
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci. , 16 February 2023
Sec. Veterinary Experimental and Diagnostic Pathology
Volume 10 - 2023 | https://doi.org/10.3389/fvets.2023.1116352
 Chihai Ji1,2,3†
Chihai Ji1,2,3† Ling Zhou1,2†
Ling Zhou1,2† Yonghui Chen1,2†
Yonghui Chen1,2† Xueen Fang4
Xueen Fang4 Yanhong Liu5
Yanhong Liu5 Mengkan Du6
Mengkan Du6 Xiandong Lu5
Xiandong Lu5 Qianniu Li1,2
Qianniu Li1,2 Heng Wang2,3
Heng Wang2,3 Yuan Sun1,2*
Yuan Sun1,2* Tian Lan1,2*
Tian Lan1,2* Jingyun Ma1,2*
Jingyun Ma1,2*Introduction: Different pathogens causing mixed infection are now threatening the pig industry in the context of the African Swine Fever (ASF) circulating especially in China, and it is crucial to achieving the early diagnosis of these pathogens for disease control and prevention.
Methods: Here we report the development of a rapid, portable, sensitive, high-throughput, and accurate microfluidic-LAMP chip detection system for simultaneous detection and differentiation of gene-deleted type and wild-type African swine fever virus (ASFV), pseudorabie virus (PRV), porcine parvovirus (PPV), porcine circovirus type 2 (PCV2), and porcine reproductive and respiratory syndrome (PRRSV).
Results and discussion: The newly developed system was shown to be sensitive with detection limits of 101 copies/μl for ASFV-MGF505-2R/P72, PPV, and PCV2, 102 copies/μl for ASFV-CD2v, PRV, and PRRSV. The system was highly specific (100%) and stable (C.V.s < 5%) in its ability to detect different pathogens. A total 213 clinical samples and 15 ASFV nucleic acid samples were collected to assess the performance of the detection system, showing highly effective diagnosis. Altogether, the developed microfluidic-LAMP chip system provides a rapid, sensitive, high-throughput and portable diagnostic tool for the accurate detection of multiple swine pathogens.
In recent decades, with the increasing demand for pork, the pig industry in China has witnessed a large-scale growth accompanied by multiple viral disease outbreaks (1). Under the intensive captivity conditions, it is common for sows to be infected simultaneously with a variety of pathogens such as African swine fever virus (ASFV), porcine reproductive and respiratory syndrome (PRRSV), porcine circovirus type 2 (PCV-2), porcine parvovirus (PPV), and pseudorabies virus (PRV) (2–4).
African swine fever (ASF) is a infections disease caused by ASFV with the mortality rates near 100% (5). Since the identification of ASF in China in August 2018, ASFV has been causing series of outbreaks which resulted in significant economic loss nationwide (6). Recently, newly emerging virulent mutant strains of ASFV with mutations, deletions, insertions, or short-fragment replacement in genomes have made the early diagnosis of ASF more difficult (7). Although artificial deletion of EP402R gene encoding the CD2v protein and multigene family 505 (MGF505) have been demonstrated to result in attenuated virulence in HLJ/18 strain and Georgia2007 isolate (8, 9), but their safety remains to be elucidated (10, 11). There is a need to identify the gene-deletion strain from the natural isolates, as the obtained phenotype of the genetically modified ASFV has some degree of uncertainty (12). This is essential to establish biosafety control measures in pig farms.
Porcine reproductive and respiratory syndrome (PRRS) is one of the most important economically diseases to the pig industry, which causes great losses to the pig industry in China (13). Pseudorabies (PR) caused by suid herpesvirus 1, is an economically important viral disease of pigs with high prevalence of its variants in China (14). PPV is one of the major etiologic agents of reproductive disorders in China, Canada and many other countries (15). PCV-2 is a widespread, economically challenging pathogen in most pig-producing countries and it has co-infections with multiple diseases including ASF and PRRS (16). The aforementioned diseases appear with similar symptoms such fever, loss of appetite, and reproductive impairments which makes it hard to diagnose at an early stage.
So far, the pathogens of swine diseases are commonly detected by single-pathogen isolation and identification, serological test, PCR and ELISA. As the same time, there are numbers of methods for detection on the basis of laboratory diagnosis. All the above-mentioned methods have been used extensively due to advantages of the accuracy and high sensitivity. Despite these merits, most of these assays are intricate and limited by specific laboratory settings and large amounts of reagents that can't be used in remote areas. Therefore, simple, rapid, reliable, and multi-targeted portable point-of-care (POC) diagnostic tools of swine pathogens are in urgent need for early disease detection and the prevention of viral transmission (17). Centrifugal microfluidic lab-on-a-chip (LOC) is an integrated and miniaturized technology that has the ability to combine sample extraction, reactions and detection on a compact disc (CD) (18, 19). This advanced technology makes detection assays more efficient and time-saving, which is calculated for Point of Care Testing (POCT) in viral detection (20).
In the present study, we integrated the loop-mediated isothermal amplification (LAMP) into the CD-like microfluidic chip for rapid multiplex detection of five pathogens in portable device. Our test shows that, the entire analysis including sample preparation and detection can be completed in 60 min, demonstrating high feasibility for the accurate, sensitive and specific detection of diseases in livestock farms in remote areas.
Clinical serum samples and tissue samples were collected from pigs that were suspected to carry fever and reproductive disorder- associated pathogens from 2016 to 2017 in South China in accordance with the recommendations of National Standards for Laboratory Animals of the People's Republic of China (GB149258-2010). All samples were preserved at −80°C from the time of original receipt until use. Fifteen ASFV positive nucleic acid samples and 50 ASFV negative serum samples were detected. The positive nucleic acid samples were provided by African Swine Fever Regional Laboratory of China (Guangzhou), and the negative serum samples were collected by our laboratory in 2018.
Individual samples including spleens, lungs, lymph nodes, tonsils, brain tissues, nose swabs, and kidney tissues were mixed and ground into homogenates with phosphate buffered saline (PBS; 20% w/v), and frozen and thawed three times, then centrifuged for 10 min at 10, 000 g. Viral nucleic acid was extracted using the Magnetic Viral DNA/RNA Kit (iGeneTech, Ningbo, China) according to the manufacturer's specifications. Total extraction time was 15 min and the products were stored at −80°C until used.
The qRT-RCR detection of ASFV, PRV, PPV, PRRSV and PCV2 was carried out with ChamQ Universal SYBR qPCR Master Mix (Vazyme, China) according to the previously described methods (21–27).
The microfluidic chip used in this study were manufactured out of PMMA using micro-injection molding technology with a diameter of 80 mm and a thickness of 2.5 mm. A kind of heat-sealing film (SC-MA2000, Suchuang Co., Ltd) was used to form an integrated chip. Four independent units compose a microfluidic chip, each of which consisted of a sample well, a vent and eight micro reaction wells (Figure 1).
