
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci. , 21 March 2023
Sec. Parasitology
Volume 10 - 2023 | https://doi.org/10.3389/fvets.2023.1145096
Cryptosporidium spp. are important enteric protozoan parasites that infect humans and other animals throughout the world. Cryptosporidium infection in cattle industry leads to substantial economic losses due to diarrhea, growth retardation, weight loss, and possibly death. Most studies have focused on C. parvum, and studies on other Cryptosporidium spp. and calf diarrhea are limited. Therefore, this study aimed to investigate the occurrence of Cryptosporidium spp. in pre-weaned calves, to determine the risk factors for Cryptosporidium spp. infection such as age and season, and to identify subtypes of C. parvum circulating in the Republic of Korea (ROK). A total of 510 fecal samples were collected from calves with diarrhea and divided by age and season. Cryptosporidium spp. were first screened using PCR targeting the small subunit (SSU) rRNA gene and further the 60-kDa glycoprotein gene for subtyping of C. parvum. Out of 510 fecal samples, 71 (13.9%) were positive for Cryptosporidium spp. in pre-weaned calves with diarrhea. C. andersoni (2.8%), C. bovis (30.9%), C. parvum (29.6%), and C. ryanae (36.6%) were identified. C. ryanae was the most predominant in calves in the ROK. Calf age was a significant risk factor for C. bovis (χ2 = 13.83, P = 0.001), C. parvum (χ2 = 7.57, P = 0.023), and C. ryanae (χ2 = 20.18, P = 0.000) occurrence. Additionally, C. parvum was detected 3.1-fold more frequently in pre-weaned calves with diarrhea in fall (95% CI: 1.23–7.81; P = 0.016) than in spring, whereas C. ryanae was 8.9-fold more frequently detected in summer (95% CI: 1.65–48.68; P = 0.011) than in spring. Three subtypes (IIaA17G4R1, IIaA18G3R1, and IIaA20G3R1) of C. parvum were identified. Of them, IIaA17G4R1 was the most common, whereas IIaA20G3R1 was not previously detected in calves in the ROK. To our knowledge, this is the first report of C. andersoni in pre-weaned calves in the ROK. The occurrence of Cryptosporidium spp. appears to be age-dependent in calves. Season had a significant effect on the occurrence of C. parvum and C. ryanae. Taken together, C. bovis and C. ryanae along with C. parvum are detected in pre-weaned calves with diarrhea and these two pathogens should not be overlooked in the diagnosis of calf diarrhea.
Cryptosporidium spp. are important protozoan parasites that affect the gastrointestinal tract in various animals, including humans. Cryptosporidium, with Giardia, is known as representative water-borne pathogen in humans (1–3). Infection with these species occurs via the fecal-oral route, either by direct contact with infected animals or by ingestion of infective oocysts from contaminated water and/or food (4). Cryptosporidium is one of the major pathogens causing diarrhea in neonatal calves, which is characterized by watery and sometimes bloody stool, loss of appetite, abdominal pain, growth retardation, weight loss, and possibly death, resulting in substantial economic losses (5–11).
To date, 44 Cryptosporidium spp. have been identified (12), and among them, four species, namely, C. andersoni, C. bovis, C. parvum, and C. ryanae, have been identified in cattle. Their prevalence has been shown to have an age-related distribution: C. parvum in pre-weaned (13), C. ryanae and C. bovis in post-weaned (14), and C. andersoni in yearlings and adult cattle (15). In addition, C. andersoni has been found in all age groups (16, 17). However, the occurrence of these species is not necessarily age-related, and a few studies performed in several countries have reported different patterns of infection (13, 16, 18–20). C. bovis, C. parvum, and C. ryanae are bovine intestinal species and often cause atrophy of the villi, shortening of the microvilli, and destruction of the intestine, resulting in diarrhea, whereas C. andersoni infects the abomasum, which causes gastritis (5, 17, 21). Most importantly, C. andersoni, C. bovis, and C. ryanae are associated with subclinical infection in cattle (22–24). Molecular methods should be used for species differentiation because of their indistinguishable sizes and shapes (24, 25).
C. parvum accounts for over 90% of Cryptosporidium infections in neonatal calves (26). Calves with diarrhea were reportedly 36.5 times more likely to be infected with C. parvum than calves without diarrhea (17). C. parvum is the most prevalent species infecting humans and calves worldwide, whereas C. andersoni and C. bovis are rarely found in humans (4, 23, 27). Recently, C. bovis and C. ryanae have been identified in pre-weaned calves (28–31); however, their pathogenicity and role in calf diarrhea remain unclear. According to a previous study performed by our group, C. ryanae was detected at a higher frequency in calves with diarrhea than C. bovis and has now emerged as a leading cause of diarrhea alongside C. parvum (31). C. parvum subtypes have been detected using the 60-kDa glycoprotein (gp60) gene, and to date, at least 14 families (IIa and IIo) have been identified (12, 27). Among them, two families, IIa and IId, are found in both humans and ruminants, and are known as having zoonotic potential. Family IIa has been identified in calves in most industrialized nations (4), and IIaA15G2R1 is the most dominant subtype reported in cattle worldwide (27). Additionally, family IId has been found in dairy calves in some countries (20, 32–36). In contrast, pre-weaned calves with diarrhea in the Republic of Korea (ROK) are mostly infected with C. parvum, almost exclusively with family IIa, especially IIaA18G3R1 (31). C. parvum oocysts in water flowing out of stables contaminate the surrounding agricultural water or infect humans or wildlife. Hence, infected cattle pose a potential threat to human health owing to environmental contamination by excreted oocysts (37).
Currently, most studies have focused on C. parvum, and reports on the association between other Cryptosporidium spp. and calf diarrhea are limited. Therefore, the objective of this study was to investigate the positivity of Cryptosporidium spp. in pre-weaned calves with diarrhea in the Republic of Korea (ROK), to identify Cryptosporidium spp. that cause diarrhea according to age group, to assess the association between age or season and Cryptosporidium spp., and to determine C. parvum subtypes circulating in calves, which act as major risk factors for zoonotic infection.
All animal procedures were conducted according to ethical guidelines for the use of animal samples, and were approved by the Jeonbuk National University (Institutional Animal Care and Use Committee Decision No. JBNU 2020-052).
