- 1Faculty of Veterinary Medicine, Chattogram Veterinary and Animal Sciences University, Chattogram, Bangladesh
- 2Institute of Epidemiology, Disease Control and Research (IEDCR), Dhaka, Bangladesh
- 3EcoHealth Alliance, New York, NY, United States
- 4School of Veterinary Medicine, Texas Tech University, Amarillo, TX, United States
- 5Queensland Alliance for One Health Sciences, School of Veterinary Science, The University of Queensland, Brisbane, QLD, Australia
- 6Antimicrobial Resistance Action Center (ARAC), Bangladesh Livestock Research Institute, Savar, Bangladesh
- 7Directorate General of Health Services, Dhaka, Bangladesh
Introduction: Antimicrobial resistance (AMR) is a growing global health threat for humans and animals. Environmental contamination of antimicrobials from human and domestic animal feces has been linked to AMR in wildlife populations, including rhesus macaques. This study aimed to describe the eco-epidemiology of AMR within Salmonella and Staphylococcus species isolated from rhesus macaques.
Methods: We followed macaque groups for 4 h per day (2 days) to observe the direct and indirect contact rate and type between macaques and people and livestock. We collected 399 freshly defecated, non-invasive fecal samples from macaques at seven sites in Bangladesh in January–June 2017. Bacterial isolation and identification were conducted using culture, biochemical characteristics, and polymerase chain reaction (PCR). An antimicrobial susceptibility test (AST) for 12 antimicrobials for each organism was conducted using the Kirby–Bauer disc diffusion method.
Results: The overall prevalence of Salmonella spp. and Staphylococcus spp. in rhesus macaques was 5% (n = 18; 95% CI: 3–7%) and 16% (n = 64; 95% CI: 13–20%), respectively. All the isolated Salmonella spp. and most of the Staphylococcus spp. (95%; 61/64; 95% CI: 86.9–99%) were resistant to at least one antimicrobial. The odds of a fecal sample having antimicrobial-resistant Salmonella spp (OR = 6.6; CI: 0.9–45.8, P = 0.05) and Staphylococcus spp. (OR = 5.6; CI: 1.2–26, P = 0.02) were significantly higher in samples collected at peri-urban sites than those collected at rural and urban sites. Salmonella spp. were most frequently resistant to tetracycline (89%), azithromycin (83%), sulfamethoxazole-trimethoprim (50%), and nalidixic acid (44%). Staphylococcus spp. were found to be highly resistant to ampicillin (93%), methicillin (31%), clindamycin (26%), and rifampicin (18%). Both bacterial species produced colonies with multidrug resistance to up to seven antimicrobials. Direct and indirect contact rates (within 20 m for at least 15 min) and resource sharing between macaques and people were higher in urban sites, while macaque-livestock contact rates were higher in rural sites.
Discussion: The study shows that resistant microorganisms are circulating in rhesus macaque, and direct and indirect contact with humans and livestock might expand the resistant organisms.
1. Introduction
Antimicrobial-resistant bacteria represent a critical health threat to people, domestic animals, and wildlife. Antimicrobial resistance (AMR) is recognized as a significant emerging public health concern by the World Health Organization (WHO) and represents a significant challenge to human and veterinary medicine due to the therapeutic failure of life-saving treatments (1). Natural ecosystems are believed to be contaminated with antimicrobial-resistant bacteria due to excessive antimicrobial use in clinical and agriculture settings (2). Antibiotics are used for therapeutic, prophylactic, and growth promotion purposes in livestock and poultry (3–5). Further, the application of manure as fertilizer for crop growth has been associated with AMR, regardless of whether the animals that produced the manure were treated with antibiotics, because the manure enriches the environment, allowing the growth of resident soil bacteria that harbor AMR elements (6). In Bangladesh, it is common for people to use antimicrobials without consulting with a registered doctor or veterinarian to treat themselves or other people and animals. Indiscriminate use of antimicrobials in multiple health sectors accelerates the development of antimicrobial resistance in bacteria. A One Health approach is important to identify the human, animal, and environmental factors associated with increasing levels of AMR (7).
Globalization has driven AMR to become a global problem. It is known that the movement of people and animals can permit the rapid spread of resistant pathogens (8). Asia is considered an epicenter of AMR, especially in densely populated South Asian countries such as Bangladesh and India (9). Multidrug-resistant genes are prevalent in many commensal and environmental bacteria, including Escherichia coli, Klebsiella pneumonia, Salmonella, Enterococcus, and Staphylococcus aureus (10). Multiple studies have revealed that bacterial resistance genes, which are frequently detected in human or animal microbiota, are spreading to environments and wild animals where antimicrobials have not previously been used (11).
Antimicrobial-resistant microorganisms in wildlife are frequently detected (12). Bacteria resistant to antimicrobials have been detected in various wild animal species (13) and wild birds (14). Wildlife has been implicated as a potential reservoir of resistant bacteria and resistance genes (15). Wild animals are rarely treated with antimicrobials. Thus, the source of antimicrobial-resistant bacteria in wildlife is not clear. Some proposed transmission routes are the consumption of human feces, household and kitchen waste, and sewage water (16). Several studies in Bangladesh have already proved the presence of resistant microorganisms in household water supply (17) and environmental effluents (7, 18). Potential exposure to resistant bacteria likely depends on host species, as some species have more frequent contact with humans, human landscapes, or domestic animals than others (19). Antimicrobial-resistant bacteria have been found in 54% of wild rodents living in proximity to livestock harbored bacteria resistant to at least one antimicrobial agent (20).
Rhesus macaques (Macaca mulatta) are synanthropic, thriving in human-altered environments, allowing them to become one of the most widely distributed and successful primates (21). Macaques are distributed throughout Bangladesh, living in urban, rural, and forested/protected areas. Feeroz et al. (22) described non-human primate habitats in the northeastern and southeastern parts of Bangladesh. One study estimated that there were 5,313 macaques in Bangladesh, with group sizes varying from 10 to 78 animals (23).
It has been estimated that up to 61% of monkeys and great apes in sub-Saharan Africa (Gabon and Ivory Coast) are infected with Staphylococcus aureus (24). Among captive rhesus macaques, infections with Staphylococcus spp. in the wild non-human primate are also common, with a prevalence of 39% rhesus macaques in the Netherlands (25) and 23.6% monkeys in Wisconsin, USA (26). Other potential human bacterial pathogens have been identified in multiple primate species. This includes human-habituated mountain gorillas (Gorilla gorilla beringei) in Uganda, with prevalences of 19, 13, and 6% for Campylobacter spp., Salmonella spp., and Shigella spp., respectively (27). Evidence of Escherichia coli sharing between people, domestic animals, and great apes has been reported in densely human-populated areas of western Uganda. Within Uganda, habituated groups of wild apes are visited daily by researchers and tourists [e.g., chimpanzees (28) and mountain gorillas at Bwindi Impenetrable National Park (29)].
Several studies have identified antimicrobial-resistant bacteria in non-human primates. A study indicated that 75% of Staphylococcus aureus isolates from captive rhesus macaques in the United States were resistant to methicillin/oxacillin (30). Campylobacter spp. were isolated from 15, 36, and 67% of cynomolgus macaques (Macaca fascicularis) in China, Cambodia, and Indonesia, respectively, and resistance to tetracycline, ciprofloxacin, and erythromycin was prevalent in China (80%) and Cambodia (75%), as well as in Indonesia (100%) (31). Rolland et al. (16) found that non-human primates (baboons) feeding on human garbage and in contact with human feces had significantly greater levels of antibiotic-resistant gut bacteria than their counterparts.
In wildlife, direct and indirect interspecies contacts are likely critical routes of transmission of antimicrobial-resistant bacteria (32). These routes include consuming food and water contaminated by antimicrobial residues or resistant bacteria within or through indirect contact with resistant bacteria in a contaminated environment (15). Wildlife at zoos and safari parks may potentially be administered antimicrobials directly. Areas with high rates of interactions between people or livestock and non-human primates are recognized as potential drivers of inter-species pathogens transmission (33).
AMR dynamics has a potential role in human, animal, and environmental health by increasing zoonotic diseases and microorganisms, but knowledge of AMR research in wildlife is limited. It has been hypothesized that the occurrence of antibiotic-resistant strains in untreated, free-ranging wild animals is a consequence of bacterial transmission with people or domestic animals which have undergone antibiotic treatment. Here we evaluated the prevalence of two bacteria (Salmonella spp. and Staphylococcus spp.) and their antimicrobial resistance levels in rhesus macaques from different districts and habitats.
2. Materials and methods
2.1. Ethical approval
Ethical approval was approved by Chattogram Veterinary and Animal Sciences University-Animal Experimentation Ethics Committee (AEEC), Chattogram, Bangladesh [permit number: CVASU/Dir (R&E) AEEC/2015/751] to conduct the study. With the help of a defined protocol, we assured the animal ethics and animal safety as well as the safety of working personnel in both field and laboratory throughout the whole study period.
