
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci. , 17 April 2023
Sec. Veterinary Infectious Diseases
Volume 10 - 2023 | https://doi.org/10.3389/fvets.2023.1155061
This article is part of the Research Topic Pathogenic and Symbiotic Bacteria in Ruminants: Antimicrobial Resistance and Microbial Homeostasis View all 10 articles
 Dong Wang1†
Dong Wang1† Haihui Gao1,2†
Haihui Gao1,2† Long Zhao1†
Long Zhao1† Changrong Lv1
Changrong Lv1 Wei Dou1
Wei Dou1 Xiuping Zhang1
Xiuping Zhang1 Yong Liu1
Yong Liu1 Xiaodong Kang2*
Xiaodong Kang2* Kangkang Guo1*
Kangkang Guo1*Introduction: Calf diarrhea is a complex disease that has long been an unsolved problem in the cattle industry. Ningxia is at the forefront of China in the scale of cattle breeding, and calf diarrhea gravely restricts the development of Ningxia's cattle industry.
Methods: From July 2021 to May 2022, we collected diarrhea stool samples from calves aged 1–103 days from 23 farms in five cities in Ningxia, and performed PCR using specific primers for 15 major reported pathogens of calf diarrhea, including bacteria, viruses, and parasites. The effect of different seasons on the occurrence of diarrhea in calves was explored, the respective epidemic pathogens in different seasons were screened, and more detailed epidemiological investigations were carried out in Yinchuan and Wuzhong. In addition, we analyzed the relationship between different ages, river distributions and pathogen prevalence.
Results: Eventually, 10 pathogens were detected, of which 9 pathogens were pathogenic and 1 pathogen was non-pathogenic. The pathogens with the highest detection rate were Cryptosporidium (50.46%), Bovine rotavirus (BRV) (23.18%), Escherichia coli (E. coli) K99 (20.00%), and Bovine coronavirus (BCoV) (11.82%). The remaining pathogens such as Coccidia (6.90%), Bovine Astrovirus (BoAstV) (5.46%), Bovine Torovirus (BToV) (4.09%), and Bovine Kobuvirus (BKoV) (3.18%) primarily existed in the form of mixed infection.
Discussion: The analysis showed that different cities in Ningxia have different pathogens responsible for diarrhea, with Cryptosporidium and BRV being the most important pathogens responsible for diarrhea in calves in all cities. Control measures against those pathogens should be enforced to effectively prevent diarrhea in calves in China.
Diarrhea is one of the most important diseases that damages the health of calves worldwide. It is considered to be one of the diseases causing the highest economic losses to the cattle industry, with losses of up to 10 million dollars due to calf diarrhea in Norway in 2006, followed by cases of varying degrees of calf diarrhea reported in the United States in 2007, South Korea in 2013, and Pakistan in 2014 (1, 2). The main causes of calf diarrhea are intricate and complex (3, 4). In addition to genetics, age, herd and farm environment, feeding practices, poor management and other complications, the most important factor is infection (5, 6). Many countries, including China, have experienced calf diarrhea outbreaks of differing degrees caused by pathogens, such as Cryptosporidium, BRV, BCoV, E. coli K99 and other pathogens (7–10). According to the annual report of Japan in 2017, the economic losses caused by BRV in the previous years were estimated to be about 1 billion yen (11). In addition to causing diarrhea, Cryptosporidium, BCoV, and E.coli K99 also have different effects on increasing mortality, reducing immunity, and reducing milk production (12, 13).
In China, calf diarrhea outbreaks have been reported in many provinces and regions (14–17), but Ningxia has few reports in the article that has comprehensively and systematically investigated the epidemic situation and pathogen distribution characteristics of calf diarrhea. Ningxia has a natural and favorable breeding environment, coupled with the government policy support for the cattle breeding industry, making it one of the important cattle breeding areas in China. With the growing scale of the cattle industry in Ningxia, diarrhea in calves has become an increasingly serious problem, such as the absence of clinical symptoms in calves carrying the pathogen, the rapid spread of the pathogen, and the effect of different environments on the occurrence of diarrhea, which have not been reported or studied.
In order to investigate the prevalence of calf diarrhea in Ningxia and clarify the main pathogens that cause calf diarrhea prevalence in different cities, and study the effects of different seasons to diarrhea in calves, calf diarrhea fecal samples were collected from 23 large-scale cattle farms in five cities of Yinchuan, Wuzhong, Shizuishan, Zhongwei and Guyuan. Pathogens that have been reported to be associated with calf diarrhea were tested, including E. coli K99 (18), Salmonella (19), Proteus mirabilis (20), Clostridium perfringens (C. perfringens) (21), Bovine Viral Diarrhea Virus (BVDV) (22), BRV (23), BCoV (23), BToV (22), BoAstV (24), BKoV (24), Bovine Norovirus (BNoV) (24), Bovine Enterovirus (BEV) (25), Cryptosporidium (26), Coccidia (27, 28), and Giardia (29). The prevalence and distribution characteristics of these pathogens were analyzed to develop a reasonable and effective treatment plan for diarrhea in calves and to provide basic data for the prevention of diarrhea in calves.
From July 2021 to May 2022, 315 calf stool samples including 220 fresh calf stool samples with diarrhea and 95 fresh normal samples from 23 large-scale cattle farms in 5 cities of Ningxia were collected. Using sterile disposable gloves to collect normal calf rectal stool samples; 4 mL fetal calf serum(FBS)-free DMEM was taken to a sterile 15 mL tube, and the diarrhea stool samples were collected into the tube and stored at 4°C. The common symptoms of diarrheal calves were dehydration, loss of appetite, watery diarrhea, and mental depression. Figure 1A shows the geographical location of the Ningxia Hui Autonomous Region, and Figure 1B shows the geographical location of the cattle farm and the total number of samples collected in each area. Table 1 shows the specific sampling numbers in Ningxia.
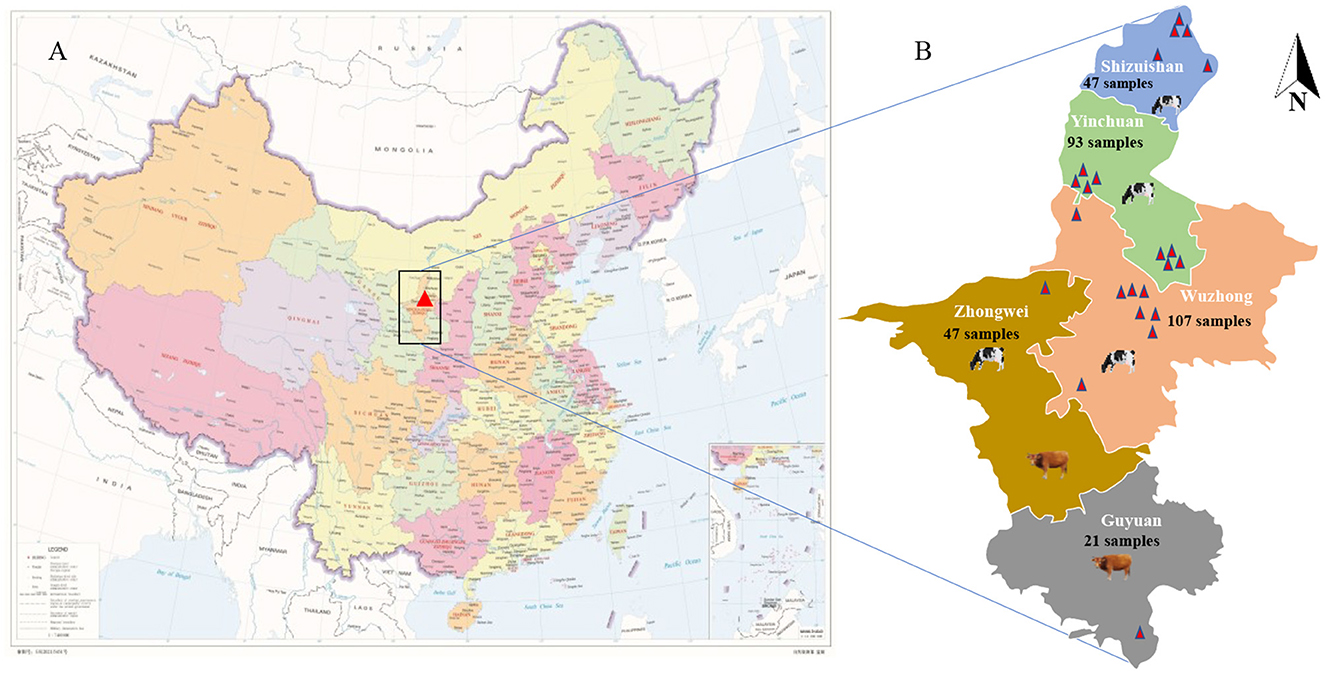
Figure 1. Cattle farm location and sampling information. (A) The geographical location of Ningxia Hui Autonomous Region (red marked as Ningxia Hui Autonomous Region, red star marked as Beijing, the capital of China). (B) The geographical location of the cattle farm and the total number of samples collected in each city (different colors represent different cities).