Between January 2021 and May 2022, a diarrhea outbreak was noticed on 181 farms in four different provinces (Gyeonggi, Gyeongbuk, Gyeongnam, and Jeonbuk) in the ROK. A total of 510 fresh fecal samples were collected directly from the rectum of individual diarrheic pre-weaned calves by a veterinarian using sterile plastic gloves. The majority of calves (n = 497) were of the Korean native breed (Hanwoo), while 13 were of the Holstein breed. All these calves were maintained indoors. The collected samples were placed in sterile plastic tubes and immediately transported to the laboratory for subsequent DNA extraction. During sampling, age, sex, sampling date, fecal consistency, and farm location were recorded. All samples were stored at 4°C until DNA extraction. Fecal samples were divided according to calf age as follows: 1–10 days (n = 251), 11–30 days (n = 205), and 31–70 days (n = 54). To investigate the association between the occurrence of Cryptosporidium spp. and season, stool samples were classified into four groups: spring (March–May; n = 206), summer (June–August; n = 80), fall (September–November; n = 158), and winter (December–February; n = 66). The average temperature and rainfall for each season were as follows: spring (13.0 ± 4.3°C, 72.4 ± 39.6 mm), summer (24.6 ± 2.0°C, 247.7 ± 117.8 mm), fall (15.6 ± 6.6°C, 97.2 ± 58.5 mm), and winter (0.7 ± 1.8°C, 14.0 ± 9.7 mm).
Genomic DNA was extracted from 200 mg of each fecal sample using an AccuPrep Stool DNA extraction kit (Bioneer, Daejeon, ROK) according to the manufacturer's instructions. Extracted DNA was stored at −20°C until use. Cryptosporidium spp. was first identified using the small subunit (SSU) rRNA gene by nested polymerase chain reaction (PCR) (38). Next, C. parvum was screened by targeting the 60-kDa glycoprotein (gp60) gene using nested PCR to determine its subtype (39, 40). Primers used in this study are listed in Table 1. The PCR conditions were as follows: 94°C for 3 min, followed by 35 cycles at 94°C for 45 s, annealing at 54°C for 45 s, 72°C for 60 s, and a final extension at 72°C for 7 min. Negative controls were included in all runs. Amplified PCR products were sepa-rated by electrophoresis on a 1.5% agarose gel and visual-ized after staining with ethidium bromide. All secondary PCR products were purified using an AccuPower PCR Purification Kit (Bioneer, Daejeon, ROK) and directly sequenced (Macrogen, Daejeon, ROK). PCR amplicons were sequenced using a BigDye Terminator 3.1 Cycle Sequencing Kit on a 3500 xL Genetic Analyzer (Applied Biosystems, Foster City, CA, USA), according to the manufacturer's instructions, using the same primer set as in the conventional PCR reaction. Among the PCR-positive samples, only samples with good sequencing results were considered positive for Cryptosporidium spp. The nucleotide sequences obtained in this study were analyzed using BioEdit (version 7.2.5) and compared with reference sequences using the Basic Local Alignment Search Tool available at the National Center for Biotechnology Information database (http://www.ncbi.nlm.nih.gov). The sequences of C. bovis, C. ryanae, and C. andersoni were compared using Geneious Prime software (version 2021.1.1; https://www.geneious.com) and analyzed via direct comparison with reference sequences from GenBank. The subtypes of gp60 were named based on the repeated numbers of TCA-(A), TCG- (G), and ACATCA-(R), as described previously (41). All nucleotide sequences generated in this study were deposited in the GenBank database under the following accession numbers; OQ001482–OQ001483 for C. andersoni, OQ001456–OQ001477 for C. bovis, OQ001429–OQ001454 for C. ryanae, and OQ025024–OQ025044 for C. parvum.
The statistical analysis was performed using the SPSS Statistics 26 software package for Windows (SPSS Inc, Chicago, IL, USA). A chi-square test was determined the association between the occurrence of each Cryptosporidium spp. and age or season. We constructed a generalized linear mixed model (GLMM) for a binomial family, with a logit link function for each Cryptosporidium species to test whether age and season affected the infection. In the model, the origin of farm and sampling year were considered as random effects. Odd ratios (OR) with 95% confidence intervals (CIs) were calculated to assess the association between age or season and occurrence of Cryptosporidium spp. The correlation between precipitation and occurrence was confirmed through correlation analysis. A P-value < 0.05 was considered statistically significant.
Of the 510 fecal samples, 71 (13.9%) were positive for Cryptosporidium spp. as determined by PCR analysis using the SSU rRNA gene. Four species were identified in the pre-weaned Korean native calves with diarrhea (Table 2; Supplementary Figure 1), of which C. ryanae was the most predominant species (36.6%, 26/71), followed by C. bovis (30.9%, 22/71), C. parvum (29.6%, 21/71), and C. andersoni (2.8%, 2/71). Co-infections with two or three species were not observed.
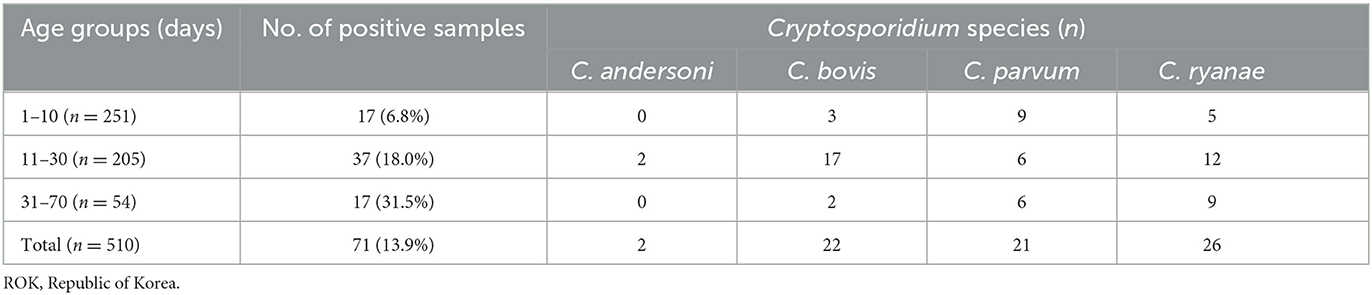
Table 2. Positivity of Cryptosporidium spp. according to age group in pre-weaned calves with diarrhea in the ROK.
The occurrence of Cryptosporidium spp. in calves showed distinct characteristics according to age (Table 2). C. bovis, C. parvum, and C. ryanae were detected in all the age groups. Positivity of C. bovis was the highest in calves aged 11–30 days, whereas positivity of C. ryanae gradually increased with age and peaked in calves aged 31–70 days. Positivity of C. parvum were the highest in calves aged 31–70 days. C. andersoni was found in only two calves aged 11–30 days. Except for C. andersoni, C. bovis (χ2 = 13.825, P = 0.001), C. parvum (χ2 = 7.570, P = 0.023), and C. ryanae (χ2 = 20.184, P = 0.000) was significantly associated with the age of the calves. Thus, the risk of contracting C. bovis and C. parvum was 7.8-fold higher in calves aged 11–30 days (95% confidence interval [CI]: 2.25–27.04; P = 0.001) and 4.2-fold higher in calves aged 31–70 days (95% CI: 1.42–12.54; P = 0.010), respectively, than in calves aged 1–10 days (Table 3). Moreover, the risk of contracting C. ryanae was 3.30-fold higher in calves aged 11–30 days and 11.38-fold higher in calves aged 31–70 days (95% CI: 3.62–35.84; P = 0.000) than in calves aged 1–10 days (Table 3).