2.2. Study areas, study design, and period
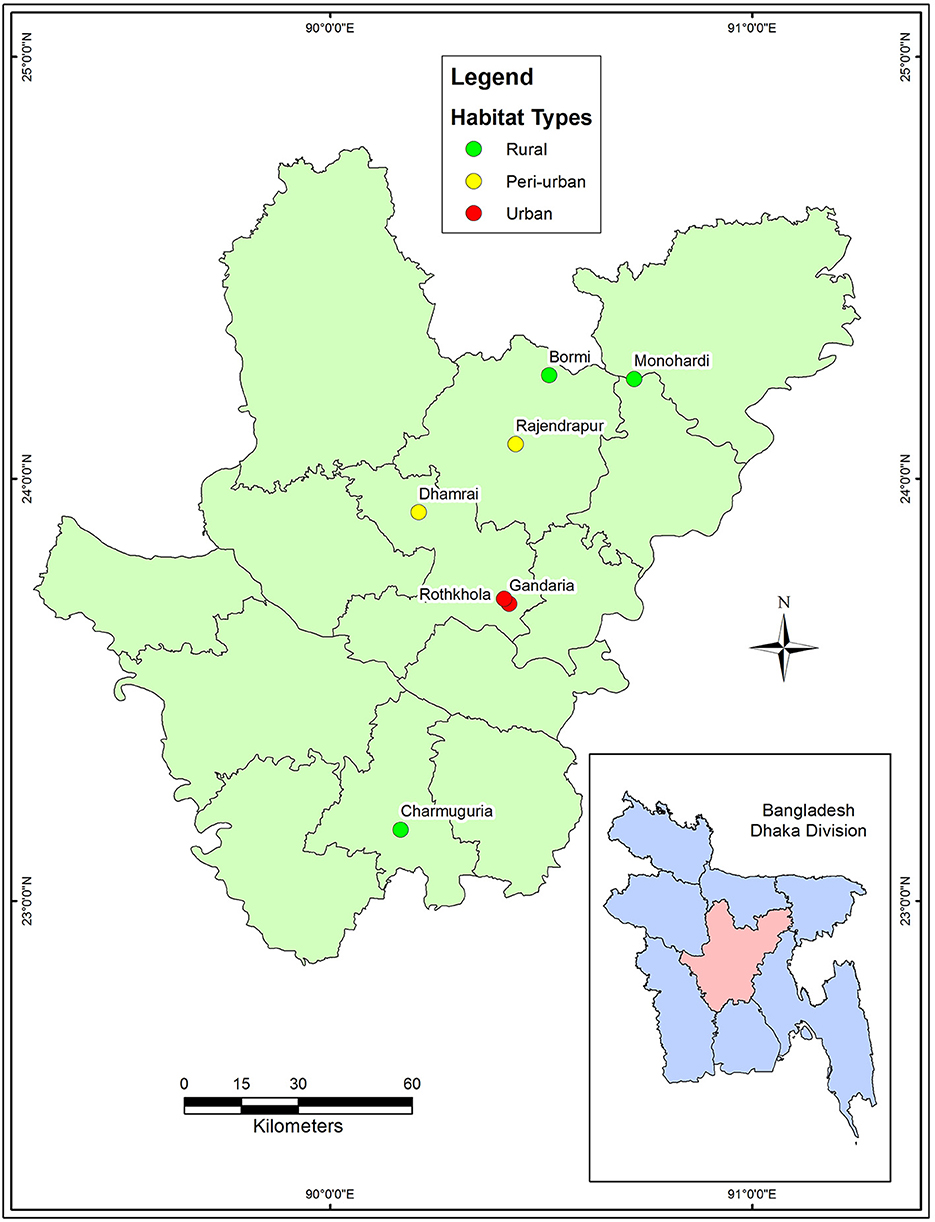
A cross-sectional study was conducted from January to June 2017. The macaque population in Bangladesh is distributed among urban, peri-urban, and rural habitats where they live close to human settlements. We selected seven sites from four districts: Dhaka (Gendaria, Rothkhola, and Dhamrai), Madaripur (Charmuguria), Gazipur (Rajendrapur and Bormi), and Narshingdi (Monohardi) (Figure 1). The distribution of rhesus macaque populations in Dhaka, Madaripur, Narshingdi, and Gazipur districts, where temples and shrines had been built from the earliest stages of civilization (34). Nevertheless, the macaque population is prevalent in these regions, thus we chose these sampling locations for our study. The sampling map was plotted using the spatial analyst tool of ArcGIS (ArcMap, version 10.2, Environmental Systems Research Institute, Redlands, California, CA, USA).
2.3. Sample collection
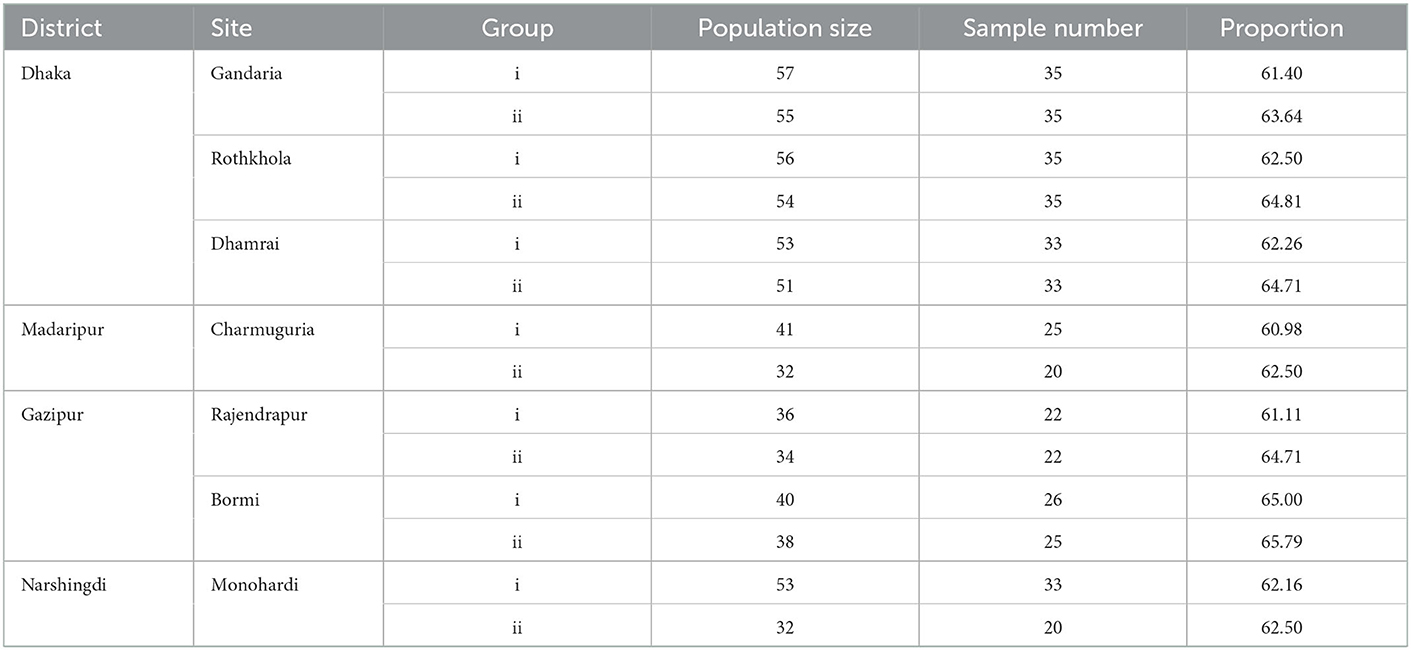
The required sample size was calculated to be 385 using the formula given by Daniel and Cross (35). We collected a total of 399 samples. Samples were collected proportionally (>60%) from each site's macaque population (Table 1). Macaques were baited by provisioning bread and bananas before sample collection. We observed defecation and immediately collected fecal samples (~1 gm) using convenience sampling. All samples were collected within 30 min to avoid re-sampling the same individual. Four trained personnel collected the samples and identified the individual animal's sex and age. Each monkey was seen defecating during the sampling period by two trained persons. To prevent duplication sampled monkey was continuously observed until the sampling was completed (21). Confirmation of the age was also done by the fecal lobe diameter (27). Fecal samples were collected by sterile swab and placed in a falcon tube (15 ml) containing buffered peptone water (Oxoid, Basingstoke, Hampshire, UK) for Salmonella spp. (36) and Mueller-Hinton broth for Staphylococcus spp. (37). Falcon tubes were placed in the ice box, and within 24 hours, they were sent to Bangladesh Livestock Research Institute (BLRI), Savar, for laboratory analysis.
2.4. Data collection
We made field observations to collect data on population structure age (adult, sub-adult, and infant), and sex and ecological data: types of habitats (rural, urban and peri-urban), season [winter (January to March), and summer (April to June)] and GPS coordinates. We also observed the interactions rhesus macaques had with people, domestic animals, and sampling sites for 2 h in the morning and 2 h in the evening for 2 days (a total of 8 h). We identified the frequency and type of both direct contact and indirect contact. Direct contact was defined as any type of inter-species touching/contact, biting, and scratching. Whereas, indirect contact was recorded if the macaques remained within 20 meters of another species for at least 15 min.
2.5. Laboratory testing
2.5.1. Isolation and identification of bacteria
Salmonella spp. was cultured in xylose lysine deoxycholate (XLD) agar (producing a black centered colony) and on brilliant green agar (BGA) media (producing red-colored colony) (38), Staphylococcus spp was cultured on mannitol salt agar producing yellow color and cocci-shaped gram-positive clusters (37). All the positive samples were confirmed using biochemical assays: triple sugar iron (TSI) agar slant (39), and carbohydrate fermentation test (40) were used for Salmonella spp., and the coagulase (41) and catalase test (42) were used for Staphylococcus spp. Finally, PCR was conducted for final confirmation of a bacterial genus Salmonella with Salmonella genus-specific primers ST-11 (5′ -AGCCAACCATTGCTAAATTGGCGCA-3′) and ST-15(5′-TGGTAGAAATTCCCAGCGGGTACTG-3′ described by Gouws et al. (43) and Staphylococcus with Staphylococcus genus-specific primers TStaG422 (5′-GGC CGT GTT GAA CGT GGT CAA ATC A-3′) and TStag765 (5′-TIA CCA TTT CAG TAC CTT CTG GTA A-3′) described by Martineau et al. (44).