After the collected fresh stool was transported back to the laboratory at 4°C, 200 g of stool were dispensed into 2 mL sterile EP tubes on a sterile clean bench, and total DNA was extracted using stool DNA kit (OMEGA, Georgia, USA), and then PCR detection was performed to detect E. coli K99, Salmonella, Proteus mirabilis, C. perfringens, Coccidia, Cryptosporidium, Giardia.
The collected fresh diarrhea stool samples were diluted with 0.9% sterile normal saline. After repeated freezing and thawing at −80°C for three times, the samples were centrifuged at 4°C, 12,000 r/min for 5 min, and the supernatant was collected. Total RNA was extracted from stool using Trizol reagent AG RNA ex Pro (Accurate Biotechnology, Hunan, China). According to the manufacturer's operating rules, 2 μg total RNA was reverse transcribed into cDNA using Evo M-MLV RT Mix kit with gDNase. The cDNA was used to detect viruses that caused bovine diarrhea such as BVDV, BRV, BCoV, BToV, BoAstV, BKoV, BNoV, and BEV.
The primers used to detect the above pathogens are shown in Table 2. The extracted RNA was measured using NanoDrop One (Thermo Fisher Scientific, Waltham, MA, USA) and the RNA concentration was in the normal range. Each sample was taken 2 μg RNA for reverse transcription to obtain the same concentration of cDNA. Nested PCR was performed to detect Cryptosporidium and Giardia using 2 × Taq Master Mix (Vazyme Biotech, Nanjing, China), the specific PCR system was 2 × Taq Master mix 10 μL, upstream and downstream primers 1 μL, template 2 μL, supplemented with ddH2O to 20 μL, the primer concentration was 10 μM. PCR amplification of other pathogens was carried out using 2 × M5 HiPer plus Taq HiFi PCR mix (Mei5 biotechnology, Beijing, China), the specific PCR system was 2 × M5 HiPer plus Taq HiFi PCR mix 10 μL, upstream and downstream primers 1 μL, template 2 μL, supplemented with ddH2O to 20 μL, the primer concentration was 10 μM.
Take 2 g stools sample of diarrheal calves (≥18 d), put it into a beaker, add 5 mL of water first, stir and mix well, add saturated saline to 60 mL, filter through a copper mesh after mixing, absorb the stool liquid, and inject it into McMaster Egg Slide Counting Chamber, after stewing for 5 min, count the number of EPG (Egg Per Gram) or OPG (Oocysts Per Gram) in the two graduated chambers under the microscope (27, 28).
The average A of the number of eggs in the two counting chambers multiplied by 200 is the number of eggs or oocysts per gram of stool. Compute the amount of EPG or OPG of oocysts per gram of stool according to the following formula:
All PCR products were visualized on a 1.0% agarose gel. All positive samples were purified and sequenced by Tsingke Biotechnology (Beijing, China). The sequence results were aligned in GenBank.
The correlation between the pathogen detection rate and the distance between the cattle farm and the river was analyzed using GraphPad, version 9.0.0. Statistical analyses of pathogen detection rates in different seasons throughout Ningxia and in different seasons in Yinchuan and Wuzhong were performed using GraphPad, version 9.0.0. Chi-square tests were performed at a 5% level of significance in SPSS 20.
PCR detection using primers designed by Xiao et al. (6), 111 (50.46%) of 220 stool samples were positive, of which 53 (24.09%) were infected by Cryptosporidium alone, and the rest were mixed infection (26.36%). The two highest proportions of mixed infections were Cryptosporidium and E. coli K99 (5.91%), followed by Cryptosporidium and BRV (5.45%), and then Cryptosporidium and Giardia (4.09%).
PCR detection using primers designed by Sulaiman (30), 30 (13.64%) of 220 stool samples were positive, of which 9 (4.09%) were infected by Giardia alone and the rest were mixed infection (9.55%). The two highest proportions of mixed infections were Giardia and Cryptosporidium (4.09%), followed by Giardia & Cryptosporidium & E. coli K99, Giardia & Cryptosporidium & BRV, Giardia & Cryptosporidium & BCoV, with a detection rate of 0.91%.
PCR detection using the primers reported by Keykhaei (18), among the 220 stool samples, 44 (20.00%) were positive, of which 14 (6.36%) were infected by E. coli K99 alone, and the rest were mixed infection (13.64%). The two highest proportions of mixed infections are E. coli K99 and Cryptosporidium (7.73%), followed by E. coli K99 and BRV (4.09%), and the proportion of simultaneous infection of E. coli K99, Cryptosporidium, and BRV is 1.82%. In the stool samples in which E. coli K99 was detected, it was only coinfected with Cryptosporidium and BRV, and no other pathogens were detected.
Using the VP6 gene primers of BRV designed by Guo (23) for PCR detection, 51 (23.18%) of 220 stools were positive, of which 16 (7.28%) were infected by BRV alone, and the rest were mixed infection (15.91%). The two highest proportions of mixed infections were BRV and Cryptosporidium (7.73%), followed by BRV and E. coli K99 (4.09%), and then BRV and BCoV (2.73%).
Using the Nsp10 gene primers in ORF1a of BCoV designed by Guo (23), 26 (11.82%) of 220 stools were positive, of which 8 (3.64%) were infected alone and 18 (8.18%) were infected with mixed infection. The two highest proportions of mixed infections were BCoV and Cryptosporidium (4.55%), followed by BCoV and BRV (0.91%), and then BCoV and E. coli K99 (0.91%).
Using the 3D gene primers of BKoV designed by Shi et al. (20) for PCR detection, 7 (3.18%) of 220 stools were positive, all of which were mixed infections. BKoV was predominantly coinfected with Cryptosporidium (1.82%) and BRV (1.36%).
PCR detection using the primers of the ORF1a gene of BoAstV reported by Shi (20) showed that 12 (5.46%) of 220 stools were positive, of which 1 was infected alone (0.45%), and the rest were mixed infection (5.00%). The two highest proportions of mixed infections were BoAstV and Cryptosporidium (2.73%), followed by BoAstV and BRV (1.36%).