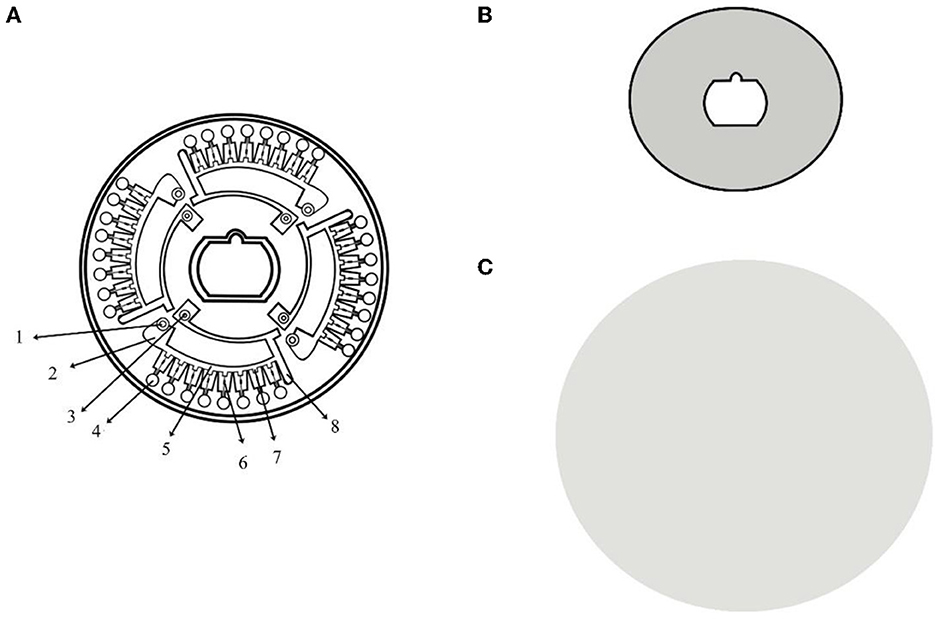
Figure 1. Structure chart of microfluidic chip. (A) The structure of the CD-like microfluidic chip. 1: sample well; 2: liquid storage chamber; 3: vent hole; 4: reaction chamber; 5: the second arc passage; 6: ball valve; 7: reservoirs; and 8: waste liquid tank; (B) Heat-sealing film of the front side; (C) Heat-sealing film of the back side.
The ball valve played a pivotal role in the entire process ensuring the reaction mixture diffuse into the adjacent pipe and the reaction wells were filled with liquid. The volume of each micro reaction well was 5 μl. The supporting microfluidic detection appliance (dimensions: length, width, and height are 280 × 200 × 135 mm) was actually a device (Product Model: AJYGene TM MA2000) equipped with the functions of constant temperature control, high-speed centrifugation and fluorescence reading and analysis, which was provided by iGeneTech (Ningbo, China). The mixture containing the sample would be equably distributed in each reaction chambers at centrifugal speeds (1,600 rpm/min, 30 s; 4,600 rpm/min, 10 s) when the microfluidic device started running. Fluorescent signals were collected every minute for each reaction well and displayed in real time on the electronic screen during the reaction period.
The reference complete genomic sequences of ASFV, PRV, PPV, PRRSV, and PCV2 were acquired from GenBank with accession numbers of MK333180.1, MH766894.2, KU056477.1, JQ249927.1, KU131565.1, and AF201897.1, respectively. To identify the gene-deleted type and wild type ASFV, we choose three genes (CD2v, MGF505-2R, and P72) as the target for the detection of ASFV. CD2v and MGF505-2R would not be detected if the sample contained gene-deleted type ASFV. The specific primers were designed based on the conserved nucleic acid fragments of ASFV CD2v/MGF505-2R/P72, PRV gB, PPV VP2, PCV2 ORF2, and PRRSV P83 genes and synthesized by Beijing Genomics institution, BGI (Shenzhen, China). To assure the success of obtaining optimal primer sets, three sets of primers were designed for each gene. All primer sequences were listed in Table 1.
All target gene fragments were synthesized and constructed into pMD19-T vector by BGI (Shenzhen, China), then were transformed into DH5α Escherichia coli cells following the manufacture's recommendations (TaKaRa, Dalian, China) and purified by Plasmid Mini Kit 2 (Omega Bio-tek, USA). The standard plasmids were tested and verified, and sent to BGI (Shenzhen, China) for sequencing. The concentration of the reconstructed standard plasmids was tested by a Qubit Fluorometer.
A volume of 5 μl primer mixture contains 0.2 μM each of outer primer (F3, B3), 1.6 μM each of inner primer (FIP, BIP), 0.8 μM each of loop primer (LF, LB), 0.3 μl trehalose and 1 μl ultrapure water. Before the start of the reaction, primer mixture was pre-immobilized in corresponding reaction chambers by heating drying. Seven micro chambers were activated for detections of seven viruses, and the remaining one was used for negative controls (Figure 2). Fifteen microliter of nucleic acid template and 10 μl premixed real-time LAMP reaction solution were transferred into the sample well by pipette and then sample wells and vent wells were sealed by the film. The preprocessed microfluidic chip was then placed in the provided microfluidic device reacted at 63.5°C for 60 min. Generally, the amplification result was regarded as positive when the chamber showed a positive signal and the negative controls showed negative signals in 40 min. A complete set of procedure was showed in Figure 2.
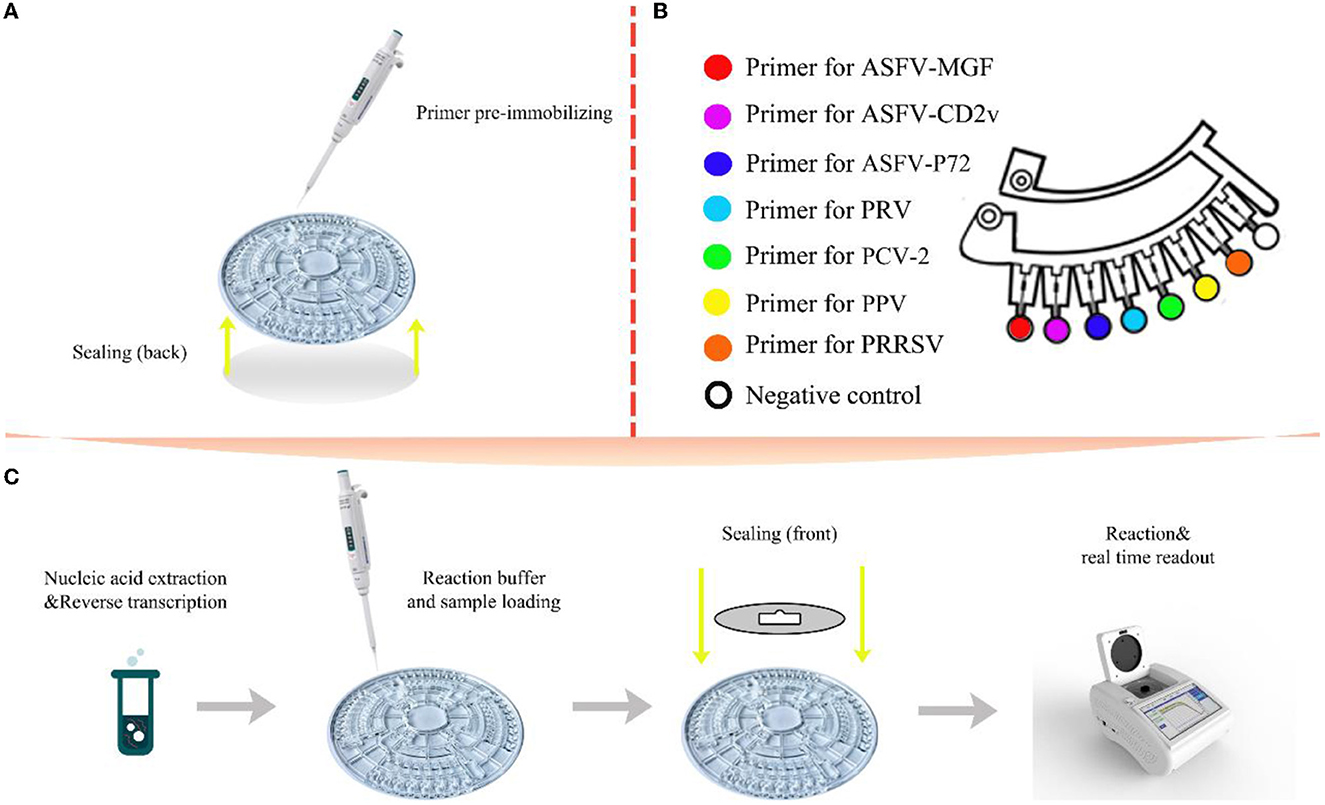
Figure 2. Schematic illustration of the multiplex detection based on microfluidic-LAMP chip system. (A) Primer pre-immobilizing and back side sealing; (B) a single section of the chip with its reaction area; (C) the entire flow of the diagnosis on microfluidic-LAMP chip detection system.