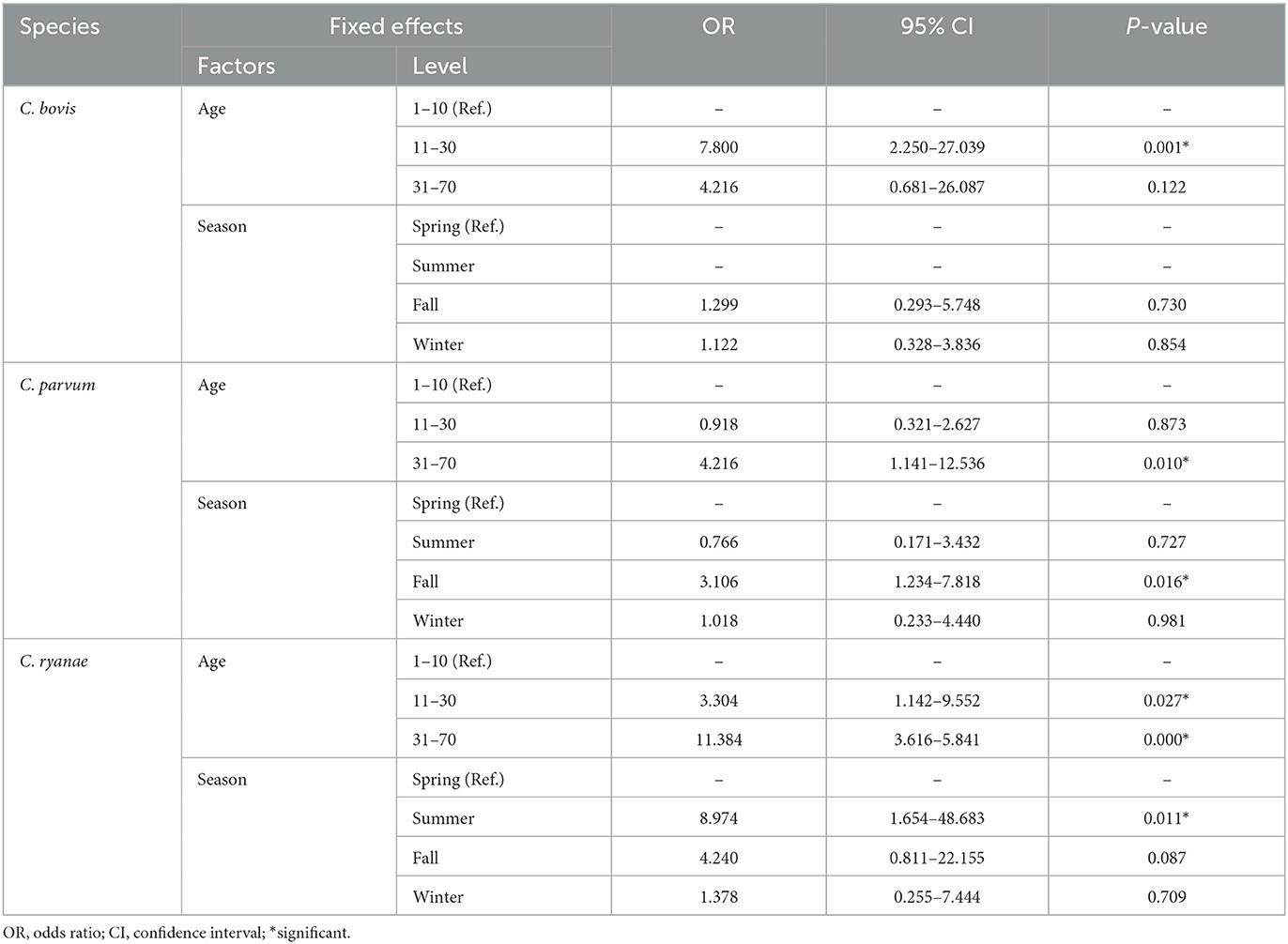
Table 3. Generalized linear mixed model results for Cryptosporidium spp. occurrence in pre-weaned calves with diarrhea in the ROK.
All four Cryptosporidium spp. were detected in spring and fall. Positivity of Cryptosporidium spp. was the highest in fall (22.2%), and there were no differences in the occurrence in any other season (Figure 1A). Of the four species, C. ryanae was found in all seasons and, unlike the other species, was exclusively detected in summer (Figure 1B). The GLMM approach was used to evaluate the association between the occurrence of Cryptosporidium spp. and season. The results showed that the occurrence of C. parvum and C. ryanae was significantly associated with season. C. parvum was detected 3.1-fold more frequently in pre-weaned calves with diarrhea in fall (95% CI: 1.23–7.81; P = 0.016) than in spring. C. ryanae was 8.9-fold more frequently detected in summer (95% CI: 1.65–48.68; P = 0.011) than in spring (Table 3). Although positivity of C. bovis was relatively high in calves in spring and winter, there was no statistically significant difference between seasons (P = 0.149).
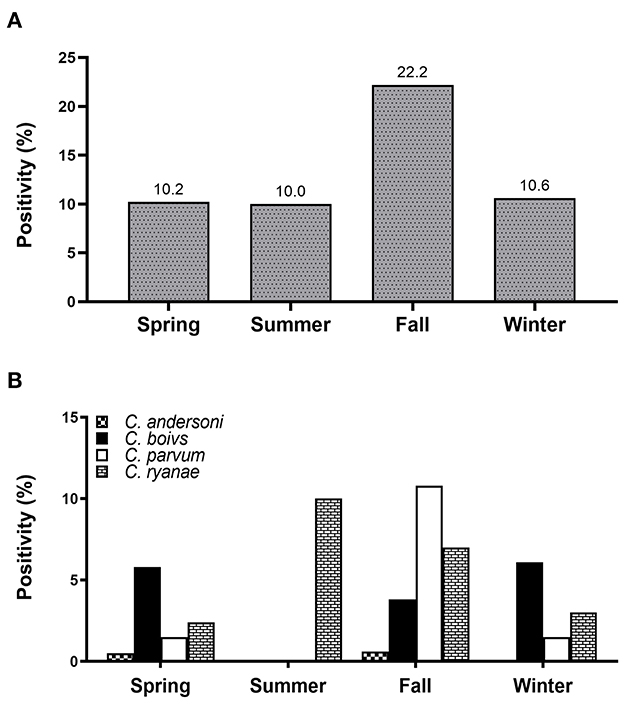
Figure 1. Seasonal positivity of Cryptosporidium spp. in pre-weaned native Korean calves with diarrhea (A). Distribution of four Cryptosporidium spp. in calves according to the season (B).
