- 1Institute of Animal Health, Guangdong Academy of Agricultural Sciences, Guangzhou, China
- 2Key Laboratory of Livestock Disease Prevention and Treatment of Guangdong Province, Guangzhou, China
- 3College of Veterinary Medicine Shanxi Agricultural University, Taigu, China
- 4Guangdong Laboratory for Lingnan Modern Agriculture, Guangzhou, China
- 5Key Laboratory for Prevention and Control of Avian Influenza and Other Major Poultry Diseases, Ministry of Agriculture and Rural Affairs, Guangzhou, China
In 2020, a chicken-origin Cluster 3 Tembusu virus (TMUV) caused outbreaks of a disease characterized by egg-drop syndrome in laying hens in China. In the present study, a TMUV strain, TMUV-GX, was isolated from tissue samples of laying hens with egg drop syndrome in south China. Phylogenetic analysis grouped TMUV-GX into TMUV Cluster 3.2, which was distinct from the prevalent TMUV Cluster 2 in duck flocks. To study the infectivity and pathogenicity of TMUV-GX in chickens and ducks, 7 day-old specific pathogen-free (SPF) chicks and SPF ducklings were infected with the same dose of the TMUV-GX. As a comparison, the duck-origin Cluster 2 strain, TMUV-JM, infection groups were set up in chicks and ducklings. Compared with the low infectivity and pathogenicity of TMUV-JM in chicks, the chicken-origin TMUV-GX displayed high replication competence in multiple tissues and caused tissues histopathological damage. In addition, the replication competence of TMUV-GX in ducklings was comparable to that of TMUV-JM. Our study revealed chicken-origin Cluster 3.2 TMUV exhibits high infectivity in chicks and ducklings, and suggested that chicken-origin Cluster 3.2 TMUV possesses a biological basis for widespread infection of chickens and ducks.
Introduction
Tembusu virus (TMUV) belongs to the genus Flavivirus in the family Flaviviridae and is an enveloped virus containing positive-sense single-stranded RNA. In 1955, the first TMUV strain, MM1775, was isolated from Culex mosquitoes in Malaysia (1). In 2000, the first avian-origin TMUV (Sitiawan virus), causing chick encephalitis and retarded growth, was isolated in the Sitiawan District of Perak State, Malaysia (2). In subsequent years, multiple fowl-origin TMUV were identified in Southeastern Asia (3). In 2010, a TMUV-caused disease, characterized by a significant decrease in egg production of laying ducks, broke out in many duck farms in Southeastern China, and spread widely in China and Southeastern Asia in following years (4–6).
The viral full genomic length is 10990 nucleotides (nt), and contains a 5' untranslated region (UTR), a large open reading frame (ORF), and a 3' UTR. The ORF encodes a polyprotein that can be processed by viral or cellular proteases into three structural proteins [capsid (C), membrane protein (M), and envelope protein (E)] and seven non-structural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5). Phylogenetic analyses based on the genes encoding E and NS5 showed that the TMUVs were divided into three clusters (Cluster 1, 2, and 3). In China, TMUV isolates that circulated predominantly in water-fowl belonged to Cluster 2. Although TMUV has been reported in a variety of birds, including chickens and sparrows, the virus did not cause large-scale epidemics in these birds (7, 8).
In China, although there were a few reports of TMUV causing egg drop syndrome in laying hens (7, 9), there was no report of the widespread of TMUV in chicken flocks. In these cases, the TMUV isolates that caused egg drop syndrome in chickens belonged to TMUV Cluster 2. However, in 2020, a TMUV-caused disease associated with egg drop syndrome occurred laying hen flocks in southwest Guangdong, China (10). In a further study, researchers revealed that the TMUV isolate causing the disease belonged to TMUV Cluster 3. Subsequently, laying hen flocks infected with Cluster 3 TMUV causing egg drop syndrome were reported sequentially in Guangxi, Fujian, Hubei, Jiangsu, and Shandong, China (11). The factors that cause Cluster 3 TMUV to spread more rapidly and widely in chicken flocks than Cluster 2 TMUV remain unclear.
In this study, we isolated a Cluster 3 TMUV strain, TMUV-GX, which was identified as the pathogen of laying hen egg drop syndrome in Guangxi, China. Furthermore, We investigated the differences in infectivity and pathogenicity between TMUV-GX and the Cluster 2.2 TMUV strain TMUV-JM in both chicks and ducklings.
Materials and methods
Sample collection and virus isolation
In 2020, a disease characterized by a decline in egg production spread through chicken farms in many regions of south China. In this study, tissues, including livers, spleens, and ovaries, of laying hens with egg drop syndrome were collected from Guangxi, China and submitted for microbial diagnosis. For virus isolation, samples were pooled and homogenized in phosphate-buffered saline (PBS) to give a 10% suspension (wt/vol). The homogenized samples were centrifuged at 8,000 × g for 10 min. The supernatant was filtered through a 0.22 μm pore-size sterile filter (Millipore, Bedford, MA, USA) and inoculated into the allantoic cavity of 10-day-old specific pathogen-free (SPF) chicken embryonated eggs. Allantoic fluid of embryos that died at 2–6 days post-inoculation were collected and centrifuged. The virus was propagated three times in SPF chicken embryonated eggs.