Viruses in this study including PRV, PRRSV, PPV, PCV2, and four other porcine viruses containing Classical swine fever virus (CSFV), foot and mouth disease virus (FMDV), seneca valley virus (SVA), and rotavirus (RV) were applied to verify the specificity of the assays. RNase Free ddH2O was also contained in the run as negative control.
All the standard plasmids were diluted serially 10-fold with RNase free ddH2O (106-100 copies/μl) as templates to define the detection limit of every amplified system, and each detection limit test was repeated three times.
For stability verification, 10-fold serial plasmids dilutions with high (106 copies/μl), medium (104 copies/μl) and low (detection limits of each microfluidic LAMP chip system) concentrations were used to assess coefficients of variation (C.V.) of the microfluidic-LAMP chip system. Synthetic ASFV DNAs and clinical positive samples of PRV, PPV, PCV-2 and PRRSV were also applied to evaluate the reproducibility of the microfluidic-LAMP chip system. The C.V. was calculated by testing microfluidic-LAMP chip system in three consecutive runs (inter-group) in different chips and four times in the same chip (intra-group). Threshold time (Tt) was estimated from the reaction time when the positive signals of a particular sample exceed the baseline during the real-time amplification.
A total of 163 swine clinical serum and tissue samples with fever and reproductive disorders, 15 ASFV positive nucleic acid samples and 50 ASFV negative serum samples were detected using the microfluid-LAMP chip system, and the diagnostic accuracy of the developed system was compared with the qRT-PCR assays.
The specificity of the microfluidic-LAMP chip system was determined with synthetic ASFV DNAs and viral nucleic acid including PRV, PPV, PRRSV, PRRSV, PCV2, CSFV, FMDV, SVA, and RV. As shown in Figure 3, only ASFV-MGF-, ASFV-CD2v-, ASFV-P72-, PRV-, PPV-, PCV2-, PRRSV-positive samples presented obvious amplification curves. Four other pathogens and the negative control did not show amplification curves, and no cross-reaction was observed during the amplification.
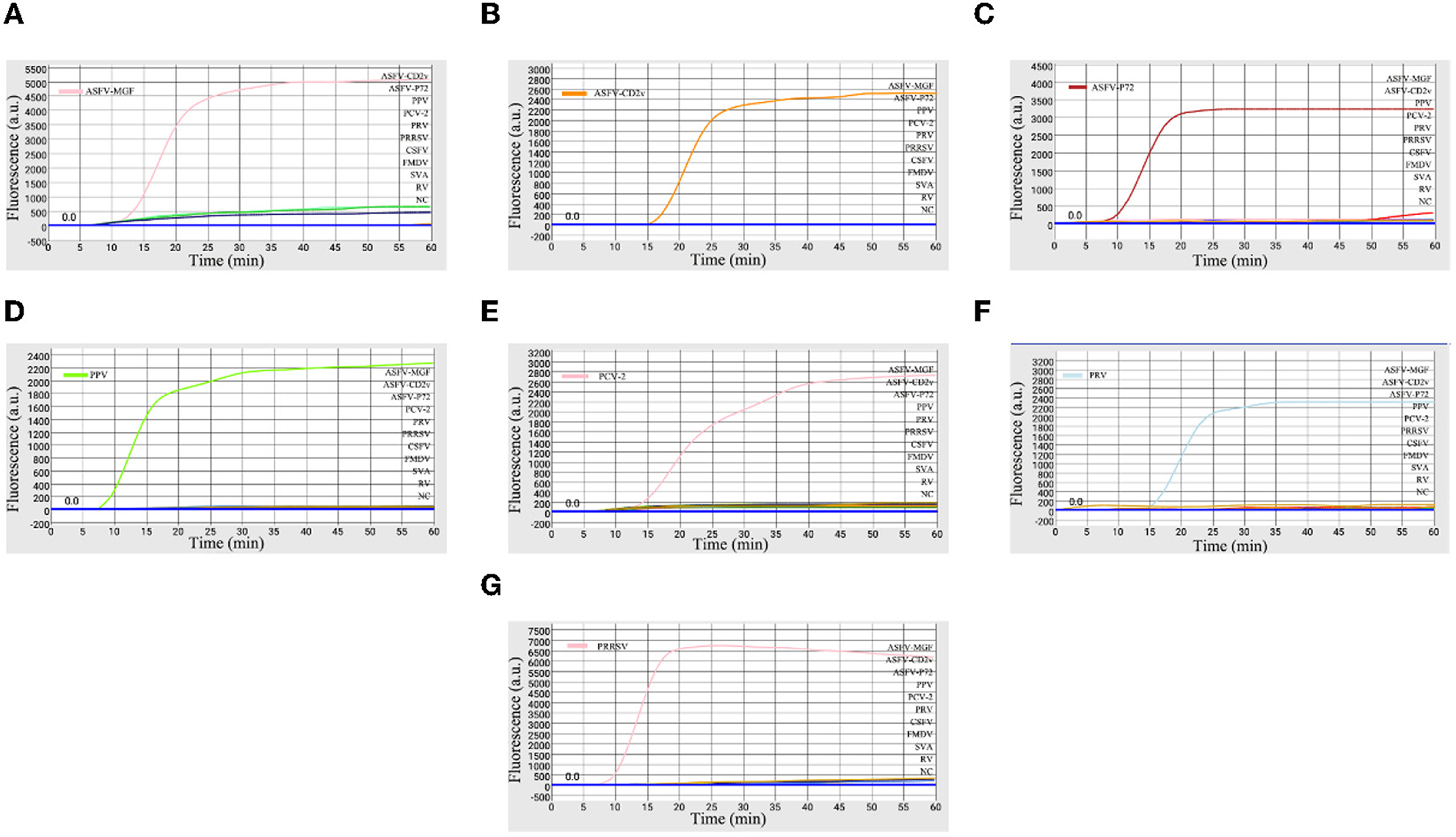
Figure 3. Specificity of microfluidic-LAMP chip detection system. (A) Specificity analysis for primers targeting to ASFV-MGF505-2R; (B) specificity analysis for primers targeting to ASFV-CD2v; (C) specificity analysis for ASFV-P72; (D) specificity analysis for PPV; (E) specificity analysis for PCV-2; (F) specificity analysis for PRV; and (G) specificity analysis for PRRSV.