Among 21 C. parvum-positive samples, three subtypes were identified using the gp60 gene (Supplementary Table 1). IIaA17G4R1 (66.7%, 14/21) was the predominant subtype, followed by IIaA18G3R1 (19.0%, 4/21), and IIaA20G3R1 (14.3%, 3/21). IIaA17G4R1 and IIaA18G3R1 were detected in pre-weaned calves of all age groups, whereas the IIaA20G3R1 subtype was identified only in calves younger than 30 days.
In the present study, we identified different Cryptosporidium spp. in pre-weaned calves with diarrhea, with a 13.9% positivity, which is higher than that reported previously in the ROK (42–44). The main reason for the difference is that past research has focused mainly on C. parvum, and the occurrence of other Cryptosporidium spp. has recently increased (29–31, 45, 46). Compared with a previous study (31), the current results showed that the occurrence of C. bovis and C. ryanae have increased noticeably in calves. C. bovis, C. parvum, and C. ryanae, but not C. andersoni, were detected at similar rates in pre-weaned calves. Although C. bovis and C. ryanae were detected in calves with diarrhea in this study, the role and clinical significance of these species in calf diarrhea remain unclear. Thus, these findings suggest that in addition to C. parvum, C. bovis and C. ryanae might be representative species that infect young calves. Notably, this is the first report of C. andersoni infection and the first time that the IIaA20G3R1 subtype of C. parvum has been described in pre-weaned calves with diarrhea in the ROK.
Four Cryptosporidium species were identified in pre-weaned calves with diarrhea: C. andersoni, C. bovis, C. parvum, and C. ryanae. In the ROK, C. andersoni infection was detected in calves aged 11–30 days for the first time. C. andersoni is more frequently found in post-weaned or adult cattle (17, 47), and its infection causes decreased milk production and poor weight gain (48–50). In particular, C. andersoni has been observed in cattle of all age groups and was reported to be the second most prevalent species in pre-weaned calves in China (51). However, to date, this has not been reported in cattle in the ROK. Although in this study, C. andersoni infection was found in only two calves, our results demonstrate that young calves can acquire C. andersoni infection at an early stage of life (20, 52). C. andersoni causes diarrhea in humans and is one of the main Cryptosporidium species found in contaminated water (53, 54). Moreover, no information is available on the occurrence of C. andersoni and its association with diarrhea in humans in the ROK. Thus, we cannot draw a conclusion regarding the infection route of C. andersoni in these calves. However, the possibility that the calves may have been infected through the ingestion of water contaminated with oocysts shed by humans infected with C. andersoni cannot be completely ruled out. These results suggest that young calves are more susceptible to Cryptosporidium spp. and may act as an infection source for other animals. Further studies are thus required to investigate the occurrence and symptoms of C. andersoni in pre-weaned calves in the ROK.
According to our results, C. ryanae was mostly detected in pre-weaned native Korean calves. These results differ from those of a previous study (31). The reason for the difference may be related to the calf management system and method used for diagnosis. Our findings revealed that positivity of C. ryanae increased with age and was highest in calves older than 31 days. Our results indicated much higher occurrence with C. ryanae (36.6%) than reported previously (8.6%) for calves of this age in the ROK (31). C. ryanae infection in the ROK showed a continuous increase in pre-weaned calves. Compared with early neonatal calves, the risk of contracting C. ryanae in pre-weaned calves increased significantly with age. The reason for the high occurrence of C. ryanae in this study is not clear. It is speculated that C. parvum may affect the occurrence of C. ryanae and C. bovis in calves, and these two species may occur early on farms with little or no C. parvum infection, resulting in increased occurrence. Another possibility is that the oocysts of C. ryanae may be more resistant to adverse environmental conditions than those of other species and survive well in high temperatures and moist conditions (55). Therefore, the infection may last for weeks or months. In addition, C. ryanae is associated with the occurrence of severe diarrhea in calves (28, 56). However, several studies have shown that C. ryanae is less frequently associated with diarrhea (24, 30, 45). However, the findings of this study indicated that C. ryanae was detected in pre-weaned calves with diarrhea, and calves older than 31 days were at higher risk.
C. bovis was the second most dominant species in pre-weaned calves with diarrhea in the ROK. This is in contrast to other studies in which C. bovis was the most common species (20, 30, 51, 57, 58) which demonstrates the differences in the occurrence of Cryptosporidium species between countries. In relation to age, positivity of C. bovis was highest in calves aged 11–30 day. This result is consistent with those of previous studies (59–61). C. bovis has also been reported in pre-weaned calves and post-weaned calves (18, 30, 45, 62) and is associated with the occurrence of severe diarrhea in calves (63, 64). However, several studies have reported that C. bovis does not cause diarrhea, similar to studies of C. ryanae (18, 29, 30, 45, 65). To date, studies on the occurrence and clinical symptoms of C. bovis in calves in the ROK are considerably scarce (31, 43) owing to the rarity of this infection in calves. Although C. bovis was detected in the diarrheic calves, currently, it cannot be asserted whether the observed diarrhea was caused by C. bovis. Accordingly, the results suggest that calves aged 11–30 days might be at high risk for C. bovis infection. Therefore, further research should be conducted to elucidate the pathogenicity of C. bovis and C. ryanae in calf diarrhea.
Interestingly, positivity of C. parvum in this study was the third lowest in pre-weaned calves in the ROK. Contrary to previous studies which showed that C. parvum mainly infects young calves (15, 59, 60, 66), the present results showed that the positivity of C. parvum were highest in calves aged 31–70 days, whereas those in calves under 30 days of age were very low. In general, C. parvum infection starts soon after birth, peaks at 2–3 weeks of age, and recovers mostly by 4 weeks (45, 65–68). The results of the present study are inconsistent with those of our previous studies, which showed no occurrence in calves aged 31–70 days (31, 42). The reason for this discrepancy cannot be explained at this time, but might be due to several factors, such as crowding, inadequate disinfection of the barn floor, environmental contamination, or the presence of other hosts or wildlife. Unfortunately, we did not consider any of these factors. Several studies have shown that C. parvum infection occurs mostly in calves aged between 31 days and 6 months (15, 17, 28). These results are somewhat consistent with ours. Wells et al. (69) reported that 80% of adult cattle were shedding C. parvum (69). This implies that adult cattle are likely a source of C. parvum infection in calves. Although C. parvum has been regarded as a major pathogen in young calves, it has not been extensively examined in post-weaning calves and adult cattle in the ROK. Considering the high positivity of C. parvum in calves aged 31–70 days, the possibility of re-infection from their dams cannot be excluded. Thus, C. parvum infection in calves appears to be more affected by the environment in which the calves are raised than by their age.