Viral whole genome sequencing
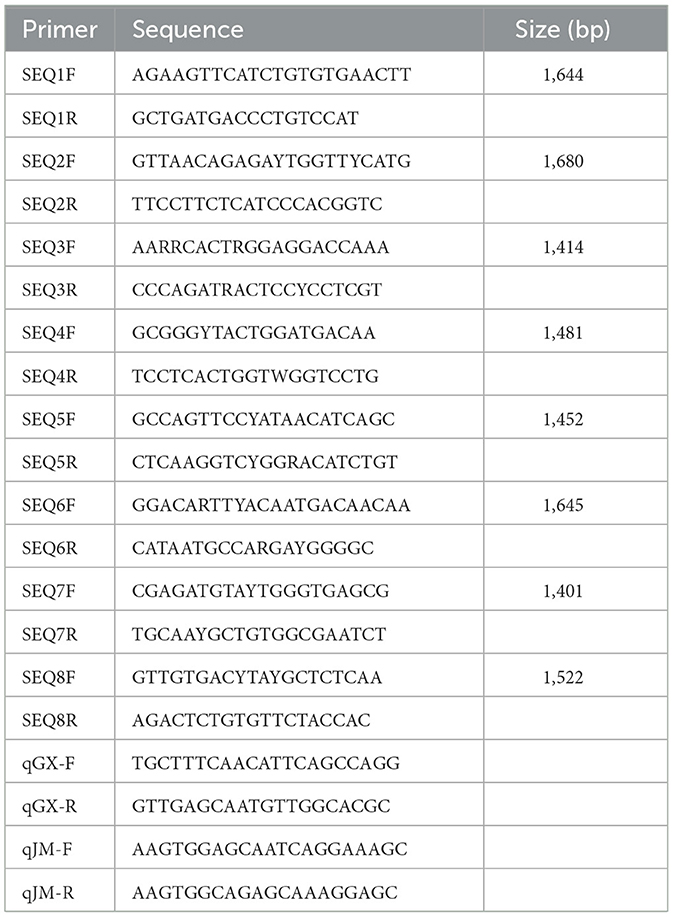
The viral RNA was extracted using a Tianlong Nucleic Acid Extraction & Purification Kit (T180H; Tianlong Technology Co., Ltd., Xi'an, China). First-strand cDNA was synthesized using Reverse Transcriptase ALV (Takara Biotechnology Co., Ltd., Dalian, China). To acquire the complete genome of the virus, eight pairs primers were designed based on published TMUVs sequences (Table 1). The eight PCR products were sequenced using the Sanger method by Shanghai Sangon Biotechnology Co., Ltd. (Shanghai, China). The 5'- and 3'- UTRs were amplified using a Takara SMARTer RACE 5′/3′ Kit. The complete genome was assembled using SeqMan of the DNASTAR package (DNASTAR Inc., Madison, WI, USA).
Phylogenetic analysis
To determine the phylogenetic relationship of the TMUV-GX strain with other reported isolates, E gene nucleotide sequences of TMUV-GX and other reference isolates were used. The nucleotide sequences were aligned, and the best nucleotide substitution model was selected using MEGA v7.0. Phylogenetic tree was generated using the maximum likelihood (ML) method implemented on MEGA v7.0 with the bootstrap method (12). The phylogenetic trees were visualized and annotated using MEGA v7.0.
Animal experiments
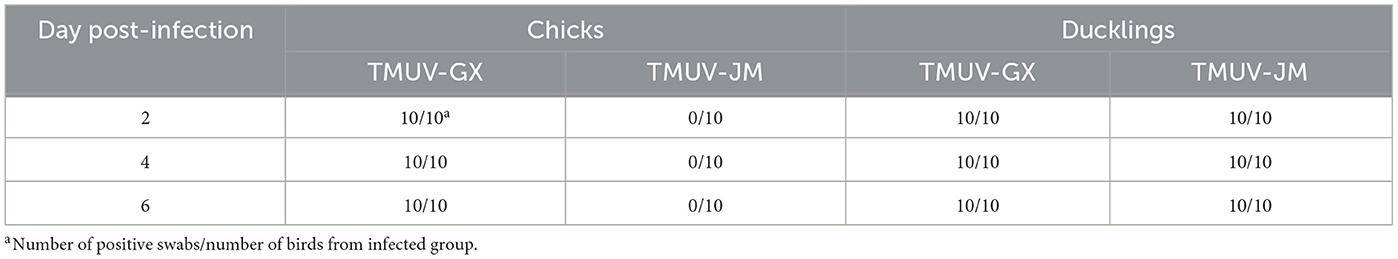
To investigated the differences in infectivity and pathogenicity between TMUV-GX and TMUV-JM in both chicks and ducklings, sixty 7 day-old SPF chicks (Xinxing Dahuanong Poultry Egg Co., Ltd., Yunfu, China) and SPF ducklings (Harbin veterinary research institute, China Academy of Agricultural Sciences) were used for infection experiments. Groups of twenty chicks or twenty ducklings were intramuscularly inoculated with 105 TCID50 (median tissue culture infectious dose) of TMUV-GX or TMUV-JM in volume of 0.1 ml. Twenty chicks and twenty ducklings were intramuscularly injected with 0.1 ml of PBS as negative controls. At 2, 4, and 6 days post-infection (dpi), three birds in each group were euthanized and liver, spleen, lung, kidney, and brain samples were collected for viral copy number detection. To investigate viral excretion, cloacal swabs and throat swabs were collected every 2 days after infection, and the mixed cloacal swabs and throat swabs samples were detected using reverse transcription quantitative real-time PCR (RT-qPCR).
For histopathological examinations, livers, kidneys, and brains were collected at 4 dpi, fixed in 4% paraformaldehyde solution, embedded in paraffin, sectioned, and stained with hematoxylin and eosin (HE).