So far, there is no safe, effective and approved ASF vaccines, and only live attenuated ASF vaccines developed through gene-deletions are the most promising vaccine candidate in the near future (21). CD2v and MGF505 genes have been considered as factors to reduce the ability of replication of the ASFV and have already been selected as the deletion target in live attenuated vaccines (8, 28). Thus, the identification of gene-deled type and wild type ASFV and other reproductive failure-associated swine pathogens including PRV, PPV, PCV2, and PRRSV becomes essential. Taken together, our system here displayed a high degree of specificity to the aimful viruses.
Serially diluted standard plasmids each with highly conserved genes of ASFV, PRV, PPV, PCV2, and PRRSV were used to investigate the detection limits of the microfluidic-LAMP chip system for those viruses. The results in Figure 4 showed that for ASFV-MGF/P72, PPV, and PCV-2 the detection limit of the established system was 101 copies/μl, and for ASFV-CD2v, PRV and PRRSV the detection limit was 102 copies/μl. The sensitivity analysis showed that the detection limit of each microfluidic-LAMP chip system was in the range of 102 to 101 copies/μl, which can be comparable to the existing detection methods (29–38). Based on the data of the sensitivity test with standard plasmids, the semi-logarithmic regression curves were established (Figure 4). The R2 values for ASFV-MGF, ASFV-CD2v, ASFV-P72, PRV, PPV, PCV-2, and PRRSV were 0.9570, 0.9587, 0.9527, 0.9589, 0.9543, 0.9643, and 0.9653, respectively and a significant linear relationship can be seen in each test, which indicate that the microfluidic- LAMP chip system has the potential for the quantitative detection.
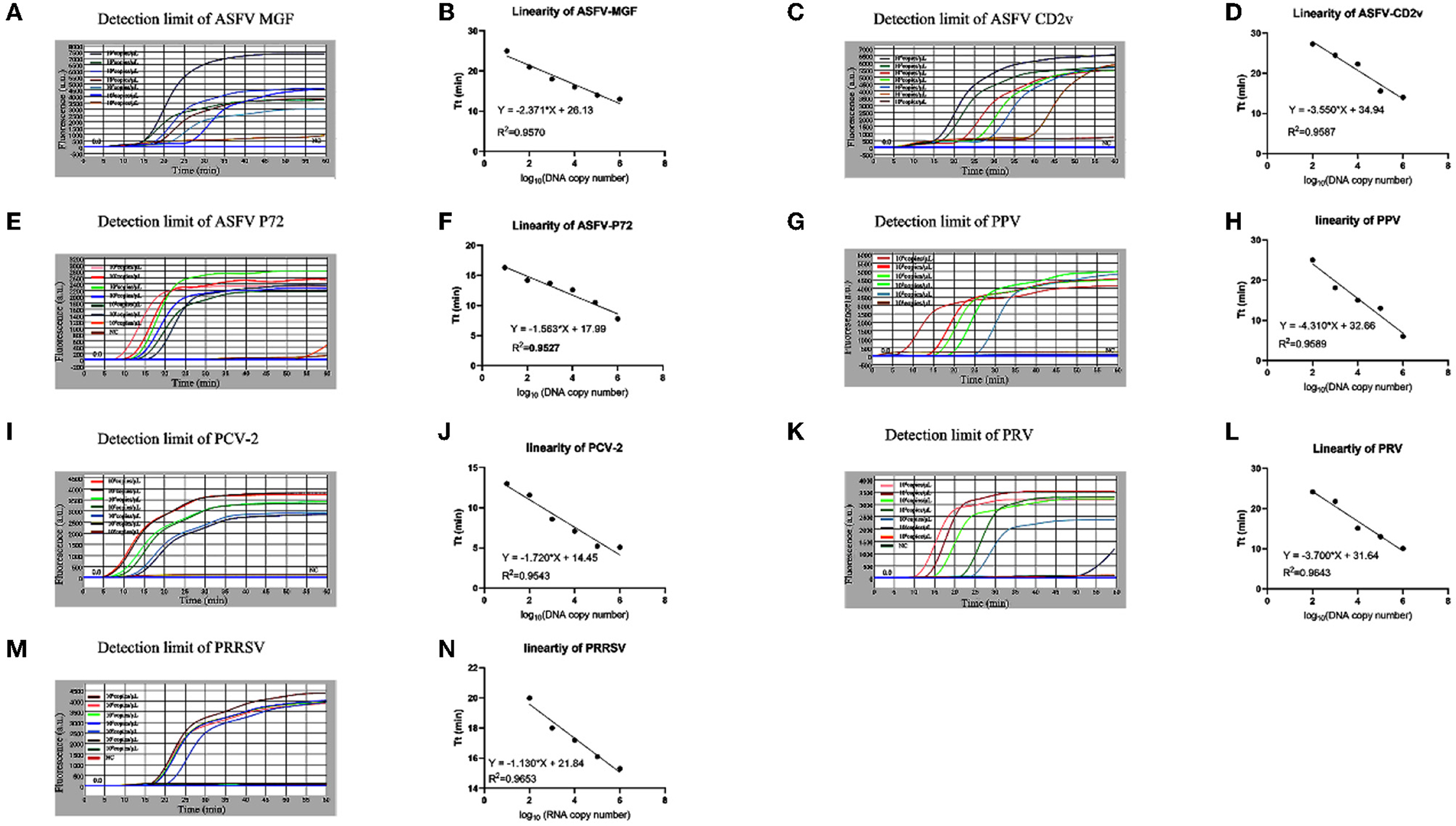
Figure 4. Sensitivity test of the microfluidic-LAMP chip detection system for five swine pathogens. (A–N) All tests were carried out using gradient-diluted standard plasmid of each pathogen. Semi-logarithmic regression between the Tt values and standards concentration by GraphPad Prism 8.3.0.
The high, medium and low detectable concentration standard plasmids were used to test in triplicate to assess the stability of the microfluidic-LAMP chip system (Figure 5). There was a clear gradient from high to low plasmid concentration group of each test, which demonstrated the stability of the established method. To further assess the results, synthetic ASFV DNAs and the nucleic acid extracted from clinical positive samples of PRV, PCV2, PRRSV, and PPV were used for evaluation (Figure 6), which showed the good reproducibility for the developed system. All the C.V. values for the stability and reproducibility tests were < 5% (Table 2), indicating that the microfluidic-LAMP chip system has good reproducibility.
