- 1Key Laboratory of Animal Genetics, Breeding and Reproduction of Shaanxi Province, College Animal Science and Technology, Northwest A&F University, Yangling, Shaanxi, China
- 2Shandong Key Lab of Animal Disease Control and Breeding, Institute of Animal Science and Veterinary Medicine, Shandong Academy of Agricultural Sciences, Jinan, China
- 3Bijie Animal Husbandry and Veterinary Science Research Institute, Bijie, China
- 4College of Life Science and Engineering, Northwest Minzu University, Lanzhou, China
Background: The hippocampal abundant transcript 1 (HIAT1) gene, also known as major facilitator superfamily domain-containing 14A (MFSD14A), encodes for a transmembrane transporter protein and has been previously shown to be associated with milk production in buffalo and sheep breeds, as well as growth traits in chicken and goats. However, tissue level distribution of the ovine HIAT1 gene, as well as its effect on body morphometric traits in sheep, has yet to be studied.
Methods: The HIAT1 mRNA expression profile of Lanzhou fat-tailed (LFT) sheep was determined by quantitative real-time PCR (qPCR). A total of 1498 sheep of three indigenous Chinese sheep breeds were PCR-genotyped for polymorphisms of HIAT1 gene. Student's t-test was used to observe the association between the genotype and sheep morphometric traits.
Results: HIAT1 was widely expressed in all examined tissues, and was particularly abundant in the testis of male LFT sheep. Additionally, a 9-bp insertion mutation (rs1089950828) located within the 5'-upstream region of HIAT1 was investigated in Luxi black-headed (LXBH) sheep and Guiqian semi-fine wool (GSFW) sheep. The wildtype allele frequency 'D' was found to be more prevalent than that of the mutant allele ‘I'. Furthermore, low genetic diversity was confirmed in all sampled sheep populations. Subsequent association analyses indicated an association between the 9-bp InDel mutation of interest and the morphometric traits of LXBH and GSFW sheep. Furthermore, yearling ewes with a heterozygous genotype (ID) demonstrated smaller body sizes, while yearling rams and adult ewes with the heterozygous genotype were found to have overall better growth performance.
Conclusion: These findings imply that functional InDel polymorphism (rs1089950828) has the potential to be utilized for marker-assisted selection (MAS) of growth traits in domestic Chinese sheep populations.
1. Introduction
The final grown performance of sheep and other domesticated animals is affected by multiple factors including environmental, nutritional, and genetic influences (1). Using high-throughput whole genome sequencing, RNA sequencing, and molecular biology techniques, a number of candidate functional genes and genetic variants linked to important growth traits in livestock were found and used for the purposes of molecular-assisted and omics-assisted breeding (2–6). Among the numerous genome variants including SNPs (single nucleotide polymorphisms), CNVs (copy number variations), and other genetic markers, numerous studies have demonstrated that InDels (insertion/deletion) function by regulating gene translation or altering a protein and are related to differences in the characteristics of individual animals (7–9).
The hippocampal abundant transcript 1 (HIAT1) gene is also known as the major facilitator superfamily domain 14A (MFSD14A) gene (10). The HIAT1 protein contains a conserved sugar transporter motif as well as 12 transmembrane alpha helices (TMs) linked by hydrophilic loops, and belongs to the major facilitator superfamily (MFS) of secondary protein transporters (11). The MFS protein superfamily is the largest secondary transporter family present in a broad variety of species from archaea to mammals and plays an essential role in the exchange of cellular materials and energy metabolism, which facilitates the transport of many substrates across both cytoplasmic and internal cell membranes (12). The expression of both MFSD1 and MFSD3 genes has previously been shown to be significantly different in both the cerebellum and brain of mice after they were subjected to starvation and high-fat diets. This suggests that these genes are regulated at the nutrient level and may be related to nutrient intake and transport (13). Two members of the MFS gene family, MFSD6 and MFSD10 were found to be related to the energy metabolism of cells. Expression levels of these two genes were found to be down-regulated when the cellular energy consumption rate increased or when cellular energy was significantly depleted (14). These findings suggest that HIAT1 may also be related to fatty acid uptake and transport.
Chinese scholars have found that HIAT1 affects buffalo milk fat synthesis mainly through regulation of cellular membrane transport functions (15). Moreover, studies have shown that the HIAT1 protein has the capacity to transport solute from the blood stream that is required for spermiogenesis, thus disruption of this gene has been shown to cause round-headed sperm and sterility in male mice (16). HIAT1 has previously been shown to be commonly expressed in multiple tissues types and highly expressed in testis, suggesting that it has extensive biological functions (17). HIAT1 is expressed in brown adipose tissue and is negatively correlated with the process in which glucose is converted to fat (18). Additionally, polymorphisms in HIAT1 have been shown to affect milk traits in buffalo as well as growth in goats (15, 19). However, to date, there are few studies that have reported on the function of HIAT1 in indigenous Chinese sheep populations.
In order to further improve the production features of native Chinese sheep populations, the current study aimed to detect HIAT1 expression features in Lanzhou fat-tailed (LFT) sheep, explore InDel variations of the HIAT1 gene in three indigenous Chinese sheep breeds, which included Luxi black-headed (LXBH) sheep from east China, Guiqian semi-fine wool (GSFW) sheep from southwest China, and LFT sheep from northwest China, as well as conduct association analyses between sheep genotypes and growth traits to provide a basis for use of the HIAT1 gene for sheep selection and breeding.