In the present study, we found a seasonal association in the occurrence of Cryptosporidium spp. Our results revealed that season had a significant effect on the occurrence of C. parvum and C. ryanae: C. parvum dominance in fall, and C. ryanae dominance in summer. However, this finding differs from previous results, which reported that the occurrence of C. bovis was dominant in summer, whereas that of C. parvum was dominant in spring and winter (58, 70). According to another study performed in the ROK, season and the occurrence of C. parvum were not associated (43). The relationship between season and the occurrence of Cryptosporidium spp. remains unclear, but it seems that their occurrence may vary depending on the farm situation in each country. Notably, levels of sunlight, temperature, humidity, and precipitation, as well as breeding density, may play a role in this variation (71, 72). One possibility is that calves are mostly born from spring to summer in the ROK, which increases the density of calves within the herd and makes calves more susceptible to Cryptosporidium spp. oocysts shed by older calves that were born earlier in the season. Especially, in the ROK, it rains heavily in summer. Heavy rain results in the overcrowding of animals and hampers the drying of the floor, thereby increasing the survival of Cryptosporidium oocysts (71). Our results showed that precipitation in the preceding month was significantly associated with the occurrence of Cryptosporidium spp. (P = 0.015) (data not shown). As previously mentioned, precipitation is highest in summer, followed by that in fall, spring, and winter. Cryptosporidium spp. oocysts are likely to easily spread through rivers or groundwater to surrounding farms due to heavy rain, which increases the opportunity for calves to be exposed. Owing to the long survival of Cryptosporidium spp. oocysts, susceptible calves with lower immunity are more frequently present from summer to fall. These results suggest that precipitation may act as an important factor in the occurrence of cryptosporidiosis. Our findings provide new insights on the seasonal occurrence of Cryptosporidium spp., and hence, their control.
Subtype analysis of C. parvum-positive samples revealed the presence of three subtypes: IIaA17G4R1, IIaA18G3R1, and IIaA20G3R1. Of these, IIaA17G4R1 was the most dominant subtype in pre-weaned calves with diarrhea in the ROK. Our results differed from those of a previous study in which IIaA18G3R1 was the predominant subtype in pre-weaned calves in the ROK (31). Contrary to a previous report (31) that found 11 subtypes, only three subtypes were identified in this study. This was mainly because of the difference in the number of positive samples. However, this could be explained by the decreased movement of humans or animals due to the COVID-19 pandemic. The IIaA17G4R1 subtype has been identified in calves and goats in several countries (31, 73–76); however, its age-related distribution in calves remains unclear. Nevertheless, IIaA17G4R1 was detected in all age groups in the present study. As mentioned earlier, the IIaA17G4R1 subtype may be shed from adult cattle and re-infected calves. Moreover, the IIaA20G3R1 subtype has been detected in humans and cattle (3, 77), and was identified for the first time in calves younger than 1 month in the ROK. This subtype is considered to be more zoonotic than IIaA17G4R1. Thus, calves may be major contributors to human cryptosporidiosis.
The study confirmed the presence of four Cryptosporidium spp., C. andersoni, C. bovis, C. parvum, and C. ryanae in pre-weaned calves with diarrhea, with C. ryanae being the most predominant species and IIaA17G4R1 being the most predominant subtype of C. parvum in the ROK. To the best of our knowledge, this is the first study to report a C. andersoni infection and a new subtype, IIaA20G3R1 of C. parvum, in calves. The occurrence of Cryptosporidium spp. was significantly associated with the season, C. parvum being dominant in fall and C. ryanae dominant in summer. Although the current study did not elucidate the association between C. bovis and C. ryanae and diarrhea, these findings improved our understanding of the epidemiology and control of bovine cryptosporidiosis in the ROK. Further research is required to investigate the association between C. bovis and C. ryanae and diarrhea according to fecal consistency and identify the risk factors affecting Cryptosporidium infection in calves.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
The animal study was reviewed and approved by the Jeonbuk National University (Institutional Animal Care and Use Committee Decision No. JBNU 2020-052). Written informed consent was obtained from the owners for the participation of their animals in this study.
KSC and JP conceived the study. DHJ, HCC, YJP, and JP performed the experiments. DHJ, HCC, and KSC analyzed the data and wrote the manuscript. All authors read and approved the final manuscript.
This research was supported by the National Research Foundation of Korea (NRF), which was funded by the Mid-Career Research Program (Grant no. NRF-2021R1A2C1011579) and this research was partly funded by the Korea Institute of Planning and Evaluation for Technology in Food, Agriculture, and Forestry (IPET) (Grant no. 122017-02-1-HD020).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2023.1145096/full#supplementary-material
Supplementary Figure 1. Nested PCR targeting SSU rRNA and gp60 genes. Amplified PCR products were sepa-rated by electrophoresis on a 1.5% agarose gel and visual-ized after staining with ethidium bromide. Lane 1: DNA ladder (1 kb); Lanes 2–5: PCR amplicons for SSU rRNA (830 bp); Lanes 6 and 7: PCR amplicons for gp60 (467 bp); and Lane 8: negative control.
Supplementary Table 1. Distribution of C. parvum subtypes in pre-weaned calves with diarrhea in the ROK.