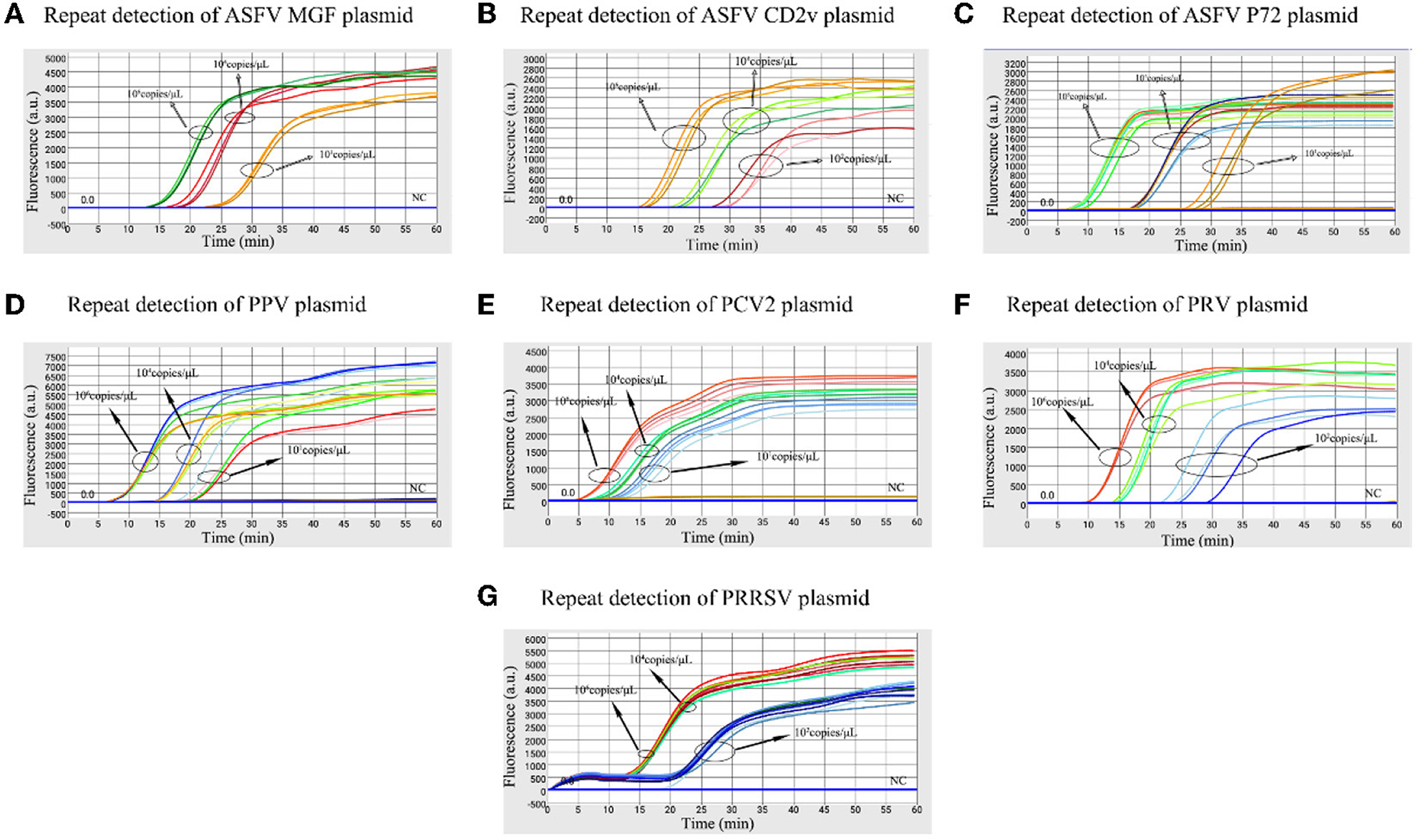
Figure 5. Stability test of the microfluidic-LAMP chip detection system for five swine pathogens. (A–G) All tests were performed using high, medium and low (detection limits) concentration standard plasmids.
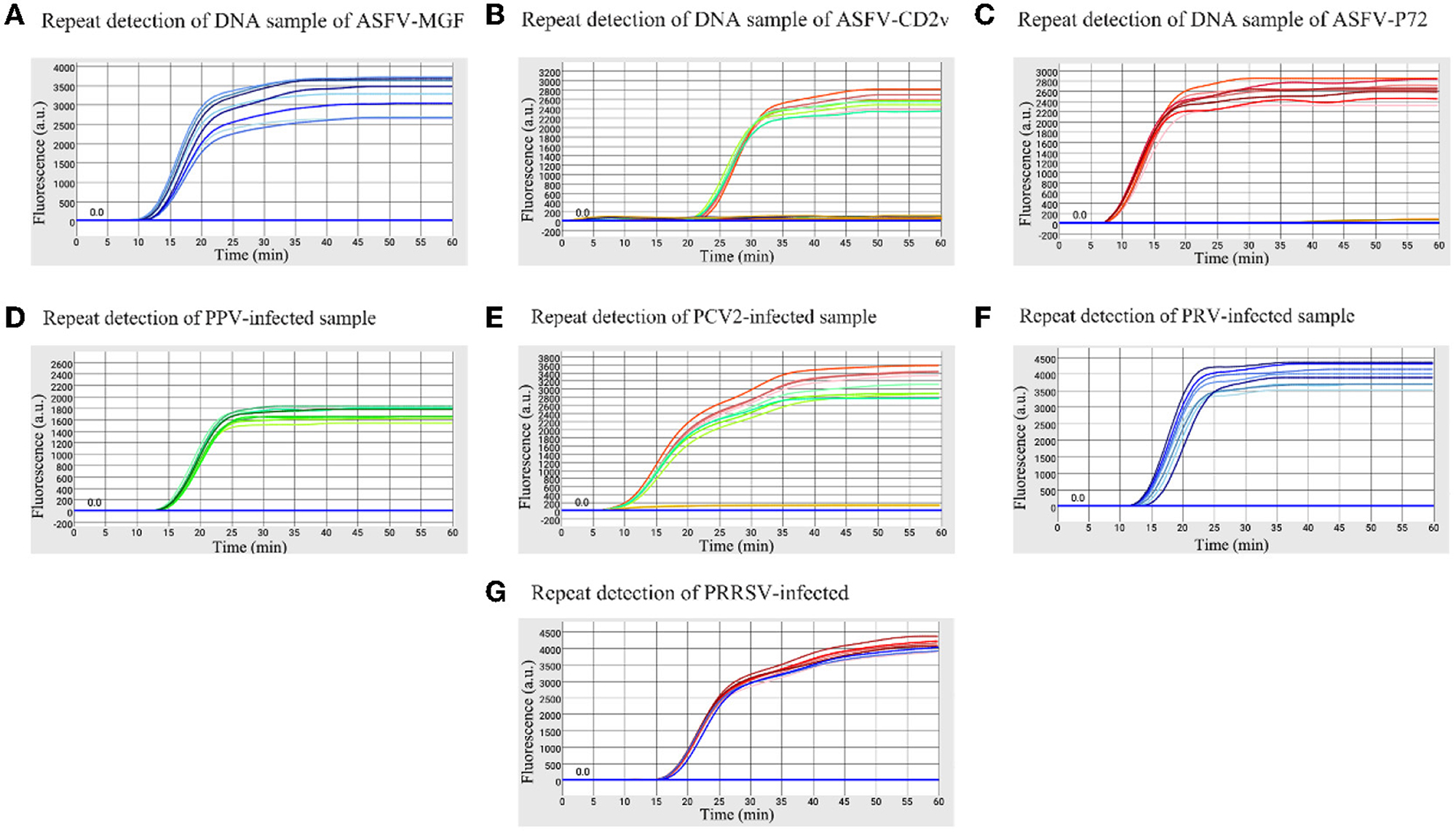
Figure 6. Reproducibility of the microfluidic-LAMP chip detection system for five swine pathogens. (A–C) Stability test of ASFV-MGF, ASFV-CD2v, and ASFV-P72 for eight replicate experiments using a DNA sample. (D–G) Stability test of PRV, PPV, PCV-2, and PRRSV for eight replicate experiments using a clinical virus-infected sample.