The detection was executed using primers designed by Park (22) for the M gene of BToV to PCR. The results showed that 9 of 220 stools (4.09%) were positive, of which 1 was a single infection (0.45%), and the rest were mixed infections (3.64%), suggesting BToV is likely coinfected with two or more pathogens. In a sense, significant diarrhea symptoms only occur when BToV is coinfected with other pathogens.
Through the McMaster Egg Slide Counting Chamber, four cases (6.90%) of 58 calves (≥18 days) were detected positive for coccidia in this study, including two cases in Wuzhong and two cases in Yinchuan. The OPG levels of the two cases were 4,100 and 8,600 in WuZhong, and the calf ages were 89 and 84 d. The OPG content of the 2 cases was 17,200 and 19,000 in Yinchuan, and the age of the calf was 28 and 27 d. The specific results are shown in Table 3.
Together, a total of nine pathogens causing diarrhea including bacteria, viruses and parasites were detected in this study. The single infection rate of the detected pathogens is shown in Figure 2. The details of a single infection are shown in Tables 4, 5 for details of a mixed infection.
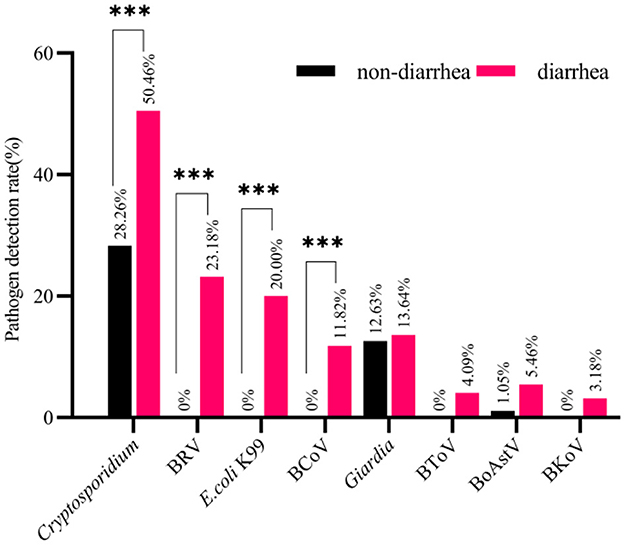
Figure 2. The Pathogens detection rate of diarrhea samples and the Pathogens detection rate of normal samples. The number of pathogen detection and non-detection in normal stool samples with the number of pathogen detection and non-detection in diarrhea stool samples were calculated, and then chi-square test was performed (mean ± standard deviation) ***p < 0.01.
In this study, 220 stool samples of calves with diarrhea and 95 normal samples of calves were detected. Among bacterial pathogens, E. coli K99 and C. perfringens were detected, and Proteus mirabilis and Salmonella were not detected. The primers reported by Jiang (21) were used to identify C. perfringens. The C. perfringens detected in this study were all type A and had no pathogenicity.
The detection rates of all pathogens from high to low are Cryptosporidium (50.46%), BRV (23.18%), E. coli K99 (20.00%), BCoV (11.82%), Giardia (13.64%), BoAstV (5.46%), BToV (4.09%), BKoV (3.18%). Comparison of detection details between diarrhea stool samples and normal stool samples by chi-square test, among them, the detection rates of Cryptosporidium (p < 0.01), BRV (p < 0.01), E. coli K99 (p < 0.01), and BCoV (p < 0.01) were significantly different between diarrhea stool samples and normal stool samples, while no significant differences were found for the other four pathogens including Giardia (p = 0.859), BoAstV (p = 0.118), BToV (p = 0.062), BKoV (p = 0.107).
Among the four diarrhea-related pathogens, BRV, E. coli K99, and BCoV were not detected in normal stool samples, but Cryptosporidium (28.26%) was detected in normal stool samples. The results showed that Cryptosporidium had a certain content in normal stool samples and diarrhea stool samples. No clinical diarrhea symptoms in normal stool samples were due to the low content of Cryptosporidium in calves. Among other pathogens, Giardia (12.63%) and BoAstV (1.05%) were also detected in normal samples and were present in the same situation as Cryptosporidium. In contrast, BToV and BKoV were not detected in normal samples, but the results were not significantly different from BToV (4.09%) and BKoV (3.18%) detection rates of diarrhea samples.
A total of 68 stool samples with diarrhea were detected in Yinchuan: E. coli K99 was detected in 19 samples (27.94%); BRV in 14 samples (20.59%); BCoV in 6 samples (8.82%); BToV in 2 samples (2.94%); BoAstV in 1 sample (1.47%); BKoV in 2 samples (2.94%); Coccidia in 2 samples (2.94%); Cryptosporidium in 28 samples (41.18%); Giardia in 6 (8.82%) samples.
The Chi-square test showed that the main epidemic cause of diarrhea happened in Yinchuan was E. coli K99 (p < 0.01), followed by BRV (p < 0.05). Although the detection rate of Cryptosporidium (41.18%) was the highest in diarrhea stool samples in Yinchuan, it was also the highest in normal stool samples, and the difference was not significant (32.00%). Other pathogens were detected in normal stool samples. The specific results are illustrated in Figure 3A.
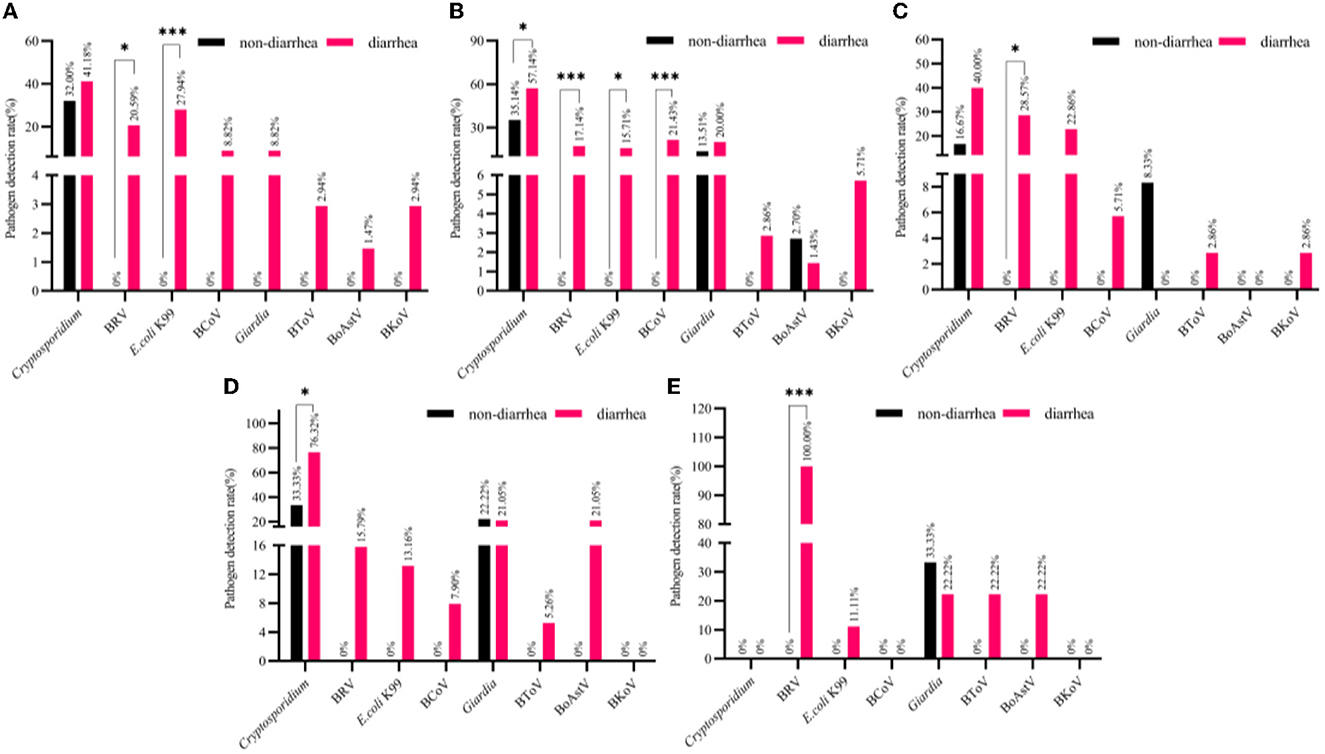
Figure 3. The detection rate of normal and diarrhea samples in different cities [(A) Yinchuan, (B) Wuzhong, (C) Shizuishan, (D) Zhongwei, (E) Guyuan]. The number of pathogen detection and non-detection in normal stool samples with the number of pathogen detection and non-detection in diarrhea stool samples in different cities were counted for chi-square test (mean ± standard deviation) ***p < 0.01, *p < 0.05.