1. Efstratiou A, Ongerth JE, Karanis P. Waterborne transmission of protozoan parasites: review of worldwide outbreaks - an update 2011-2016. Water Res. (2017) 114:14–22. doi: 10.1016/j.watres.2017.01.036
2. Huang DB, White AC. An updated review on Cryptosporidium and Giardia. Gastroenterol Clin North Am. (2006) 35:291–314. doi: 10.1016/j.gtc.2006.03.006
3. Nolan MJ, Jex AR, Koehler AV, Haydon SR, Stevens MA, Gasser RB. Molecular-based investigation of Cryptosporidium and Giardia from animals in water catchments in southeastern Australia. Water Res. (2013) 47:1726–40. doi: 10.1016/j.watres.2012.12.027
4. Xiao L. Molecular epidemiology of cryptosporidiosis: an update. Exp Parasitol. (2010) 124:80–9. doi: 10.1016/j.exppara.2009.03.018
5. de Graaf DC, Vanopdenbosch E, Ortega-Mora LM, Abbassi H, Peeters JE, A. review of the importance of cryptosporidiosis in farm animals. Int J Parasitol. (1999) 29:1269–87. doi: 10.1016/S0020-7519(99)00076-4
6. Niine T, Dorbek-Kolin E, Lassen B, Orro T. Cryptosporidium outbreak in calves on a large dairy farm: effect of treatment and the association with the inflammatory response and short-term weight gain. Res Vet Sci. (2018) 117:200–8. doi: 10.1016/j.rvsc.2017.12.015
7. Brar APS, Sood NK, Kaur P, Singla LD, Sandhu BS, Gupta K, et al. Periurban outbreaks of bovine calf scours in Northern India caused by Cryptosporidium in association with other enteropathogens. Epidemiol Infect. (2017) 145:2717–26. doi: 10.1017/S0950268817001224
8. Brook E, Hart CA, French N, Christley R. Prevalence and risk factors for Cryptosporidium spp. infection in young calves. Vet Parasitol. (2008) 152:46–52. doi: 10.1016/j.vetpar.2007.12.003
9. Cui Z, Wang R, Huang J, Wang H, Zhao J, Luo N, et al. Cryptosporidiosis caused by Cryptosporidium parvum subtype IIdA15G1 at a dairy farm in Northwestern China. Parasit Vectors. (2014) 7:529. doi: 10.1186/s13071-014-0529-z
10. Klein P, Kleinova T, Volek Z, Simunek J. Effect of Cryptosporidium parvum infection on the absorptive capacity and paracellular permeability of the small intestine in neonatal calves. Vet Parasitol. (2008) 152:53–9. doi: 10.1016/j.vetpar.2007.11.020
11. Tzipori S, Ward H. Cryptosporidiosis: biology, pathogenesis and disease. Microbes Infect. (2002) 4:1047–58. doi: 10.1016/S1286-4579(02)01629-5
12. Ryan U, Feng Y, Fayer R, Xiao L. Taxonomy and molecular epidemiology of Cryptosporidium and Giardia- a 50 year perspective (1971-2021). Int J Parasitol. (2021) 51:1099–119. doi: 10.1016/j.ijpara.2021.08.007
13. Feng Y, Ortega Y, He G, Das P, Xu M, Zhang X, et al. Wide geographic distribution of Cryptosporidium bovis and the deer-like genotype in bovines. Vet Parasitol. (2007) 144:1–9. doi: 10.1016/j.vetpar.2006.10.001
14. Fayer R, Santin M, Trout JM, Greiner E. Prevalence of species and genotypes of Cryptosporidium found in 1-2-year-old dairy cattle in the Eastern United States. Vet Parasitol. (2006) 135:105–12. doi: 10.1016/j.vetpar.2005.08.003
15. Santin M, Trout JM, Xiao L, Zhou L, Greiner E, Fayer R. Prevalence and age-related variation of Cryptosporidium species and genotypes in dairy calves. Vet Parasitol. (2004) 122:103–17. doi: 10.1016/j.vetpar.2004.03.020
16. Wang R, Ma G, Zhao J, Lu Q, Wang H, Zhang L, et al. Cryptosporidium andersoni is the predominant species in post-weaned and adult dairy cattle in China. Parasitol Int. (2011) 60:1–4. doi: 10.1016/j.parint.2010.09.002
17. Huetink RE, van der Giessen JW, Noordhuizen JP, Ploeger HW. Epidemiology of Cryptosporidium spp. and Giardia duodenalis on a dairy farm. Vet Parasitol. (2001) 102:53–67. doi: 10.1016/S0304-4017(01)00514-3
18. Fan Y, Wang T, Koehler AV, Hu M, Gasser RB. Molecular investigation of Cryptosporidium and Giardia in pre- and post-weaned calves in Hubei province, China. Parasit Vectors. (2017) 10:519. doi: 10.1186/s13071-017-2463-3
19. Budu-Amoako E, Greenwood SJ, Dixon BR, Barkema HW, McClure JT. Occurrence of Cryptosporidium and Giardia on beef farms and water sources within the vicinity of the farms on Prince Edward Island, Canada. Vet Parasitol. (2012) 184:1–9. doi: 10.1016/j.vetpar.2011.10.027
20. Silverlas C, Naslund K, Bjorkman C, Mattsson JG. Molecular characterisation of Cryptosporidium isolates from Swedish dairy cattle in relation to age, diarrhoea and region. Vet Parasitol. (2010) 169:289–95. doi: 10.1016/j.vetpar.2010.01.003
21. Mahmoudi M, Hazrati Tapeh K, Abasi E, Sayyadi H, Aminpour A. Prevalence and genetic characterization of Cryptosporidium in pre-weaned cattle in Urmia (Northwestern Iran). J Infect Dev Ctries. (2021) 15:422–7. doi: 10.3855/jidc.12122
22. Fayer R, Santin M, Trout JM. Prevalence of Cryptosporidium species and genotypes in mature dairy cattle on farms in Eastern United States compared with younger cattle from the same locations. Vet Parasitol. (2007) 145:260–6. doi: 10.1016/j.vetpar.2006.12.009
23. Ryan U, Hijjawi N. New Developments in Cryptosporidium research. Int J Parasitol. (2015) 45:367–73. doi: 10.1016/j.ijpara.2015.01.009
24. Fayer R, Santin M, Trout JM. Cryptosporidium ryanae n. sp (Apicomplexa: Cryptosporidiidae) in cattle (Bos Taurus). Vet Parasitol. (2008) 156:191–8. doi: 10.1016/j.vetpar.2008.05.024
25. Fayer R, Santin M, Xiao L. Cryptosporidium bovis n. sp (Apicomplexa: Cryptosporidiidae) in cattle (Bos Taurus). J Parasitol. (2005) 91:624–9. doi: 10.1645/GE-3435
26. Thomson S, Hamilton CA, Hope JC, Katzer F, Mabbott NA, Morrison LJ, et al. Bovine cryptosporidiosis: impact, host-parasite interaction and control strategies. Vet Res. (2017) 48:42. doi: 10.1186/s13567-017-0447-0
27. Ryan U, Fayer R, Xiao L. Cryptosporidium species in humans and animals: current understanding and research needs. Parasitology. (2014) 141:1667–85. doi: 10.1017/S0031182014001085
28. Follet J, Guyot K, Leruste H, Follet-Dumoulin A, Hammouma-Ghelboun O, Certad G, et al. Cryptosporidium infection in a veal calf cohort in France: molecular characterization of species in a longitudinal study. Vet Res. (2011) 42:116. doi: 10.1186/1297-9716-42-116