Total RNA isolation and RT-qPCR
Total RNA isolation from tissues and swabs was performed using RNAiso plus (Takara) according to the manufacturer's protocol. The RNA was then reverse transcribed to cDNA. To detect the copy number of TMUV RNA in samples, the cDNA was processed using the quantitative real-time PCR (qPCR) step of the RT-qPCR protocol using two pairs of qPCR primers designed for TMUV-JM and TMUV-GX NS2A genes, respectively (Table 1). Standard plasmids pMD18T-JMNS2A and pMD18T-GXNS2A, containing viral NS2A genes, were constructed and used in the assay to generate a standard curve to quantify the viral copy number. The qPCR process was carried out using a Roche LightCycler96 System (Roche Diagnostics, Basel, Switzerland) and a ChamQ SYBR qPCR Master Mix (Vazyme, Nanjing, China).
Statistical analysis
In the figures, all data in are expressed as the mean ± standard deviation (SD). Data were analyzed using an independent-sample t-test in Graph Prism software (GraphPad Software Inc., San Diego, CA, USAS). Statistical significance was set at p < 0.05.
Results
Virus isolation and genome sequencing
The tissues samples collected from laying hens with egg drop syndrome were homogenized, and the supernatant tested negative for avian influenza virus, Newcastle disease virus, fowl adenovirus serotype-4, egg drop syndrome 76 virus, and infectious bursal disease virus, but tested positive for TMUV, as determined using PCR or RT-PCR assays. The virus was isolated in 10-day-old SPF chicken embryos, in which embryo death was observed. The dead embryos showed severely congestion and hemorrhage (Figure 1). The virus was propagated three times in SPF chicken embryonated eggs for purification and named as TMUV-GX.
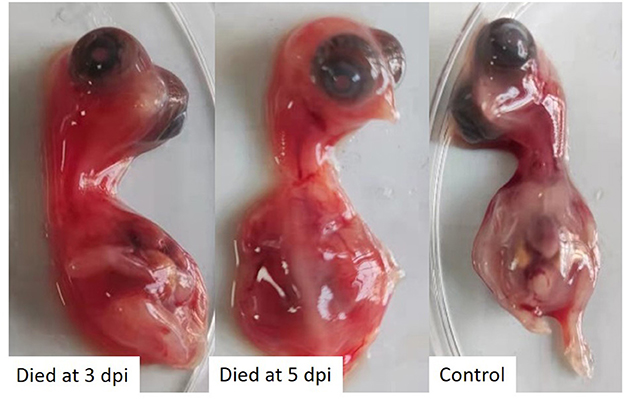
Figure 1. SPF chicken embryos infected with TMUV-GX died and showed severely congestion and hemorrhage.
To acquire the complete viral genome, a set of primers were used to amplify the viral genome using RT-PCR (Table 1). PCR amplicons were then sequenced. The complete genome comprised 10994 nt.
Phylogenetic analysis