2. Materials and methods
2.1. Animals and ethics statement
Twelve tissue samples were collected from three 6-month-old male Lanzhou fat-tailed sheep and include heart, liver, spleen, lung, kidney, testis, longissimus dorsi muscle, tail adipose, perirenal fat, subcutaneous fat, small intestine, and rennet stomach (Yongjing county, Gansu Province). A total of 1498 indigenous Chinese sheep were used for DNA experiments and included Luxi black-headed sheep (n = 616) raised in Liaocheng city, Shandong province; Guiqian semi-fine wool sheep (n = 824) raised in Bijie city, Guizhou province; and Lanzhou fat-tailed sheep (n = 58) provided by Gansu Ruilin Science and Technology Breeding Company (Yongjing county, Gansu province). All tissue samples were frozen in liquid nitrogen and stored at −80°C for RNA and DNA isolation. Relevant body morphometric traits were recorded, including body weight, chest circumference, height at hip cross, body height, body length, cannon circumference, hip width, abdominal circumference, chest depth, and chest width. Animal experiments were conducted in line with the guidelines for the care and use of animals and approved by the Animal Ethics Committee of Northwest A&F University.
2.2. DNA isolation, primer design, and InDel genotyping
Genomic DNA was isolated following previously established protocols (20, 21). Total RNA and genomic DNA concentrations were determined using Nanodrop 2000. DNA was diluted to a concentration of 20 ng/μL and stored at −40°C. Thirty DNA samples were randomly selected from each breed to construct three genomic DNA pools for investigation of HIAT1 variations.
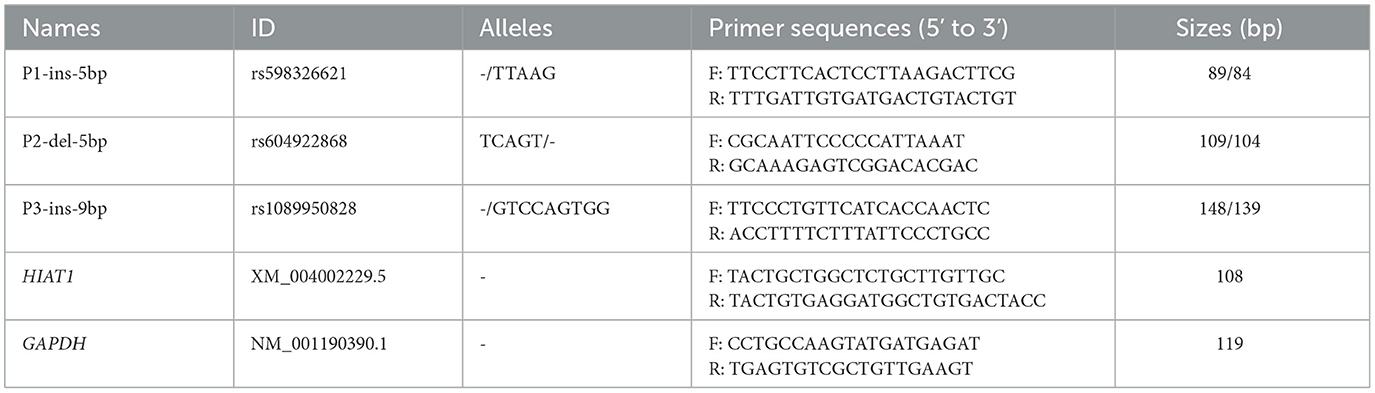
Three upstream gene variants were screened from the Ensembl database, and then several primers (Table 1) were designed using Primer Premier 5 software and synthesized by Sangon Biotech (Xi'an, China) based on reference sequences in ovine HIAT1 (NC 056054.1). Polymorphic fragments were amplified using the polymerase chain reaction (PCR) method and utilized a touch-down program and reaction volumes as described in previous studies (8, 22). PCR products were then separated by electrophoresis on a 3.5% agarose gel stained with GoldView. The genotype of each individual sheep was analyzed.
2.3. RNA isolation, cDNA synthesis, and quantitative real-time PCR
Total RNA was extracted from tissue samples using TRIzol reagent, and cDNA was synthesized according to manufactures' instructions using an Evo M-MLV RT Kit with gDNA Clean for qPCR II (Accurate biology Co., Ltd, Changsha, China). A LightCycler 96 real-time PCR system (Roche, Switzerland) was used to run the qPCR program. qPCR reactions (10 μL) contained 5 μL 2 × ChamQ SYBR qPCR Master Mix (Vazyme, Nanjing, China), 0.2 μL of each primer, and 4.6 μL cDNA (1/100 dilution). qPCR conditions were as follows: 95°C for 180 s, followed by 40 amplification cycles lasting 40 s each (95°C for 10 s and 60°C for 30 s). Gene expression was quantitated using the 2−ΔΔCt method.
2.4. Bioinformatics and statistical analysis
Differing amino acid sequences alignment were made using DNAMAN 6.0 software. The percent similarity of amino acid sequences was analyzed by NCBI-Blast online software. Using AliBaba2.1 (http://gene-regulation.com/pub/programs/alibaba2/), predictions of transcription factor binding to the mutant ovine HIAT1 gene sequence were generated (23). Population genetic indices, as well as allelic and genotypic frequencies, were determined. χ2 test was performed to determine whether the gene variants are within Hardy-Weinberg equilibrium (HWE) and analyze the difference in genotype distributions amongst each sheep population. The statistical model used was Yij = μ + Gi + εij, where Yij = observed growth traits, μ = population mean, Gi = fixed effect of the genotype, εij = random error. If there were only two genotypes, student's t-test was performed to determine the effect of different genotypes on economic traits.