2.5.2. Antimicrobial susceptibility test
A panel of 12 antibiotics was used for each organism: chloramphenicol (CPL) (30 μg), gentamicin (GEN) (10 μg), sulfamethoxazole-trimethoprim (SXT) (25 μg), and tetracycline (TET) (30 μg) were used for both genera. In addition, amoxicillin-clavulanic acid (AMC) (30 μg), azithromycin (AZM) (15 μg), cefixime (CFM) (5 μg), cefotaxime (CTX) (30 μg), ceftriaxone (CTR) (30 μg), ciprofloxacin (CIP) (5 μg), imipinem (IPM) (10 μg), nalidixic acid (NA) (30 μg) were used for Salmonella spp. For Staphylococcus, we used: ampicillin (AMP) (10 μg), clindamycin (CDA) (2 μg), linezolid (LZD) (30 μg), methicillin (MET) (5 μg), oxacillin (OXA) (1 μg), rifampicin (RMP) (5 μg), streptomycin (STP) (10 μg), tigecyclin (TGC) (15 μg). We used the Kirby-Bauer disc diffusion method for antimicrobial susceptibility testing (45). Zone diameters were calculated and interpreted as resistant, intermediate, and sensitive according to the size of inhibition following the Clinical and Laboratory Standards Institute (CLSI) guideline, 25th edition (46).
2.6. Statistical evaluation
Field and laboratory data were entered into MS Excel (Excel 2013, Microsoft Corporation, USA). After checking data integrity, it was exported to STATA/IC13 (StataCorp 4905, Lakeway Drive, College Station, Texas 77845, USA) for epidemiological analysis. The prevalence of the bacteria and antimicrobial-resistant bacteria were expressed as a percentage with a 95% confidence interval (CI). If any microorganism was found to be resistant to a single antimicrobial, it was considered resistant. Antimicrobial susceptibility was expressed as a percentage according to resistance, intermediate, and sensitivity. Variables were assessed for collinearity. A univariate Chi-square test was done to identify potential risk factors for the presence of AMR. The significant risk factors (P < 0.2) were included in multiple logistic regression models, but the district was omitted for Salmonella spp. due to collinearity. Model adequacy was verified by the lowest AIC after dropping variables from the model. Confounders were confirmed by observing the variation in the coefficient (β > 10%). The final model was assessed using a receiver operating characteristic curve (ROC) and goodness of fit tests (47). The results were expressed as odds ratios (ORs) and 95% CI.
3. Results
We sampled 399 macaques. The overall prevalence of Salmonella spp. and Staphylococcus spp. in macaques were 5% (n = 18; 95%CI: 2.7–7.1%) and 16% (n = 64; 95%CI: 12.6–20%), respectively. All of the Salmonella spp. and 95% of the Staphylococcus spp. (n = 61; 95%CI: 86.9–99%) isolated in the study were resistant to at least one antimicrobial.
3.1. Descriptive statistics for macaques carrying antimicrobial-resistant Salmonella spp.
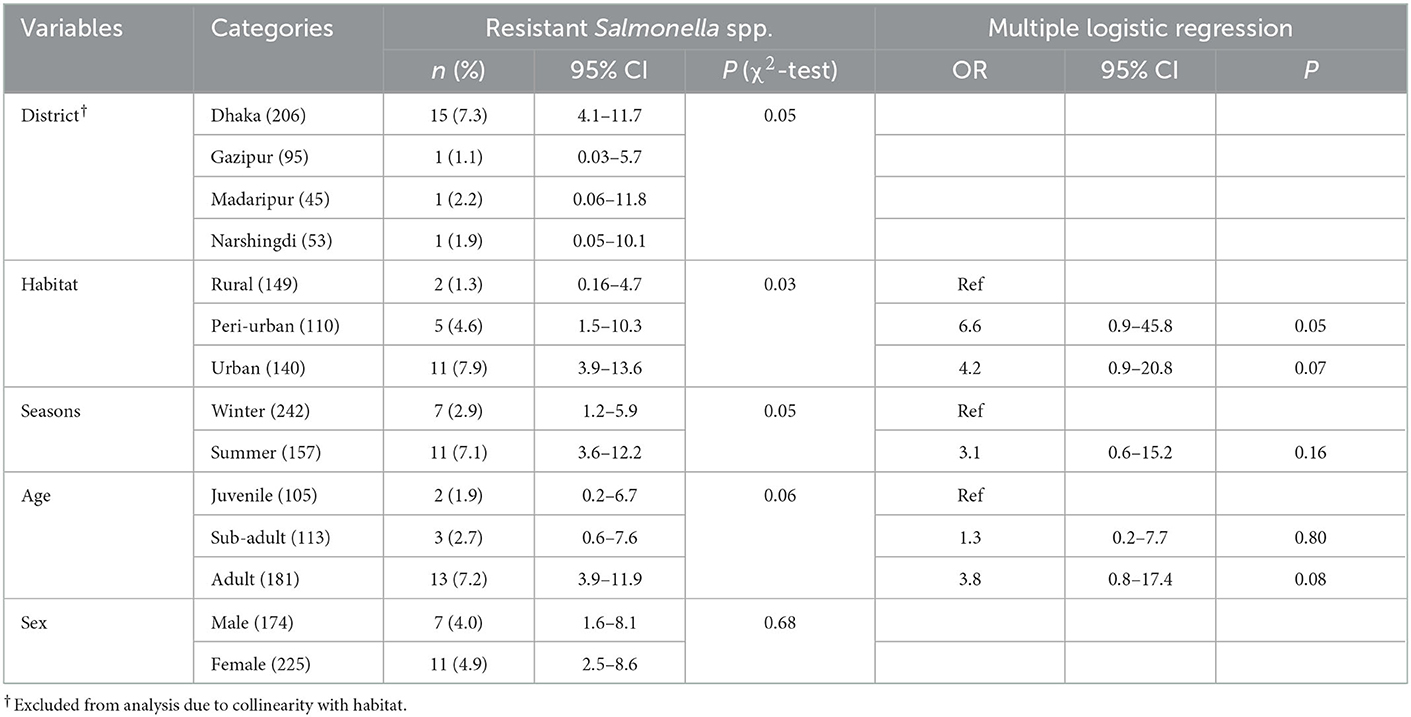
Among all macaques, the prevalence of resistant Salmonella spp. was highest in macaques in the Dhaka District (n = 15; 7.3%; 95%CI: 4.1–11.7), whereas the lowest prevalence (n = 1; 1.1%; 95%CI: 0.03–5.7) was found in Gazipur (see Table 2 for all descriptive statistics). Habitat, season, and age were included in the multivariable regression, and none of the variables were significant in the multiple logistic regression (Table 2).
3.2. Descriptive statistics and risk factors for macaques carrying antimicrobial-resistant Staphylococcus spp.
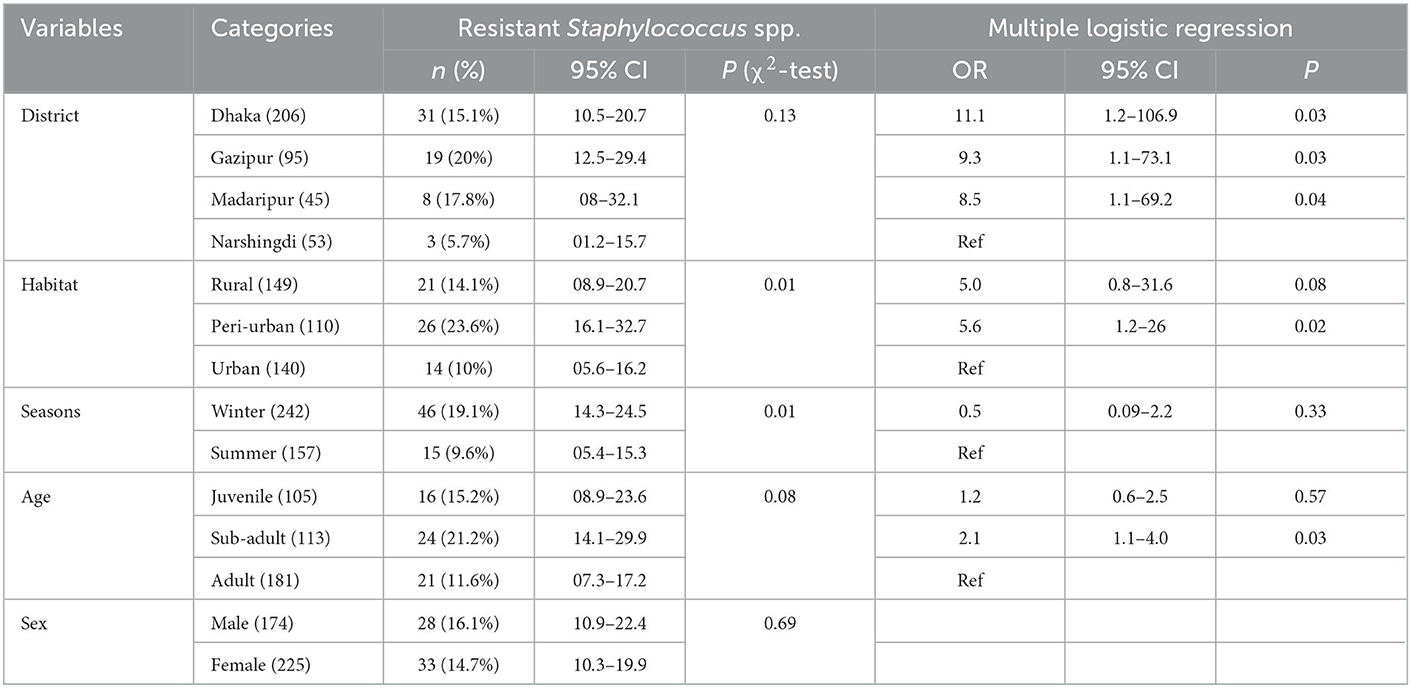
In univariate analysis, only sex did not meet the selection criteria to be included in the multivariable analysis. In the multivariable analysis, we found that macaques sampled in Dhaka (OR = 11.1, CI: 1.2–106.9, P = 0.03), Gazipur (OR = 9.3, CI: 1.1–73.1, P = 0.03) and Madaripur (OR = 8.5, CI: 1.1–69.2, P = 0.04) had significantly higher odds of carrying antimicrobial resistant Staphylococcus spp, compared to animals living in Narshingdi (Table 3). Additionally, macaques sampled in a peri-urban habitat (OR = 5.6, CI: 1.2–26, P = 0.02) and those that were sub-adults (OR = 2.1, CI: 1.1–4.0, P = 0.03) had higher odds of infection (Table 3).
