mediated gene transfer in zebrafish (27). Teratorn, too, was originally isolated as causative DNA fragments for several medaka spontaneous mutants (Figure 1). First, Kondo et al. analyzed rs-3 mutant, which shows almost complete loss of scales, and demonstrated that a transposon-like sequence (which later turned out to be Teratorn) was inserted in the first intron of ectodysplasin-A receptor (EDAR) (28) (Figure 1A). Similarly, Hashimoto et al. reported that the same sequence was inserted in the fourth intron of the pc/glis3 gene, causing polycystic kidney disease (29) (Figure 1B). Our group also isolated the same insertions from two medaka spontaneous mutants; abcdef [(30)] and Da [(31)]. In abcdef, the DNA fragment is inserted at the 47th intron of the pkd1l1 gene, causing randomized left–right organ placement (30) (Figure 1C), while in Da, it is inserted in the regulatory region of zic1/zic4 gene. The latter insertion impairs one of the tissue-specific enhancers of zic1/zic4 gene, which results in specific loss of their expression in somites with the expression in the neural tube maintained, leading to ventralization of the dorsal part of the trunk morphology (e.g., fin morphology, pigmentation pattern, lateral line distribution, and body shape; Figure 1D) (31, 32). In the initial study, sequencing of fosmid clones at the zic1/zic4 locus was performed and partial sequences of Teratorn were identified (about 20 kb long regions at both ends) (31). From the sequence information, we assumed that Teratorn is a DNA transposon that moves in a “cut-and-paste” manner, since it contains 18 bp terminal inverted repeats (TIR) at its termini, and is flanked by target site duplications (TSD) (16). Notably, this anticipated transposon was unique in size (at least 41 kb long) and in gene number (at least six genes), since most cut-and-paste DNA transposons are small (less than 10 kb long) and encode a limited number of genes (transposase) (31). We named this transposon Teratorn after the largest bird of prey that inhabited the North American continent. However, its complete sequence failed to be provided in the initial study, as it could not be deduced from the previously published medaka draft genome; the medaka genome has multicopy of the sequence as repetitive ones [ref. Kasahara et al., 2007 (33)].
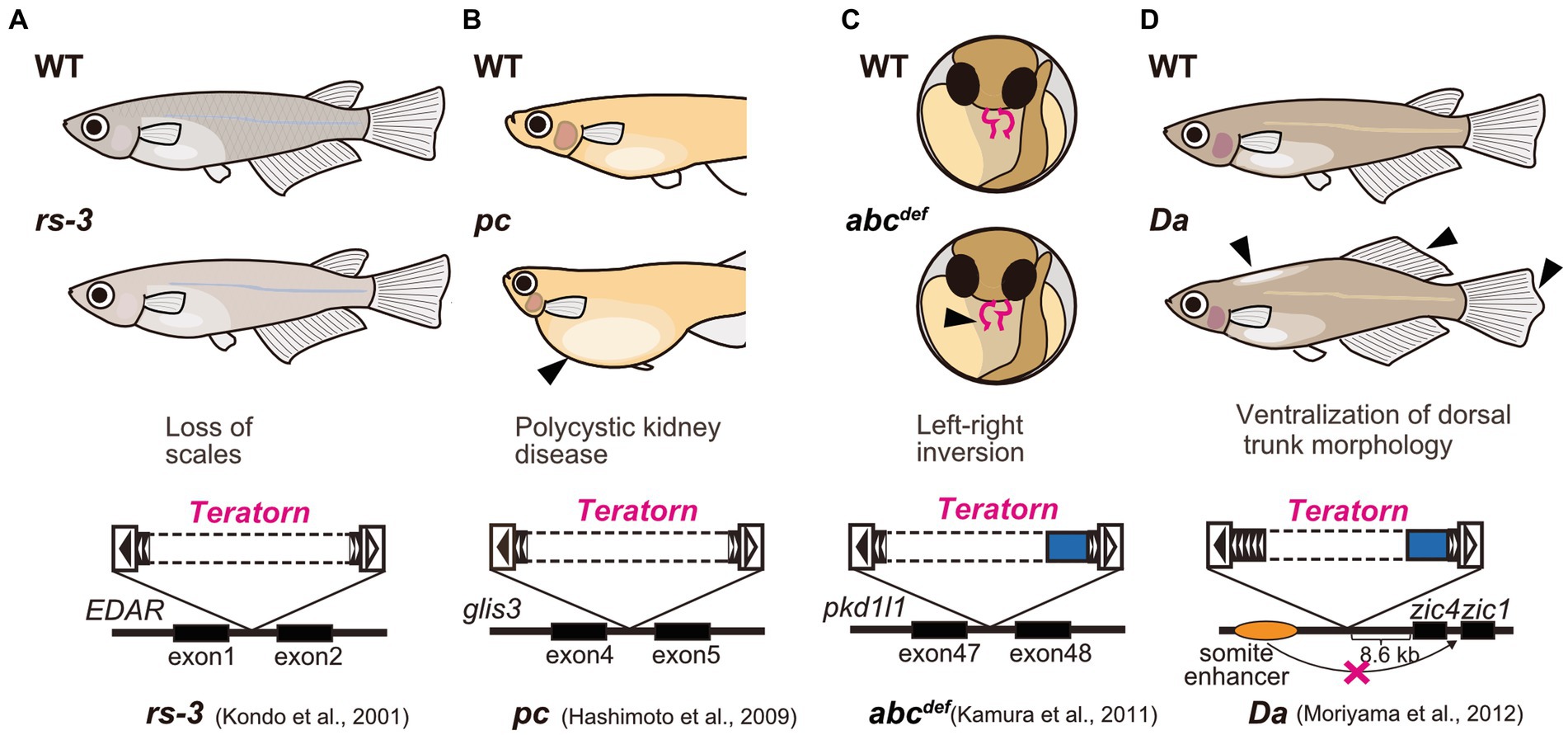
Figure 1. Medaka spontaneous mutants caused by Teratorn insertion. (A) rs-3 mutant [loss of scales, (28)]. (B) pc mutant [polycystic kidney disease, (29)] (C) abcdef mutant [defect in left–right axis formation, (30)]. (D) Da mutant [ventralization of dorsal trunk morphology, (31)]. Solid and dotted boxes inside Teratorn indicate the sequence-determined and undetermined region, respectively.
Teratorn: a 180 kb long giant DNA transposon created by a fusion of a piggyBac-like DNA transposon and a herpesvirus
To determine the full-length sequence of Teratorn, we screened a BAC library of Hd-rR strain to obtain clones containing the entire Teratorn. Using PacBio long-read sequencing, we found that Teratorn was ~180 kb long, quite larger than any other transposon reported at that time (Figure 2A). Further genomic survey indicated that there are two subtypes of Teratorn, which show ~88% sequence identity with each other, and their copy numbers were predicted to be 30 to 40 copies for subtype 1 and ~ 5 for subtype 2 in the Hd-rR strain medaka genome. Gene annotation inside Teratorn revealed two important points. First, a transposase gene, which is homologous to that of piggyBac superfamily DNA transposon, was identified (Figure 2B, red). piggyBac is one of the major “cut-and-paste” DNA transposon superfamilies and is widely distributed among eukaryotes, especially in metazoans (11). In addition, the sequence composition of terminal inverted repeats (TIRs) and target site duplications (TSDs) at its termini follows the rule of the piggyBac superfamily; they possess TIRs, 12–19 bp in length beginning with a “CCYT” motif, and preferentially target TTAA motif to give rise to TSDs. Furthermore, we experimentally showed that Teratorn transposase retains the transposition activity as demonstrated by in vitro assay with human culture cells; i.e., recognition of the TIRs, excision, and re-integration of the internal DNA sequence into chromosomal DNA. These characteristics indicate that Teratorn belongs to piggyBac superfamily and is still active as a transposon. Secondly, and quite surprisingly, besides the transposase gene, Teratorn encodes at least ~90 putative genes, 17 of which show sequence similarity to those of herpesviruses. Those include genes essential for herpesvirus life cycles, such as DNA polymerase, primase, UL21 homolog DNA helicase, capsid maturation protease, DNA packaging terminase, major capsid protein (MCP), subunit 2 capsid triplex protein, and envelope glycoprotein (Figure 2B, blue). Herpesviruses are double-stranded DNA viruses that infect a wide variety of vertebrate species and some molluscan species. Their genomes are relatively large, ranging from 124 to 295 kb, and contain 70 to 200 genes (34, 35). Phylogenetic analyses indicate that Teratorn belongs to Alloherpesviridae (infecting fish and amphibians), one of the three families of the order Herpesvirales (Figure 2C). Alloherpesviruses are known as several important pathogens in the field of aquaculture. Those include Cyprinid herpesvirus 3 (CyHV-3, known as koi herpesvirus) and Ictalurid herpesvirus 1 (IcHV-1, known as channel catfish virus), which caused massive outbreaks in the past (35). The existence of all 13 genes conserved among all alloherpesvirus species, as well as its size (~180 kb), strongly suggests that Teratorn contains the whole genome of a herpesvirus. Besides the essential genes, ~20 genes show no sequence similarity to those of other herpesviruses but are found in other organisms (Figure 2B, yellow). Most of them appear to function in regulation of immune response or cell proliferation in hosts. Given that alloherpesviruses primarily infect epithelial cells and cause pathologies such as epidermal cell necrosis, hyperplasia and lesions, branchial hyperplasia, papillomas and renal adenocarcinomas (35, 36), those genes could be utilized for the infection processes. Taken together, Teratorn is a complete fusion of a piggyBac-like DNA transposon and a whole herpesvirus genome.