Wuzhong detected E. coli K99 in 11 (15.71%) of 70 diarrhea stool samples; BRV in 12 (17.14%) samples; BCoV in 15 (21.43%) samples; BToV in 2 (2.86%) samples; BoAstV in 1 (1.43%) sample; BKoV in 4 (5.71%) samples; Coccidia in 2 (2.68%) samples; Cryptosporidium in 40 (57.14%) samples; Giardia in 14 (20.00%) samples.
The Chi-square test showed that the main epidemic pathogen causing diarrhea in Wuzhong calves was BCoV (p < 0.01), followed by BRV (p < 0.01), E. coli K99 (p < 0.05), Cryptosporidium (p < 0.05). Only Cryptosporidium was detected in both diarrhea and normal stool samples, and the other three pathogens were not detected in normal stool samples. Although the detection rate of Giardia in Wuzhong diarrhea stool samples was higher (20.00%), and it (13.51%) was second only to Cryptosporidium (35.14%) in normal stool samples, and the detection rate of Giardia in normal stool samples and diarrhea stool samples showed no difference. BToV and BKoV were not detected in normal samples, the detection rates in diarrhea stool samples were low (2.86%, 5.71%), and the difference was not significant. The detection rate of BoAstV in normal stool samples (2.70%) was even higher than that in diarrhea stool samples (1.43%). The results are detailed in Figure 3B.
In Shizuishan, detected 35 diarrhea stool samples including eight samples (22.86%) of E. coli K99; 10 samples of BRV (28.57%); two samples of BCoV (5.71%); one sample of BToV (2.86%); one sample of BKoV (2.86%); 14 samples of Cryptosporidium (40.00%).
The Chi-square test showed that the main epidemic pathogen causing diarrhea in Shizuishan calves was BRV (p < 0.05). Although Cryptosporidium (40.00%) and E. coli K99 (22.86%) had higher detection rates in diarrhea stool samples, there was no significant difference between them and normal stool samples. In particular, Giardia was not detected in diarrhea stool samples, but its detection rate in normal stool samples (8.33%) was second only to Cryptosporidium (16.67%). The results are detailed in Figure 3C.
A total of 38 stool samples with diarrhea were detected in Zhongwei: five samples (13.16%) of E. coli K99; four samples of BRV (15.79%); three samples of BCoV (7.90%); two samples of BToV (5.26%); eight samples of BoAstV (21.05%); Cryptosporidium 29 (76.32%) samples; Giardia 8 (21.05%) samples; BKoV and Coccidia were not detected.
The Chi-square test showed that the main epidemic pathogen causing diarrhea in Zhongwei calves was Cryptosporidium (p < 0.05), with a detection rate of 76.32%. The detection rates of other pathogens between diarrhea and normal stool samples were showed no significant difference. The detection rate of Giardia in normal stool samples (22.22%) was even higher than that in diarrhea stool samples (21.05%). The results are detailed in Figure 3D.
In Guyuan, a total of nine diarrhea stool samples were detected in E. coli K99 in 1 (11.11%); BRV in 9 (100%); BToV in 2 (22.22%); BoAstV in 2 (22.22%); Giardia in 2 (22.22%).
The Chi-square test showed that the main epidemic pathogen causing diarrhea in Guyuan calves was BRV (p < 0.01). Other pathogens were not significantly different. The detection rate of Giardia in normal stool samples (33.33%) was higher than that in diarrhea stool samples (22.22%), which was similar to the detection of Giardia in Zhongwei and Shizuishan. The results are detailed in Figure 3E.
The Chi-square test was performed on the number of cattle farms collected in different seasons and the pathogen detection rate of diarrhea fecal samples in each cattle farm. The correlation between the four main pathogens with the significant difference in detection rate in each season and diarrheal calves was analyzed.
The results showed that the dominant pathogens of diarrhea in spring in Ningxia were BCoV (30.50%), E. coli K99 (27.98%), BRV (27.05%) and Cryptosporidium (25.38%). In summer, the dominant pathogens of diarrhea were Cryptosporidium (59.63%), BRV (35.56%), E. coli K99 (31.44%) and BCoV (13.39%). In autumn, the dominant pathogens of diarrhea were Cryptosporidium (63.57%), BRV (29.61%), BCoV (19.55%) and E. coli K99 (15.56%). In winter, the dominant pathogens of diarrhea were Cryptosporidium (75.04%), BRV (53.57%), E. coli K99 (16.70%) and BCoV (13.39%). The detail results are illustrated in Figure 4.
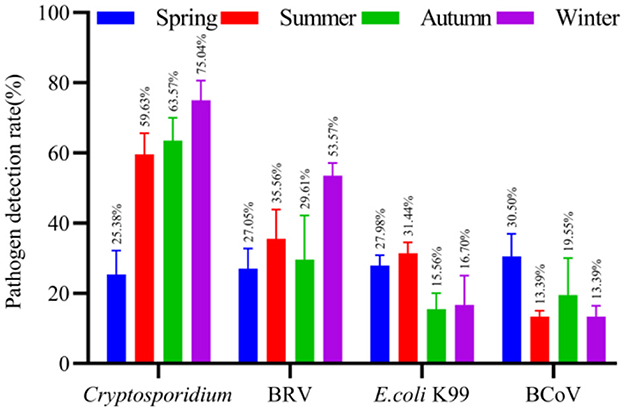
Figure 4. The detection rate of normal and diarrhea samples in different seasons. The number of cattle farms in Ningxia where samples were collected at different seasons, and the detection rates of Cryptosporidium, BRV, E. coli K99, and BCoV in each cattle farm were counted.
After the chi-square test of the entire Ningxia, the pathogens of calf diarrhea that were prevalent in each season have been obtained. Yinchuan and Wuzhong are the concentrated breeding areas of cattle in Ningxia. Analyzing the correlation between the significant pathogens in Yinchuan and Wuzhong in each season and diarrheal calves is more important. Based on the detection rate of different pathogens in the cattle farms of Yinchuan and Wuzhong in different seasons, the average detection rate of pathogens in each cattle farm was calculated, and the epidemic diarrhea pathogens in Yinchuan and Wuzhong in different seasons were obtained.