3.3. AMR patterns and multidrug resistance of Salmonella spp. in rhesus macaque
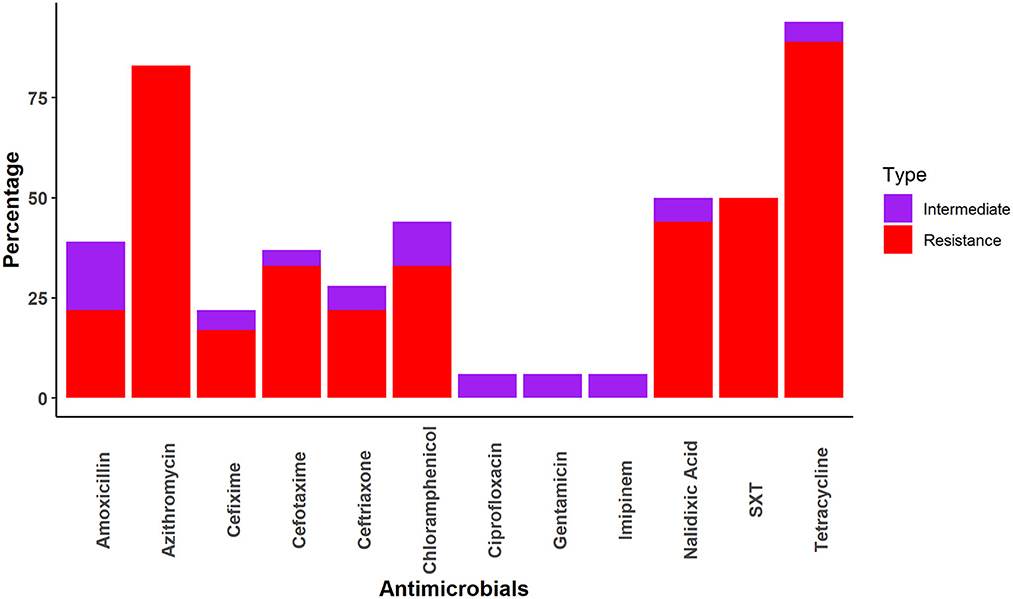
Salmonella spp. was most frequently resistant to tetracycline (TET) (89%), followed by azithromycin (AZM) (83%) and cefixime (17%). However, Salmonella spp. demonstrated no resistance to ciprofloxacin, gentamicin, or imipenem (Figure 2).
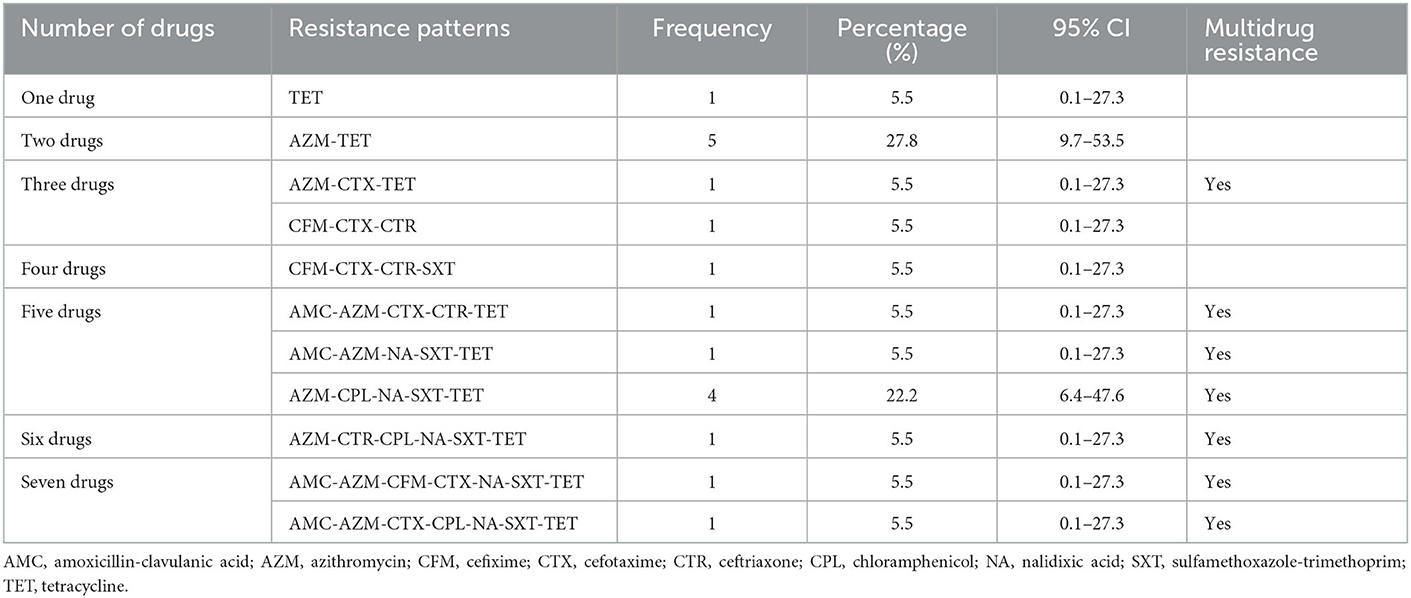
A total of seven combinations of drugs were found to be resistant to Salmonella spp. Two combinations of multiple drugs were found most frequently for Salmonella spp.; AZM-TET (27.8%; 95%CI: 9.7–53.5) and AZM-CPL-NA-SXT-TET (22.2%; 95%CI: 6.4–47.6). All other combinations of multidrug resistance were detected in a single sample (Table 4).
3.4. AMR patterns and multidrug resistance of Staphylococcus spp. in rhesus macaque
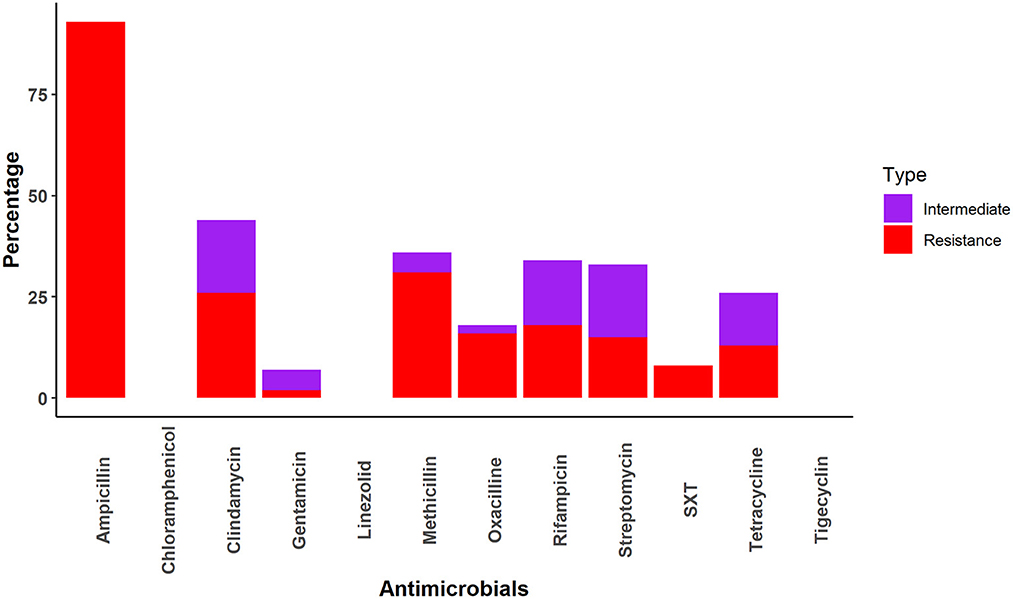
All Staphylococcus samples were resistant to at least one drug. The Staphylococcus samples were most frequently resistant to ampicillin (93%), followed by methicillin (31%), clindamycin (26%), rifampicin (18%), oxacillin (16%), streptomycin (15%), tetracycline (13%) and sulfamethoxazole-trimethoprim (8%). No resistance was identified against chloramphenicol, linezolid, or tetracycline (Figure 3).