The evolutionary relationship between TMUV-GX and reference strains was investigated using a phylogenetic tree based on the viral E genes (Figure 2). The phylogenetic tree showed that the TMUVs were divided into three clusters. Most of the Cluster 1 TMUV strains were isolated from Southeast Asia. Cluster 2 was divided into Cluster 2.1 and Cluster 2.2, and contained most of the waterfowl-origin TMUVs in the Chinese mainland. Cluster 3 was further divided into Cluster 3.1 and Cluster 3.2. Cluster 3.1 contained mosquito-origin, duck-origin, and chicken-origin TMUVs found in Southeast Asia, but not found in the Chinese mainland. The TMUV-GX was grouped into Cluster 3.2, which contained most of the mosquito-origin TMUVs. Specifically, TMUV-GX is closely related to the TMUV CTLN strain, which causes egg-drop in chickens.
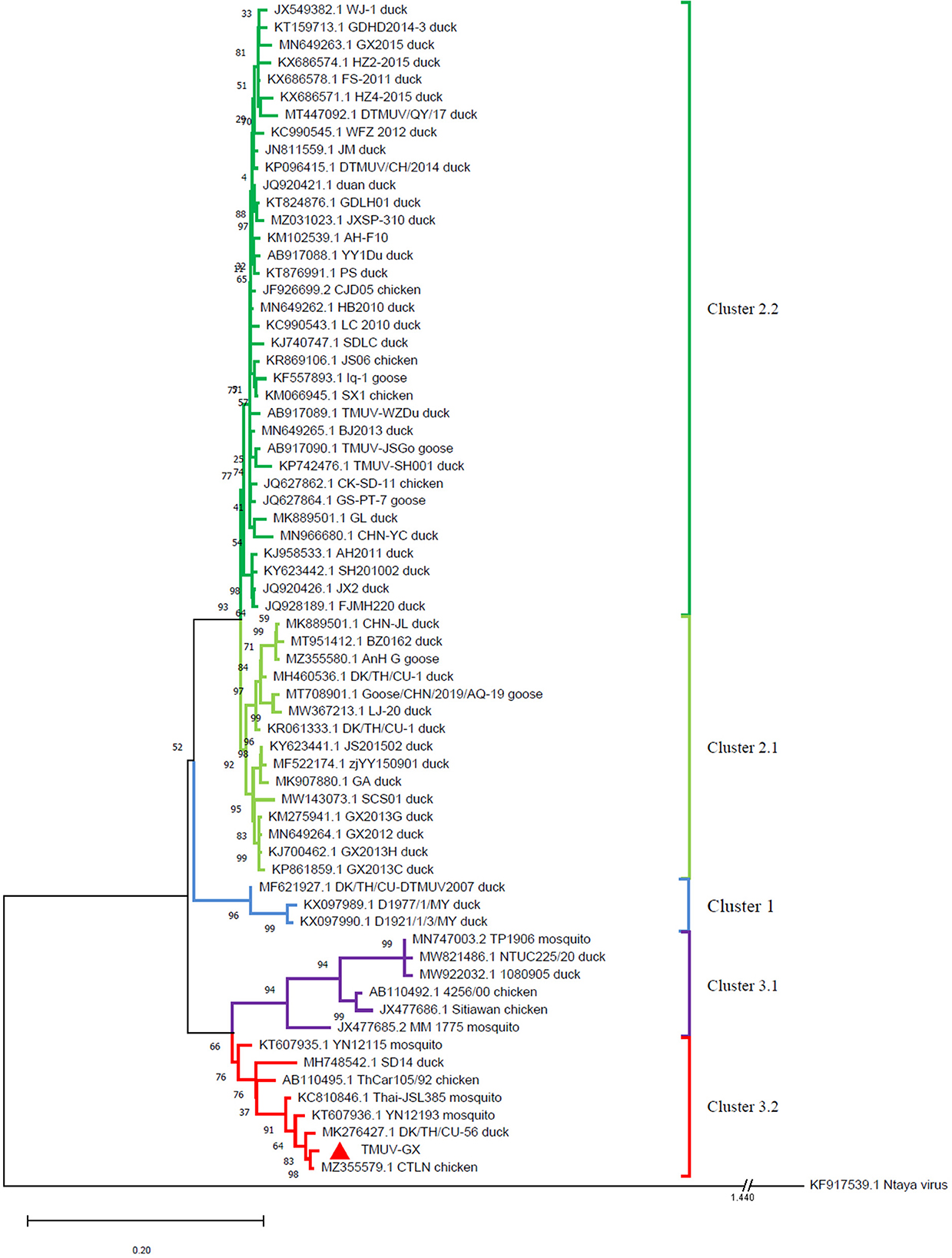
Figure 2. Phylogenetic analysis based on the E genes of TMUVs. The phylogenetic tree was generated via the ML method implement on MEGA v.7.0. Bootstrap values are shown at the nodes. The Ntaya isolate was used as the outgroup.
Comparison of pathogenicity between TMUV-GX and TMUV-JM
In the chick infection experiments, chicks in the TMUV-GX infection group did not die, but showed very mild diarrhea, moderate lack of appetite, and very mild depression (Table 2). The mean body weight in the TMUV-GX-infected chicks decreased by 20–25% compared with that in the control group at 4–7 dpi (Figure 3A). In the TMUV-JM infection group, very mild lack of appetite but no others obvious clinical signs were observed.
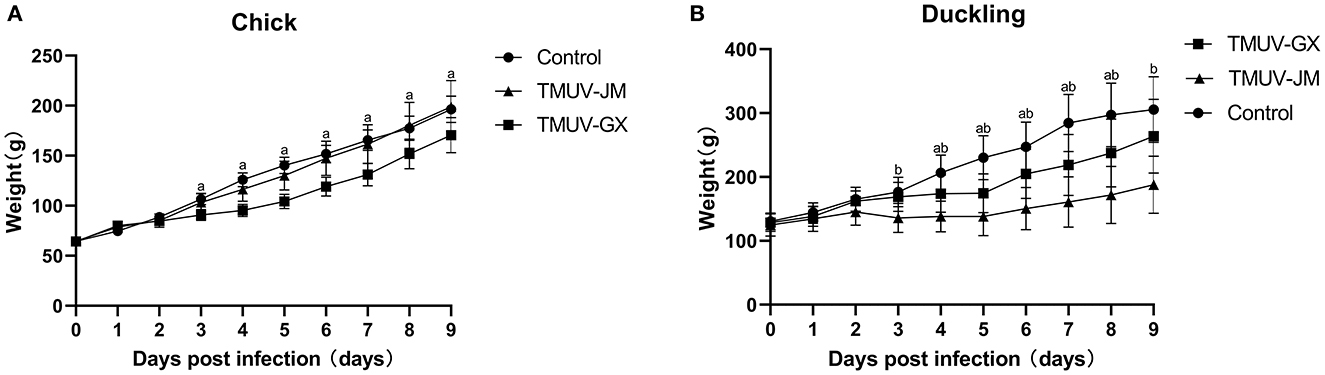
Figure 3. Weight changes of chicks (A) and ducklings (B) after infection with TMUV. The error bars indicate the means ± standard deviation (SD). Different lowercase letters over the bars denote statistically significant differences (p < 0.05) between the infection groups and the control group [(a) TMUV-GX infection group vs. control group; (b) TMUV-JM infection group vs. control group].
In the duckling infection experiments, diarrhea, lack of appetite, and depression were observed in both the TMUV-GX and TMUV-JM infection groups (Table 2). No ducks died during the experiment. However, the clinical signs were more severe in the TMUV-JM infection group than in the TMUV-GX infection group. Compared with that in the control group, the body weights gain in TMUV-infected ducklings decreased significantly after infection. Specifically, compared with that in the control group, the mean body weight of the TMUV-GX-infected ducklings decreased by 15–25% at 4–8 dpi, and the mean body weight of the TMUV-JM-infected ducklings decreased by 33–45% at 4–9 dpi (Figure 3B).