3. Results
3.1. Multiple sequence alignment and mRNA expression
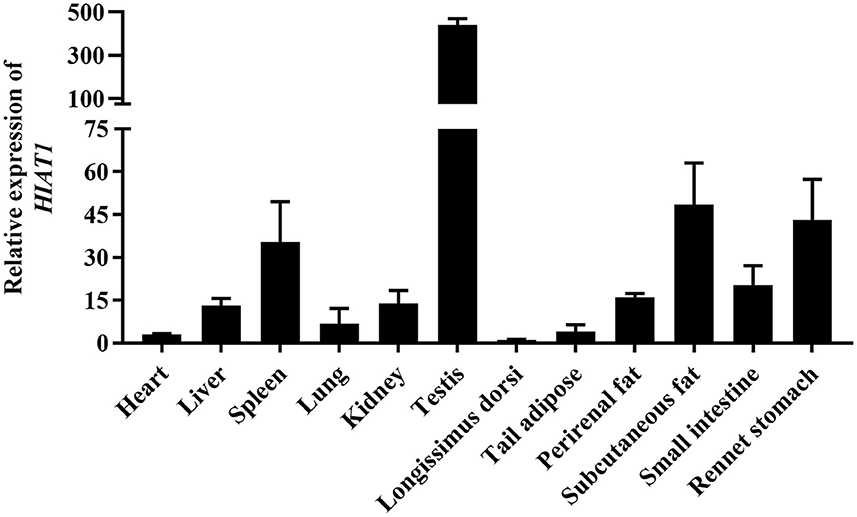
The results of NCBI-BLASTP sequence analysis showed that the HIAT1 in Ovis aries (Genbank: XP_004002278.1) shares 100%, 100%, 99.80%, 99.80% and 99.59% similarity with that of Capra hircus (XP_017901305.1), Bos taurus (NP_001095508.1), Mus musculus (NP_032272.2), Sus scrofa (XP_013852805.2) and Homo sapiens (NP_149044.2), respectively (Figure 1). Additionally, qPCR results revealed that HIAT1 was expressed in most sheep tissues, especially in the testis of male LFT sheep (Figure 2), which coincided with expression and distribution in human tissues.

Figure 1. Amino acid sequence alignment of HIAT1 in different mammalian species. The red boxes denote the parts of the sequence that are unable to be matched with the Ovis aries sequence. Note: • denotes the same amino acid.
3.2. Detection and genetic parameter analysis of InDel polymorphisms within the HIAT1 gene
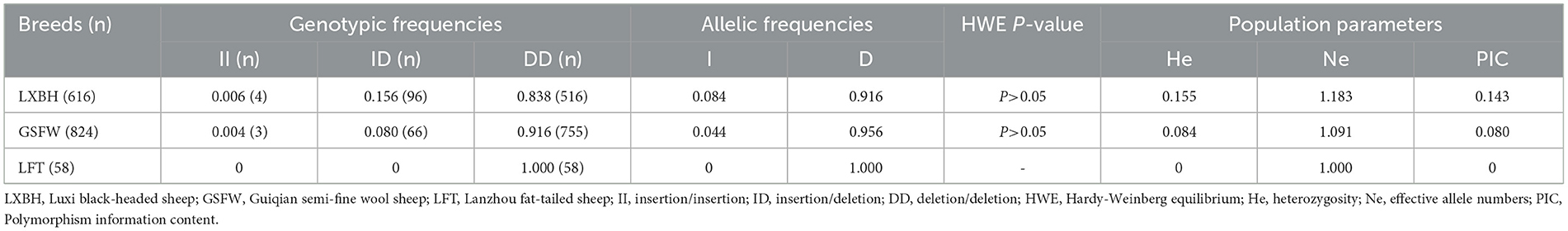
Electrophoresis patterning and sequence mapping of the P3-ins-9bp InDel showed polymorphisms in the 5'-upstream region of HIAT1 for both Luxi black-headed sheep and Guiqian semi-fine wool sheep. There were no detected HIAT1 polymorphisms found in Lanzhou fat-tailed sheep (Figure 3). Genotyping analyses showed that the HIAT1 P3-ins-9bp locus contained three genotypes (the homozygous insertion type, II, 148 bp; the homozygous deletion type, DD, 139 bp; and the heterozygous mutation type, ID, 148 bp and 139 bp) for both Luxi black-headed sheep and Guiqian semi-fine wool sheep; however, only one genotype (DD) was discovered in the Lanzhou fat-tailed breed.