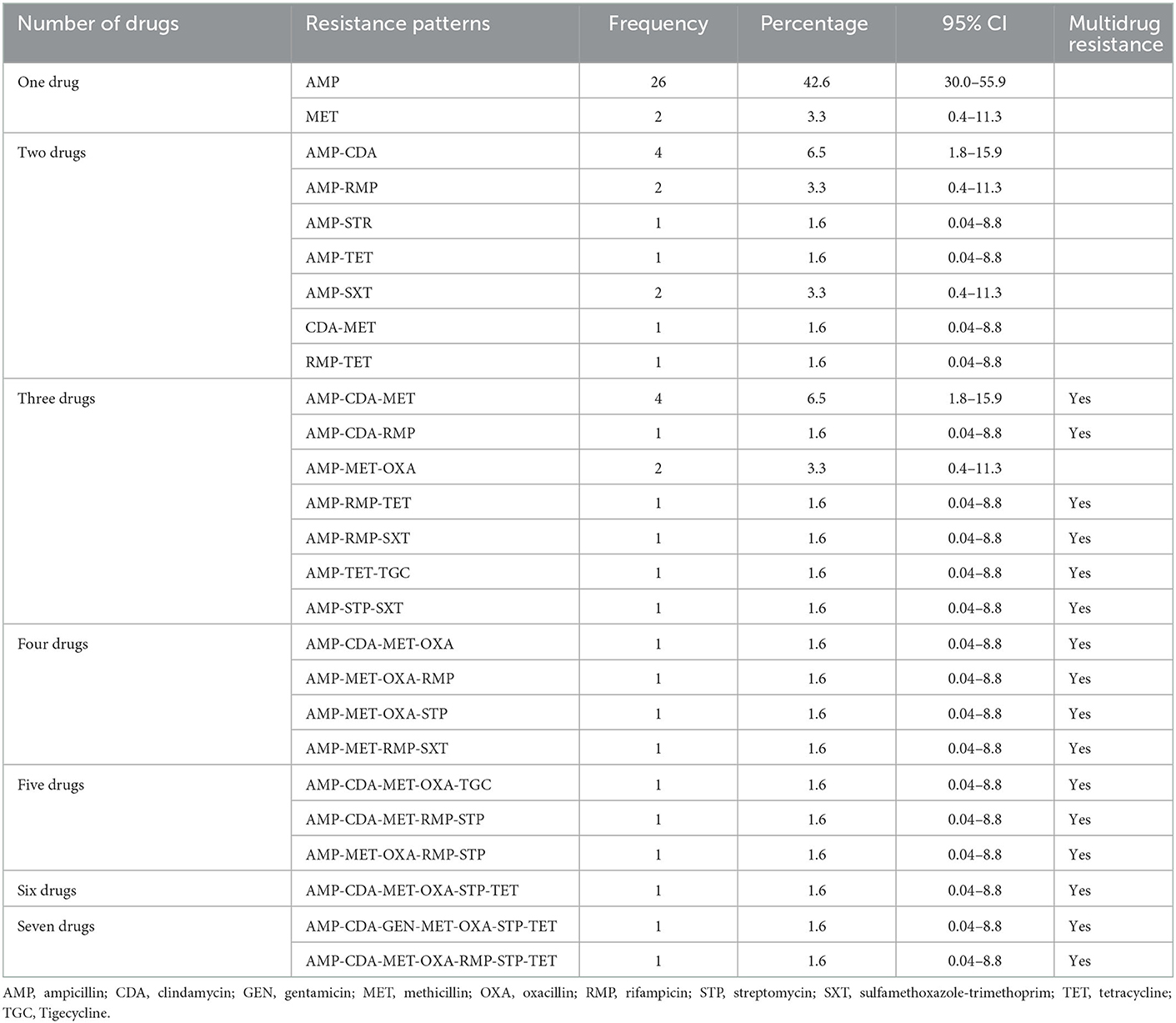
Nearly half of the Staphylococcus cultures were resistant to only one drug (AMP [42.6%] and MET [3.3%]; Table 5). Four samples had resistance patterns of each AMP-CDA (6.5%) and AMP-CDA-MET (6.5%) combination (Table 5). Multidrug combinations of one, two, three, four, five, six, and seven drugs were identified (Table 5); however, most combinations were only found in a single sample.
3.5. Inter-species interaction in different locations
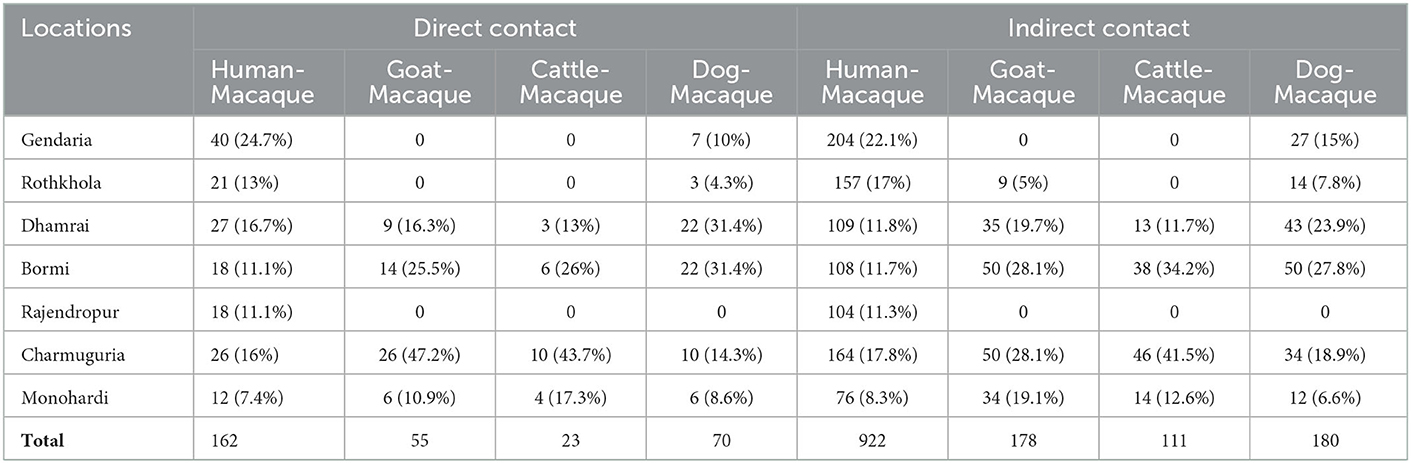
We observed direct and indirect interactions between macaques and people, cattle, goats, and dogs (Table 6). The most frequently observed inter-species interactions were between people and macaques (Table 6). We observed the highest number of human-macaque contacts in Gendaria, Dhaka, with 40 observations of direct contact and 204 observations of indirect contact. In contrast, Monohardi and Narshingdi had the lowest number of observed direct (12) and indirect (76) contact occurrences. No direct contact was observed between macaques, goats, and cattle in Gendaria and Rothkhola in Dhaka. No direct or indirect contact was observed between macaques and any domestic animals in Rajendropur, Gazipur (Table 6).
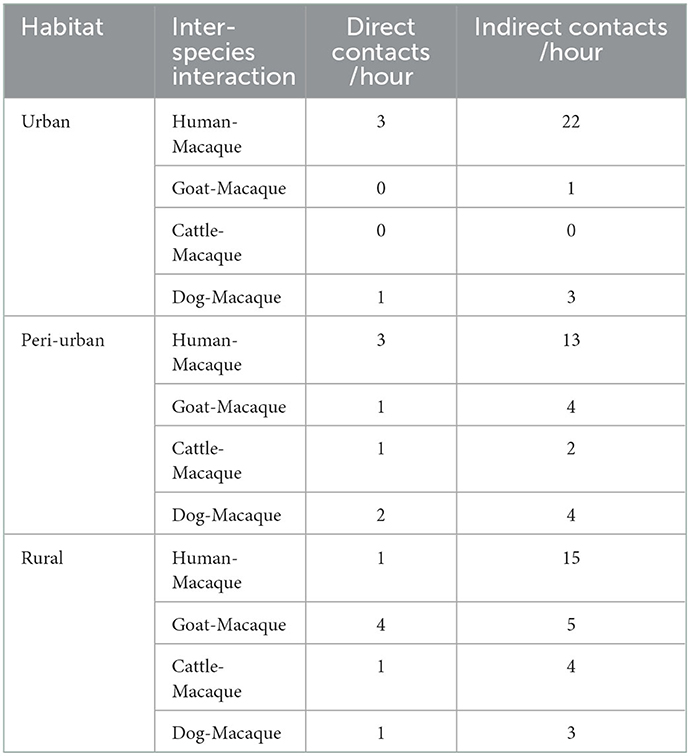
The contact rate between macaques and people was high in urban habitats for both direct contacts per hour (3) and indirect contacts per hour (22) (Table 7).
4. Discussion
We found evidence of antimicrobial-resistant bacteria in free-ranging macaque populations. This suggests that macaques are infected through contact with infected people or animals or environmental contamination. We observed direct and indirect contact between macaques and people and domestic animals at all sites where samples were collected, providing a potential transmission pathway for antimicrobial resistant bacteria, especially considering that free-ranging macaques in Bangladesh are not treated with antimicrobials.
We found that the prevalence of Salmonella spp. was low (5%). Several other studies have found similarly low rates of salmonella infection in other species of non-human primates, including a prevalence of 3% in Salmonella spp. in captive primate populations at the national center for primate biology (48). A longitudinal study in Thailand also found a similar prevalence of (7%) Salmonella serotypes in captive wildlife, including non-human primates (49). Another study from Bwindi and Mgahinga in Uganda reported that 13% of free-range mountain gorillas (Gorilla gorilla beringei) had Salmonella spp. (27).
We found that the macaques we sampled had Salmonella spp. most frequently resistant to tetracycline (89%). This is in line with a previous study where 95% of Salmonella sp. collected samples from south America, Africa, and Southeast Asia to the National Center for Primate Biology were resistant to tetracycline (48). Other studies found lower levels of tetracycline resistance in Salmonella spp. isolated from captive wildlife (29%) in Trinidad, which contains mammals (21%), birds (25%), and reptiles (40%) (49) and non-human primates (38.3%) in the USA (50). It may be that the prevalence of tetracycline resistant Salmonella spp. is associated with the usage of tetracycline to treat livestock. In Bangladesh, tetracycline is commonly used to treat livestock and does not require a prescription. Salmonella resistance to azithromycin was the second highest as it was detected in 83% of Salmonella spp. isolates in the present study. A previous study in Bangladesh found similarly high (95%) rates of azithromycin resistance Salmonella spp. in human patients (51). The authors proposed that the high rate of azithromycin resistance in Salmonella spp. may be due to the frequent misuse of this antibiotic to treat people for enteric fever and the common cold and possible transmission to macaques via environmental contamination through excrement (52). In addition, 50% of Salmonella spp. samples were also found to be resistant to sulfamethoxazole-trimethoprim. This result differs slightly with a recent study conducted in Bangladesh, in which 68 and 58% of Salmonella enterica Serovar Typhi samples were resistant to sulfamethoxazole and trimethoprim, respectively (53). In our investigation, the prevalence of nalidixic acid-resistant Salmonella was 44%, which is lower than the prevalence reported in several previous studies of human patients (73–93%) (53–55). Few other AMR studies have been conducted in other wild species (wild birds, squirrels, deer) in Bangladesh and found higher antimicrobial resistance in Salmonella, E. coli, and Staphylococcus (14, 56, 57).