Histopathology revealed that the chicks and ducklings inoculated with TMUV-GX presented significant lesions in the liver, brain, and kidney. In the liver, multifocal lymphoid infiltration was frequently observed in the TMUV-GX-infected chicks, while lymphoid infiltration was less frequently observed in the TMUV-JM-infected chicks (Figures 4A, D). In TMUV-infected ducklings, there were lesions in the liver characterized by diffuse hydropic degeneration of hepatocytes and some lymphoid infiltration (Figures 4G, J). In the brain, we observed neuronal pyknosis, enhanced basophilic cytoplasm, and increase of microglia in the TMUV-GX-infected chicks. Increased microglia were also observed in the TMUV-GX-infected ducklings, whereas no obvious lesions were found in the TMUV-JM-infected chicks and ducklings (Figures 4B, E, H, K). In the kidney, renal tubular atrophy, luminal stenosis, multiple epithelial cell necrosis, nuclear pyknosis, lymphoid infiltration, and hydropic degeneration of renal tubular epithelial cells were observed in the TMUV-GX-infected chicks and ducklings (Figures 4C, I). There were lesions characterized by renal tubular atrophy, luminal stenosis, epithelial cell necrosis, nuclear pyknosis in the kidneys of the TMUV-JM-infected ducklings, whereas less hydropic degeneration of renal tubular epithelial cells was observed in the TMUV JM infected chicks (Figures 4F, L).
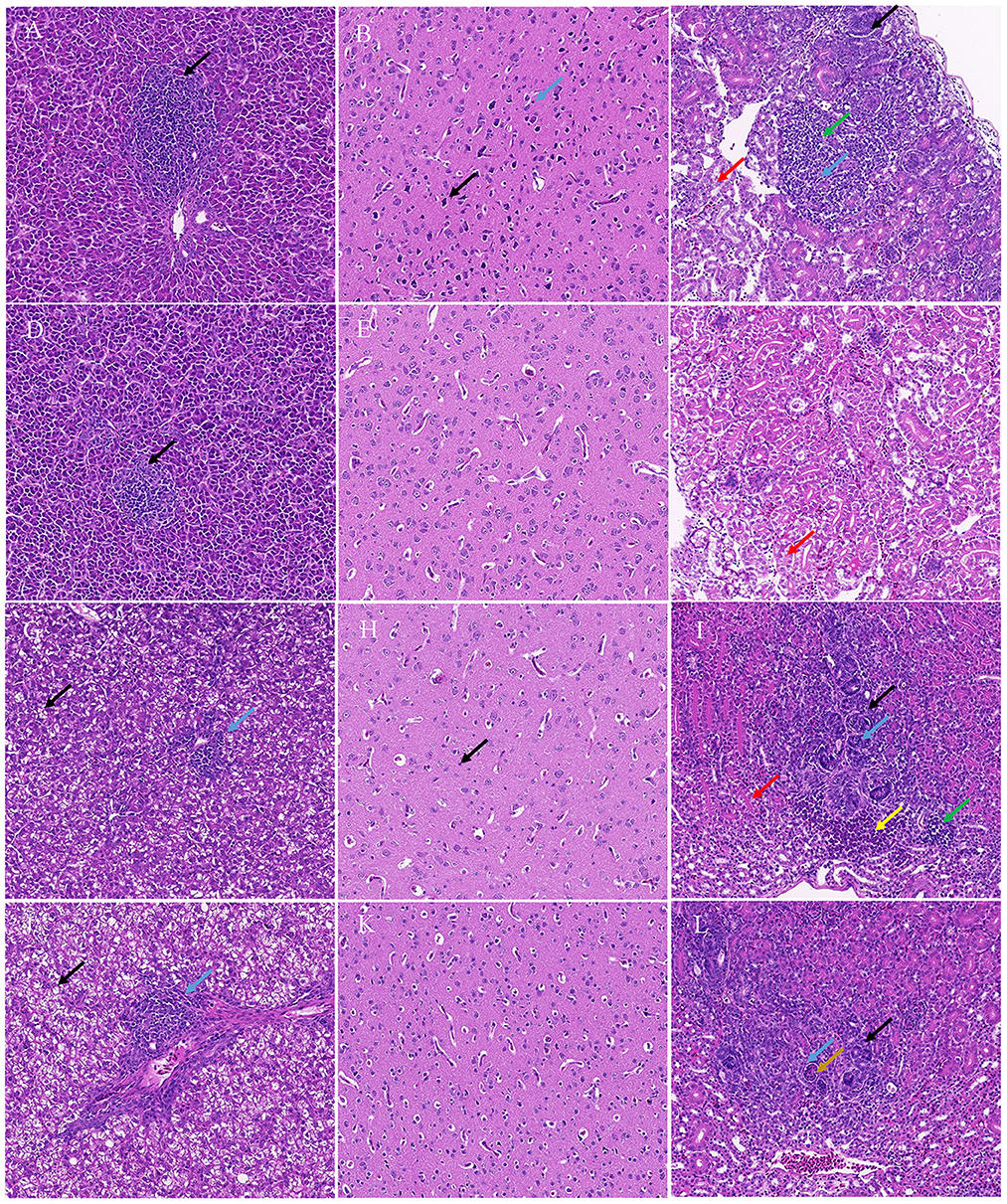
Figure 4. Histopathological lesions. (A) Liver, TMUV-GX-infected chicks: lymphoid infiltration (black arrow); (B) Brain, TMUV-GX-infected chicks: neuronal pyknosis, enhanced basophilic cytoplasm (black arrow) and increase in microglia (blue arrow); (C) Kidney, TMUV-GX-infected chicks: renal tubular atrophy, luminal stenosis (black arrow), multiple epithelial cell necrosis, nuclear pyknosis (blue arrow), lymphoid infiltration (green arrow), and hydropic degeneration of renal tubular epithelial cells (red arrow); (D) Liver, TMUV-JM-infected chicks: lymphoid infiltration (black arrow); (E) Brain, TMUV-JM-infected chicks; (F) Kidney, TMUV-JM-infected chicks: some hydropic degeneration of renal tubular epithelial cells (red arrow); (G, J) Liver, TMUV-infected ducklings: diffuse hydropic degeneration of hepatocytes (black arrow) and some lymphoid infiltration (blue arrow); (H) Brain, TMUV-GX-infected ducklings: increase in microglia (black arrow); (I) Kidney, TMUV-GX-infected ducklings: renal tubular atrophy, luminal stenosis (black arrow), multiple epithelial cell necrosis, nuclear pyknosis (blue arrow), lymphoid infiltration (green arrow), hydropic degeneration of renal tubular epithelial cells (red arrow), and hemorrhage (yellow arrow); (K) Brain, TMUV-JM-infected ducklings; (L) Kidney, TMUV-JM-infected ducklings: renal tubular atrophy, luminal stenosis (black arrow), multiple epithelial cell necrosis, nuclear pyknosis (blue arrow), and necrotic cell mass (brown arrow).