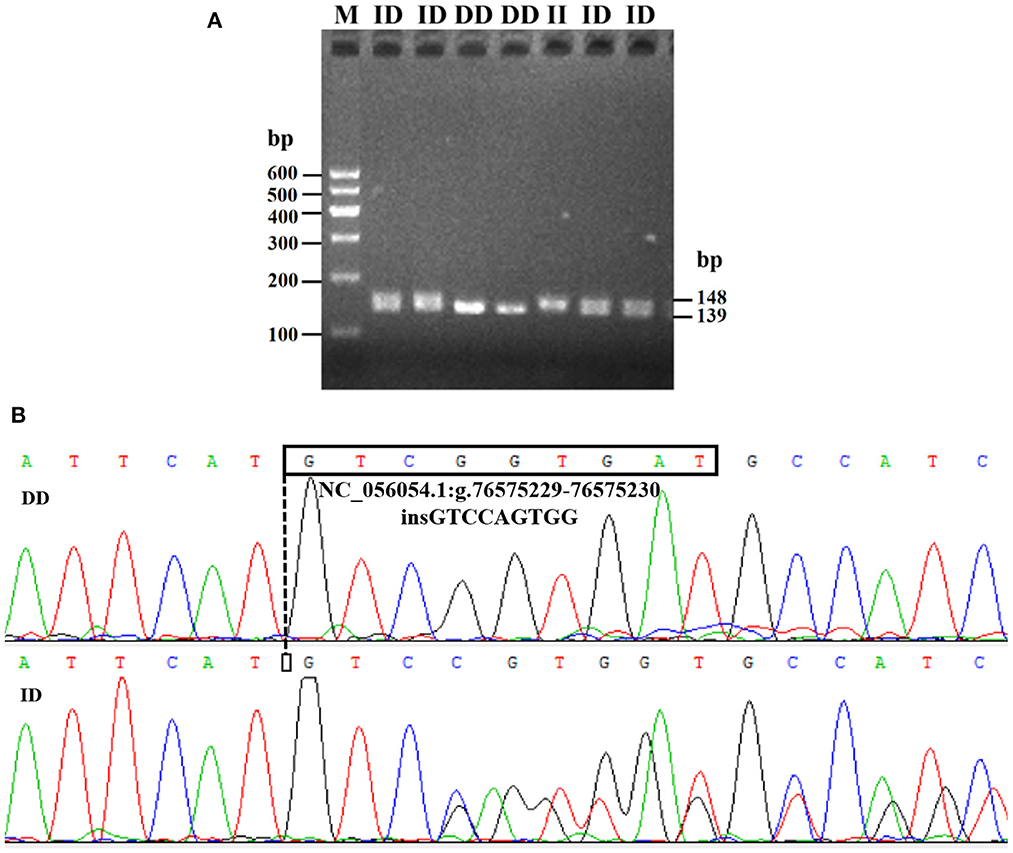
Figure 3. Electrophoresis gel (A) and sequence chromatograms (B) of the P3-ins-9bp InDel within the ovine HIAT1 gene.
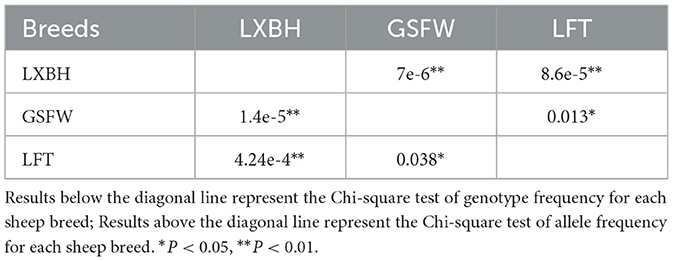
The genotypic frequency, allelic frequency, and population genetic indices were subsequently calculated and shown in Table 2. The DD genotype frequency was found to be much higher than both ID and II genotypes. Amongst all sheep breeds tested, the P3-ins-9bp locus was found to have low genetic polymorphism (PIC < 0.25) while maintaining Hardy-Weinberg equilibrium (P > 0.05). χ2 testing revealed that there were significant differences both in the genotypic and allelic frequency distributions of the P3-ins-9bp locus amongst all three populations (P < 0.05) (Table 3).
3.3. Association between the P3-ins-9bp InDel and growth traits
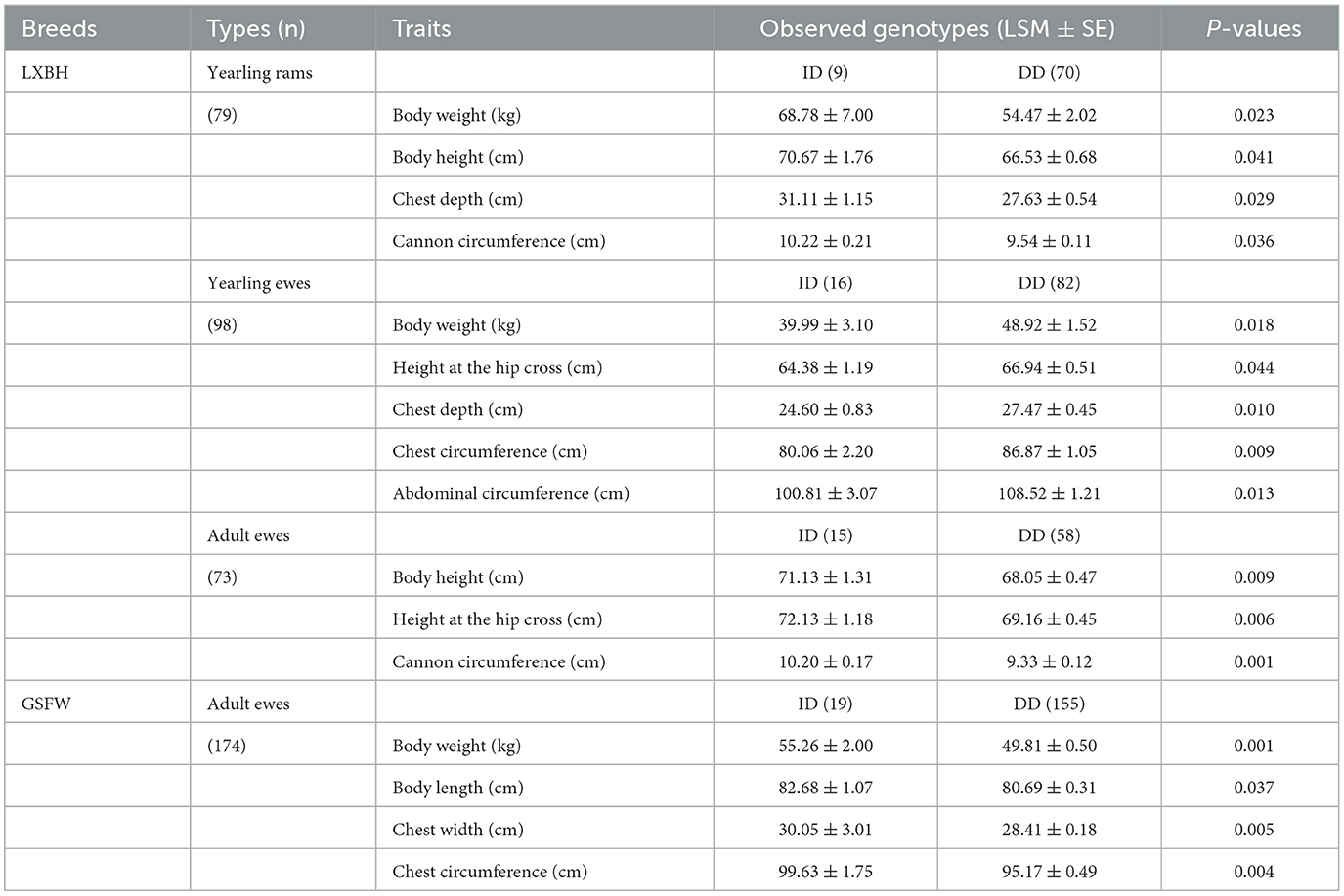
The associations between the P3-ins-9bp InDel of the ovine HIAT1 gene and growth performance were analyzed (Table 4). For LXBH sheep, t-tests results confirmed that the P3-ins-9bp InDel had no remarkable influence on growth traits for lambs (P > 0.05) (not shown). Yearling rams with the homozygous DD genotype were found to have lower chest depth, body weight, cannon circumference, and body height (P < 0.05). Yearling ewes with the homozygous DD genotype had significantly higher body weight, chest depth, chest circumference, height at the hip cross and abdominal circumference than those with the heterozygous ID genotype (P < 0.05). Additionally, adult ewes exhibited significant differences in height at the hip cross, body height, and cannon circumference (P < 0.01), with the heterozygous ID genotype being the dominant individual. For GSFW sheep, adult ewes with the heterozygous ID genotype showed superior performance compared to those with the homozygous DD genotype with regards to body weight, body length, chest width, and chest circumference.
4. Discussion
Recently, a study by Liu et al. (24) identified a quantitative trait locus (QTL) that affects milk traits in buffalo breed and includes SLC35A3 (Solute carrier family 35 member A3) and HIAT1 (24). The SLC35A3 gene is located adjacent to the HIAT1 gene and is involved in the transport of glucose, vitamins and other substances (25). A genome-wide association study (GWAS) on milk production traits determined that SLC35A3 may also play a significant role in sheep milk traits (26). Moreover, a missense mutation in bovine SLC35A3 has been found to lead to complex vertebral malformation (25). A recent study by Ye (15) found that HIAT1 regulates milk fat synthesis in mammary epithelial cells through the PPAR signaling pathway, and that two SNPs within HIAT1 are significantly associated with both