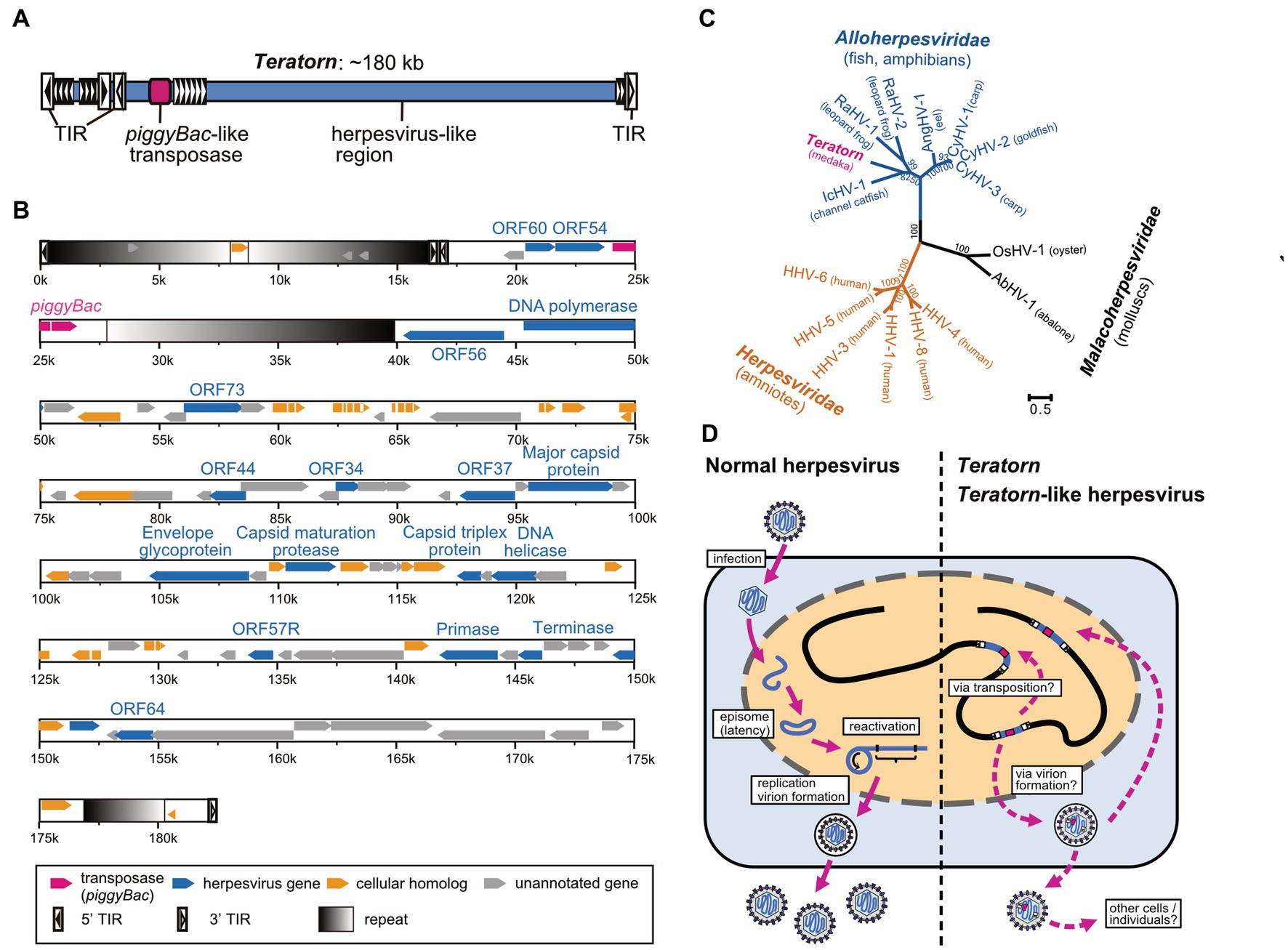
Figure 2. Sequence characteristics of Teratorn and its proposed life cycle. (A) The overall structure of Teratorn. (B) Gene map of the subtype 1 Teratorn copy. Predicted genes are classified into four categories depicted by colored arrowheads; magenta, PiggyBac-like transposase gene; blue, herpesvirus-like genes; yellow, cellular homologs; gray, unannotated genes. Terminal inverted repeats (TIRs) of PiggyBac-like transposon are depicted by squares with black-to-white gradients. (C) Phylogeny of Teratorn and representative herpesvirus species, based on the maximum-likelihood analysis. Amino acid sequences of the DNA packaging terminase, the only gene confidently conserved among all herpesvirus species, were used. Bootstrap values of branching are indicated at the nodes. (D) Comparison of life cycles of normal herpesviruses (left) and Teratorn (right).
Giant TEs in eukaryotes; unique to Teratorn, or broadly existed?
One of the unique characteristics of Teratorn is its large size as compared with other transposons, since eukaryotic TEs are generally small (less than 10 kb long) and encode only a few genes (11, 16, 37). The exceptions to this rule were the Ty3/Gypsy family LTR retrotransposons Ogre in pea (~25 kb) (38) and Burro in the planarian (exceeding 30 kb) (39), Ac transposons in maize (up to 22 kb) (40–42), Penelope-like retrotransposons Terminons in rotifers (up to 40 kb) (43), Helitrons (up to 40 kb) (44, 45), and Polintons/Mavericks (up to 40 kb) (46, 47). However, TEs which exceed 50 kb were not reported until Teratorn was identified. One of the reasons why a few cases of large TEs had been reported could be the size limitation of TEs; the efficiency of reverse-transcription (for retrotransposons) and transposition (for cut-and-paste DNA transposons) decreases as its size increases (48). Teratorn could overcome the size limitation by acquiring the replication machinery of a herpesvirus genome (discussed later). The other reason could be technical difficulties in identifying large TEs from the draft genome, since repetitive sequences were hardly assembled from the whole-genome short-read data. However, recent advances in long-read sequencing technologies, as well as the wealth of genomic data of closely related species and strains, allow us to identify large TEs. For example, a series of studies reported a novel family of giant TEs, named Starships (reaching ~400 kb in size), in fungal genomes (49–51). Initially, members of the Starship elements were independently identified by several groups as sequences associated with specific traits of the hosts. Those include HEPHAESTUS, a ~ 85 kb long element containing many genes involved in tolerance to metal ions in Paecilomyces vcariotii (50), and Enterprise, reaching up to 247 kb, some of which contain a meiotic drive gene of the Spoks (spore killing) (51). A comprehensive genomic survey across Ascomycetes fungi genomes was later performed and proposed that HEPHAESTUS and Enterprise belong to the same family of giant mobile element Starships (49). Starships share characteristic genes such as tyrosine recombinase-like gene DUF3435 presumably responsible for transposition, a novel gene containing DUF3723 domain, ferric reductases (FREs), patatin-like phosphatases (PLPs), and NOD-like receptors (NLRs). However, they comprise only a few copies in the host genomes (~5 copies at most), have a large variation in size (from 27 to 393 kb), and contain accessory genes (e.g., metal tolerance genes in HEPHAESTUS, meiotic drive genes in Spok block, and virulence genes in ToxA), providing specific traits to the hosts. Thus, Starship elements are akin to bacterial integrative and conjugative elements (ICEs) (52) and are suggested to play key roles in horizontal gene transfer of beneficial genes (49–51). This highly contrasts with Teratorn, in that it comprises a large number of copies (~40 copies) in the medaka genome and acts as a selfish genetic element. As such, further research is expected to provide new examples of giant TEs in the near future.
The life cycle of Teratorn: a new life form of a herpesvirus?
A second unique character of Teratorn is that it exists as a chromosomally-integrated form, which is quite different from other herpesviruses. Generally, herpesviruses establish latent infection after entry into host cells, maintaining their genomes as an episomal form in the nucleus, and recurrently activate under some circumstances (53) (Figure 2D, left). Thus, chromosomal integration is not a general phenomenon for most herpesviruses. To our knowledge, among the >100 herpesvirus species reported so far, there are only two chromosomally-integrated herpesviruses other than Teratorn, human herpesvirus 6 (HHV-6) (54) and tarsier endogenous herpesvirus (55). Both of them belong to the Betaherpesvirinae subfamily of the Herpesviridae family, and are integrated into the telomeric region of the host chromosome through their own telomeric repeats (TMRs) at their termini (54, 56). However, it remains unclear whether these chromosomally-integrated herpesviruses are genuine genomic parasites. Tarsier endogenous herpesvirus is thought to be an already fossilized endogenous viral element (EVE), since nearly all genes have lost coding capacity accumulating deleterious mutations (55). On the other hand, chromosomally-integrated HHV-6 is estimated to exist in approximately 1% of the human population (56), and horizontal transfer is still the major way of HHV-6 transmission. In contrast, Teratorn exists in the genomes of several medaka-related species retaining viral genes as intact forms, and the phylogeny of Teratorn is the same as that of host species, suggesting long-term vertical inheritance among the genus Oryzias (19). We reason that chromosomal integration is an adaptive consequence to escape from host immune systems and ensure stable, vertical transmission of their progenies across host generations. Herpesviruses are successful pathogens in establishing long-term infection without chromosomal integration (53). However, since virus molecules can be detected by pathogen recognition receptors (PRRs) evoking the host immune system, herpesviruses need to take various strategies to overcome them, especially during viral replication (57). piggyBac acquisition of the chromosomal integration system could provide an alternative way of transmission (vertical inheritance), in addition to infection.