In Yinchuan, the dominant pathogens of diarrhea in spring were BRV (39.55%), E. coli K99 (36.82%), Cryptosporidium (19.55%) and BCoV (4.55%). In summer, the dominant diarrhea pathogens were Cryptosporidium (53.46%), E. coli K99 (30.39%), BRV (10.00%), and BCoV was not detected. In autumn, the dominant diarrhea pathogens were Cryptosporidium (18.18%), BRV (9.09%), BCoV (9.09%), and E. coli K99 was not detected. In winter, the dominant diarrhea pathogens were Cryptosporidium (92.86%), BRV (28.57%), E. coli K99 (16.67%), BCoV (7.15%). The results are detailed in Figure 5A.
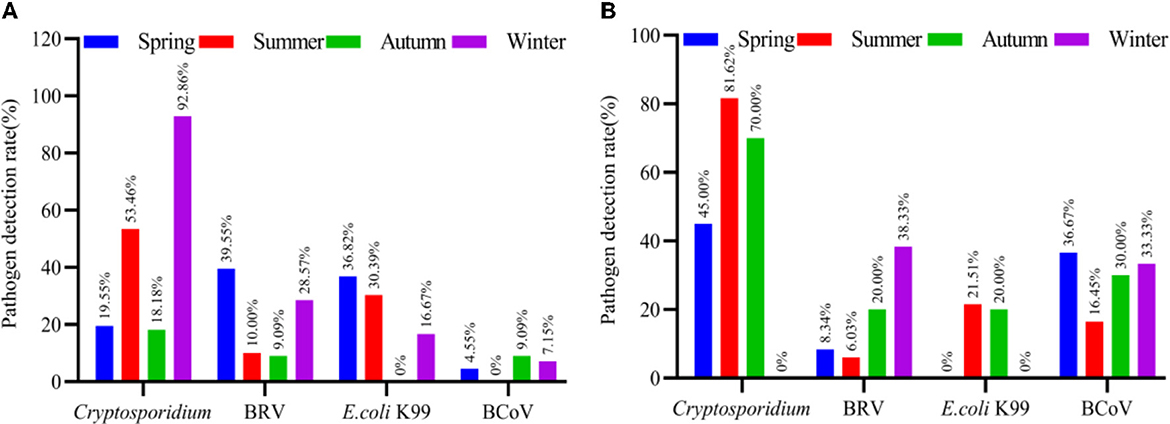
Figure 5. The detection rate of normal and diarrhea samples in different seasons [(A) Yinchuan, (B) Wuzhong]. The number of cattle farms in Yinchuan and Wuzhong, where samples were collected at different seasons, and the detection rates of Cryptosporidium, BRV, E. coli K99, and BCoV in each cattle farm were counted.
In Wuzhong, the dominant pathogens of diarrhea in spring were Cryptosporidium (45.00%), BCoV (36.67%), BRV (8.34%), and E. coli K99 was not detected. In summer, the dominant diarrhea pathogens were Cryptosporidium (81.62%), E. coli K99 (21.51%), BCoV (16.45%), BRV (6.03%). In autumn, the dominant diarrhea pathogens were Cryptosporidium (70.00%), BCoV (30.00%), BRV (20.00%), E. coli K99 (20.00%). In winter, the dominant diarrhea pathogens were BRV (38.33%), BCoV (33.33%), Cryptosporidium and E. coli K99 were not detected. The results are detailed in Figure 5B.
The earliest onset time and the common age of nine pathogens were illustrated in Figure 6. The earliest onset age of Cryptosporidium was 4 days, and the frequent onset age was 5–18 days. The earliest onset age of BRV was 4 days, and the frequent onset age was 7–30 days. The earliest onset age of E. coli K99 was 1 day and the common onset age was 8–15 days The earliest onset age of Giardia was 7 days, and the most frequent age was 11–30 days. The earliest onset age of BCoV was 2 days, and the most frequent age was 9–26 days. The earliest onset age of BoAstV was 8 days, and the most frequent age was 8-30 days. The earliest onset age of BToV was 8 days, and the most frequent age was 8–44 days. The earliest onset age of BKoV was 10 days, and the most frequent age was 10–26 days. The earliest onset age of Coccidia was 27 days.
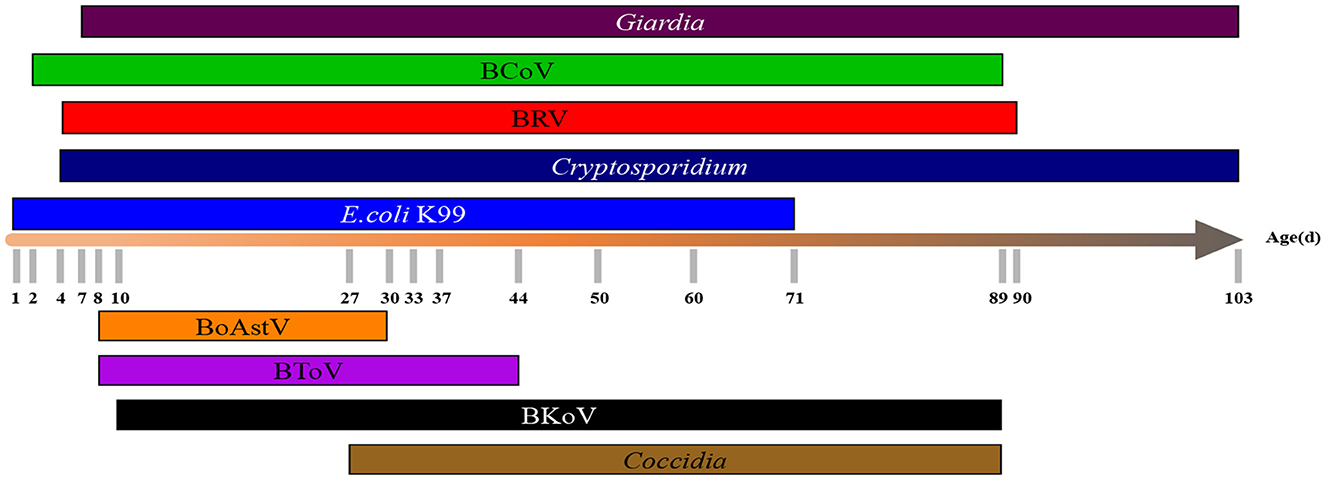
Figure 6. The distribution of different pathogens in different ages. The distribution of 9 pathogens in calves of different ages (1–103 d) was counted.
Ningxia is a province through which the Yellow River flows, with a length of about 397 km. There are two other tributaries, the Qingshui River and the Kushui River. Among the 23 large-scale cattle farms in this study, 18 cattle farms were close to the river, and the average number of Cryptosporidium detected per farm was 7.22, of which 5 cattle farms detected Cryptosporidium number ≥10. Among the five cattle farms where no Cryptosporidium were detected and where E. coli K99, BRV, and BCoV were the main diarrhea pathogens, three cattle farms were not surrounded by a river and one cattle farm was relatively far from a river.
This suggests Cryptosporidium is the main diarrhea pathogen in cattle farms, < 500 m from the water source. However, the detection rate of Cryptosporidium was positively correlated with the distance from cattle farms to rivers, but not significant (r = 0.1941), while the detection rates of E.coli K99, BRV, and BCoV were not correlated with the distance from cattle farms to rivers. The specific analysis results are detailed in Figure 7.