milk fat and milk protein rates in buffalo (15). A more surprising finding is that both HIAT1 and SLC35A3 have the potential to affect both carcass and growth traits in chickens (27). In this study, we found that HIAT1 has high amino acid sequence conservation, and that its tissue expression pattern is similar to that of humans. These results suggest that HIAT1 might have a similar function amongst multiple types of mammals. Moreover, HIAT1 mRNA expression was detected in perirenal fat, subcutaneous fat, and the livers of LFT sheep, suggesting that HIAT1 may be related to fat deposition in sheep. These finding suggest HIAT1 is a key candidate gene for regulating important economic traits in livestock.
Body morphology traits reflect the growth and development of different parts of the body and are also important economic and breeding indicators for both meat and meat-wool sheep breeds (28, 29). Existing studies have showed that the HIAT1 gene influenced both growth and meat production traits in goats, and that a 15-bp insertion in HIAT1 was associated with body morphology in Shaanbei white cashmere goats (19, 30). In this study, a 9-bp insertion mutation located in the 5'-upstream region of the ovine HIAT1 gene (rs1089950828 -/GTCCAGTGG) was elucidated. It is well known that variations in the 5'-upstream region of genes has the potential to affect gene transcription and mRNA expression levels through changing transcription factor binding sites (TFBSs) and can also influence an individual's phenotype (31, 32). Thus, this research studied the association between the P3-ins-9bp InDel and sheep growth traits. Furthermore, the P3-ins-9bp InDel was strongly associated with body measurement traits in both LXBH and GSFW sheep based on association analysis. For yearling LXBH sheep, ewes with a heterozygous ID genotype demonstrated overall lower body size, while rams with a heterozygous ID genotype had better growth performance. These results indicate that the 9-bp insertion mutation may have a negative regulatory effect on both growth and development of LXBH yearling ewes as well as promote growth in yearling rams. Additionally, a heterozygous ID genotype in ewes was found to inhibit body size development in yearling sheep as well as promote growth in adult sheep. This may be due to the effect of the 9-bp InDel on the differential expression of HIAT1 at different growth stages throughout the sheep lifecycle (33). Furthermore, the PIC value is an indicator of molecular marker quality in genetic studies. Our InDel locus of interest showed low polymorphism (PIC < 0.25) amongst all tested sheep according to the PIC value, indicating that there was sufficient space in which artificial selection can take place (34, 35).