What is the entire life cycle of Teratorn? Existence of several medaka spontaneous mutants caused by Teratorn insertion, as well as polymorphism in integration sites among individuals of Hd-rR medaka inbred strain, indicated that it behaves at least as an active transposon in vivo. However, the presence of a set of intact herpesvirus-related genes suggests that those genes are also utilized for the propagation of Teratorn. In HHV-6, a chromosomally-integrated form of the virus was suggested to be reactivated in some situations (58, 59), accompanied by the formation of circular viral DNA molecules via excision of the telomeric t-loop (56, 60). It would be intriguing if Teratorn also undergoes similar processes during reactivation; excision mediated by the piggyBac-like transposase, formation of a circular form of DNA, followed by genome replication and virion formation (Figure 2D, right). However, we have not been successful in identifying virions or virus-like particles derived from Teratorn in medaka. Treatment of medaka fibroblast with 5′-azacytidine, a DNA methylation inhibitor, only caused moderate upregulation of viral genes, and we failed to detect any virus particles by electron microscopy or viral proteins by western blot under this condition (19). This could be due to the robust silencing of Teratorn by epigenetic mechanisms (61), since transposon silencing generally involves multiple mechanisms, such as DNA methylation, repressive histone modifications, KRAB zinc-finger proteins, and small RNAs (62, 63). Thus, further experiments are necessary to uncover the repression mechanisms and unlock them to fully reactivate Teratorn.
Widespread distribution of Teratorn-like elements in teleost; generality of piggyBac-herpesvirus fusion
There was an intriguing question as to whether transposon-herpesvirus fusion is a rare accidental event or a general phenomenon in nature. We previously performed a comprehensive survey of Teratorn-like sequences against a publicly available vertebrate genome dataset (Tblastn search of 13 herpesvirus core genes) and identified Teratorn-like elements in at least 22 of the 77 teleost fish species (Figure 3A) (20). In contrast, we did not get any significant hits against other vertebrate genomes, indicating that Teratorn-like elements were distributed only in teleosts. To date, there have been four genera in Alloherpesviridae; Batrachovirus, Cyprinivirus, Ictalurivirus, and Salmonivirus (65). However, phylogenetic analysis showed that Teratorn-like elements are quite distantly related to all those genera and form a single cluster among Alloherpesviridae (Figure 3B). Thus, we proposed that Teratorn-like elements (hereafter call Teratorn-like herpesviruses) represent a new genus of alloherpesvirus, characterized by a high tendency for genomic integration. Several lines of evidence support the idea of multiple integration events among teleosts, i.e., the patchy distribution of Teratorn-like herpesviruses among teleost fishes, and little correlation between the phylogeny of Teratorn-like herpesviruses and host genomes (Figures 3A,B).
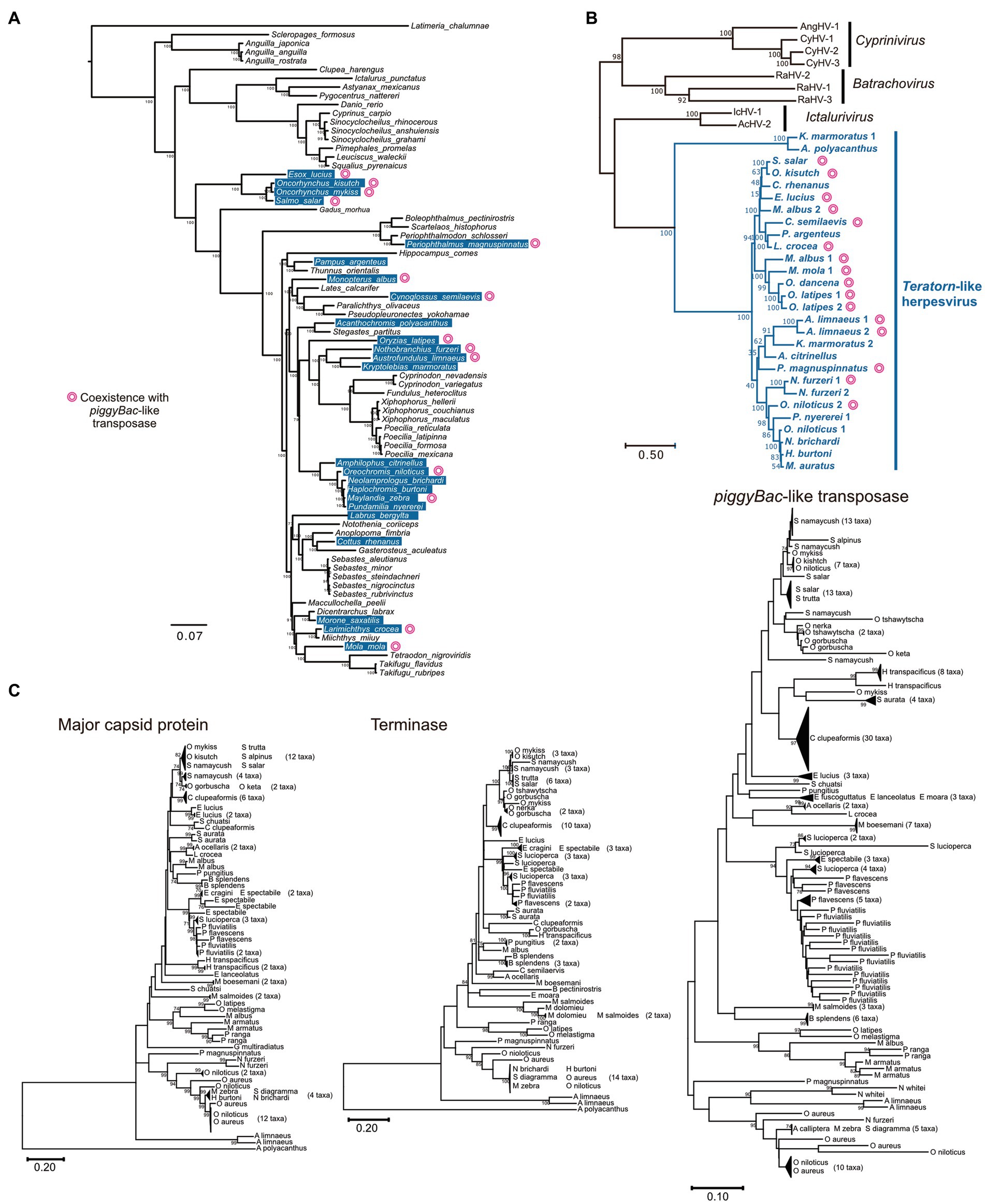
Figure 3. Widespread distribution of Teratorn-like herpesviruses in teleosts. (A) Distribution of Teratorn-like herpesviruses in the genomes of teleost fish species. Species that seem to contain Teratorn-like herpesvirus are highlighted in blue (greater than nine of the 13 herpesvirus core genes, tblastn E-value <10−3). The phylogenetic tree was constructed by bayesian inference, based on the concatenated nucleotide sequence of nine nuclear genes (64). Species in which Teratorn-like herpesviruses are adjacent to piggyBac-like transposase genes are marked. (B) Phylogeny of Teratorn-like herpesviruses and other alloherpesvirus species, based on the maximum-likelihood analysis. The concatenated amino acid sequences of five herpesvirus genes (major capsid protein, DNA helicase, DNA polymerase, capsid triplex protein, and DNA packaging terminase) were used. (C) Maximum-likelihood trees based on the nucleotide sequences of major capsid protein, DNA packaging terminase, and piggyBac-like transposase genes inside re-screened Teratorn-like herpesvirus sequences. General time reversible model was used as a substitution model, considering evolutionary rate differences among sites by discrete Gamma distribution with five categories. Total of 2,878 positions (major capsid protein), 756 positions (DNA packaging terminase), and 558 positions (piggyBac-like transposase) were used in the final dataset. Nodes of phylogenetic trees were collapsed if the sequence similarity was greater than 98% for major capsid protein and terminase, and greater than 96% for piggyBac-like transposase. Non-collapsed trees are shown in Supplementary Figure 1. The scale bars represent the number of substitutions per site.