To compared the detection performances of the microfluidic chip system and qRT-PCR assays, 213 clinical serum samples and tissue samples (spleens, lungs, lymph nodes, tonsils, brain tissues, nose swabs and kidney tissues) and 15 ASFV positive nucleic acid samples were detected by microfluidic-LAMP chip system and qRT-PCR assay, respectively (Table 3). Testing of clinical samples with the microfluidic-LAMP chip system showed that, among the 146 serum samples, eight samples were positive for PCV-2 and 38 were positive for PRRSV, and there were no co-infections with PCV-2 and PRRSV. Among the 34 lung tissues samples, 14 were positive for PRV and two were positive for PCV-2 and there was no co-infection observed in these 16 PRV and PCV-2 samples. Testing of 10 tonsil tissues showed that six were positive for PRV, one was positive for PCV-2 and one was co-infected with PRV and PCV-2. PRV was detected in remaining 23 samples, accounting for 2 (2/7), 2 (2/3), 3 (3/8), 1 (1/5) of lymph nodes, brain tissues, nose swabs and kidney tissues, respectively. Fifteen nucleic acid samples tested positive for ASFV, no samples had confirmed PPV infection. Of these 92/228 (40.4%) tested positive by the microfluidic-LAMP chip system, which was slightly lower compared to 94/228 (41.2%) positive samples detected by the existing qRT-PCR assay. Except for the PRRSV (95%), the coincidence rate of the detection results of ASFV, PRV, PPV, and PCV-2 between the microfluidic-LAMP chip system and the existing qRT-PCR assay were all 100%. The clinical sample test revealed that PRV, PCV-2 ASFV, and PRRSV can be identified by the established system. The data suggested that the overall prevalence of positive samples was 40.4% (92/228), and PRV was detected in 29.3%, PCV-2 in 12% and PRRSV in 41.3% of the positive samples. The co-infection rate of PCV-2 and PRRSV was 8.7%. The positive detection rate of ASFV samples was 16.3%. The co-infection rate of PCV-2 and PRV was 3.3%. Despite no infection of PPV, these five viruses are still the common pathogens in swine production industry. Notably, the results for all of the 15 ASFV positive nucleic acid samples showed that an amplification curve could only be obtained for the P72 channel, indicating that these positive samples were ASFV wild-type strains. This result was expected since the attenuated strains are not yet used as vaccines (Table 3). The coincidence rate of the detection results of PRV, ASFV, PPV, PCV-2, and PRRSV between the developed system and the existing qRT-PCR assay was 97.2%, indicating the new system can be applied to diagnosis of swine pathogens.
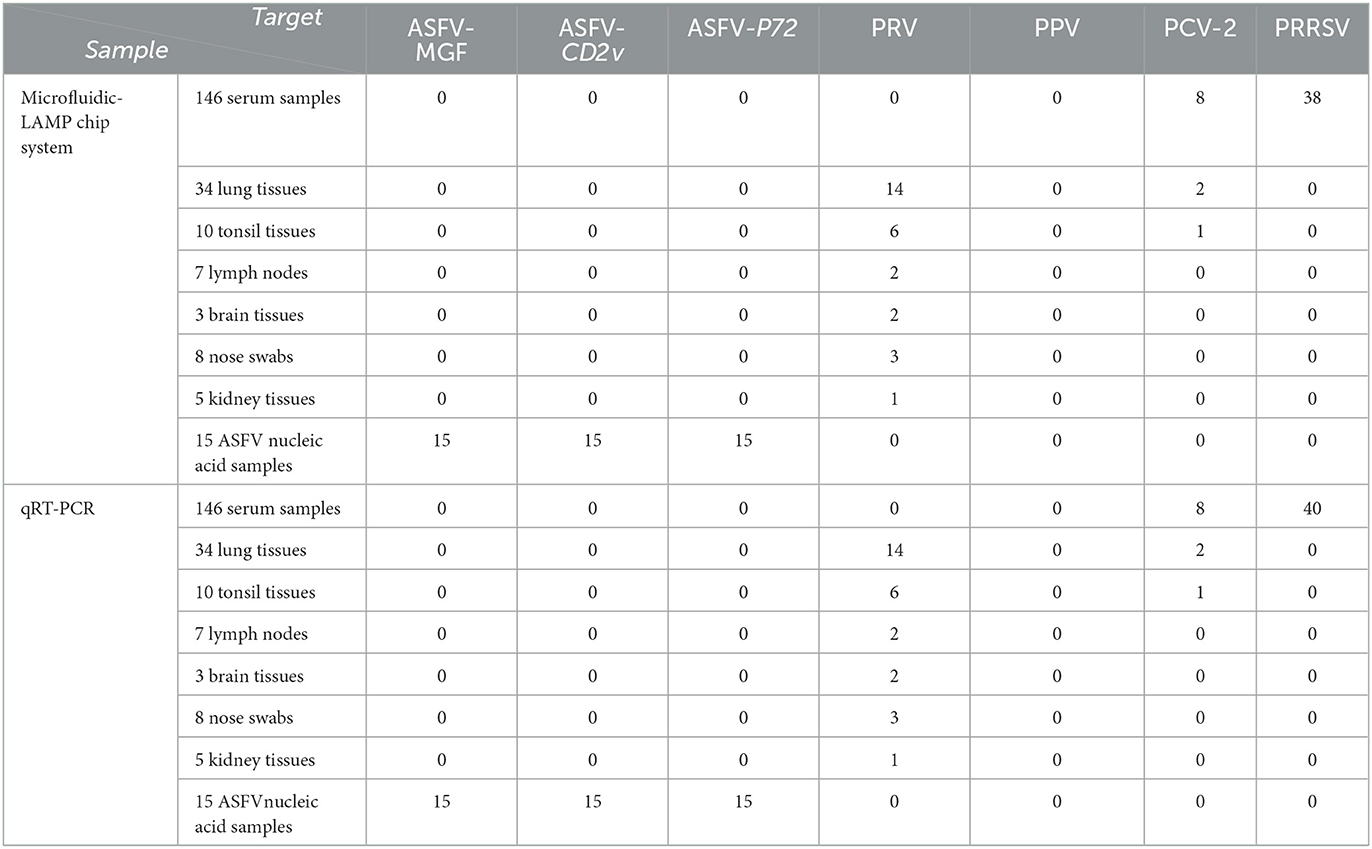
Table 3. Comparisons among the microfluidic- LAMP chip system from detection of PRV, PPV, PCV-2, ASFV, and PRRSV with the existing qRT-PCR assay using clinical samples.