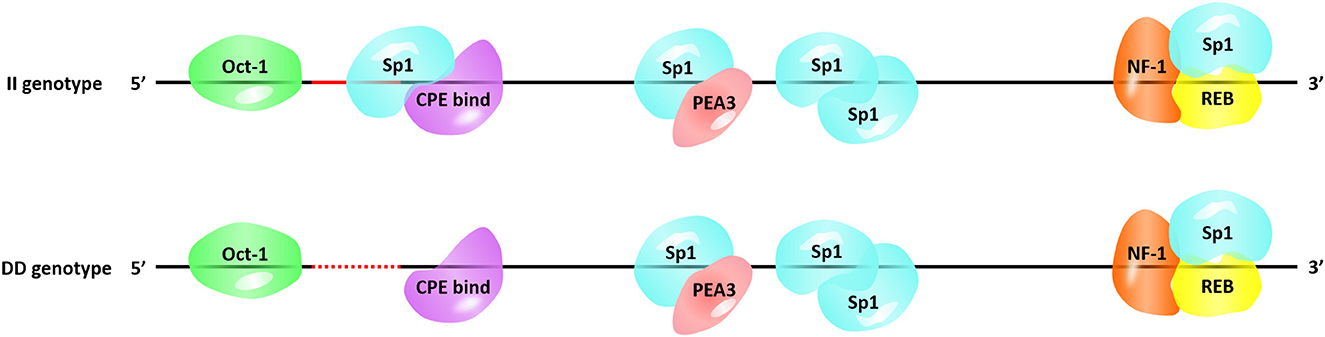
Next, we searched for the putative transcription factor binding sites that contained our InDel of interest. The P3-ins-9bp locus was found 624-bp upstream of the HIAT1 transcriptional start site (TSS), which created an additional binding site for the transcription factor Sp1 (Specificity protein 1) falling upstream of the four existing Sp1 binding sites (Figure 4). SP1 is a transcription factor of the SP/KLF family. The regulation of SP1 with regards to target genes is achieved by binding of the DNA binding domain to GC-rich motifs in the promoter region of the target gene, thereby regulating cellular processes such as autophagy, apoptosis, proliferation, differentiation and angiogenesis (36, 37). Chen et al. (38) found that Sp1 binding sites in the core promoter region are essential for positive regulation of FGF21 (Fibroblast growth factor 21) through gene transcription in both hepatic and adipose tissue (38). Furthermore, a recent study found that an SNP g.133A>C located within the SCD (Stearoyl CoA desaturase) promoter generated overall higher Sp1 binding affinity to the SCD promoter, which consequently affected gene expression (39). Moreover, a T > C mutation in the promoter region of IGF1 (Insulin-like growth factor 1) was significantly associated with the litter size of Yunshang black goats. This mutation was found to create a new binding site for SP1, thereby promoting goat granulosa cell proliferation by regulating the expression of IGF1 (40). Therefore, the interaction between Sp1 and HIAT1 leads to changes in gene expression at both the mRNA and protein levels, which in turn has the potential to affect the growth traits of sheep. In future studies this interaction is worthy of further in-depth research and exploration. Furthermore, the molecular mechanism of action and associated allelic consequences should be further validated in future studies.
5. Conclusion
This study investigated both the mRNA expression profile and genetic variations of the ovine HIAT1 gene. Our results demonstrated a high mRNA expression level of HIAT1 in the testis of Lanzhou fat-tailed sheep. Moreover, a 9-bp insertion mutation (rs1089950828) in the 5'-upstream region of ovine HIAT1 was detected. The frequency of the mutant allele “I” was found to be low. Furthermore, the 9-bp InDel locus was found to be closely associated with growth traits of LXBH and GSFW sheep, indicating that this InDel has the potential to be used as a genetic marker for assisted selection programs in domestic sheep.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding authors.
Ethics statement
All animal experiments were conducted in accordance with the the China national standard of Guidelines on Welfare and Ethical Review for Laboratory Animals (GB/T 35892-2018). Our study was approved by Institutional Animal Care and Use Committee (IACUC) of Northwest A&F University.
Author contributions
YL, XL, and HX came up with idea and revised the manuscript. YL wrote the manuscript and performed the experiments. CM, FJ, ZG, and HX collected the sheep samples and isolated the genomic DNA. YL and ZA analyzed the data. All authors approved the final version of the manuscript for submission, contributed to the article, and approved the submitted version.
Funding
This work was supported by the National Natural and Science Foundation of China (32060741).
Acknowledgments
We thank Shaanxi Key Laboratory of Molecular Biology for Agriculture and the Life Science Research Core Services (LSRCS) of Northwest A&F University (Northern Campus) for their cooperation and support. We would like to thank the staff of Shandong Academy of Agricultural Sciences, Guizhou Xinwumeng Ecological Animal Husbandry Development Co., Ltd., and Gansu Ruilin Science and Technology Breeding Company for providing samples and phenotypic traits data for us.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Li ZH Li H, Zhang H, Wang SZ, Wang QG, Wang YX. Identification of a single nucleotide polymorphism of the insulin-like growth factor binding protein 2 gene and its association with growth and body composition traits in the chicken. J Anim Sci. (2006) 84:2902–6. doi: 10.2527/jas.2006-144
2. Bakhtiarizadeh MR, Alamouti AA. RNA-Seq based genetic variant discovery provides new insights into controlling fat deposition in the tail of sheep. Sci Rep. (2020) 10:13525. doi: 10.1038/s41598-020-70527-8
3. Gu B, Sun R, Fang X, Zhang J, Zhao Z, Huang D, et al. Genome-wide association study of body conformation traits by whole genome sequencing in Dazu Black Goats. Animals. (2022) 12:548. doi: 10.3390/ani12050548