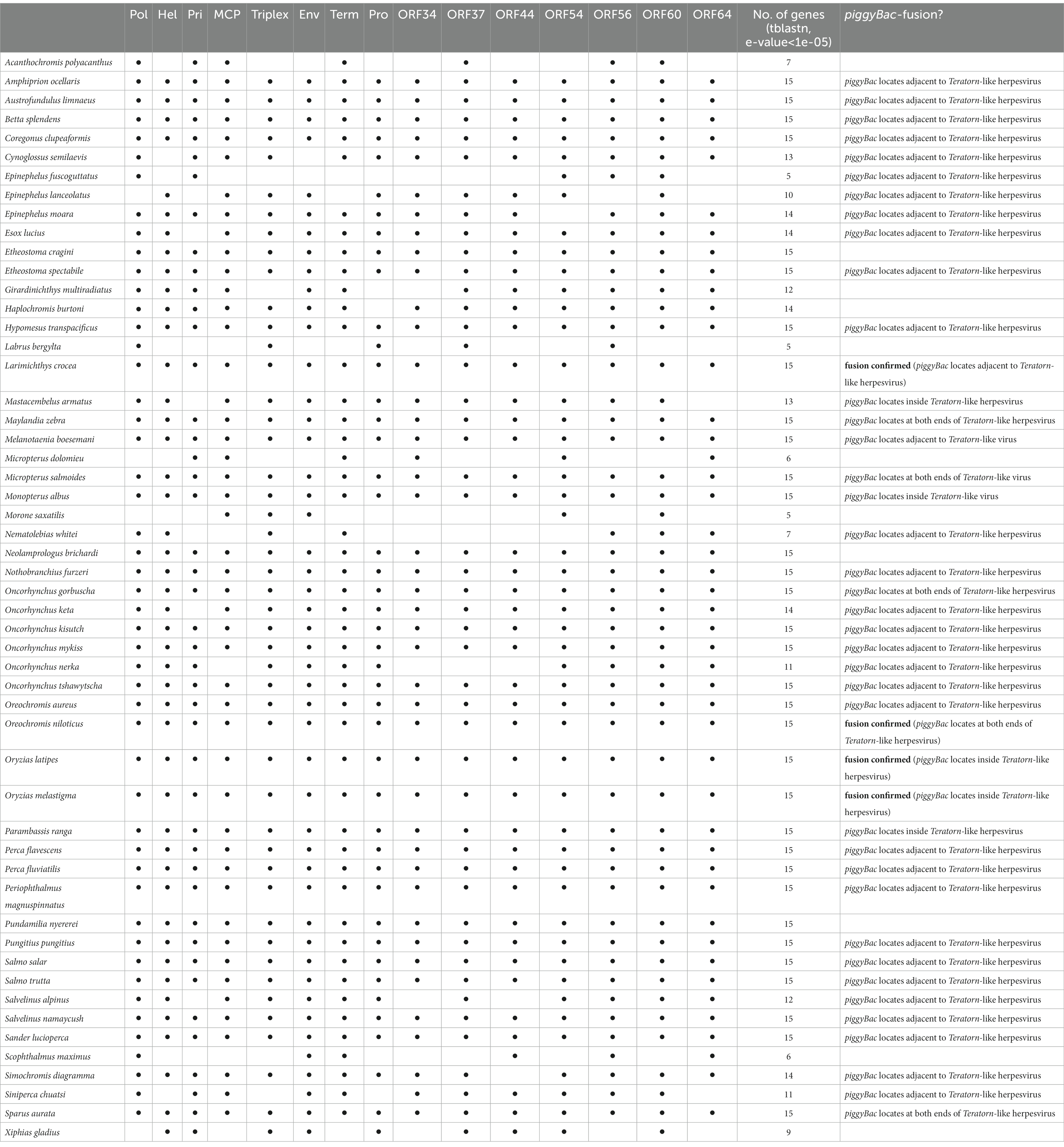
We previously found that at least 9 of the 22 Teratorn-like herpesviruses have a configuration of piggyBac-herpesvirus fusion, and suggested the co-evolution of piggyBac and herpesvirus sequences, based on the same topology of phylogenetic trees between them (20). We reperformed a comprehensive BLAST search of Teratorn-like herpesviruses against publicly available teleost genomes (NCBI RefSeq Representative Genomes). This time we identified Teratorn-like herpesviruses in at least 53 of the 138 teleost fish species (tblastn, e-value <1e-05, more than five of the 15 herpesvirus genes). Importantly, we found that piggyBac-like transposons exist close to or inside Teratorn-like herpesviruses in at least 42 of the 53 species (Table 1; Supplementary Table S1), indicating that fusion with piggyBac-like transposon is common for Teratorn-like herpesviruses. Furthermore, the topology of the phylogenetic tree of piggyBac-like transposase genes inside Teratorn-like herpesviruses is again nearly identical to that of herpesvirus genes (Figure 3C; Supplementary Figure S1). These data strongly suggest that piggyBac-like transposon and herpesvirus genome have behaved as a single unit for a long evolutionary timescale and that the fusion has allowed Teratorn-like herpesviruses for widespread habitation among teleost genomes. Structures of the representative Teratorn-like herpesviruses are shown in Figure 4A.
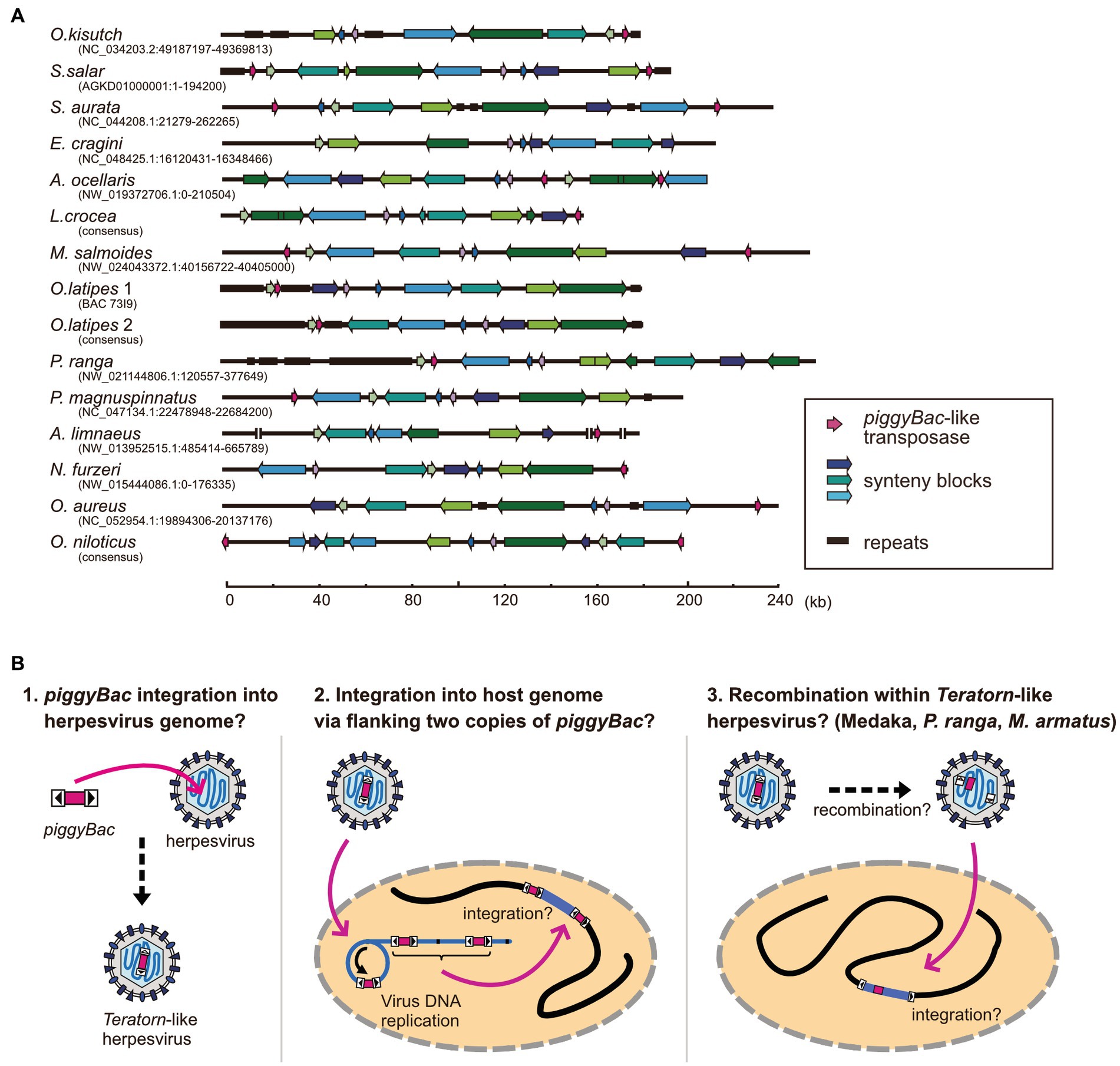
Figure 4. Structures of Teratorn-like herpesviruses and a model of the evolutionary process of piggyBac-herpesvirus fusion. (A) Structures of Teratorn-like herpesvirus in several teleost fish species. Conserved synteny blocks are depicted by the same colors. Magenta arrows indicate piggyBac-like transposase. (B) A model of the process of piggyBac-herpesvirus fusion. In this scenario, the initial event is the transposition of a piggyBac-like transposon into a herpesvirus genome (left). After formation of concatemeric virus DNA during replication, the internal herpesvirus genome was excised, with the two copies of piggyBac-like transposons as its boundary, and was integrated into the host genome (middle). A subgroup of Teratorn-like herpesviruses (those in medaka, M. albus, P. ranga, and M. armatus) might have undergone further recombination, resulting in piggyBac transposase gene located in the middle while TIRs at both ends of Teratorn-like herpesvirus (right).
Process of piggyBac-herpesvirus fusion; piggyBac transposition into herpesvirus genome, followed by recombination?