Viral RNA copy number in tissues and viral excretion
The virus loads in the different tissues of TMUV-infected chicks and ducklings were determined using RT-qPCR (Figure 5). In the chick infection experiments (Figures 5A–C), the viral RNA could be detected in all investigated tissues of the TMUV-GX infection group at 2 and 4 dpi, whereas only the spleens and kidneys of the TMUV-JM infection group were positive for viral RNA at 2 and 4 dpi. Viral RNA copy numbers in all investigated tissues obtained from the TMUV-GX infection group were significantly higher than those in the TMUV-JM infection group. The maximum viral RNA copy numbers were achieved at 4 dpi, and higher viral RNA copy numbers were found in the kidneys and spleen than in other tissues. Furthermore, viral RNA copy numbers in the brains of the TMUV-GX infection group were generally higher at 4 and 6 dpi, whereas viral RNA was not detected in the brain of most TMUV-JM-infected chicks. In the duckling infection experiments (Figures 5D-5F), the viral RNA could be detected in all investigated tissues of the TMUV-infected ducklings at 2, 4, and 6 dpi, with the maximum viral RNA copy numbers being achieved at 2 dpi. Furthermore, viral RNA copy numbers in the liver, kidney and brain were lower in the TMUV-GX infection group than in the TMUV-JM infection group at 2 dpi. No viral RNA was detected in the control groups. Generally, the viral copy numbers in the investigated tissues from the TMUV-GX-infected chicks were higher than those in the TMUV-JM-infected chicks, but did not differ between the two duckling infected groups.
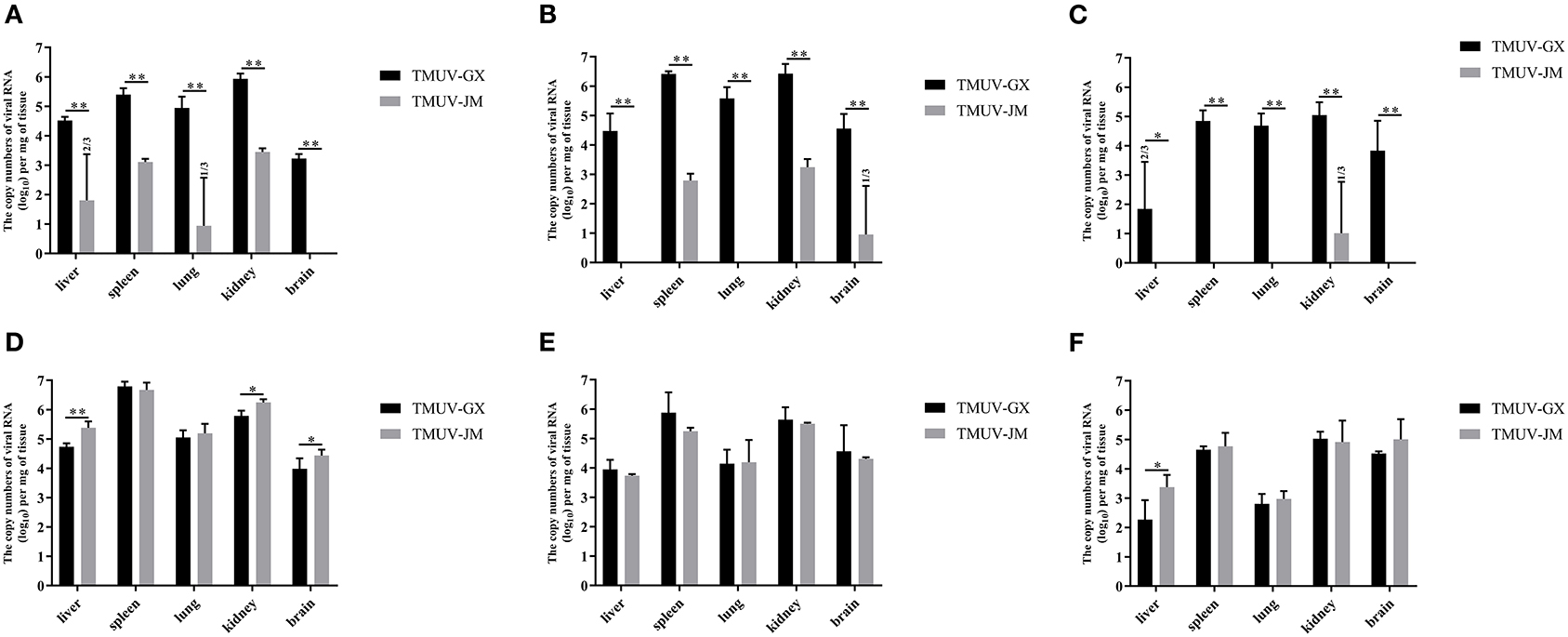
Figure 5. Viral RNA copy numbers in different tissues of experimentally infected animals. (A–C) show the viral RNA copy numbers in tissue samples of the TMUV-infected chicks at 2, 4, and 6 dpi, respectively. (D–F) show the viral RNA copy numbers in tissue samples of the TMUV-infected ducklings at 2, 4, and 6 dpi, respectively. The statistical differences between both groups were evaluated using an independent-sample t-test. *p < 0.05, **p < 0.01.