4. Salek Ardestani S, Jafarikia M, Sargolzaei M, Sullivan B, Miar Y. Genomic prediction of average daily gain, back-fat thickness, and loin muscle depth using different genomic tools in Canadian Swine populations. Front Genet. (2021) 12:665344. doi: 10.3389/fgene.2021.665344
5. Wang K, Liu X, Qi T, Hui Y, Yan H, Qu L, et al. Whole-genome sequencing to identify candidate genes for litter size and to uncover the variant function in goats (Capra hircus). Genomics. (2021) 113:142–50. doi: 10.1016/j.ygeno.2020.11.024
6. Ren F, Yu S, Chen R, Lv XY, Pan CY. Identification of a novel 12-bp insertion/deletion (indel) of iPS-related Oct4 gene and its association with reproductive traits in male piglets. Anim Reprod Sci. (2017) 178:55–60. doi: 10.1016/j.anireprosci.2017.01.009
7. Gamarra D, Aldai N, Arakawa A, de Pancorbo MM, Taniguchi M. Effect of a genetic polymorphism in SREBP1 on fatty acid composition and related gene expression in subcutaneous fat tissue of beef cattle breeds. Anim Sci J. (2021) 92:e13521. doi: 10.1111/asj.13521
8. Wang Z, Zhang X, Jiang E, Yan H, Zhu H, Chen H, et al. InDels within caprine IGF2BP1 intron 2 and the 3'-untranslated regions are associated with goat growth traits. Anim Genet. (2020) 51:117–21. doi: 10.1111/age.12871
9. Wang SH, Liu SR, Yuan TT, Sun XZ. Genetic effects of FTO gene insertion/deletion (InDel) on fat-tail measurements and growth traits in Tong sheep. Anim Biotechnol. (2021) 32:229–39. doi: 10.1080/10495398.2019.1680379
10. Fredriksson R, Nordström KJV, Stephansson O, Hägglund MGA, Schiöth HB. The solute carrier (SLC) complement of the human genome: phylogenetic classification reveals four major families. FEBS Lett. (2008) 582:3811–6. doi: 10.1016/j.febslet.2008.10.016
11. Collins S, Martin TL, Surwit RS, Robidoux J. Genetic vulnerability to diet-induced obesity in the C57BL/6J mouse: physiological and molecular characteristics. Physiol. Behav. (2004) 81:243–8. doi: 10.1016/j.physbeh.2004.02.006
12. Dittami SM, Barbeyron T, Boyen C, Cambefort J, Collet G, Delage L, et al. Genome and metabolic network of “Candidatus Phaeomarinobacter ectocarpi” Ec32, a new candidate genus of Alphaproteobacteria frequently associated with brown algae. Front Genet. (2014) 5:241. doi: 10.3389/fgene.2014.00241
13. Perland E, Hellsten SV, Lekholm E, Eriksson MM, Arapi V, Fredriksson R. The novel membrane-bound proteins MFSD1 and MFSD3 are putative SLC transporters affected by altered nutrient Intake. J Mol Neurosci. (2017) 61:199–214. doi: 10.1007/s12031-016-0867-8
14. Bagchi S, Perland E, Hosseini K, Lundgren J, Al-Walai N, Kheder S, et al. Probable role for major facilitator superfamily domain containing 6 (MFSD6) in the brain during variable energy consumption. Int J Neurosci. (2020) 130:476–89. doi: 10.1080/00207454.2019.1694020
15. Ye T. Identification of Milk Fat Regulation Gene MFSD14A and Its Mechanism in Buffalo. Wuhan: Huazhong Agricultural University. (2022).
16. Doran J, Walters C, Kyle V, Wooding P, Hammett-Burke R, Colledge WH. Mfsd14a (Hiat1) gene disruption causes globozoospermia and infertility in male mice. Reproduction. (2016) 152:91–9. doi: 10.1530/REP-15-0557
17. Sreedharan S, Stephansson O, Schioth HB, Fredriksson R. Long evolutionary conservation and considerable tissue specificity of several atypical solute carrier transporters. Gene. (2011) 478:11–8. doi: 10.1016/j.gene.2010.10.011
18. Pravenec M, Saba LM, Zídek V, Landa V, Mlejnek P, Šilhavý J, et al. Systems genetic analysis of brown adipose tissue function. Physiol Genomics. (2018) 50:52–66. doi: 10.1152/physiolgenomics.00091.2017
19. Gao JY, Song XY, Wu H, Tang Q, Wei ZY, Wang XY, et al. Detection of rs665862918 (15-bp Indel) of the HIAT1 gene and its strong genetic effects on growth traits in goats. Animals. (2020) 10:358–66. doi: 10.3390/ani10020358
20. Aljanabi SM, Martinez I. Universal and rapid salt-extraction of high quality genomic DNA for PCR-based techniques. Nucleic Acids Res. (1997) 25:4692–3. doi: 10.1093/nar/25.22.4692
21. Lan XY, Pan CY, Chen H, Zhang CL Li JY, Zhao M, Lei CZ, et al. An AluI PCR-RFLP detecting a silent allele at the goat POU1F1 locus and its association with production traits. Small Ruminant Res. (2007) 73:8–12. doi: 10.1016/j.smallrumres.2006.10.009
22. Li X, Jiang E, Zhang K, Zhang S, Jiang F, Song E, et al. Genetic variations within the bovine CRY2 gene are significantly associated with carcass traits. Animals. (2022) 12:1616. doi: 10.3390/ani12131616
23. Zhang S, Kang Z, Sun X, Cao X, Pan C, Dang R, et al. Novel lncRNA lncFAM200B: molecular characteristics and effects of genetic variants on promoter activity and cattle body measurement traits. Front Genet. (2019) 10:968. doi: 10.3389/fgene.2019.00968
24. Liu JJ, Liang AX, Campanile G, Plastow G, Zhang C, Wang Z, et al. Genome-wide association studies to identify quantitative trait loci affecting milk production traits in water buffalo. J Dairy Sci. (2018) 101:433–44. doi: 10.3168/jds.2017-13246