We also found that Salmonella spp. isolates were susceptible to ciprofloxacin, gentamicin, and imipenem. These findings were similar to those reported by previous studies in which Salmonella spp. isolated from human samples in Montenegro, Bangladesh, and India were sensitive to ciprofloxacin (99.5%) (58), gentamicin (100%) (59), imipenem (100%) (60), respectively.
We found the Staphylococcus spp. prevalence was 16%, which was much lower than a previous study (61%) on non-human primates in Africa (24). In our study, the prevalence of resistant Staphylococcus spp. was higher (15%) than that of Salmonella spp. However, it was much lower than what was detected in Macaca mulatta in Spain (80%) (61), the Netherlands (39%) (25), and Wisconsin, USA (23.6%) (26). We found that Staphylococcus spp. in rhesus macaques was highly resistant (93%) to ampicillin, which contrasts a study that found that S. aureus isolates from African non-human primates was 2.9% resistant to penicillin (24). The increased interaction between synanthropic rhesus macaques and humans in South Asia is likely contributing to the higher ampicillin resistance and transfer of resistance genes in South Asia compared to African regions (62). Taylor and Grady reported 75% resistance to methicillin/oxacillin in Staphylococcus spp. in vitro (30).
Staphylococcus spp. were highly sensitive to chloramphenicol, gentamycin, linezolid, oxacillin, tigecycline, and trimethoprim-sulfamethoxazole in our study. This sensitivity is similar to that found in human clinical specimens in the USA, where more than 90% of multidrug-resistant Staphylococcus spp. were sensitive to trimethoprim-sulfamethoxazole, linezolid, and vancomycin (63). The higher sensitivity of Staphylococcus spp. to these antimicrobials may be due to infection of macaque-specific Staphylococcus aureus (25). Another reason may be infrequent exposure of humans and animals to chloramphenicol, linezolid, Oxacillin, etc. (64).
Rhesus macaques from the Dhaka District had a higher odd (OR = 11.1) of harboring resistant Staphylococcus spp. We suspect this may be associated with the high human population density levels in Dhaka City and improper waste disposal management (65). Moreover, household drinking water supply contains diversified microorganisms that easily increase the chance of getting resistant organisms (17). A similar reason may be corresponded with the higher odds of resistance to Salmonella spp. in the urban areas compared to the rural areas. Macaques from the peri-urban areas also had a high odd (OR = 5.6) of having resistant Staphylococcus spp. Peri-urban regions have a decentralized waste management system (66) and transmission of pathogenic bacteria into the river through sewage systems (67, 68). As in the peri-urban area, rural areas were more likely to experience Staphylococcus resistance, possibly due to the transmission of pathogenic bacteria into rural via the river; additionally, waste disposal is poor in the rural area.
The age-dependent differences in AMR can also be useful for monitoring the trend of antibiotic-resistant strains spreading at high-risk interfaces in Bangladesh. In reality, the behavior of individuals of the same species can vary depending on their age and gender. It is believed that young are less exposed to antibiotics than adults throughout their lifetimes, and younger animals may therefore have a lower prevalence of AMR isolates than older animals. However, our results revealed higher odds of Staphylococcus resistance in the sub-adult compared to the adult. On the other hand, no significant differences in relation to animal age for Salmonella resistance. Similar to this study, the insignificant difference in MDR has been reported in wild micromammals from Northern, Italy (69). The age-dependent AMR phenomena in cattle and pigs are detected regardless of geographic region and production methods. However, the nature of the AMR phenomenon is currently unexplained (70).
We discovered that macaques and people had more direct and indirect contact per hour in urban and peri-urban areas. These are most likely significant risk factors for transmitting resistant organisms/genes from humans to macaques and vice versa. Microorganism transmission from humans to non-human primates has been observed for approximately three decades (71) and happening frequently e.g., SARS-CoV-2 infection in a gorilla at a zoo in California (72). A recent qualitative study suggests that interviewees from some of these regions in Bangladesh believe that recent human encroachment into the natural habitat of macaques may have increased the amount of contact between people and macaques (73). Hasan et al. reported that the distribution of macaques is high in densely populated urban areas, such as old Dhaka (23). Macaques can also be found in temples and shrines where people frequently leave food (34). As a result, the rhesus macaque may acquire resistant microorganisms from humans. As expected, the contact rate between livestock and macaques was higher in rural areas, given the abundance of livestock in these areas. According to one study, many disease-causing bacteria in livestock in Bangladesh were resistant to available antibiotics, most likely due to improper antimicrobial use (3). Contact between livestock and macaque can potentially transmit resistant organisms and/or genes. Further, macaques frequently take waste from dustbins, snatch household food and clothes, and damage crops (74). Our study detected that interaction between livestock, dogs, and people with macaques frequently occurs during the conflict, crop raiding, and when people provide food (75). Stray dogs eat waste from dustbins and the roadside and are infected with microorganisms resistant to the majority of commonly used antibiotics (76).
The exceedingly high rates of AMR that we found in Salmonella spp. (100%) and Staphylococcus spp. (95%) suggest that macaque exposure to antimicrobial resistant bacteria at all the sampling sites was likely very high. Antibiotics are commonly used in treating humans, livestock, and poultry in Bangladesh; therefore, antibiotic residues and resistant organisms could have been passed through environmental sources and/or interspecies interaction with wild species. Rhesus macaque and other wildlife are rarely treated with antibiotics, but present study showed a relatively high frequency of antimicrobial resistance in Salmonella and Staphylococcus in the rhesus macaque. So, Nationwide programs are important to control the irrational use of antibiotics and improve the waste management and sewage system and reduce human-wildlife and livestock-wildlife interaction to decrease antimicrobial resistance among humans and animals in the future.
5. Conclusions
AMR is a global concern and public health threat, and the fact that it is spilling over into wildlife indicates potential widespread environmental contamination. The study identified the prevalence of 5% Salmonella spp. and 16% Staphylococcus spp. in rhesus macaque, nearly all of which were resistant (100 and 95%, respectively) in different locations in Bangladesh. The isolated organisms showed various levels of resistance to different antimicrobials. Multidrug resistance patterns in Salmonella spp. and Staphylococcus spp. were identified, including cultures resistant to up to seven antimicrobials. The high level of resistance could be attributed to interactions and transmission of resistant microorganisms between macaques, humans, and livestock. Human-macaque interactions were more common in urban habitats, while livestock-macaque interactions were more common in peri-urban and rural habitats. Even though rhesus macaques are not directly treated with antimicrobials, this study found an alarming level of resistant organisms in macaques. These findings should be viewed as an indicator of significant environmental contamination and an urgent call for stricter controls of the use of antimicrobials.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Ethics statement
The animal study was reviewed and approved by Animal Experimentation Ethics Committee (AEEC) [permit number: CVASU/Dir (R&E) AEEC/2015/751], Chattogram Veterinary and Animal Sciences University, Chattogram, Bangladesh.
Author contributions
MRa, AI, MRo, JE, and MH: conceptualization. MRa, AI, and MU: methodology. MRa and AI: software, formal analysis, and writing—original draft preparation. AI, SI, MRa, EH, MS, MRo, JE, and MH: validation. AI and MH: investigation. MRa, AI, and MH: resources. MRa, SI, and MU: data curation. AI, SI, MRa, EH, MRo, MS, and MH: writing—review and editing. MU and MS: laboratory support. AI, MF, and MH: supervision and project administration. JE and AI: funding acquisition. All authors have read and agreed to the published version of the manuscript.
Funding
This study was made possible by the generous support of the United States Agency for International Development (USAID) Emerging Pandemic Threats PREDICT (cooperative agreement number AID-OAA-A-14-00102).
Acknowledgments
We are thankful to Pitu Biswas, Gafur Sheikh, Abdul hai, Abdullah Al Mamun and Md. Mizanur Rahman for their technical support and field assistance. We are also grateful to the laboratory stuffs of the Bangladesh Livestock and Research Institute (BLRI).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. World Health Organization. Antimicrobial Resistance: Global Report on Surveillance. World Health Organization (2014). Available online at: http://apps.who.int/iris/bitstream/10665/112642/1/9789241564748_eng.pdf (accessed January 13, 2021).