To investigate viral excretion, cloacal swabs and throat swabs were collected at 2, 4, and 6 dpi, and the mixed cloacal swabs and throat swab samples were detected using RT-qPCR (Table 3). Swabs collected from TMUV-GX-infected chicks and ducklings were detected as 100% positive for viral RNA at 2, 4, and 6 dpi. Viral RNA was also detected in swabs collected from TMUV-JM-infected ducklings at 2, 4, and 6 dpi. However, swabs collected from TMUV-JM-infected chicks were negative for viral RNA at 2, 4, and 6 dpi. No positive viral RNA was recorded in the control birds in this study.
Discussion
Generally, TMUV has been considered as highly contagious in water-fowl (13). Although TMUV-induced egg-drop sporadic cases in laying hens have been reported, there has been no report widespread of TMUV infections in chicken flocks. Since 2020, Cluster 3 TMUV infection causing laying hen egg drop syndrome occurred successively in laying hen flocks in many provinces of China. In this study, a Cluster 3 TMUV isolate, TMUV-GX, was isolated from samples of laying hens with egg drop syndrome.
The E gene plays an important role in viral antigenicity, tissue tropism, transmissibility, and virulence (14–16). The result of phylogenetic analysis based on the E gene showed that the TMUVs were divided into 3 clusters. Cluster 1 and 2 mainly contained water-fowl origin isolates. Although Cluster 2 TMUVs were sporadically isolated from other birds (including chickens, sparrows, and pigeons), they did not cause an epidemic in these birds. The TMUV-GX, isolated in this study, was phylogenetically grouped into Cluster 3.2, which contained most of the mosquito-origin TMUVs. Furthermore, TMUV-GX was most closely related to the CTLN strain, which was isolated from laying hen flocks displaying clinical egg drop syndrome in Guangdong, China, in 2020. We believe that TMUV-GX and CTLN are different isolates that caused the same epidemic that occurred successively in many regions of China. In addition, TMUV-GX was also clustered with the mosquito-origin YN12193 strain and the duck-origin DK/TH/CU-56 strain. Therefore, we conjectured that the Cluster 3.2 TMUV, which spread widely in chicken flocks recently, could become a potential threat for both fowl and water-fowl flocks.
We compared the infectivity and pathogenicity of TMUV-GX with that of duck-origin Cluster 2.2 strain TMUV-JM in chicks and ducks. TMUV-GX-infected chicks displayed clinical signs, consisting of diarrhea, lack of appetite, depression, and growth retardation, which were not observed in the TMUV-JM-infected chicks. The same clinical signs were observed in TMUV-GX-infected and TMUV-JM-infected ducklings. The histological lesions observed in the TMUV-GX-infected ducklings were similar to those of TMUV-JM-infected ducklings, except for the brain. However, the lesions in chicks attributed to TMUV-GX infection were more serious and more numerous, especially in the brain, compared with those in TMUV-JM infection. The clinical signs and microscopic lesions indicated that the TMUV-GX is more pathogenic for chicks than TMUV-JM. The results of tissue viral RNA load determination showed significantly higher viral copy numbers in all investigated tissues of TMUV-GX-infected chicks in comparison with TMUV-JM-infected chicks, demonstrating that the replication competence of TMUV-GX in chicks was stronger than that of TMUV-JM. However, the levels of tissue viral RNA load of TMUV-GX-infected and TMUV-JM-infected ducklings were almost identical, indicating that the replication competence of TMUV-GX in ducklings is comparable to that of TMUV-JM.
Conclusion
The results of the present study demonstrated that the chicken-origin TMUV-GX has high infectivity and certain pathogenicity toward chicks, whereas duck-origin cluster 2 TMUV did not have these characteristics. In addition, we revealed that chicken-origin Cluster 3.2 TMUV has high infectivity and certain pathogenicity toward ducklings. Our findings suggested that Cluster 3.2 TMUV, which has spread widely in chicken flocks recently, could be a potential threat to both chicken and duck flocks.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
Ethics statement
The animal study was reviewed and approved by the Committee on the Ethics of Animal Experiments of Institute of Animal Health, Guangdong Academy of Agricultural Sciences Experimental Animal Welfare Ethics Committee (Approve ID: 2018-007).