25. Thomsen B, Horn P, Panitz F, Bendixen E, Petersen AH, Holm LE, et al. missense mutation in the bovine SLC35A3 gene, encoding a UDP-N-acetylglucosamine transporter, causes complex vertebral malformation. Genome Res. (2006) 16:97–105. doi: 10.1101/gr.3690506
26. Rezvannejad E, Asadollahpour Nanaei H, Esmailizadeh A. Detection of candidate genes affecting milk production traits in sheep using whole-genome sequencing analysis. Vet Med Sci. (2022) 8:1197–204. doi: 10.1002/vms3.731
27. Zhang H, Shen LY, Xu ZC, Kramer LM Yu JQ, Zhang XY, Na W, et al. Haplotype-based genome-wide association studies for carcass and growth traits in chicken. Poult Sci. (2020) 99:2349–61. doi: 10.1016/j.psj.2020.01.009
28. Al-Mamun HA, Kwan P, Clark SA, Ferdosi MH, Tellam R, Gondro C. Genome-wide association study of body weight in Australian Merino sheep reveals an orthologous region on OAR6 to human and bovine genomic regions affecting height and weight. Genet Selection Evol. (2015) 47:66. doi: 10.1186/s12711-015-0142-4
29. Macé T, González-García E, Pradel J, Parisot S, Carrière F, Douls S, et al. Genetic analysis of robustness in meat sheep through body weight and body condition score changes over time. J Anim Sci. (2018) 96:4501–11. doi: 10.1093/jas/sky318
30. Zhang B, Chang L, Lan XY, Asif N, Guan FL, Fu DK, et al. Genome-wide definition of selective sweeps reveals molecular evidence of trait-driven domestication among elite goat (Capra species) breeds for the production of dairy, cashmere, and meat. GigaScience. (2018) 7:giy105. doi: 10.1093/gigascience/giy105
31. Glanzmann B, Lombard D, Carr J, Bardien S. Screening of two indel polymorphisms in the 5'UTR of the DJ-1 gene in South African Parkinson's disease patients. J Neural Transm. (2014) 121:135–8. doi: 10.1007/s00702-013-1094-x
32. Oner Y, Keskin A, Ustuner H, Soysal D, Karakaş V. Genetic diversity of the 3' and 5' untranslated regions of the HSP70. 1 gene between native Turkish and Holstein Friesian cattle breeds South African. J Anim Sci. (2017) 47:424–39. doi: 10.4314/sajas.v47i4.2
33. Cui Y, Yan H, Wang K, Xu H, Zhang X, Zhu H, et al. Insertion/Deletion within the KDM6A gene is significantly associated with litter size in goat. Front Genet. (2018) 9:91. doi: 10.3389/fgene.2018.00091
34. Serrote CML, Reiniger LRS, Silva KB, Rabaiolli SMDS, Stefanel CM. Determining the polymorphism information content of a molecular marker. Gene. (2020) 726:144175–8. doi: 10.1016/j.gene.2019.144175
35. Zhao HD, He S, Wang SH, Zhu YJ, Xu HW, Luo RY, et al. Two new insertion/deletion variants of the PITX2 gene and their effects on growth traits in sheep. Anim Biotechnol. (2018) 29:276–82. doi: 10.1080/10495398.2017.1379415
36. Guido C, Panza S, Santoro M, Avena P, Panno ML, Perrotta I, et al. Estrogen receptor beta (ERβ) produces autophagy and necroptosis in human seminoma cell line through the binding of the Sp1 on the phosphatase and tensin homolog deleted from chromosome 10 (PTEN) promoter gene. Cell cycle. (2012) 11:2911–21. doi: 10.4161/cc.21336
37. Vellingiri B, Iyer M, Devi Subramaniam M, Jayaramayya K, Siama Z, Giridharan B, et al. Understanding the role of the transcription factor Sp1 in ovarian cancer: from theory to practice. Int J Mol Sci. (2020) 21:1153. doi: 10.3390/ijms21031153
38. Chen S, Li H, Zhang J, Jiang S, Zhang M, Xu Y, et al. Identification of Sp1 as a transcription activator to regulate fibroblast growth factor 21 gene expression. BioMed Res Int. (2017) 29:8402035. doi: 10.1155/2017/8402035
39. Gu M, Cosenza G, Iannaccone M, Macciotta NPP, Guo Y, Di Stasio L, et al. The single nucleotide polymorphism g. 133A>C in the stearoyl CoA desaturase gene (SCD) promoter affects gene expression and quali-quantitative properties of river buffalo milk. J Dairy Sci. (2019) 102:442–51. doi: 10.3168/jds.2018-15059
Keywords: sheep, HIAT1 gene, insertion/deletion (InDel), growth traits, marker-assisted selection (MAS)
Citation: Luo Y, Akhatayeva Z, Mao C, Jiang F, Guo Z, Xu H and Lan X (2023) The ovine HIAT1 gene: mRNA expression, InDel mutations, and growth trait associations. Front. Vet. Sci. 10:1134903. doi: 10.3389/fvets.2023.1134903
Received: 31 December 2022; Accepted: 27 March 2023;
Published: 17 April 2023.
Edited by:
Ibrar Muhammad Khan, Fuyang Normal University, ChinaReviewed by:
Cemal Ün, Ege University, TürkiyeMuhammad Zahoor, University of Agriculture, Dera Ismail Khan, Pakistan
Copyright © 2023 Luo, Akhatayeva, Mao, Jiang, Guo, Xu and Lan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongwei Xu, eHVob25nd2VpQHhibXUuZWR1LmNu; Xianyong Lan, bGFueGlhbnlvbmc3OUBud3N1YWYuZWR1LmNu
 Yunyun Luo1
Yunyun Luo1 Zhanerke Akhatayeva
Zhanerke Akhatayeva Fugui Jiang
Fugui Jiang Xianyong Lan
Xianyong Lan