The underlying mechanism for the fusion of an ancestral herpesvirus and a piggyBac-like transposon to create Teratorn-like herpesvirus is largely unknown, but based on the configuration of Teratorn-like herpesviruses in teleosts, we propose the following scenario. The first event of the fusion would be that a piggyBac transposon was jumped from the host genome into the herpesvirus genome during viral infection (Figure 4B left). Indeed, previous studies reported that TEs can transpose into the genomes of dsDNA viruses of various families, such as Baculoviruses (66–69), Poxviruses (70), Iridoviruses (69), and Pandraviruses (71), possibly whereby acting as vectors of horizontal transposon transfer across species (72). Although transposon insertion has not yet been reported so far for herpesviruses, it could occur, from the host genome into the virus DNA, since herpesviruses usually establish a latent infection with their genomic DNA floating in the nucleus of host cells. Once DNA transposon is transposed into the herpesvirus DNA, integration of the whole virus genome into the host chromosome is theoretically possible. During herpesvirus replication, viral genomic DNA becomes circular and DNA synthesis proceeds in a rolling-circle manner, giving rise to concatenated virus DNA (73). Thus, a single unit of viral genomic DNA, flanked by two copies of piggyBac-like DNA transposons, could be integrated into host genomes via transposition (Figure 4B middle). This scenario is supported by the fact that the majority of Teratorn-like herpesviruses contain piggyBac-like transposon at their termini (Figure 4A; Supplementary Table S1). However, a clade of Teratorn-like herpesviruses (those in medaka, M. albus, P. ranga, and M. armatus) must have undergone further recombination, resulting in the piggyBac transposase gene located in the middle while TIRs at both ends of Teratorn (Figures 4A,B right). Of course, to test the above scenario, a further genomic survey of Teratorn-like herpesviruses in teleost genomes, as well as identification of the exogenous form of Teratorn-like herpesviruses, will definitely be required.
Recombination as a key driver of life cycle shift of MGEs and viruses?
Due to the lack of chromosomal integration step in the life cycles, it was generally thought that nearly all viruses except for retroviruses have lost propagation capacity after genomic integration (14). Indeed, except for ERVs, there have been only a few reports of EVEs that have increased the copy number inside host genomes. Such examples are endogenous pararetroviruses and some single-stranded DNA viruses. The former constitutes up to ~1% of the genome of some plants (74), although the mechanism underlying chromosomal integration and propagation is unknown. The latter is present in tens to one thousand copies in some species (e.g., geminiviruses in plants and fungi, circoviruses and parvoviruses in animals) and is thought to integrate via RC-Rep endonuclease, an enzyme that functions in replication of virus DNA, although not experimentally confirmed (75–77). Thus, we propose that Teratorn-like herpesviruses are the first endogenous non-retroviral element that shifted into the intragenomic lifestyle to promote propagation in host genomes. Recently, some giant (up to ~2000 kb) dsDNA viruses belonging to Mimiviruses and Phycodnaviruses were found to be integrated into the genomes of diverse green algae species (78). Although the mechanism of integration is unknown again, this study raised the possibility that integration of large dsDNA viruses into host genomes widely occurs, some of which might integrate and propagate in the host genomes with the help of DNA transposons.
So far, Teratorn is the first example of the fusion between cut-and-paste DNA transposon and DNA virus (Figure 5A). However, there are several examples of the fusion of two or more distinct mobile elements in the network of eukaryotic MGEs and viruses. The most famous and classical example is the transition from LTR retrotransposons to retroviruses (14, 18) and vice versa (81) (Figure 5B). The former transition is thought to be mediated by the gain of envelope genes from other viruses such as baculovirus and herpesvirus (18), while the latter could be a result of chromosomal integration into germline cells followed by propagation inside host genomes (81). Recent phylogenetic analyses suggest that such recombination events are rather common for all MGEs and viruses and have contributed to their diversification (82). The first example is Polintons (also known as Mavericks), a group of replicative large DNA transposons (10–40 kb) widely distributed in eukaryotes. Polintons were initially regarded as transposons but were later found to encode two capsid protein genes (double jerry roll major capsid protein (DJR-MCP) and minor capsid protein) (83), suggesting that they are bona fide viruses. Recent phylogenetic studies suggested that Polintons have evolutionary links with various viruses and MGEs (e.g., adenoviruses, nucleocytoplasmic large DNA viruses (NCLDVs), virophages, Polinton-like viruses, linear plasmids, and tectivirus-like bacteriophages), characterized by the shared genes between groups (e.g., DJR-MCP, minor capsid protein, protein-primed DNA polymerase (pPolB), Ulp1-like cysteine protease, A32-like genome packaging ATPase, and retrovirus-like integrase (RVE-INT)) (79, 83–86). Based on the phylogenetic analyses, the following scenario was proposed for the evolution of those viruses and MGEs. First, Polinton emerged from a tectivirus-like bacteriophage via the acquisition of a retrovirus-like integrase gene and cysteine protease gene from a certain DNA transposon related to Ginger 1, at the emergence of eukaryotes. Then, some of them returned to virus life forms, concurrent with the loss of integrase genes and/or exchange of DNA polymerase genes, which enabled the expansion of their genomes to form a large part of current eukaryotic DNA viruses such as adenoviruses and NCLDVs (79, 83–85) (Figure 5C). However, more recently, comprehensive surveys of Polinton-like viruses against metagenomic samples were performed and found that Polintons are polyphyletic among Polinton-like viruses. This suggests that the evolution of this group of elements is more complex than previously thought, and the transition from a transposon to a virus and vice versa is massively ongoing for this group of elements (85, 87). Another example is the evolution of ssDNA viruses, which also seemed to be driven by multiple recombination events between different classes of elements. In this scenario, eukaryotic ssDNA is thought to originate from rolling-circle replicating plasmids by the acquisition of single jelly-roll capsid protein-encoding genes from ssRNA viruses (15, 80) (Figure 5D). Such recombination events are suggested to be ongoing even now. This is supported by the finding of a novel group of ssDNA viruses named chimeric viruses (CHIVs), which are created by the acquisition of a capsid gene from another family of ssRNA viruses (tombusviruses) by several families of ssDNA viruses (geminiviruses, circoviruses, and nanoviruses) (88) (Figure 5D). The most extreme example of recombination is bidnavirus, a single-stranded DNA virus, which is composed of sequences derived from four different families of viruses and transposons [Polinton-derived DNA polymerase gene (pPolB), parvovirus-derived capsid gene, and additional two genes derived from reovirus and baculovirus (89)]. Thus, the recombination and fusion between mobile elements of distinct classes occur more frequently than previously thought, in the process of the diversification of MGEs and viruses (Figure 5E). So far, Teratorn has provided the most concrete evidence for this concept. Further identification of novel and peculiar MGEs and viruses will provide insights into mechanisms underlying their diversification and evolution.
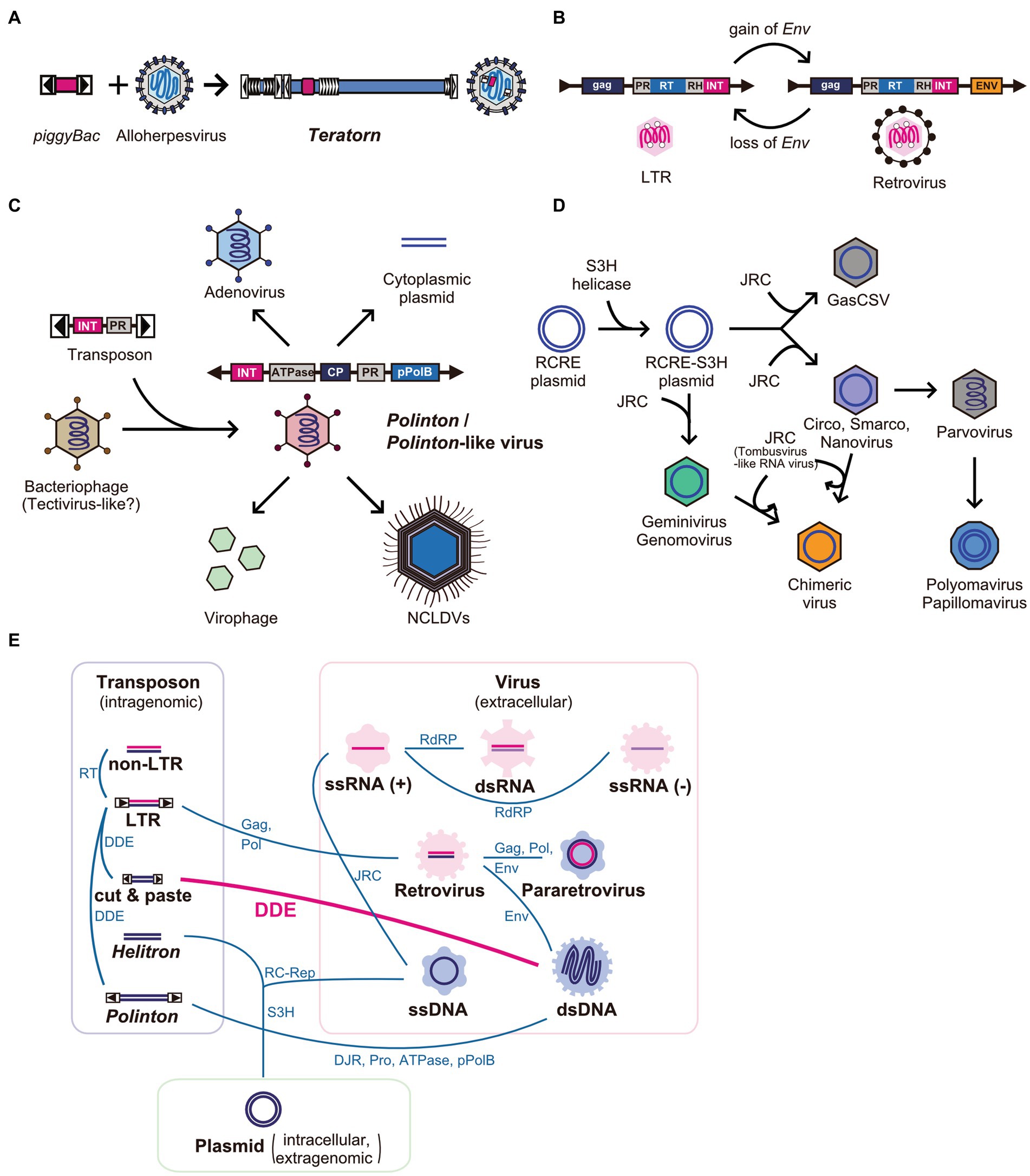
Figure 5. Network-like evolutionary relationships in eukaryotic mobile genetic elements and viruses. (A–D) Examples of the emergence of novel mobile genetic elements by recombination between different class of elements. (A) Teratorn (B) LTR retrotransposons and retroviruses (C) Polintons and several dsDNA viruses, adapted from (79) (D) ssDNA viruses, adapted from (80). Abbreviations; PR, protease; RH, RNA helicase; RT, reverse-transcriptase; Env, envelope protein; INT, retrovirus-like integrase; CP, capsid protein; pPolB, protein-primed family B DNA polymerase; NCLDV, nucleocytoplasmic large DNA virus; RCRE, rolling-circle replication endonuclease; JRC, jerry-roll capsid. (E) Network-like evolutionary relationships in eukaryotic mobile genetic elements and viruses. Evolutionary relationships between different classes of mobile elements are connected by lines. Shared genes are represented next to the lines. Abbreviations; ATPase, DNA packaging ATPase; DDE, DDE transposase; DJR, double jelly-roll capsid; Env, envelope protein; Gag, group-specific antigen; JRC, jelly-roll capsid; pPolB, protein-primed family B DNA polymerase; Pro, Ulp1-like cysteine protease; RC-Rep, rolling-circle replication initiation endonuclease; RdRP, RNA-dependent RNA polymerase; RT, reverse-transcriptase; S3H, superfamily 3 helicase.