This study describes the development of a low-cost, rapid, field deployable, microfluidic-LAMP chip system for one-step-distribution of samples and real-time loop-mediated isothermal amplification, enabling the simultaneous detection and differentiation of ASFV, PRV, PPV, CSFV, and PCV-2 within 60 min. Pre-immobilization of different primers targeting to different pathogens allow for a broad range of microfluidic-LAMP chip purposefully customized for specific diagnosis, which has implications for clinical application for early detection of swine pathogens. The minimum equipment of our proposed system required only pipettes, reagent tubes, microfluidic chips and a portable detection device, which has great potential for clinical diagnosis outside of the laboratory.
Herein, we have developed a microfluidic-LAMP chip system for synchronous detection and identification of gene-deleted and wild-type ASFV, PRV, PPV, PCV2, and PRRSV on a single chip. The developed system demonstrates the detection limit in the range from 102 to 101 copies/μl, requires < 60 min for the entire analysis and is accurate in differential diagnosis of swine pathogens. Importantly, the microfluidic-LAMP chip system detection time was within 60 min, and the test results could be seen in 40 min at the fastest. The microfluidic-LAMP chip system possessed outstanding advantages in high-throughput, rapid detection, high sensitivity and simplified manipulation that shows its application potential of porcine pathogens diagnosis and control. This is crucial to the minute-by-minute battle in pandemic surveillance.
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding authors.
The animal study was reviewed and approved by South China Agricultural University Experimental Animal Welfare Ethics Committee.
Conceived and designed the experiments: JM, TL, and YS. Performed the experiments: CJ, LZ, YC, and XF. Sample collection: XF, YL, MD, XL, QL, and HW. Analyzed the data: CJ, LZ, YC, YS, and YL. Contributed to the writing: CJ and YC. All authors have read and approved the final manuscript.
In support of this work, the Provincial Key Field Research and Development Plan funded the Research and Application of Accurate Detection Technology of African Swine Fever (No. 201902112019B020211005), the Guangdong Major Project of Basic and Applied Basic Research (No. 2020B0301030007), and the Natural Science Foundation of Guangdong Province (Nos. 2022A1515012473, 2020A1515010295, and 2020A1515010220).
YL and XL were employed by the company Ningbo iGene Technology Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Kedkovid R, Sirisereewan C, Thanawongnuwech R. Major swine viral diseases: an Asian perspective after the African swine fever introduction. Porcine Health Manag. (2020) 6:20. doi: 10.1186/s40813-020-00159-x
2. VanderWaal K, Deen J. Global trends in infectious diseases of swine. Proc Natl Acad Sci U S a. (2018) 115:11495–500. doi: 10.1073/pnas.1806068115
3. Chen NH, Ye MX, Xiao YZ Li S, Huang YC Li XD, et al. Development of universal and quadruplex real-time RT-PCR assays for simultaneous detection and differentiation of porcine reproductive and respiratory syndrome viruses. Transbound Emerg Dis. (2019) 66:2271–78. doi: 10.1111/tbed.13276
4. Lefebvre RC. Fetal mummification in the major domestic species: current perspectives on causes and management. Vet Med. (2015) 6:233–44. doi: 10.2147/VMRR.S59520
5. Gaudreault NN, Madden DW, Wilson WC, Trujillo JD, Richt JA. African swine fever virus: an emerging DNA arbovirus. Front Vet Sci. (2020) 7:215. doi: 10.3389/fvets.2020.00215
6. Tao D, Sun D, Liu Y, Wei S, Yang Z, An T, et al. One year of African swine fever outbreak in China. Acta Trop. (2020) 211:105602. doi: 10.1016/j.actatropica.2020.105602
7. Sun E, Zhang Z, Wang Z, He X, Zhang X, Wang L, et al. Emergence and prevalence of naturally occurring lower virulent African swine fever viruses in domestic pigs in China in 2020. Sci China Life Sci. (2021) 64:752–65. doi: 10.1007/s11427-021-1904-4
8. O'Donnell V, Holinka LG, Gladue DP, Sanford B, Krug PW, Lu XQ, et al. African swine fever virus georgia isolate harboring deletions of MGF360 and MGF505 genes is attenuated in swine and confers protection against challenge with virulent parental virus. J Virol. (2015) 89:6048-56. doi: 10.1128/JVI.00554-15
9. Chen WY, Zhao DM, He XJ, Liu RQ, Wang ZL, Zhang XF, et al. A seven-gene-deleted African swine fever virus is safe and effective as a live attenuated vaccine in pigs. Sci China Life Sci. (2020) 63:623–34. doi: 10.1007/s11427-020-1657-9
10. Borca MV, O'Donnell V, Holinka LG, Risatti GR, Ramirez-Medina E, Vuono EA, et al. Deletion of CD2-like gene from the genome of African swine fever virus strain Georgia does not attenuate virulence in swine. Sci Rep. (2020) 10:494. doi: 10.1038/s41598-020-57455-3
11. O'Donnell V, Holinka LG, Sanford B, Krug PW, Carlson J, Pacheco JM, et al. African swine fever virus Georgia isolate harboring deletions of 9GL and MGF360/505 genes is highly attenuated in swine but does not confer protection against parental virus challenge. Virus Res. (2016) 221:8-14. doi: 10.1016/j.virusres.2016.05.014
12. Gladue DP, O'Donnell V, Ramirez-Medina E, Rai A, Pruitt S, Vuono EA, et al. Deletion of CD2-Like (CD2v) and C-type lectin-LIKE (EP153R) genes from African swine fever virus georgia-9gl abrogates its effectiveness as an experimental vaccine. Viruses. (2020) 12:1185. doi: 10.3390/v12101185
13. Guo ZH, Chen XX, Li R, Qiao SL, Zhang GP. The prevalent status and genetic diversity of porcine reproductive and respiratory syndrome virus in China: a molecular epidemiological perspective. Virol J. (2018) 15:2. doi: 10.1186/s12985-017-0910-6
14. Sun Y, Luo YZ, Wang CH, Yuan J, Li N, Song K, et al. Control of swine pseudorabies in China: opportunities and limitations. Vet Microbiol. (2016) 183:119–24. doi: 10.1016/j.vetmic.2015.12.008
15. Meszaros I, Olasz F, Csagola A, Tijssen P, Zadori Z. Biology of porcine parvovirus (ungulate parvovirus 1). Viruses. (2017) 9:393. doi: 10.3390/v9120393
16. Opriessnig T, Karuppannan AK, Castro A, Xiao CT. Porcine circoviruses: current status, knowledge gaps and challenges. Virus Res. (2020) 286:198044. doi: 10.1016/j.virusres.2020.198044
17. Gervais L, de Rooij N, Delamarche E. Microfluidic chips for point-of-care immunodiagnostics. Adv Mater. (2011) 23:H151–76. doi: 10.1002/adma.201100464