2. Levy SB, Marshall B. Antibacterial resistance worldwide: causes, challenges and responses. Nat Med. (2004) 10:S122. doi: 10.1038/nm1145
3. Roess AA, Winch PJ, Akhter A, Afroz D, Ali NA, Shah R, et al. Household animal and human medicine use and animal husbandry practices in rural Bangladesh: risk factors for emerging zoonotic disease and antibiotic resistance. Zoonoses Public Health. (2015) 62:569–78. doi: 10.1111/zph.12186
4. Akond MA, Hassan S, Alam S, Shirin M. Antibiotic resistance of Escherichia coli isolated from poultry and poultry environment of Bangladesh. Am J Environ Sci. (2009) 5:47–52. doi: 10.3844/ajessp.2009.47.52
5. Faruk M, Ali M, Patwary Z. Evaluation of the status of use of chemicals and antibiotics in freshwater aquaculture activities with special emphasis to fish health management. J Bangladesh Agric Univ. (2008) 6:381–90. doi: 10.3329/jbau.v6i2.4838
6. Agga GE, Arthur TM, Durso LM, Harhay DM, Schmidt JW. Antimicrobial-resistant bacterial populations and antimicrobial resistance genes obtained from environments impacted by livestock and municipal waste. PLoS ONE. (2015) 10:e0132586. doi: 10.1371/journal.pone.0132586
7. Ahaduzzaman M, Hassan MM, Alam M, Islam S, Uddin I. Antimicrobial resistance pattern against Staphylococcus aureus in environmental effluents. Res J Vet Pract. (2014) 2:13–6. doi: 10.14737/journal.rjvp/2014/2.1.13.16
8. Finley RL, Collignon P, Larsson DJ, McEwen SA, Li X-Z, Gaze WH, et al. The scourge of antibiotic resistance: the important role of the environment. Clin Infect Dis. (2013) 57:704–10. doi: 10.1093/cid/cit355
9. Kang CI, Song JH. Antimicrobial resistance in Asia: current epidemiology and clinical implications. Infect Chemother. (2013) 45:22–31. doi: 10.3947/ic.2013.45.1.22
10. Robinson TP, Bu D, Carrique-Mas J, Fèvre EM, Gilbert M, Grace D, et al. Antibiotic resistance is the quintessential one health issue. Trans R Soc Trop Med Hyg. (2016) 110:377–80. doi: 10.1093/trstmh/trw048
11. Martínez J. Antibiotics and antibiotic resistance genes in natural environments. Science. (2008) 321:365–7. doi: 10.1126/science.1159483
12. Carroll D, Wang J, Fanning S, McMahon BJ. Antimicrobial resistance in wildlife: implications for public health. Zoonoses Public Health. (2015) 62:534–42. doi: 10.1111/zph.12182
13. Faruq A, Hassan MM, Uddin MM, Rahman ML, Rakib TM, Alam M, et al. Prevalence and multidrug resistance pattern of Salmonella isolated from resident wild birds of Bangladesh. Int J One Health. (2016) 2:35–41. doi: 10.14202/IJOH.2016.35-41
14. Hasan B, Sandegren L, Melhus Å, Drobni M, Hernandez J, Waldenström J, et al. Antimicrobial drug–resistant Escherichia coli in wild birds and free-range poultry, Bangladesh. Emerg Infect Dis. (2012) 18:2055. doi: 10.3201/eid1812.120513
15. Sayah RS, Kaneene JB, Johnson Y, Miller R. Patterns of antimicrobial resistance observed in Escherichia coli isolates obtained from domestic-and wild-animal fecal samples, human septage, and surface water. Appl Environ Microbiol. (2005) 71:1394–404. doi: 10.1128/AEM.71.3.1394-1404.2005
16. Rolland R, Hausfater G, Marshall B, Levy S. Antibiotic-resistant bacteria in wild primates: increased prevalence in baboons feeding on human refuse. Appl Environ Microbiol. (1985) 49:791–4. doi: 10.1128/aem.49.4.791-794.1985
17. Talukdar PK, Rahman M, Rahman M, Nabi A, Islam Z, Hoque MM, et al. Antimicrobial resistance, virulence factors and genetic diversity of Escherichia coli isolates from household water supply in Dhaka, Bangladesh. PLoS ONE. (2013) 8:e61090. doi: 10.1371/journal.pone.0061090
18. Khan SA, Imtiaz MA, Sayeed MA, Shaikat AH, Hassan MM. Antimicrobial resistance pattern in domestic animal-wildlife-environmental niche via the food chain to humans with a Bangladesh perspective; a systematic review. BMC Vet Res. (2020) 16:1–13. doi: 10.1186/s12917-020-02519-9
19. Cristóbal-Azkarate J, Dunn JC, Day JM, Amábile-Cuevas CF. Resistance to antibiotics of clinical relevance in the fecal microbiota of Mexican wildlife. PLoS ONE. (2014) 9:e107719. doi: 10.1371/journal.pone.0107719
20. Kozak GK, Boerlin P, Janecko N, Reid-Smith RJ, Jardine C. Antimicrobial resistance in Escherichia coli isolates from swine and wild small mammals in the proximity of swine farms and in natural environments in Ontario, Canada. Appl Environ Microbiol. (2009) 75:559–66. doi: 10.1128/AEM.01821-08
21. Islam S, Rahman MK, Uddin MH, Rahman MM, Chowdhury MN, Hassan MM, et al. Prevalence and diversity of gastrointestinal parasites in free-ranging rhesus macaques (Macaca mulatta) in different land gradients of Bangladesh. Am J Primatol. (2022) 84:e23345. doi: 10.1002/ajp.23345
22. Feeroz MM, Soliven K, Small CT, Engel GA, Andreina Pacheco M, Yee JL, et al. Population dynamics of rhesus macaques and associated foamy virus in Bangladesh. Emerg Microbes Infect. (2013) 2:1–14. doi: 10.1038/emi.2013.23
23. Hasan MK, Aziz MA, Alam SR, Kawamoto Y, Jones-Engel L, Kyes RC, et al. Distribution of Rhesus Macaques (Macaca mulatta) in Bangladesh: inter-population variation in group size and composition. Primate Conserv. (2013) 26:125–33. doi: 10.1896/052.026.0103
24. Schaumburg F, Alabi A, Köck R, Mellmann A, Kremsner P, Boesch C, et al. Highly divergent Staphylococcus aureus isolates from African non-human primates. Environ Microbiol Rep. (2012) 4:141–6. doi: 10.1111/j.1758-2229.2011.00316.x
25. Van Den Berg S, Van Wamel WJ, Snijders SV, Ouwerling B, de Vogel CP, Boelens HA, et al. Rhesus macaques (Macaca mulatta) are natural hosts of specific Staphylococcus aureus lineages. PLoS ONE. (2011) 6:e26170. doi: 10.1371/journal.pone.0026170
26. Kokan N, Bergdoll M. Detection of low-enterotoxin-producing Staphylococcus aureus strains. Appl Environ Microbiol. (1987) 53:2675–6. doi: 10.1128/aem.53.11.2675-2676.1987
27. Nizeyi JB, Innocent RB, Erume J, Kalema GR, Cranfield MR, Graczyk TK. Campylobacteriosis, salmonellosis, and shigellosis in free-ranging human-habituated mountain gorillas of Uganda. J Wildl Dis. (2001) 37:239–44. doi: 10.7589/0090-3558-37.2.239
28. Goldberg TL, Gillespie TR, Rwego IB, Wheeler E, Estoff EL, Chapman CA. Patterns of gastrointestinal bacterial exchange between chimpanzees and humans involved in research and tourism in western Uganda. Biol Conserv. (2007) 135:511–7. doi: 10.1016/j.biocon.2006.10.048
29. Rwego IB, Isabirye-basuta G, Gillespie TR, Goldberg TL. Gastrointestinal bacterial transmission among humans, mountain gorillas, and livestock in Bwindi Impenetrable National Park, Uganda. Conserv Biol. (2008) 22:1600–7. doi: 10.1111/j.1523-1739.2008.01018.x
30. Taylor WM, Grady AW. Catheter-tract infections in rhesus macaques (Macaca mulatta) with indwelling intravenous catheters. Comp Med. (1998) 48:448–54.
31. Koga T, Aoki W, Mizuno T, Wakazono K, Ohno J, Nakai T, et al. Antimicrobial resistance in Campylobacter coli and Campylobacter jejuni in cynomolgus monkeys (Macaca fascicularis) and eradication regimens. J Microbiol Immunol Infect. (2017) 50:75–82. doi: 10.1016/j.jmii.2014.12.006
32. Tashiro Y, Yawata Y, Toyofuku M, Uchiyama H, Nomura N. Interspecies interaction between Pseudomonas aeruginosa and other microorganisms. Microbes Environ. (2013) 28:13–24. doi: 10.1264/jsme2.ME12167
33. Devaux CA, Mediannikov O, Medkour H, Raoult D. Infectious disease risk across the growing human-non human primate interface: a review of the evidence. Front Public Health. (2019) 7:305. doi: 10.3389/fpubh.2019.00305
34. Neha SA, Hasan MAU, Baki MA, Sehrin S. Rhesus Macaque Macaca mulatta (Mammalia: Primates: Cercopithecidae) in a human-modified landscape: population, activity budget, and societal perceptions in Bangladesh. J Threat Taxa. (2021) 13:19203–11. doi: 10.11609/jott.7073.13.9.19203-19211
35. Daniel WW, Cross CL. Cross, Biostatistics: A Foundation for Analysis in the Health Sciences. Wiley (2018).
36. Putturu R, Thirtham M, Eevuri TR. Eevuri, Antimicrobial sensitivity and resistance of Salmonella enteritidis isolated from natural samples. Vet World. (2013) 6:185–8. doi: 10.5455/vetworld.2013.185-188
37. Kateete DP, Kimani CN, Katabazi FA, Okeng A, Okee MS, Nanteza A, et al. Identification of Staphylococcus aureus: DNase and Mannitol salt agar improve the efficiency of the tube coagulase test. Ann Clin Microbiol Antimicrob. (2010) 9:23. doi: 10.1186/1476-0711-9-23
38. Nesa M, Khan M, Alam M. Isolation, identification and characterization of salmonella serovars from diarrhoeic stool samples of human. Bangladesh J Vet Med. (2011) 9:85–93. doi: 10.3329/bjvm.v9i1.11218
39. Pao S, Patel D, Kalantari A, Tritschler JP, Wildeus S, Sayre BL. Detection of Salmonella strains and Escherichia coli O157: H7 in feces of small ruminants and their isolation with various media. Appl Environ Microbiol. (2005) 71:2158–61. doi: 10.1128/AEM.71.4.2158-2161.2005
40. Hossain M, Chowdhury E, Islam M, Haider M, Hossain M. Avian salmonella infection: isolation and identification of organisms and histopathological study. Bangladesh J Vet Med. (2006) 4:7–12. doi: 10.3329/bjvm.v4i1.1518
41. Graham PL III, Lin SX, Larson EL. A US population-based survey of Staphylococcus aureus colonization. Ann Intern Med. (2006) 144:318. doi: 10.7326/0003-4819-144-5-200603070-00006
42. Hogan S, Gonzalez R, Harmon J, Nickerson S, Oliver S, Pankey J, et al. Laboratory Handbook on Bovine Mastitis. WD Hoard, Fort Atkinson: National Mastitis Council, Inc. (1999).
43. Gouws PA, Visser M, Brözel VS. A polymerase chain reaction procedure for the detection of Salmonella spp. within 24 hours. J Food Prot. (1998) 61:1039–42. doi: 10.4315/0362-028X-61.8.1039
44. Martineau F, Picard FJ, Ke D, Paradis S, Roy PH, Ouellette M, et al. Development of a PCR assay for identification of staphylococci at genus and species levels. J Clin Microbiol. (2001) 39:2541–7. doi: 10.1128/JCM.39.7.2541-2547.2001
45. Biemer JJ. Antimicrobial susceptibility testing by the Kirby-Bauer disc diffusion method. Ann Clin Lab Sci. (1973) 3:135–40.