Conclusion
In summary, Teratorn has the following unique features; (I) It is very large (~180 kb long) and active transposon, (II) created by a fusion of a piggyBac-like DNA transposon and the whole genome of a herpesvirus, (III) together with related herpesviruses, forms a new genus among Alloherpesviridae, characterized by a high tendency for intragenomic propagation. Thus, Teratorn provides new insights into not only transposon and herpesvirus biology but also into the mechanisms of how novel MGEs and viruses emerge. However, many questions remain unanswered. For example, the genuine amplification mechanism of Teratorn is still unclear. Secondly, the impacts of Teratorn on host species are not fully investigated. Given its large size, Teratorn insertion could alter the 3D chromatin conformation around its inserted region, which frequently impacts gene regulation and causes serious phenotypes. Thirdly, uncovering the arms race between Teratorn and host defense systems (e.g., immune systems and epigenetic silencing) is of interest. Lastly, the evolutionary process of piggyBac-herpesvirus fusion, and the generality of DNA transposon-DNA virus fusion is unclear. Answering the above questions will require combinatorial approaches covering virology, genome science, and evolutionary biology.
Methods
Search for Teratorn–like herpesviruses in teleost fish species
A tblastn search of 15 herpesvirus genes of Teratorn (DNA polymerase, DNA helicase, primase, ATPase subunit of terminase, major capsid protein, membrane glycoprotein, capsid triplex protein, capsid maturation protease, ORF34, ORF37, ORF44, ORF54, ORF56, ORF60, ORF64) was carried out against all available teleost genomes (NCBI RefSeq Representative Genomes, 138 species in total) with default parameters. Sequences of Teratorn-like herpesvirus were obtained as follows; First, locations of the 15 herpesvirus genes were identified by tblastn. After merging the genomic loci of each herpesvirus gene which are within 40 kb of one another, sequences of the defined region and the flanking 40 kb region were extracted from the draft genomes using BEDtool (90). The list of Teratorn-like herpesviruses is shown in Supplementary Table S1.
Phylogenetic analysis
Nucleotide sequences of major capsid protein, DNA packaging terminase, and piggyBac-like transposase were obtained by tblastn search against Teratorn-like herpesvirus sequences. Nucleotide alignments were constructed using MUSCLE in MEGA11 (91), followed by removal of poorly aligned regions using trimAl (92) with a –strict option. Maximum-likelihood analysis was performed for each element using MEGA11 with 100 bootstraps (general time reversible model, discrete gamma distribution with five rate categories). All positions with less than 70% site coverage were eliminated; i.e., fewer than 30% alignment gaps, missing data, and ambiguous bases were allowed at any position (partial deletion option). Multiple alignment data was attached in Supplementary Text 1.
Author contributions
YI prepared the figures and performed a search of Teratorn-like herpesviruses and phylogenetic analyses. YI and HT wrote the manuscript. All authors contributed to the article and approved the submitted version.
Acknowledgments
This work was supported by JSPS KAKENHI grant number JP19K16036.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2023.1158023/full#supplementary-material
References
1. Siefert, JL. Defining the Mobilome. Methods Mol Biol. (2009) 532:13–27. doi: 10.1007/978-1-60327-853-9
2. Suttle, C. Marine viruses--major players in the global ecosystem. Nat Rev Microbiol. (2007) 5:801–12. doi: 10.1038/nrmicro1750
3. Lander, ES, Linton, LM, Birren, B, Nusbaum, C, Zody, MC, Baldwin, J, et al. Initial sequencing and analysis of the human genome. Nature. (2001) 409:860–921. doi: 10.1038/35057062
4. International Rice Genome Sequencing ProjectSchnable, PS, Ware, D, Fulton, RS, Stein, JC, Wei, F, et al. The B73 maize genome: complexity, diversity, and dynamics. Nature. (2005) 436:1112, 793–5, 800. doi: 10.1038/nature03895
5. Levin, HL, and Moran, JV. Dynamic interactions between transposable elements and their hosts. Nat Rev Genet. (2011) 12:615–27. doi: 10.1038/nrg3030
6. tenOever, BR. The evolution of antiviral defense systems. Cell Host Microbe. (2016) 19:142–9. doi: 10.1016/j.chom.2016.01.006
7. Forterre, P, and Prangishvili, D. The great billion-year war between ribosome-and capsid-encoding organisms (cells and viruses) as the major source of evolutionary novelties. Ann N Y Acad Sci. (2009) 1178:65–77. doi: 10.1111/j.1749-6632.2009.04993.x
8. Feschotte, C. Transposable elements and the evolution of regulatory networks. Nat Rev Genet. (2008) 9:397–405. doi: 10.1038/nrg2337
9. Lynch, VJ, Leclerc, RD, May, G, and Wagner, GP. Transposon-mediated rewiring of gene regulatory networks contributed to the evolution of pregnancy in mammals. Nat Genet. (2011) 43:1154–9. doi: 10.1038/ng.917
10. Chuong, EB, Elde, NC, and Feschotte, C. Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science. (2016) 351:1083–7. doi: 10.1126/science.aad5497
11. Feschotte, C, and Pritham, EJ. DNA transposons and the evolution of eukaryotic genomes. Annu Rev Genet. (2007) 41:331–68. doi: 10.1146/annurev.genet.40.110405.090448
12. Strand, MR, and Burke, GR. Polydnavirus-wasp associations: evolution, genome organization, and function. Curr Opin Virol. (2013) 3:587–94. doi: 10.1016/j.coviro.2013.06.004
13. Ono, R, Nakamura, K, Inoue, K, Naruse, M, Usami, T, Wakisaka-Saito, N, et al. Deletion of Peg10, an imprinted gene acquired from a retrotransposon, causes early embryonic lethality. Nat Genet. (2006) 38:101–6. doi: 10.1038/ng1699
14. Feschotte, C, and Gilbert, C. Endogenous viruses: insights into viral evolution and impact on host biology. Nat Rev Genet. (2012) 13:283–96. doi: 10.1038/nrg3199
15. Koonin, EV, Dolja, VV, Krupovic, M, Varsani, A, Wolf, YI, Yutin, N, et al. Global organization and proposed Megataxonomy of the virus world. Microbiol Mol Biol Rev. (2020) 84. doi: 10.1128/MMBR.00061-19
16. Wicker, T, Sabot, F, Hua-Van, A, Bennetzen, JL, Capy, P, Chalhoub, B, et al. A unified classification system for eukaryotic transposable elements. Nat Rev Genet. (2007) 8:973–82. doi: 10.1038/nrg2165-c4
17. King, AMQ, Lefkowitz, E, Adams, MJ, and Carstens, EB. Virus Taxonomy; Ninth Report of the International Committee on Taxonomy of Viruses. Elsevier (2011):1338. doi: 10.1016/B978-0-12-384684-6.00115-4
18. Malik, HS, Henikoff, S, and Eickbush, TH. Poised for contagion: evolutionary origins of the infectious abilities of invertebrate retroviruses. Genome Res. (2000) 10:1307–18. doi: 10.1101/gr.145000
19. Inoue, Y, Saga, T, Aikawa, T, Kumagai, M, Shimada, A, Kawaguchi, Y, et al. Complete fusion of a transposon and herpesvirus created the Teratorn mobile element in medaka fish. Nat Commun. (2017) 8:551. doi: 10.1038/s41467-017-00527-2
20. Inoue, Y, Kumagai, M, Zhang, X, Saga, T, Wang, D, Koga, A, et al. Fusion of piggyBac-like transposons and herpesviruses occurs frequently in teleosts. Zoological Lett. (2018) 4:6. doi: 10.1186/s40851-018-0089-8
21. Aswad, A, and Katzourakis, A. A novel viral lineage distantly related to herpesviruses discovered within fish genome sequence data. Virus Evol. (2017) 3:1–10. doi: 10.1093/ve/vex016
22. Takeda, H, and Shimada, A. The art of medaka genetics and genomics: what makes them so unique? Annu Rev Genet. (2010) 44:217–41. doi: 10.1146/annurev-genet-051710-151001
23. Ichikawa, K, Tomioka, S, Suzuki, Y, Nakamura, R, Doi, K, Yoshimura, J, et al. Centromere evolution and CpG methylation during vertebrate speciation. Nat Commun. (2017) 8:1833. doi: 10.1038/s41467-017-01982-7
24. Koga, A, Inagaki, H, Bessho, Y, and Hori, H. Insertion of a novel transposable element in the tyrosinase gene is responsible for an albino mutation in the medaka fish, Oryzias latipes. Mol Gen Genet. (1995) 249:400–5. doi: 10.1007/BF00287101
25. Koga, A, Suzuki, M, Inagaki, H, Bessho, Y, and Hori, H. Transposable element in fish. Nature. (1996) 383:30. doi: 10.1038/383030a0
26. Akihiko, K, and MHH, S. Transposable elements in Medaka fish. Zool Sci. (2002) 19:1–6. doi: 10.2108/zsj.19.1
27. Kawakami, K. Tol2: A versatile gene transfer vector in vertebrates. Genome Biol. (2007) 8:S7. doi: 10.1186/gb-2007-8-s1-s7
28. Shu, K, Yoshikazu, K, Mariko, K, Kiyoshi, N, Nobuyoshi, S, and Akihiro, S. The medaka rs-3 locus required for scale development encodes ectodysplasin-a receptor. Curr Biol. (2001) 11:1202–6.