29. Aberg M, Emanuelson U, Troell K, Bjorkman C. Infection dynamics of Cryptosporidium bovis and Cryptosporidium ryanae in a Swedish dairy herd. Vet Parasitol. (2019) 276S:100010. doi: 10.1016/j.vpoa.2019.100010
30. Aberg M, Emanuelson U, Troell K, Bjorkman C. A single-cohort study of Cryptosporidium bovis and Cryptosporidium ryanae in dairy cattle from birth to calving. Vet Parasitol Reg Stud Reports. (2020) 20:100400. doi: 10.1016/j.vprsr.2020.100400
31. Jang DH, Cho HC, Shin SU, Kim EM, Park YJ, Hwang S, et al. Prevalence and distribution pattern of Cryptosporidium spp. among pre-weaned diarrheic calves in the Republic of Korea. PLoS ONE. (2021) 16:e0259824. doi: 10.1371/journal.pone.0259824
32. Muhid A, Robertson I, Ng J, Ryan U. Prevalence of and management factors contributing to Cryptosporidium sp. infection in pre-weaned and post-weaned calves in Johor, Malaysia. Exp Parasitol. (2011) 127:534–8. doi: 10.1016/j.exppara.2010.10.015
33. Vieira PM, Mederle N, Lobo ML, Imre K, Mederle O, Xiao L, et al. Molecular characterisation of Cryptosporidium (Apicomplexa) in children and cattle in Romania. Folia Parasitol. (2015) 62:1–4. doi: 10.14411/fp.2015.002
34. Ibrahim MA, Abdel-Ghany AE, Abdel-Latef GK, Abdel-Aziz SA, Aboelhadid SM. Epidemiology and public health significance of Cryptosporidium isolated from cattle, buffaloes, and humans in Egypt. Parasitol Res. (2016) 115:2439–48. doi: 10.1007/s00436-016-4996-3
35. Bjorkman C, Lindstrom L, Oweson C, Ahola H, Troell K, Axen C. Cryptosporidium infections in suckler herd beef calves. Parasitology. (2015) 142:1108–14. doi: 10.1017/S0031182015000426
36. Amer S, Zidan S, Adamu H, Ye J, Roellig D, Xiao L, et al. Prevalence and characterization of Cryptosporidium spp. in dairy cattle in Nile River delta provinces, Egypt. Exp Parasitol. (2013) 135:518–23. doi: 10.1016/j.exppara.2013.09.002
37. Imre K, Morar A, Ilie MS, Plutzer J, Imre M, Emil T, et al. Survey of the occurrence and human infective potential of Giardia duodenalis and Cryptosporidium spp. in wastewater and different surface water sources of Western Romania. Vector Borne Zoonotic Dis. (2017) 17:685–91. doi: 10.1089/vbz.2017.2155
38. Xiao L, Escalante L, Yang C, Sulaiman I, Escalante AA, Montali RJ, et al. Phylogenetic analysis of Cryptosporidium parasites based on the small-subunit rRNA gene locus. Appl Environ Microbiol. (1999) 65:1578–83. doi: 10.1128/AEM.65.4.1578-1583.1999
39. Alves M, Xiao L, Sulaiman I, Lal AA, Matos O, Antunes F. Subgenotype analysis of Cryptosporidium isolates from humans, cattle, and zoo ruminants in Portugal. J Clin Microbiol. (2003) 41:2744–7. doi: 10.1128/JCM.41.6.2744-2747.2003
40. Peng MM, Matos O, Gatei W, Das P, Stantic-Pavlinic M, Bern C, et al. A comparison of Cryptosporidium subgenotypes from several geographic regions. J Eukaryot Microbiol. (2001) 48:28S−31S. doi: 10.1111/j.1550-7408.2001.tb00442.x
41. Sulaiman IM, Hira PR, Zhou L, Al-Ali FM, Al-Shelahi FA, Shweiki HM, et al. Unique endemicity of cryptosporidiosis in children in Kuwait. J Clin Microbiol. (2005) 43:2805–9. doi: 10.1128/JCM.43.6.2805-2809.2005
42. Lee YJ, Ryu JH, Shin SU, Choi KS. Prevalence and molecular characterization of Cryptosporidium and Giardia in pre-weaned native calves in the Republic of Korea. Parasitol Res. (2019) 118:3509–17. doi: 10.1007/s00436-019-06482-9
43. Lee SH, VanBik D, Kim HY, Lee YR, Kim JW, Chae M, et al. Multilocus typing of Cryptosporidium spp. in young calves with diarrhea in Korea. Vet Parasitol. (2016) 229:81–9. doi: 10.1016/j.vetpar.2016.09.019
44. Lee SH, Kim HY, Choi EW, Kim D. Causative agents and epidemiology of diarrhea in Korean native calves. J Vet Sci. (2019) 20:e64. doi: 10.4142/jvs.2019.20.e64
45. Cai M, Guo Y, Pan B, Li N, Wang X, Tang C, et al. Longitudinal monitoring of Cryptosporidium species in pre-weaned dairy calves on five farms in Shanghai, China. Vet Parasitol. (2017) 241:14–9. doi: 10.1016/j.vetpar.2017.05.005
46. Qi MZ, Fang YQ, Wang XT, Zhang LX, Wang RJ, Du SZ, et al. Molecular characterization of Cryptosporidium spp. in pre-weaned calves in Shaanxi province, North-Western China. J Med Microbiol. (2015) 64:111–6. doi: 10.1099/jmm.0.079327-0
47. Enemark HL, Ahrens P, Lowery CJ, Thamsborg SM, Enemark JM, Bille-Hansen V, et al. Cryptosporidium andersoni from a Danish cattle herd: identification and preliminary characterisation. Vet Parasitol. (2002) 107:37–49. doi: 10.1016/S0304-4017(02)00083-3
48. Liang N, Wu Y, Sun M, Chang Y, Lin X, Yu L, et al. Molecular epidemiology of Cryptosporidium spp. in dairy cattle in Guangdong province, South China. Parasitology. (2019) 146:28–32. doi: 10.1017/S0031182018001129
49. Ralston B, Thompson RC, Pethick D, McAllister TA, Olson ME. Cryptosporidium andersoni in Western Australian feedlot cattle. Aust Vet J. (2010) 88:458–60. doi: 10.1111/j.1751-0813.2010.00631.x
50. Esteban E, Anderson BC. Cryptosporidium muris: prevalence, persistency, and detrimental effect on milk production in a drylot dairy. J Dairy Sci. (1995) 78:1068–72. doi: 10.3168/jds.S0022-0302(95)76723-6
51. Zhang W, Wang R, Yang F, Zhang L, Cao J, Zhang X, et al. Distribution and genetic characterizations of Cryptosporidium spp. in pre-weaned dairy calves in Northeastern China's Heilongjiang province. PLoS ONE. (2013) 8:e54857. doi: 10.1371/journal.pone.0054857
52. Kvac M, Kouba M, Vitovec J. Age-related and housing-dependence of Cryptosporidium infection of calves from dairy and beef herds in South Bohemia, Czech Republic. Vet Parasitol. (2006) 137:202–9. doi: 10.1016/j.vetpar.2006.01.027
53. Jiang Y, Ren J, Yuan Z, Liu A, Zhao H, Liu H, et al. Cryptosporidium andersoni as a novel predominant Cryptosporidium species in outpatients with diarrhea in Jiangsu province, China. BMC Infect Dis. (2014) 14:555. doi: 10.1186/s12879-014-0555-7