Author contributions
SG, MS, and ML supervised this project, designed the experiments. YH, ZH, JD, LL, JZ, and RK collected samples and performed experiments. YH analyzed data and prepared manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (No. 31902272; No. 32102691), Guangdong Basic and Applied Basic Research Foundation (2021A1515012071), Guangdong Laboratory of Lingnan Modern Agriculture Project (NZ2021015), National Key R&D Program of China (2022YFD1801000), Jiangmen Basic and Applied Basic Research Foundation (2021030102450004518).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2023.1152802/full#supplementary-material
References
1. Kuno G, Chang GJ, Tsuchiya KR, Karabatsos N, Cropp CB. Phylogeny of the genus Flavivirus. J Virol. (1998) 72:73–83. doi: 10.1128/JVI.72.1.73-83.1998
2. Kono Y, Tsukamoto K, Abd HM, Darus A, Lian TC, Sam LS, et al. Encephalitis and retarded growth of chicks caused by Sitiawan virus, a new isolate belonging to the genus Flavivirus. Am J Trop Med Hyg. (2000) 63:94–101. doi: 10.4269/ajtmh.2000.63.94
3. Thontiravong A, Ninvilai P, Tunterak W, Nonthabenjawan N, Chaiyavong S, Angkabkingkaew K, et al. Tembusu-related Flavivirus in ducks, Thailand. Emerg Infect Dis. (2015) 21:2164–7. doi: 10.3201/eid2112.150600
4. Su J, Li S, Hu X, Yu X, Wang Y, Liu P, et al. Duck egg-drop syndrome caused by BYD virus, a new Tembusu-related Flavivirus. PLoS One. (2011) 6:e18106–e18106. doi: 10.1371/journal.pone.0018106
5. Cao Z, Zhang C, Liu Y, Liu Y, Ye W, Han J, et al. Tembusu virus in ducks, China. Emerg Infect Dis. (2011) 17:1873–5. doi: 10.3201/eid1710.101890
6. Ninvilai P, Limcharoen B, Tunterak W, Prakairungnamthip D, Oraveerakul K, Banlunara W, et al. Pathogenesis of Thai duck Tembusu virus in Cherry Valley ducks: The effect of age on susceptibility to infection. Vet Microbiol. (2020) 243:108636. doi: 10.1016/j.vetmic.2020.108636
7. Liu M, Chen S, Chen Y, Liu C, Chen S, Yin X, et al. Adapted Tembusu-like virus in chickens and geese in China. J Clin Microbiol. (2012) 50:2807–9. doi: 10.1128/JCM.00655-12
8. Yu G, Lin Y, Tang Y, Diao Y. Evolution of Tembusu virus in ducks, chickens, geese, sparrows, and mosquitoes in northern China. Viruses. (2018) 10:485. doi: 10.3390/v10090485
9. Xu T, Liu X, Liu Q, Han K, Liu Y, Jang J, et al. Effect of experimental infections of various Tembusu virus strains isolated from geese, ducks and chickens on ducklings. Pol J Vet Sci. (2018) 21:389–96. doi: 10.24425/122613
10. Yu Z, Ren H, Sun M, Xie W, Sun S, Liang N, et al. Tembusu virus infection in laying chickens: Evidence for a distinct genetic cluster with significant antigenic variation. Transbound Emerg Dis. (2022) 69:e1130–41. doi: 10.1111/tbed.14402
11. Yan D, Li X, Wang Z, Liu X, Dong X, Fu R, et al. The emergence of a disease caused by a mosquito origin Cluster 32 Tembusu virus in chickens in China. Vet Microbiol. (2022) 272:109500. doi: 10.1016/j.vetmic.2022.109500
12. Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 70 for bigger datasets. Mol Biol Evolut. (2016) 33:1870. doi: 10.1093/molbev/msw054
13. Li X, Shi Y, Liu Q, Wang Y, Li G, Teng Q, et al. Airborne transmission of a novel tembusu virus in ducks. J Clin Microbiol. (2015) 53:2734–6. doi: 10.1128/JCM.00770-15
14. Sun M, Dong J, Li L, Lin Q, Sun J, Liu Z, et al. Recombinant Newcastle disease virus (NDV) expressing Duck Tembusu virus (DTMUV) pre-membrane and envelope proteins protects ducks against DTMUV and NDV challenge. Vet Microbiol. (2018) 218:60–9. doi: 10.1016/j.vetmic.2018.03.027
15. Li L, Zhang Y, Dong J, Zhang J, Zhang C, Sun M, et al. The truncated E protein of DTMUV provide protection in young ducks. Vet Microbiol. (2020) 240:108508. doi: 10.1016/j.vetmic.2019.108508
Keywords: tembusu virus, egg-drop syndrome, infectivity, pathogenicity, chicks
Citation: Huang Y, Hu Z, Dong J, Li L, Zhang J, Kuang R, Gao S, Sun M and Liao M (2023) Chicken-origin Cluster 3.2 Tembusu virus exhibits higher infectivity than duck-origin Cluster 2 Tembusu virus in chicks. Front. Vet. Sci. 10:1152802. doi: 10.3389/fvets.2023.1152802
Received: 28 January 2023; Accepted: 03 March 2023;
Published: 24 March 2023.
Edited by:
Jianzhu Liu, Shandong Agricultural University, ChinaCopyright © 2023 Huang, Hu, Dong, Li, Zhang, Kuang, Gao, Sun and Liao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shimin Gao, c21nYW8mI3gwMDA0MDsxMjYuY29t; Minhua Sun, c21oMjAwMnNtaCYjeDAwMDQwOzE2My5jb20=; Ming Liao, bWxpYW8mI3gwMDA0MDtzY2F1LmVkdS5jbg==
 Yunzhen Huang
Yunzhen Huang Ziqiang Hu
Ziqiang Hu Jiawen Dong
Jiawen Dong Linlin Li
Linlin Li Junqin Zhang
Junqin Zhang Ruihuan Kuang
Ruihuan Kuang Shimin Gao3*
Shimin Gao3* Minhua Sun
Minhua Sun Ming Liao
Ming Liao