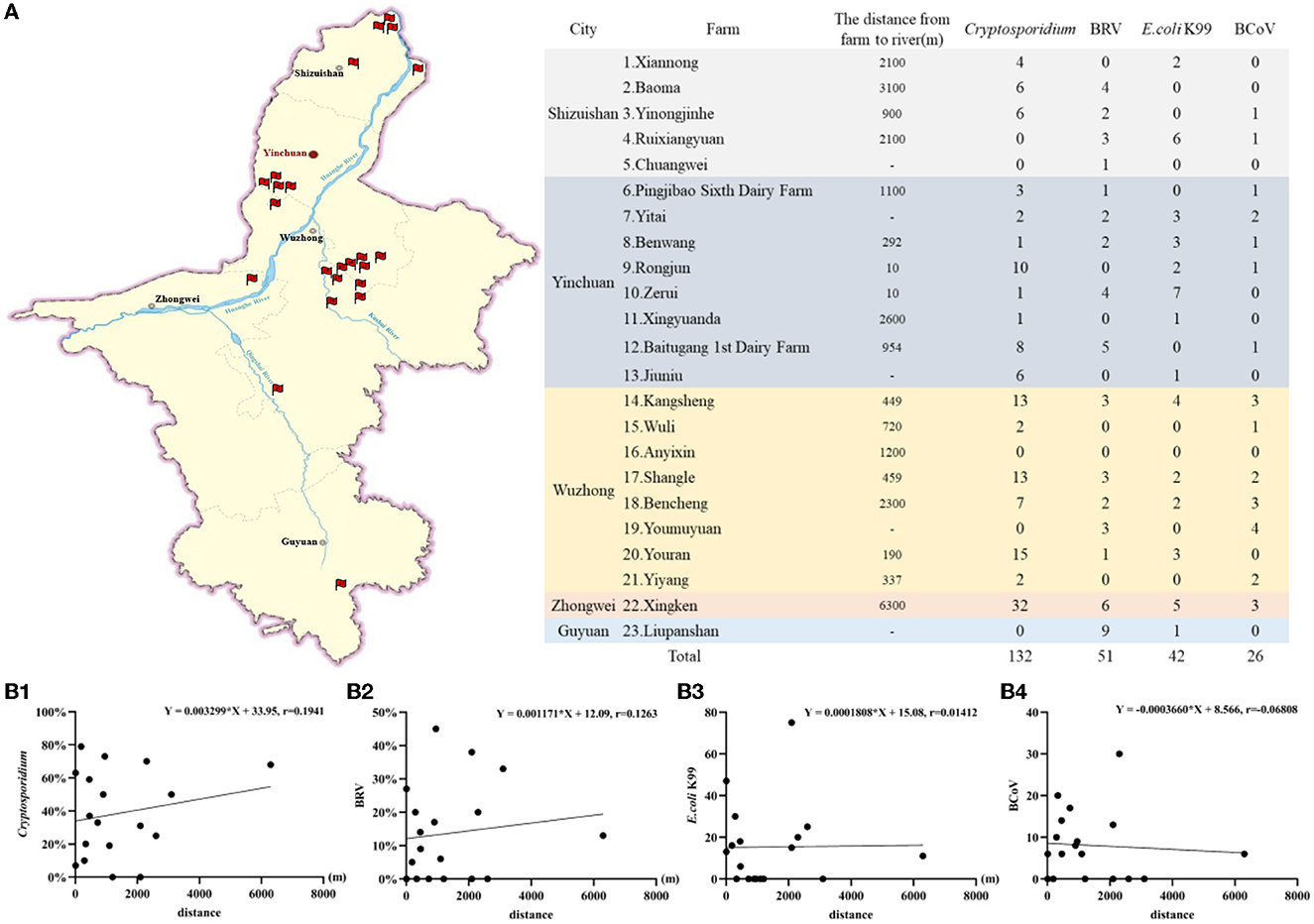
Figure 7. The correlation analysis of main pathogens causing calf diarrhea and river distribution. (A) The number of Cryptosporidium, BRV, E. coli K99, and BCoV detected in cattle farms and distribution of sampling cattle farms and rivers. (B1) The correlation between the Cryptosporidium detection rate and the distance from cattle farm to river. (B2) The correlation between the BRV detection rate and the distance from cattle farm to river. (B3) The correlation between the E.coli K99 detection rate and the distance from cattle farm to river. (B4) The correlation between the BCoV detection rate and the distance from cattle farm to river.
Wuzhong is the city with the most types of pathogens and the highest average detection rate, followed by Yinchuan. Because the etiology of calf diarrhea is more complex, in addition to other environmental factors, it is predominantly caused by pathogens [viruses (31), bacteria (1), parasites (32)], especially Cryptosporidium, which is principally transmitted by fecal-oral transmission (33). Therefore, calf density is one of the important factors affecting its transmission rate, and Wuzhong, Yinchuan, and some counties in Shizuishan and Zhongwei are the location of cattle breeding areas in Ningxia, and the density of calf herds is extremely high more than other cities. Consistently, like the results reported in other studies, Cryptosporidium is an important cause of diarrhea in Ningxia calves (33–35). In this study, a total of 315 stool samples were collected from all five cities in Ningxia, and 137 stool samples (43.49%) were positive for Cryptosporidium, including diarrhea samples (50.46%) and normal samples (27.37%). In 2015, researchers reported on Cryptosporidium infection in Ningxia and Gansu (35), 150 positive samples (5.09%) were detected in 2,945 stools in both diarrhea and normal calves. The detection rate of our study is significantly higher than 5.09%, which suggests that the infection rate of Cryptosporidium in Ningxia is rising year by year. Since December 2011, the detection rate of Cryptosporidium in Ningxia has shown a significant increase, from 1.68% (23/1,366) (33) to 50.46% (111/220). The infection of Cryptosporidium in calves with diarrhea and normal calves also coexist in this study, which is consistent with the results of the above studies.
At present, the treatment measures for Cryptosporidium are only preventive, and there is no effective commercial vaccine on the market to prevent long-term infection in cattle. The increased prevalence is one of the serious problems faced by researchers. Therefore, it is important to take care of deworming cattle in all growth stages and pay attention to biological safety measures.
BRV is the main pathogen that causes calf diarrhea worldwide. It has been also reported in many regions of China. Rotaviruses are a major causative pathogen of diarrhea in humans and animals, involving the deaths of 200,000 children in developing countries and causing economic losses in the livestock industry globally. In this study, the detection rate of BRV in Ningxia from 2021 to 2022 (23.18 %) was lower than the average detection rate of Ningxia over the years (32%), which was lower than the pooled prevalence of BRV in China 46% (6,635/10,677) (36). This is greatly related to the fact that the Ningxia agricultural department pays more attention to the impact of viruses on the cattle industry.
Compared with Cryptosporidium and BRV, the infection rates of E. coli K99 in Ningxia were relatively low. However, compared with other pathogens, E. coli K99 and other pathogenic Escherichia coli are still important pathogens causing calf diarrhea. The detection rate of BoAstV in Zhongwei (21.05%) was significantly higher than that in other cities, but it was not the main cause of diarrhea in Zhongwei calves, the reason may be that the BoAstV detected in this study was neurotype rather than diarrhea type. Evolutionary analyses showed that astrovirus strains from bovine brain tissue were closely related to astrovirus strains from humans, pigs, sheep and other animals with neurological symptoms, indicating that cross-species transmission may occur.