29. Hashimoto, H, Miyamoto, R, Watanabe, N, Shiba, D, Ozato, K, Inoue, C, et al. Polycystic kidney disease in the medaka (Oryzias latipes) pc mutant caused by a mutation in the Gli-similar3 (glis3) gene. PLoS One. (2009) 4:e6299. doi: 10.1371/journal.pone.0006299
30. Kamura, K, Kobayashi, D, Uehara, Y, Koshida, S, Iijima, N, Kudo, A, et al. Pkd1l1 complexes with Pkd2 on motile cilia and functions to establish the left-right axis. Dev. (2011) 138:1121–9. doi: 10.1242/dev.058271
31. Moriyama, Y, Kawanishi, T, Nakamura, R, Tsukahara, T, Sumiyama, K, Suster, ML, et al. The medaka zic1/zic4 mutant provides molecular insights into teleost caudal fin evolution. Curr Biol. (2012) 22:601–7. doi: 10.1016/j.cub.2012.01.063
32. Kawanishi, T, Kaneko, T, Moriyama, Y, Kinoshita, M, Yokoi, H, Suzuki, T, et al. Modular development of the teleost trunk along the dorsoventral axis and zic1/zic4 as selector genes in the dorsal module. Development. (2013) 140:1486–96. doi: 10.1242/dev.088567
33. Kasahara, M, Naruse, K, Sasaki, S, Nakatani, Y, Qu, W, Ahsan, B, et al. The medaka draft genome and insights into vertebrate genome evolution. Nature. (2007) 447:714–9. doi: 10.1038/nature05846
34. van Beurden, S, and Engelsma, M. “Herpesviruses of Fish, Amphibians and Invertebrates”. In: Herpesviridae - A Look Into This Unique Family of Viruses. eds. George D. Magel, S. Tyring Intech Open (2012):217–42. doi: 10.5772/1207
35. Hanson, L, Dishon, A, and Kotler, M. Herpesviruses that infect fish. Viruses. (2011) 3:2160–91. doi: 10.3390/v3112160
36. Boutier, M, Morvan, L, Delrez, N, Origgi, F, Doszpoly, A, and Vanderplasschen, A. “Fish and Amphibian Alloherpesviruses (Herpesviridae)”, In: Encyclopedia of Virology 4th Edition. eds. Bamford D, Zuckerman M. Elsevier (2021):306–15. doi: 10.1016/b978-0-12-809633-8.20931-x
37. Kojima, KK. Structural and sequence diversity of eukaryotic transposable elements. Genes Genet Syst. (2019) 94:233–52. doi: 10.1266/ggs.18-00024
38. Neumann, P, Požárková, D, and Macas, J. ighly abundant pea LTR retrotransposon Ogre is constitutively transcribed and partially spliced. Plant Mol Biol. (2003). 53:399–410. doi: 10.1023/b:plan.0000006945.77043.ce
39. Grohme, MA, Schloissnig, S, Rozanski, A, Pippel, M, Young, GR, Winkler, S, et al. The genome of Schmidtea mediterranea and the evolution of core cellular mechanisms. Nature. (2018) 554:56–61. doi: 10.1038/nature25473
40. Huang, JT, and Dooner, HK. Macrotransposition and other complex chromosomal restructuring in maize by closely linked transposons in direct orientation. Plant Cell. (2008) 20:2019–32. doi: 10.1105/tpc.108.060582
41. Yu, C, Zhang, J, Pulletikurti, V, Weber, DF, and Peterson, T. Spatial configuration of transposable element ac termini affects their ability to induce chromosomal breakage in maize. Plant Cell. (2010) 22:744–54. doi: 10.1105/tpc.109.070052
42. Wang, D, Yu, C, Zhang, J, and Peterson, T. Excision and reinsertion of ac macrotransposons in maize. Genetics. (2022) 221:iyac067. doi: 10.1093/genetics/iyac067
43. Arkhipova, IR, Yushenova, IA, and Rodriguez, F. Giant reverse transcriptase-encoding transposable elements at telomeres. Mol Biol Evol. (2017) 34:2245–57. doi: 10.1093/MOLBEV/MSX159
44. Kapitonov, VV, and Jurka, J. Rolling-circle transposons in eukaryotes. Proc Natl Acad Sci U S A. (2001) 98:8714–9. doi: 10.1073/pnas.151269298
45. Du, C, Fefelova, N, Caronna, J, He, L, and Dooner, HK. The polychromatic Helitron landscape of the maize genome. Proc Natl Acad Sci U S A. (2009) 106:19916–21. doi: 10.1073/pnas.0904742106
46. Kapitonov, VV, and Jurka, J. Self-synthesizing DNA transposons in eukaryotes. Proc Natl Acad Sci U S A. (2006) 103:4540–5. doi: 10.1073/pnas.0600833103
47. Pritham, EJ, Putliwala, T, and Feschotte, C. Mavericks, a novel class of giant transposable elements widespread in eukaryotes and related to DNA viruses. Gene. (2007) 390:3–17. doi: 10.1016/j.gene.2006.08.008
48. Arkhipova, IR, Yushenova, IA, and Angert, E. Giant transposons in eukaryotes: is bigger better? Genome Biol Evol. (2019) 11:906–18. doi: 10.1093/gbe/evz041
49. Gluck-Thaler, E, Ralston, T, Konkel, Z, Ocampos, CG, Ganeshan, VD, Dorrance, AE, et al. Giant starship elements mobilize accessory genes in fungal genomes. Mol Biol Evol. (2022) 39:msac109. doi: 10.1093/molbev/msac109
50. Urquhart, AS, Chong, NF, Yang, Y, and Idnurm, A. A large transposable element mediates metal resistance in the fungus Paecilomyces variotii. Curr Biol. (2022) 32:937–950.e5. doi: 10.1016/j.cub.2021.12.048
51. Vogan, AA, Ament-Velásquez, SL, Bastiaans, E, Wallerman, O, Saupe, SJ, Suh, A, et al. The Enterprise, a massive transposon carrying Spok meiotic drive genes. Genome Res. (2021) 31:789–98. doi: 10.1101/GR.267609.120
52. Johnson, CM, and Grossman, AD. Integrative and conjugative elements (ICEs): what they do and how they work. Annu Rev Genet. (2015) 49:annurev-genet-112414-055018:577–601. doi: 10.1146/annurev-genet-112414-055018
53. Arvin, A, Campadelli-Fiume, G, Mocarski, E, Moore, PS, Roizman, B, Whitley, R, et al. eds. Human Herpesviruses: Biology, Therapy, and Immunoprophylaxis. ed. K. Yamanishi, Cambridge: Cambridge University Press (2007), 1408.