54. Leoni F, Amar C, Nichols G, Pedraza-Diaz S, McLauchlin J. Genetic analysis of Cryptosporidium from 2414 humans with diarrhoea in England between 1985 and 2000. J Med Microbiol. (2006) 55:703–7. doi: 10.1099/jmm.0.46251-0
55. King BJ, Monis PT. Critical processes affecting Cryptosporidium oocyst survival in the environment. Parasitology. (2007) 134:309–23. doi: 10.1017/S0031182006001491
56. Li N, Wang R, Cai M, Jiang W, Feng Y, Xiao L. Outbreak of cryptosporidiosis due to Cryptosporidium parvum subtype IIdA19G1 in neonatal calves on a dairy farm in China. Int J Parasitol. (2019) 49:569–77. doi: 10.1016/j.ijpara.2019.02.006
57. Wang R, Wang H, Sun Y, Zhang L, Jian F, Qi M, et al. Characteristics of Cryptosporidium transmission in preweaned dairy cattle in Henan, China. J Clin Microbiol. (2011) 49:1077–82. doi: 10.1128/JCM.02194-10
58. Feng Y, Gong X, Zhu K, Li N, Yu Z, Guo Y, et al. Prevalence and genotypic identification of Cryptosporidium spp, Giardia duodenalis and Enterocytozoon bieneusi in pre-weaned dairy calves in Guangdong, China. Parasit Vectors. (2019) 12:41. doi: 10.1186/s13071-019-3310-5
59. Feng Y, Xiao L. Molecular epidemiology of cryptosporidiosis in China. Front Microbiol. (2017) 8:1701. doi: 10.3389/fmicb.2017.01701
60. Rieux A, Paraud C, Pors I, Chartier C. Molecular characterization of Cryptosporidium isolates from pre-weaned calves in Western France in relation to age. Vet Parasitol. (2013) 197:7–12. doi: 10.1016/j.vetpar.2013.05.001
61. Wang R, Zhao G, Gong Y, Zhang L. Advances and perspectives on the epidemiology of bovine Cryptosporidium in China in the past 30 years. Front Microbiol. (2017) 8:1823. doi: 10.3389/fmicb.2017.01823
62. Deksne G, Mateusa M, Cvetkova S, Derbakova A, Keidane D, Troell K, et al. Prevalence, risk factor and diversity of Cryptosporidium in cattle in Latvia. Vet Parasitol Reg Stud Reports. (2022) 28:100677. doi: 10.1016/j.vprsr.2021.100677
63. Ayinmode AB, Olakunle FB, Xiao L. Molecular characterization of Cryptosporidium spp. in native calves in Nigeria. Parasitol Res. (2010) 107:1019–21. doi: 10.1007/s00436-010-1972-1
64. Lichtmannsperger K, Harl J, Freudenthaler K, Hinney B, Wittek T, Joachim A. Cryptosporidium parvum, Cryptosporidium ryanae, and Cryptosporidium bovis in samples from calves in Austria. Parasitol Res. (2020) 119:4291–5. doi: 10.1007/s00436-020-06928-5
65. Qi M, Zhang K, Huang M, Wang S, Xu C, Wang T, et al. Longitudinal detection of Cryptosporidium spp. in 1-10-week-old dairy calves on a farm in Xinjiang, China. Parasitol Res. (2020) 119:3839–44. doi: 10.1007/s00436-020-06904-z
66. Santin M, Trout JM, Fayer R. A longitudinal study of cryptosporidiosis in dairy cattle from birth to 2 years of age. Vet Parasitol. (2008) 155:15–23. doi: 10.1016/j.vetpar.2008.04.018
67. Soltane R, Guyot K, Dei-Cas E, Ayadi A. Cryptosporidium parvum (Eucoccidiorida: Cryptosporiidae) in calves: results of a longitudinal study in a dairy farm in Sfax, Tunisia. Parasite. (2007) 14:309–12. doi: 10.1051/parasite/2007144309
68. Thomson S, Innes EA, Jonsson NN, Katzer F. Shedding of Cryptosporidium in calves and dams: evidence of re-infection and shedding of different gp60 subtypes. Parasitology. (2019) 146:1404–13. doi: 10.1017/S0031182019000829
69. Wells B, Shaw H, Hotchkiss E, Gilray J, Ayton R, Green J, et al. Prevalence, species identification and genotyping Cryptosporidium from livestock and deer in a catchment in the Cairngorms with a history of a contaminated public water supply. Parasit Vectors. (2015) 8:66. doi: 10.1186/s13071-015-0684-x
70. Szonyi B, Bordonaro R, Wade SE, Mohammed HO. Seasonal variation in the prevalence and molecular epidemiology of Cryptosporidium infection in dairy cattle in the New York City watershed. Parasitol Res. (2010) 107:317–25. doi: 10.1007/s00436-010-1864-4
71. Maurya PS, Rakesh RL, Pradeep B, Kumar S, Kundu K, Garg R, et al. Prevalence and risk factors associated with Cryptosporidium spp. infection in young domestic livestock in India. Trop Anim Health Prod. (2013) 45:941–6. doi: 10.1007/s11250-012-0311-1
72. Yang F, Ma L, Gou JM, Yao HZ, Ren M, Yang BK, et al. Seasonal distribution of Cryptosporidium spp, Giardia duodenalis and Enterocytozoon bieneusi in Tibetan sheep in Qinghai, China. Parasit Vectors. (2022) 15:394. doi: 10.1186/s13071-022-05442-0
73. Al-Habsi K, Yang R, Williams A, Miller D, Ryan U, Jacobson C. Zoonotic Cryptosporidium and Giardia shedding by captured rangeland goats. Vet Parasitol Reg Stud Reports. (2017) 7:32–5. doi: 10.1016/j.vprsr.2016.11.006
74. Holzhausen I, Lendner M, Gohring F, Steinhofel I, Daugschies A. Distribution of Cryptosporidium parvum gp60 subtypes in calf herds of Saxony, Germany. Parasitol Res. (2019) 118:1549–58. doi: 10.1007/s00436-019-06266-1
75. Mercado R, Pena S, Ozaki LS, Fredes F, Godoy J. Multiple Cryptosporidium parvum subtypes detected in a unique isolate of a Chilean neonatal calf with diarrhea. Parasitol Res. (2015) 114:1985–8. doi: 10.1007/s00436-015-4364-8
76. Avendano C, Ramo A, Vergara-Castiblanco C, Sanchez-Acedo C, Quilez J. Genetic uniqueness of Cryptosporidium parvum from dairy calves in Colombia. Parasitol Res. (2018) 117:1317–23. doi: 10.1007/s00436-018-5818-6
Keywords: Cryptosporidium spp., diarrhea, pre-weaned calves, age, season
Citation: Jang D-H, Cho H-C, Park Y-J, Park J and Choi K-S (2023) First report of Cryptosporidium andersoni and risk factors associated with the occurrence of Cryptosporidium spp. in pre-weaned native Korean calves with diarrhea. Front. Vet. Sci. 10:1145096. doi: 10.3389/fvets.2023.1145096
Received: 15 January 2023; Accepted: 02 March 2023;
Published: 21 March 2023.
Edited by:
Calin Mircea Gherman, University of Agricultural Sciences and Veterinary Medicine of Cluj-Napoca, RomaniaReviewed by:
Kalman Imre, Banat University of Agricultural Sciences and Veterinary Medicine, RomaniaCopyright © 2023 Jang, Cho, Park, Park and Choi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kyoung-Seong Choi, a3NjaG9pM0BrbnUuYWMua3I=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.