To date, Cryptosporidium, E. coli K99, BRV and BCoV have been identified as important pathogens prevalent in calf diarrhea in China. In addition, previous studies have demonstrated that BRV can be transmitted to humans directly or through recombination during the evolution of the strain and Cryptosporidium and E. coli K99, and is typical zoonosis (37). Thus, the in-depth investigation of the above calf diarrhea pathogens is the basis for the prevention and treatment of calf diarrhea, and how to avoid the mixed infection caused by multiple pathogens is of clinical significance. Thus, more efforts should be taken to block the spread of these pathogens in cattle farms and reduce the external factors leading to calf diarrhea. In total, it is possible to reduce the incidence of calf diarrhea.
The area around the reiver is a high-frequency area for parasite reproduction and transmission, and many parasites, including Cryptosporidium, can be transmitted through water (38, 39). Cryptosporidium in its oocyst stage can remain infectious for many months under cool, moist conditions such as rivers, lakes and ponds (40), and in a relatively dry environment, it is more suitable for the growth of viruses and bacteria (24, 41). The distribution of calf diarrhea pathogens in Ningxia also showed similar characteristics in this study, and how to prevent the spread of the pathogen due to geographic environmental factors is one of the issues the researchers have been facing.
In this study, Cryptosporidium can be detected in both diarrheal calves and normal calves, and other pathogens are a mixed infection of two or more pathogens in the same or different calves. Together, Cryptosporidium, BRV, E. coli K99 and BCoV are the main pathogens causing calf diarrhea in Ningxia, the remaining four pathogens are mainly infected in the form of mixed infection.
From June 2021 to May 2022, the main pathogens causing calf diarrhea in Yinchuan were E. coli K99 and BRV; the main pathogens causing calf diarrhea in Wuzhong are Cryptosporidium, BCoV, BRV and E. coli K99; BRV was the main pathogen causing calf diarrhea in Shizuishan; Cryptosporidium was the main pathogen causing calf diarrhea in Zhongwei; BRV was the main pathogen causing calf diarrhea in Guyuan.
Different seasons had a more obvious effect on the detection rate of calf diarrhea-related pathogens. In addition, the rivers had an effect on the detection rate of Cryptosporidium. In conclusion, the distribution of diarrhea pathogens in Ningxia calves is associated with geographical and environmental factors.
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding authors.
The animal study was reviewed and approved by Executive Committee of Laboratory Animal Management and Ethics Inspection of Northwest A&F University, Xianyang, China. Written informed consent was obtained from the owners for the participation of their animals in this study.
KG, XK, and HG designed the experiments. DW, LZ, and WD carried out the experiments. HG and CL collected samples. DW wrote the manuscript. XZ and YL contributed to data analysis and helped complete the experiments. All authors discussed the results and commented on the manuscript.
This work was supported by the Key Research and Development Program of Ningxia under Grant [Project No. 2021BEF03005].
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Ali A, Liaqat S, Tariq H, Abbas S, Arshad M, Li WJ, et al. Neonatal calf diarrhea: A potent reservoir of multi-drug resistant bacteria, environmental contamination and public health hazard in Pakistan. Sci Total Environ. (2021) 799:149450. doi: 10.1016/j.scitotenv.2021.149450
2. Hur J, Jeon BW, Kim YJ, Oh IG, Lee JH. Escherichia coli isolates from calf diarrhea in Korea and their virulent genetic characteristics. J Vet Med Sci. (2013) 75:519–22. doi: 10.1292/jvms.12-0378
3. Cho YI, Yoon KJ. An overview of calf diarrhea - infectious etiology, diagnosis, and intervention. J Vet Sci. (2014) 15:1–17. doi: 10.4142/jvs.2014.15.1.1
5. McGuirk SM. Disease management of dairy calves and heifers. The Veterinary clinics of North America. Food Animal Pract. (2008) 24:139–53. doi: 10.1016/j.cvfa.2007.10.003
6. Windeyer MC, Leslie KE, Godden SM, Hodgins DC, Lissemore KD, LeBlanc SJ. (2014). Factors associated with morbidity, mortality, and growth of dairy heifer calves up to 3 months of age. Prevent Vet Med. 113:231–240. doi: 10.1016/j.prevetmed.2013.10.019
7. Caffarena RD, Casaux ML, Schild CO, Fraga M, Castells M, Colina R, et al. Causes of neonatal calf diarrhea and mortality in pasture-based dairy herds in Uruguay: a farm-matched case-control study. Brazilian J Microbiol. (2021) 52:977–88. doi: 10.1007/s42770-021-00440-3
8. Chae JB, Kim HC, Kang JG, Choi KS, Chae JS Yu DH, et al. The prevalence of causative agents of calf diarrhea in Korean native calves. J Animal Sci Technol. (2021) 63:864–71. doi: 10.5187/jast.2021.e63
9. Dall Agnol AM, Lorenzetti E, Leme RA, Ladeia WA, Mainardi RM, Bernardi A, et al. Severe outbreak of bovine neonatal diarrhea in a dairy calf rearing unit with multifactorial etiology. Brazilian J Microbiol. (2021) 52:2547–53. doi: 10.1007/s42770-021-00565-5
10. Geng HL, Ni HB, Li JH, Jiang J, Wang W, Wei XY, et al. Prevalence of Cryptosporidium spp. in Yaks (Bos grunniens) in China: a systematic review and meta-analysis. Front Cell Infect Microbiol. (2021) 11:770612. doi: 10.3389/fcimb.2021.770612
11. Odagiri K, Yoshizawa N, Sakihara H, Umeda K, Rahman S, Nguyen SV, et al. Development of genotype-specific anti-bovine rotavirus a immunoglobulin yolk based on a current molecular epidemiological analysis of bovine rotaviruses a collected in Japan during 2017-2020. Viruses. (2020) 12:1386. doi: 10.3390/v12121386
12. Lombardelli JA, Tomazic ML, Schnittger L, Tiranti KI. Prevalence of Cryptosporidium parvum in dairy calves and GP60 subtyping of diarrheic calves in central Argentina. Parasitol Res. (2019) 118:2079–86. doi: 10.1007/s00436-019-06366-y
13. Burimuah V, Sylverken A, Owusu M, El-Duah P, Yeboah R, Lamptey J, et al. Molecular-based cross-species evaluation of bovine coronavirus infection in cattle, sheep and goats in Ghana. BMC Vet Res. (2020) 16:405. doi: 10.1186/s12917-020-02606-x
14. Guo Z, He Q, Yue H, Zhang B, Tang C. First detection of Nebovirus and Norovirus from cattle in China. Arch Virology. (2018) 163:475–8. doi: 10.1007/s00705-017-3616-6
15. Li N, Wang R, Cai M, Jiang W, Feng Y, Xiao L. Outbreak of cryptosporidiosis due to Cryptosporidium parvum subtype IIdA19G1 in neonatal calves on a dairy farm in China. Int J Parasitol. (2019) 49:569–577. doi: 10.1016/j.ijpara.2019.02.006