54. Morissette, G, and Flamand, L. Herpesviruses and chromosomal integration. J Virol. (2010) 84:12100–9. doi: 10.1128/JVI.01169-10
55. Aswad, A, and Katzourakis, A. The first endogenous herpesvirus, identified in the tarsier genome, and novel sequences from primate Rhadinoviruses and Lymphocryptoviruses. PLoS Genet. (2014) 10:e1004332. doi: 10.1371/journal.pgen.1004332
56. Kaufer, BB, and Flamand, L. Chromosomally integrated HHV-6: impact on virus, cell and organismal biology. Curr Opin Virol. (2014) 9:111–8. doi: 10.1016/j.coviro.2014.09.010
57. Bhowmik, D, and Zhu, F. Evasion of intracellular DNA sensing by human herpesviruses. Front Cell Infect Microbiol. (2021) 11:647992. doi: 10.3389/fcimb.2021.647992
58. Strenger, V, Caselli, E, Lautenschlager, I, Schwinger, W, Aberle, SW, Loginov, R, et al. Detection of HHV-6-specific mRNA and antigens in PBMCs of individuals with chromosomally integrated HHV-6 (ciHHV-6). Clin Microbiol Infect. (2014) 20:1027–32. doi: 10.1111/1469-0691.12639
59. Prusty, BK, Siegl, C, Hauck, P, Hain, J, Korhonen, SJ, Hiltunen-Back, E, et al. Chlamydia trachomatis infection induces replication of latent HHV-6. PLoS One. (2013) 8:e61400. doi: 10.1371/journal.pone.0061400
60. Huang, Y, Hidalgo-Bravo, A, Zhang, E, Cotton, VE, Mendez-Bermudez, A, Wig, G, et al. Human telomeres that carry an integrated copy of human herpesvirus 6 are often short and unstable, facilitating release of the viral genome from the chromosome. Nucleic Acids Res. (2014) 42:315–27. doi: 10.1093/nar/gkt840
61. Nakamura, R, Tsukahara, T, Qu, W, Ichikawa, K, Otsuka, T, Ogoshi, K, et al. Large hypomethylated domains serve as strong repressive machinery for key developmental genes in vertebrates. Development. (2014) 141:2568–80. doi: 10.1242/dev.108548
62. Slotkin, RK, and Martienssen, R. Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet. (2007) 8:272–85. doi: 10.1038/nrg2072
63. Imbeault, M, Helleboid, P-Y, and Trono, D. KRAB zinc-finger proteins contribute to the evolution of gene regulatory networks. Nature. (2017) 543:550–4. doi: 10.1038/nature21683
64. Near, TJ, Eytan, RI, Dornburg, A, Kuhn, KL, Moore, JA, Davis, MP, et al. Resolution of ray-finned fish phylogeny and timing of diversification. Proc Natl Acad Sci. (2012) 21:13698–703. doi: 10.1073/pnas.1206625109
65. Waltzek, TB, Kelley, GO, Alfaro, ME, Kurobe, T, Davison, AJ, and Hedrick, RP. Phylogenetic relationships in the family Alloherpesviridae. Dis Aquat Org. (2009) 84:179–94. doi: 10.3354/dao02023
66. Fraser, MJ, Smith, GE, and Summers, MD. Acquisition of Host Cell DNA sequences by Baculoviruses: relationship between host DNA insertions and FP mutants of Autographa californica and galleria mellonella nuclear Polyhedrosis viruses. J Virol. (1983) 47:287–300. doi: 10.1128/jvi.47.2.287-300.1983
67. Gilbert, C, Chateigner, A, Ernenwein, L, Barbe, V, Bézier, A, Herniou, EA, et al. Population genomics supports baculoviruses as vectors of horizontal transfer of insect transposons. Nat Commun. (2014) 5:3348. doi: 10.1038/ncomms4348
68. Gilbert, C, Peccoud, J, Chateigner, A, Moumen, B, Cordaux, R, and Herniou, EA. Continuous influx of genetic material from host to virus populations. PLoS Genet. (2016) 12:e1005838. doi: 10.1371/journal.pgen.1005838
69. Loiseau, V, Peccoud, J, Bouzar, C, Guillier, S, Fan, J, Gueli Alletti, G, et al. Monitoring insect transposable elements in large double-stranded DNA viruses reveals host-to-virus and virus-to-virus transposition. Mol Biol Evol. (2021) 38:3512–30. doi: 10.1093/molbev/msab198
70. Piskurek, O, and Okada, N. Poxviruses as possible vectors for horizontal transfer of retroposons from reptiles to mammals. Proc Natl Acad Sci U S A. (2007) 104:12046–51. doi: 10.1073/pnas.0700531104
71. Sun, C, Feschotte, C, Wu, Z, and Mueller, RL. DNA transposons have colonized the genome of the giant virus Pandoravirus salinus. BMC Biol. (2015) 13:38. doi: 10.1186/s12915-015-0145-1
72. Schaack, S, Gilbert, C, and Feschotte, C. Promiscuous DNA: horizontal transfer of transposable elements and why it matters for eukaryotic evolution. Trends Ecol Evol. (2010) 25:537–46. doi: 10.1016/j.tree.2010.06.001
73. Weller, SK, and Coen, DM. Herpes simplex viruses: mechanisms of DNA replication. Cold Spring Harb Perspect Biol. (2012) 4:a013011–1. doi: 10.1101/cshperspect.a013011
74. Geering, ADW, Maumus, F, Copetti, D, Choisne, N, Zwickl, DJ, Zytnicki, M, et al. Endogenous florendoviruses are major components of plant genomes and hallmarks of virus evolution. Nat Commun. (2014) 5:5269. doi: 10.1038/ncomms6269
75. Liu, H, Fu, Y, Li, B, Yu, X, Xie, J, Cheng, J, et al. Widespread horizontal gene transfer from circular single-stranded DNA viruses to eukaryotic genomes. BMC Evol Biol. (2011) 11:276. doi: 10.1186/1471-2148-11-276
76. Krupovic, M, and Forterre, P. Single-stranded DNA viruses employ a variety of mechanisms for integration into host genomes. Ann N Y Acad Sci. (2015) 1341:41–53. 1341:n/a-n/a. doi: 10.1111/nyas.12675
77. Sharma, V, Lefeuvre, P, Roumagnac, P, Filloux, D, Teycheney, PY, Martin, DP, et al. Large-scale survey reveals pervasiveness and potential function of endogenous geminiviral sequences in plants. Virus Evol. (2020) 6:veaa071. doi: 10.1093/ve/veaa071
78. Moniruzzaman, M, Weinheimer, AR, Martinez-Gutierrez, CA, and Aylward, FO. Widespread endogenization of giant viruses shapes genomes of green algae. Nature. (2020) 588:141–5. doi: 10.1038/s41586-020-2924-2
79. Krupovic, M, and Koonin, EV. Polintons: a hotbed of eukaryotic virus, transposon and plasmid evolution. Nat Rev Microbiol. (2015) 13:105–15. doi: 10.1038/nrmicro3389
80. Kazlauskas, D, Varsani, A, Koonin, EV, and Krupovic, M. Multiple origins of prokaryotic and eukaryotic single-stranded DNA viruses from bacterial and archaeal plasmids. Nat Commun. (2019) 10:3425. doi: 10.1038/s41467-019-11433-0
81. Magiorkinis, G, Gifford, RJ, Katzourakis, A, de Ranter, J, and Belshaw, R. Env-less endogenous retroviruses are genomic superspreaders. Proc Natl Acad Sci U S A. (2012) 109:7385–90. doi: 10.1073/pnas.1200913109
82. Koonin, EV, and Dolja, VV. Virus world as an evolutionary network of viruses and capsidless selfish elements. Microbiol Mol Biol Rev. (2014) 78:278–303. doi: 10.1128/MMBR.00049-13
83. Krupovic, M, Bamford, DH, and Koonin, EV. Conservation of major and minor jelly-roll capsid proteins in Polinton (maverick) transposons suggests that they are bona fide viruses. Biol Direct. (2014) 9:6. doi: 10.1186/1745-6150-9-6
84. Yutin, N, Raoult, D, and Koonin, EV. Virophages, polintons, and transpovirons: a complex evolutionary network of diverse selfish genetic elements with different reproduction strategies. Virol J. (2013) 10:158. doi: 10.1186/1743-422X-10-158
85. Yutin, N, Shevchenko, S, Kapitonov, V, Krupovic, M, and Koonin, EV. A novel group of diverse Polinton-like viruses discovered by metagenome analysis. BMC Biol. (2015) 13:95. doi: 10.1186/s12915-015-0207-4
86. Fischer, MG, and Suttle, CA. A virophage at the origin of large DNA transposons. Science. (2011) 332:231–4. doi: 10.1126/science.1199412
87. Bellas, CM, and Sommaruga, R. Polinton-like viruses are abundant in aquatic ecosystems. Microbiome. (2021) 9:13. doi: 10.1186/s40168-020-00956-0
88. Roux, S, Enault, F, Bronner, G, Vaulot, D, Forterre, P, and Krupovic, M. Chimeric viruses blur the borders between the major groups of eukaryotic single-stranded DNA viruses. Nat Commun. (2013) 4:2700. doi: 10.1038/ncomms3700
89. Krupovic, M, and Koonin, EV. Evolution of eukaryotic single-stranded DNA viruses of the Bidnaviridae family from genes of four other groups of widely different viruses. Sci Rep. (2014) 4:5347. doi: 10.1038/srep05347
90. Quinlan, AR, and Hall, IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. (2010) 26:841–2. doi: 10.1093/bioinformatics/btq033
91. Tamura, K, Stecher, G, and Kumar, S. MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol. (2021) 38:3022–7. doi: 10.1093/molbev/msab120
Keywords: mobile genetic element, transposable element, virus, piggyBac , herpesvirus, teleost, recombination
Citation: Inoue Y and Takeda H (2023) Teratorn and its relatives – a cross-point of distinct mobile elements, transposons and viruses. Front. Vet. Sci. 10:1158023. doi: 10.3389/fvets.2023.1158023
Edited by:
Pavulraj Selvaraj, Louisiana State University, United StatesReviewed by:
Ganesan Muthusamy, Periyar University, IndiaDafang Wang, Hofstra University, United States
Copyright © 2023 Inoue and Takeda. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yusuke Inoue, eS1pbm91ZUBicy5zLnUtdG9reW8uYWMuanA=; Hiroyuki Takeda, aHRha2VkYUBicy5zLnUtdG9reW8uYWMuanA=
 Yusuke Inoue
Yusuke Inoue Hiroyuki Takeda*
Hiroyuki Takeda*