18. Chen YQ, Zhu YZ, Shen MJ, Lu Y, Cheng J, Xu YC. Rapid and automated detection of six contaminants in milk using a centrifugal microfluidic platform with two rotation axes. Anal Chem. (2019) 91:7958–64. doi: 10.1021/acs.analchem.9b01998
19. Lee SK Yi GR, Yang SM. High-speed fabrication of patterned colloidal photonic structures in centrifugal microfluidic chips. Lab Chip. (2006) 6:1171–77. doi: 10.1039/b606448e
20. Zhuang JJ, Yin JX, Lv SW, Wang B, Mu Y. Advanced “lab-on-a-chip” to detect viruses - current challenges and future perspectives. Biosens Bioelectron. (2020) 163:112291. doi: 10.1016/j.bios.2020.112291
21. McIntosh KA, Tumber A, Harding J, Krakowka SK, Ellis JA, Hill JE. Development and validation of a SYBR green real-time PCR for the quantification of porcine circovirus type 2 in serum, buffy coat, feces, and multiple tissues. Vet Microbiol. (2009) 133:23–33. doi: 10.1016/j.vetmic.2008.06.010
22. Bian PY, Ye C, Zheng XY, Yang J, Ye W, Wang Y, et al. Mesenchymal stem cells alleviate Japanese encephalitis virus-induced neuroinflammation and mortality. Stem Cell Res Ther. (2017) 8:38. doi: 10.1186/s13287-017-0486-5
23. Gil S, Sepulveda N, Albina E, Leitao A, Martins C. The low-virulent African swine fever virus (ASFV/NH/P68) induces enhanced expression and production of relevant regulatory cytokines (IFN alpha, TNF alpha and IL12p40) on porcine macrophages in comparison to the highly virulent ASFV/L60. Arch Virol. (2008) 153:1845–54 doi: 10.1007/s00705-008-0196-5
24. Huang J, Qi YH, Wang AT, Huang C, Liu X, Yang X, et al. Porcine beta-defensin 2 inhibits proliferation of pseudorabies virus in vitro and in transgenic mice. Virol J. (2020) 17:18. doi: 10.1186/s12985-020-1288-4
25. Sun P, Bai CX, Zhang D, Wang J, Yang KK, Cheng BZ, et al. SYBR Green-based real-time polymerase chain reaction assay for detection of porcine parvovirus 6 in pigs. Pol J Vet Sci. (2020) 23:197–202. doi: 10.24425/pjvs.2020.132766
26. Zheng HH, Zhang SJ, Cui JT, Zhang J, Wang L, Liu F, et al. Simultaneous detection of classical swine fever virus and porcine circovirus 3 by SYBR green I-based duplex real-time fluorescence quantitative PCR. Mol Cell Probes. (2020) 50:101524. doi: 10.1016/j.mcp.2020.101524
27. Zheng LL, Chai LY, Tian RB, Zhao Y, Chen HY, Wang ZY. Simultaneous detection of porcine reproductive and respiratory syndrome virus and porcine circovirus 3 by SYBR Green capital I, Ukrainian-based duplex real-time PCR. Mol Cell Probes. (2020) 49:101474. doi: 10.1016/j.mcp.2019.101474
28. Teklue T, Wang T, Luo YZ, Hu RL, Sun Y, Qiu HJ. Generation and evaluation of an African swine fever virus mutant with deletion of the CD2v and UK genes. Vaccines. (2020) 8:763. doi: 10.3390/vaccines8040763
29. Lin YX, Cao CF, Shi WJ, Huang CH, Zeng SL, Sun J, et al. Development of a triplex real-time PCR assay for detection and differentiation of gene-deleted and wild-type African swine fever virus. J Virol Methods. (2020) 280:113875. doi: 10.1016/j.jviromet.2020.113875
30. Chen N, Huang Y, Ye M, Li S, Xiao Y, Cui B, et al. Co-infection status of classical swine fever virus (CSFV), porcine reproductive and respiratory syndrome virus (PRRSV) and porcine circoviruses (PCV2 and PCV3) in eight regions of China from 2016 to 2018. Infect Genet Evol. (2019) 68:127–35. doi: 10.1016/j.meegid.2018.12.011
31. Cheng TY, Henao-Diaz A, Poonsuk K, Buckley A, van Geelen A, Lager K, et al. Pseudorabies (Aujeszky's disease) virus DNA detection in swine nasal swab and oral fluid specimens using a gB-based real-time quantitative PCR. Prev Vet Med. (2021) 189:105308. doi: 10.1016/j.prevetmed.2021.105308
32. Henriques AM, Duarte M, Barros SC, Fagulha T, Ramos F, Luis T, et al. Development and validation of a real-time PCR for the detection and quantification of porcine circovirus type 2. Virusdisease. (2018) 29:355–61. doi: 10.1007/s13337-018-0476-y
33. Kim SC, Jeong CG, Nazki S, Lee SI, Baek YC, Jung YJ, et al. Evaluation of a multiplex PCR method for the detection of porcine parvovirus types 1 through 7 using various field samples. PloS ONE. (2021) 16:e0245699. doi: 10.1371/journal.pone.0245699
34. Mee PT, Wong SN, O'Riley KJ, Da Conceicao F, Jong J, Phillips DE, et al. Field verification of an African swine fever virus loop-mediated isothermal amplification (LAMP) assay during an outbreak in timor-leste. Viruses. (2020) 12:1444. doi: 10.3390/v12121444
35. Wang DG, Yu JH, Wang YZ, Zhang M, Li P, Liu M, et al. Development of a real-time loop-mediated isothermal amplification (LAMP) assay and visual LAMP assay for detection of African swine fever virus (ASFV). J Virol Methods. (2020) 276:113775. doi: 10.1016/j.jviromet.2019.113775
36. Yuan XF, Lv JZ, Lin XM, Zhang CY, Deng JH, Wang CX, et al. Multiplex detection of six swine viruses on an integrated centrifugal disk using loop-mediated isothermal amplification. J Vet Diagn Invest. (2019) 31:415–25. doi: 10.1177/1040638719841096
37. Zhou DN, Wang SS, Yang KL, Liu X, Liu W, Guo R, et al. Rapid and simultaneous detection of Japanese encephalitis virus by real-time nucleic acid sequence-based amplification. Microb Pathog. (2021) 150:104724. doi: 10.1016/j.micpath.2020.104724
Keywords: multiplex detection, microfluidic chip, loop-mediated isothermal amplification (LAMP), portable diagnostic tool, pathogens
Citation: Ji C, Zhou L, Chen Y, Fang X, Liu Y, Du M, Lu X, Li Q, Wang H, Sun Y, Lan T and Ma J (2023) Microfluidic-LAMP chip for the point-of-care detection of gene-deleted and wild-type African swine fever viruses and other four swine pathogens. Front. Vet. Sci. 10:1116352. doi: 10.3389/fvets.2023.1116352
Received: 04 January 2023; Accepted: 01 February 2023;
Published: 16 February 2023.
Edited by:
Bui Thi To Nga, Vietnam National University of Agriculture, VietnamReviewed by:
Tereza Cristina Cardoso, Universidade Estadual de São Paulo, BrazilCopyright © 2023 Ji, Zhou, Chen, Fang, Liu, Du, Lu, Li, Wang, Sun, Lan and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jingyun Ma,  bWFqeTI0MDBAc2NhdS5lZHUuY24=; Tian Lan,
bWFqeTI0MDBAc2NhdS5lZHUuY24=; Tian Lan,  bGFudGlhbjIwMTZAc2NhdS5lZHUuY24=; Yuan Sun,
bGFudGlhbjIwMTZAc2NhdS5lZHUuY24=; Yuan Sun,  c3VueXVhbkBzY2F1LmVkdS5jbg==
c3VueXVhbkBzY2F1LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.