16. Liu X, Yan N, Yue H, Wang Y, Zhang B, Tang C. Detection and molecular characteristics of bovine rotavirus A in dairy calves in China. J Vet Sci. 22:e69. doi: 10.4142/jvs.2021.22.e69
17. Wei X, Wang W, Dong Z, Cheng F, Zhou X, Li B, et al. Detection of infectious agents causing neonatal calf diarrhea on two large dairy farms in Yangxin County, Shandong Province, China. Front Vet Sci. (2021) 7:589126. doi: 10.3389/fvets.2020.589126
18. Keykhaei N, Salari S, Rashki A. Frequency of k99, stx1, and stx2 virulence factors in escherichia coli isolated from diarrheic and clinically healthy suckling calves in Sistan and Baluchistan Province, Iran. Arch Razi Inst. (2021) 76:283−91. doi: 10.22092/ari.2019.124040.1268
19. Zishiri OT, Mkhize N, Mukaratirwa S. Prevalence of virulence and antimicrobial resistance genes in Salmonella spp isolated from commercial chickens and human clinical isolates from South Africa and Brazil. Onderstepoort J Vet Res. (2016) 83:1067. doi: 10.4102/ojvr.v83i1.1067
20. Sanches MS, Rodrigues da Silva C, Silva LC, Montini VH, Lopes Barboza MG, Migliorini Guidone GH, et al. Proteus mirabilis from community-acquired urinary tract infections (UTI-CA) shares genetic similarity and virulence factors with isolates from chicken, beef and pork meat. Microbial Pathogenesis. (2021) 158:105098. doi: 10.1016/j.micpath.2021.105098
21. Jiang Y, Ma Y, Liu Q, Li T, Li Y, Guo K, et al. Tracing Clostridium perfringens strains from beef processing of slaughter house by pulsed-field gel electrophoresis, and the distribution and toxinotype of isolates in Shaanxi province, China. Food Microbiol. (2022) 101:103887. doi: 10.1016/j.fm.2021.103887
22. Park SI, Jeong C, Kim HH, Park SH, Park SJ, Hyun BH, et al. Molecular epidemiology of bovine noroviruses in South Korea. Vet Microbiol. (2007) 124:125–33. doi: 10.1016/j.vetmic.2007.03.010
23. Guo Z, He Q, Zhang B, Yue H, Tang C. Detection molecular characteristics of neboviruses in dairy cows in China. J Gen Virol. (2019) 100:35–45. doi: 10.1099/jgv.0.001172
24. Shi Z, Wang W, Chen C, Zhang X, Wang J, Xu Z, et al. First report and genetic characterization of bovine torovirus in diarrhoeic calves in China. BMC Vet Res. (2020) 16:272. doi: 10.1186/s12917-020-02494-1
25. He H, Tang C, Chen X, Yue H, Ren Y, Liu Y, et al. Isolation and characterization of a new enterovirus F in yak feces in the Qinghai-Tibetan Plateau. Arch Virol. (2017) 162:523–7. doi: 10.1007/s00705-016-3119-x
26. Xiao L, Morgan UM, Limor J, Escalante A, Arrowood M, Shulaw W, et al. Genetic diversity within Cryptosporidium parvum and related Cryptosporidium species. Appl Environ Microbiol. (1999) 65:3386–91. doi: 10.1128/AEM.65.8.3386-3391.1999
27. Dong H, Li C, Zhao Q, Li J, Han H, Jiang L, et al. Prevalence of Eimeria infection in yaks on the Qinghai-Tibet Plateau of China. J Parasitol. (2012) 98:958–62. doi: 10.1645/GE-3079.1
28. Dong H, Zhao Q, Han H, Jiang L, Zhu S, Li T, et al. Prevalence of coccidial infection in dairy cattle in Shanghai, China. J Parasitol. (2012) 98:963–6. doi: 10.1645/GE-2966.1
29. Sulaiman IM, Fayer R, Bern C, Gilman RH, Trout JM, Schantz PM, et al. Triosephosphate isomerase gene characterization and potential zoonotic transmission of Giardia duodenalis. Emerg Infect Dis. (2003) 9:1444–52. doi: 10.3201/eid0911.030084
30. Shimoda T, Okubo T, Enoeda Y, Yano R, Nakamura S, Thapa J, et al. Effect of thermal control of dry fomites on regulating the survival of human pathogenic bacteria responsible for nosocomial infections. PLoS ONE. (2019) 14:e0226952. doi: 10.1371/journal.pone.0226952
31. Lotfollahzadeh S, Madadgar O, Reza Mohebbi M, Reza Mokhber Dezfouli M, George Watson D. Bovine coronavirus in neonatal calf diarrhoea in Iran. Vet Med Sci. (2020) 6:686–94. doi: 10.1002/vms3.277
32. Li N, Zhao W, Song S, Ye H, Chu W, Guo Y, et al. Diarrhoea outbreak caused by coinfections of Cryptosporidium parvum subtype IIdA20G1 and rotavirus in pre-weaned dairy calves. Transbound Emerg Dis. (2022) 69:e1606–17. doi: 10.1111/tbed.14496
33. Huang J, Yue D, Qi M, Wang R, Zhao J, Li J, et al. Prevalence and molecular characterization of Cryptosporidium spp. and Giardia duodenalis in dairy cattle in Ningxia, northwestern China. BMC Vet Res. (2014) 10:292. doi: 10.1186/s12917-014-0292-6
34. Cui Z, Wang R, Huang J, Wang H, Zhao J, Luo N, et al. Cryptosporidiosis caused by Cryptosporidium parvum subtype IIdA15G1 at a dairy farm in Northwestern China. Parasites Vectors. (2014) 7:529. doi: 10.1186/s13071-014-0529-z
35. Zhang XX, Tan QD, Zhou DH, Ni XT, Liu GX, Yang YC, et al. Prevalence and molecular characterization of Cryptosporidium spp. in dairy cattle, northwest China. Parasitology Res. (2015) 114:2781–7. doi: 10.1007/s00436-015-4537-5
36. Chen S, Zhang W, Zhai J, Chen X, Qi Y. Prevalence of bovine rotavirus among cattle in mainland China: a meta-analysis. Microb Pathog. (2022) 170:105727. doi: 10.1016/j.micpath.2022.105727
37. Doan YH, Nakagomi T, Aboudy Y, Silberstein I, Behar-Novat E, Nakagomi O, et al. (2013). Identification by full-genome analysis of a bovine rotavirus transmitted directly to and causing diarrhea in a human child. J Clini Microbiol. 51, 182–189. doi: 10.1128/JCM.02062-12
38. Bajer A, Toczylowska B, Bednarska M, Sinski E. Effectiveness of water treatment for the removal of Cryptosporidium and Giardia spp. Epidemiol Infect. (2012) 140:2014–22. doi: 10.1017/S0950268811002780
39. Omarova A, Tussupova K, Berndtsson R, Kalishev M, Sharapatova K. Protozoan parasites in drinking water: a system approach for improved water, sanitation and hygiene in developing countries. Int J Res Public Health. (2018) 15:495. doi: 10.3390/ijerph15030495
40. Fayer R. (2004). Cryptosporidium: a water-borne zoonotic parasite. Vet Parasitol. 126:37–56. doi: 10.1016/j.vetpar.2004.09.004
Keywords: diarrhea, calf, epidemic investigation, Ningxia, pathogens
Citation: Wang D, Gao H, Zhao L, Lv C, Dou W, Zhang X, Liu Y, Kang X and Guo K (2023) Detection of the dominant pathogens in diarrheal calves of Ningxia, China in 2021–2022. Front. Vet. Sci. 10:1155061. doi: 10.3389/fvets.2023.1155061
Received: 31 January 2023; Accepted: 30 March 2023;
Published: 17 April 2023.
Edited by:
Yi Yang, Yangzhou University, ChinaReviewed by:
Tianle Xu, Yangzhou University, ChinaCopyright © 2023 Wang, Gao, Zhao, Lv, Dou, Zhang, Liu, Kang and Guo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaodong Kang, a2FuZ3hkMTk3MUAxMjYuY29t; Kangkang Guo, Z3Vva2syMDA3QG53c3VhZi5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.