- 1Institute of Zoology, Bahauddin Zakariya University, Multan, Pakistan
- 2Centro Nacional de Investigación Disciplinaria en Salud Animal e Inocuidad, Instituto Nacional de Investigaciones Forestales, Agrícolas y Pecuarias (INIFAP), Jiutepec, Morelos, Mexico
- 3Department of Animal Production, College of Food and Agriculture Sciences, King Saud University, Riyadh, Saudi Arabia
- 4Institute of Molecular Biology and Biotechnology, Bahauddin Zakariya University, Multan, Pakistan
- 5Shaheed Benazir Bhuto University Sheringal, District Dir, Khyber Pakhtunkhwa, Pakistan
- 6Department of Botany and Zoology, Bacha Khan University, Charsadda, Khyber Pakhtunkhwa, Pakistan
Background: Majority of Pakistani population lives in rural areas and raising animals, especially the small ruminants, is their primary source of income. Anaplasma ovis is known to infect small ruminants globally and causing significant economic losses to livestock owners, however prevalence of Anaplasma ovis has been least investigated from Pakistan despite having a huge sheep population.
Methods: The present study was conducted from June 2021 till December 2021 to report the PCR based prevalence of Anaplasma ovis in the blood samples of sheep (n = 239) that were collected from District Dera Ghazi Khan in Pakistan.
Results: Out of 239 samples, 30 (12.5%) amplified a 347 bp fragment specific for the msp4 gene of Anaplasma ovis. Represented partial msp4 gene sequences were confirmed by Sanger sequencing and deposited to GenBank (OP620757-59). None of the studied epidemiological factors (age, sex, breed, size of herd, dogs with herd, and composition of herd) showed an association (P > 0.05) with the Anaplasma ovis infection in enrolled sheep. Analysis of the amplified partial mSP4 sequence of Anaplasma ovis revealed that this gene is highly conserved as all three sequences were identical and phylogenetically resembled with the msp4 sequences amplified from small ruminants in China, Kenya, and Germany, Turkey, Portugal, Tunisia and India. In conclusion, for the first time, we are reporting a moderate prevalence of Anaplasma ovis prevalence in Pakistani sheep and this data will help in developing the integrated control policies against this newly reported tick-borne disease that is infecting our sheep breeds.
Introduction
Diseases, including metabolic, infectious and vector bone, are a major constraint to small ruminants and livestock industry as diseases impose financial risks to the livestock owners due to morbidity and mortality of their animals (1, 2). Ecto parasites, especially ticks, are the major source of pathogen transmission to sheep (3, 4). Tick population as seen a substantial increase globally as well as in Pakistan in recent years due to global weather changes and due to introduction of the animal species in environments/countries where they did not exist before (4). This rise in tick population has led to an increase in the incidence of tick borne diseases (TBDs) and hence the economic losses (5).
Anaplasma (A.) ovis, an intra erythrocytic gram negative rickettsial bacterium, that belongs to the genus Anaplasma, family Anaplasmataceae that infect sheep, goats and some wild ruminants causing anaplasmosis (6). Anaplasma ovis is frequently reported to be transmitted to sheep by ticks belonging to Ixodes, Dermacentor, Rhipicephalus, and Amblyomma genera (7). In sheep, infection due to Anaplasma ovis is very common and chances of infection increases when the weather is hot and dry or when the sheep are already co infected with some other parasite (8). In extreme cases, infection with A. ovis can lead to the death of infected animal (6). The most marked clinical signs of anaplasmosis are anemia and jaundice. Usually a persistent infection is developed in those animals that survive the acute phase of disease (9). Doxycycline is the treatment of choice for the treatment of anaplasmosis and all other tick borne rickettsial diseases (10).
Although Dera Ghazi Khan is known for its large sheep population but ovine anaplasmosis has never been reported from this District. Hence the present study was designed to report the molecular prevalence of A. ovis in enrolled sheep breeds and to report the association of this infection with the epidemiological risk factors, if any.
Materials and methods
Sample and data collection
Randomly selected herds in Dera Ghazi Khan District were targeted to collect 239 blood samples from apparently healthy sheep during June 2021 till December 2021. Enrolled sheep belonged to four breeds: Mundri (N = 171), Kajli (N = 44), Latti (N = 14), and Baluchi (N = 10). Following the informed consent from the owners, around 3–5 ml of blood sample was collected by pricking the Jugular vein of each sheep with a disposable syringe into a tube containing 0.5 M EDTA as an anticoagulant. This blood was later on used for the DNA extraction. In order to find out the epidemiological factors that are associated with A. ovis infect in enrolled sheep, a questionnaire was filled on the sampling site. Data regarding sex, age, herd size, composition of herd, and dogs present in herd was collected.