46. Clinical and Laboratory Standards Institute. M100-S25 performance standards for antimicrobial susceptibility testing; twenty-fifth informational supplement. CLSI. (2015) 35:1–240.
48. Good RC, May BD, Kawatomari T. Enteric pathogens in monkeys. J Bacteriol. (1969) 97:1048–55. doi: 10.1128/jb.97.3.1048-1055.1969
49. Gopee NV, Adesiyun AA, Caesar K. Caesar, retrospective and longitudinal study of salmonellosis in captive wildlife in Trinidad. J Wildl Dis. (2000) 36:284–93. doi: 10.7589/0090-3558-36.2.284
50. Kim J, Coble DJ, Salyards GW, Bower JK, Rinaldi WJ, Plauche GB, et al. Antimicrobial use for and resistance of zoonotic bacteria recovered from nonhuman primates. Comp Med. (2017) 67:79–86.
51. Lina TT, Rahman SR, Gomes DJ. Multiple-antibiotic resistance mediated by plasmids and integrons in uropathogenic Escherichia coli and Klebsiella pneumoniae. Bangladesh J Microbiol. (2007) 24:19–23. doi: 10.3329/bjm.v24i1.1231
52. Hooda Y, Sajib MS, Rahman H, Luby SP, Bondy-Denomy J, Santosham M, et al. Molecular mechanism of azithromycin resistance among typhoidal Salmonella strains in Bangladesh identified through passive pediatric surveillance. PLoS Negl Trop Dis. (2019) 13:e0007868. doi: 10.1371/journal.pntd.0007868
53. Chiou C-S, Lauderdale T-L, Phung DC, Watanabe H, Kuo J-C, Wang P-J, et al. Antimicrobial resistance in Salmonella enterica serovar Typhi isolates from Bangladesh, Indonesia, Taiwan, and Vietnam. Antimicrob Agents Chemother. (2014) 58:6501–7. doi: 10.1128/AAC.03608-14
54. Siddique MA, Shamsuzzaman SM, Khandakar AR, Khondaker FA, Sumi SA, Jahan R. Detection of mutated gyra gene from nalidixic acid resistant Salmonella typhi and paratyphi a isolated from enteric fever patients in a tertiary care hospital of Bangladesh. J Med Microbiol. (2016) 10:3–7. doi: 10.3329/bjmm.v10i1.31431
55. Afroj S, Ilias M, Islam M, Saha SK. Prevalence of ciprofloxacin and nalidixic acid resistant Salmonella enterica serovar Typhi in Bangladesh. Bangladesh J Microbiol. (2011) 28:7–11. doi: 10.3329/bjm.v28i1.11802
56. Sarker MS, Ahad A, Ghosh SK, Mannan MS, Sen A, Islam S, et al. Antibiotic-resistant Escherichia coli in deer and nearby water sources at safari parks in Bangladesh. Vet World. (2019) 12:1578. doi: 10.14202/vetworld.2019.1578-1583
57. Jalal MS, Islam MZ, Dutta A, Dhar PK, Das A, Hasan MM, et al. Antibiotic resistant zoonotic bacteria in Irrawaddy squirrel (Callosciurus pygerythrus). Vet Med Sci. (2019) 5:260–8. doi: 10.1002/vms3.138
58. Mijović G. Antibiotic susceptibility of Salmonella spp: a comparison of two surveys with a 5 years interval. J IMAB–Ann Proceed Sci Papers. (2012) 18:216–9. doi: 10.5272/jimab.2012181.216
59. Hassan MM, Amin KB, Ahaduzzaman M, Alam M, Faruk MS, Uddin I. Antimicrobial resistance pattern against E. coli and Salmonella in layer poultry. Res J Vet Pract. (2014) 2:30–5. doi: 10.14737/journal.rjvp/2014/2.2.30.35
60. Singh L, Cariappa M. Blood culture isolates and antibiogram of Salmonella: experience of a tertiary care hospital. Med J Armed Forces India. (2016) 72:281–4. doi: 10.1016/j.mjafi.2015.07.007
61. Woods SE, Lieberman MT, Lebreton F, Trowel E, de la Fuente-Núñez C, Dzink-Fox J, et al. Characterization of multi-drug resistant Enterococcus faecalis isolated from cephalic recording chambers in research macaques (Macaca spp). PLoS ONE. (2017) 12:e0169293. doi: 10.1371/journal.pone.0169293
62. Priston NE, McLennan MR. Managing humans, managing macaques: human–macaque conflict in Asia and Africa. In: The Macaque Connection. New York, NY: Springer (2013). p. 225–50.
63. Styers D, Sheehan DJ, Hogan P, Sahm DF. Laboratory-based surveillance of current antimicrobial resistance patterns and trends among Staphylococcus aureus: 2005 status in the United States. Ann Clin Microbiol Antimicrob. (2006) 5:2. doi: 10.1186/1476-0711-5-2
64. Biswas M, Roy MN, Manik MIN, Hossain MS, Tapu STA, Moniruzzaman M, et al. Self medicated antibiotics in Bangladesh: a cross-sectional health survey conducted in the Rajshahi City. BMC Public Health. (2014) 14:1–7. doi: 10.1186/1471-2458-14-847
65. Parvin M, Begum A. Organic solid waste management and the urban poor in Dhaka City. Int J Waste Res. (2018) 8:2. doi: 10.4172/2252-5211.1000320
66. Parkinson J, Tayler K. Decentralized wastewater management in peri-urban areas in low-income countries. Environ Urban. (2003) 15:75–90. doi: 10.1177/095624780301500119
67. Saha ML, Khan MR, Ali M, Hoque S. Bacterial load and chemical pollution level of the River Buriganga, Dhaka, Bangladesh. Bangladesh J Botany. (2009) 38:87–91. doi: 10.3329/bjb.v38i1.5128
68. Das M, Ahmed MK, Begum F, Parveen S, Islam MM, Begum M. Microbial load in tannery and textile effluents and their receiving rivers of Dhaka. Dhaka Univ J Biol Sci. (2010) 19:73–81. doi: 10.3329/dujbs.v19i1.8946
69. Zanardi G, Iemmi T, Spadini C, Taddei S, Cavirani S, Cabassi CS. Wild micromammals as bioindicators of antibiotic resistance in ecopathology in Northern Italy. Animals. (2020) 10:1184. doi: 10.3390/ani10071184
70. Gaire TN, Scott HM, Sellers L, Nagaraja TG, Volkova VV. Age dependence of antimicrobial resistance among fecal bacteria in animals: a scoping review. Front Vet Sci. (2021) 7:622495. doi: 10.3389/fvets.2020.622495
71. ASM American Society for Microbiology. Green Monkeys Acquired Staphylococcus aureus from Humans. ScienceDaily. (2016). Available online at: https://www.sciencedaily.com/releases/2016/07/160729132845.htm (accessed January 9, 2021).
72. USDA. Confirmation of COVID-19 in Gorillas at a California Zoo. (2011). Available online at: https://www.aphis.usda.gov/aphis/newsroom/stakeholder-info/sa_by_date/sa-2021/sa-01/ca-gorillas-SARS-CoV-2 (accessed January 11, 2021).
73. Shano S, Islam A, Hagan E, Rostal MK, Martinez S, Al Shakil A, et al. Environmental change and zoonotic disease risk at human-macaque interfaces in Bangladesh. EcoHealth. (2021) 18:487–99. doi: 10.1007/s10393-021-01565-5
74. Imam E, Ahmad A. Population status of rhesus monkey (Macaca mulatta) and their menace: a threat for future conservation. Int J Environ Sci. (2013) 3:1279. doi: 10.6088/ijes.2013030400007
75. Ahsan M, Uddin M. Human-rhesus monkey conflict at Rampur village under Monohardi upazila in Narsingdi district of Bangladesh. J Threat Taxa. (2014) 6:5905–8. doi: 10.11609/JoTT.o3818.5905-8
Keywords: rhesus macaque, antimicrobial resistance, Salmonella, Staphylococcus, Bangladesh
Citation: Rahman MK, Hassan MM, Islam S, Rostal MK, Uddin MH, Hagan E, Samad MA, Flora MS, Epstein JH and Islam A (2023) Characterization and epidemiology of antimicrobial resistance patterns of Salmonella spp. and Staphylococcus spp. in free-ranging rhesus macaque (Macaca mulatta) at high-risk interfaces with people and livestock in Bangladesh. Front. Vet. Sci. 10:1103922. doi: 10.3389/fvets.2023.1103922
Received: 21 November 2022; Accepted: 04 January 2023;
Published: 30 January 2023.
Edited by:
Ihab Habib, United Arab Emirates University, United Arab EmiratesReviewed by:
Maja Velhner, Scientific Veterinary Institute Novi Sad, SerbiaPawin Padungtod, Food and Agriculture Organization of the United Nations, Vietnam
Copyright © 2023 Rahman, Hassan, Islam, Rostal, Uddin, Hagan, Samad, Flora, Epstein and Islam. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ariful Islam,  YXJpZkBlY29oZWFsdGhhbGxpYW5jZS5vcmc=
YXJpZkBlY29oZWFsdGhhbGxpYW5jZS5vcmc=
 Md. Kaisar Rahman
Md. Kaisar Rahman Mohammad Mahmudul Hassan
Mohammad Mahmudul Hassan Shariful Islam
Shariful Islam Melinda K. Rostal
Melinda K. Rostal Md. Helal Uddin
Md. Helal Uddin Emily Hagan3
Emily Hagan3 Mohammed Abdus Samad
Mohammed Abdus Samad Meerjady Sabrina Flora
Meerjady Sabrina Flora Jonathan H. Epstein
Jonathan H. Epstein Ariful Islam
Ariful Islam