DNA extraction and PCR amplification
DNA was extracted from the collected blood by using a non-organic method as reported by Grimberg et al. (11). A pair of primers, AOF 5'-TGAAGGGAGCGGGGTCATGGG-3' and AOR 5'-GAGTAATTGCAGCCAGGCACTCT-3' was used to amplify a 347 bp fragment specific for msp4 gene of Anaplasma ovis as previously reported by Torina et al. (12). A master mixture of 25 μl was prepared containing 3 mM MgCl2, 10X PCR buffer, 5 μl of template DNA, 0.2 mM deoxy ribonucleotide triphosphates, 2 U of Taq DNA Polymerase (Parstous, Iran) and 0.5 mM of each primer. Reaction conditions comprised of an initial denaturation at 95°C for 5 min, 30 cycles of denaturation for 30 s at 94°C, annealing for 30 s at 62°C and extension for 30 s at 72°C and a final elongation for 5 min at 72°C (12).
DNA sequencing and phylogenetic analysis
Randomly selected PCR products (n = 3) were sent to First Base Sequencing Service (Malaysia) for purification and DNA sequencing. The resultant partial msp4 sequences from A. ovis isolates were deposited at GenBank under the accession numbers OP620757- OP620759. Anaplasma ovis sequences (having 96–100% similarity to the ones generated in present investigation) were download from the GenBank database (https://www.ncbi.nlm.nih.gov/) and all sequences were trimmed to 309 bp to be used in phylogeny. Maximum Likelihood method was applied in MEGA version 11 for evolutionary analysis (13). Kimura 2-parameter model with invariant sites was the top ranking substitution model according to lowest Bayesian Information Criterion score (14). Also, a bootstrap analysis with 1,000 replicates was used for the tree construction. Anaplasma phagocytophilum's msp4 partial gene sequence was used as an outgroup. Sequence alignment was performed by using ClustalW and visualizated with BioEdit (15).
Statistical analysis
Minitab (version 17, USA) was used for data analysis. P ≤ 0.05 was selected as significant level. Comparison of A. ovis prevalence between various sheep breeds was made by applying one way analysis of variance (ANOVA). Association between A. ovis occurrence and various risk factors was assessed through the Fisher's exact test (for 2 × 2 tables). Tajima's D Fu and Li's D values were estimated with DnaSP v5 (16).
Results
Molecular investigation and risk factors' analysis
Analysis of the results revealed that PCR amplified a 347 base pair fragment specific for msp4 gene of A. ovis in 30 out of 239 (12.5%) collected sheep blood samples during present molecular survey. When prevalence of A. ovis was compared between enrolled sheep breed, one way ANOVA results revealed that the bacterium prevalence was not restricted to a particular sheep breed (P = 0.3; Table 1). Fisher Exact test results revealed that all the epidemiological parameters investigated during this survey were not associated (P > 0.05) with the A. ovis infection in sheep (Table 2).
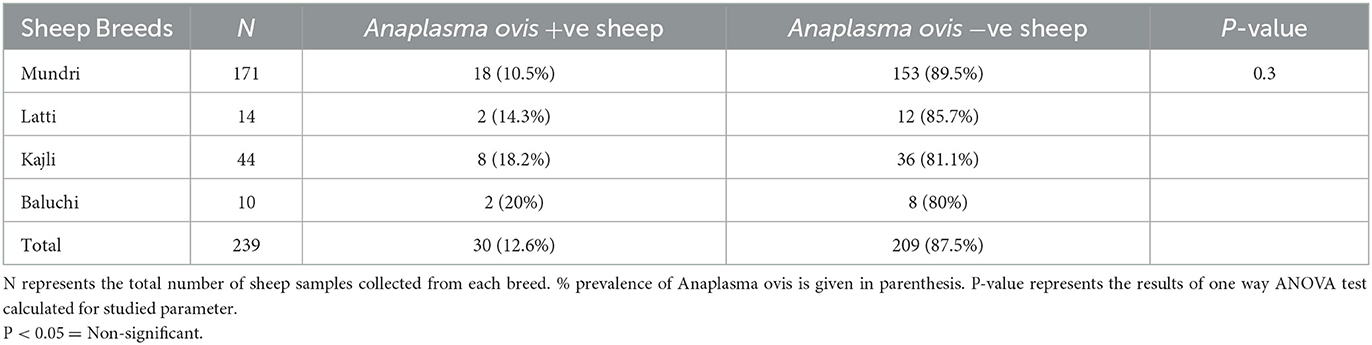
Table 1. Comparison of Anaplasma ovis prevalence in blood samples of various sheep breeds enrolled from District Dera Ghazi Khan during present study.
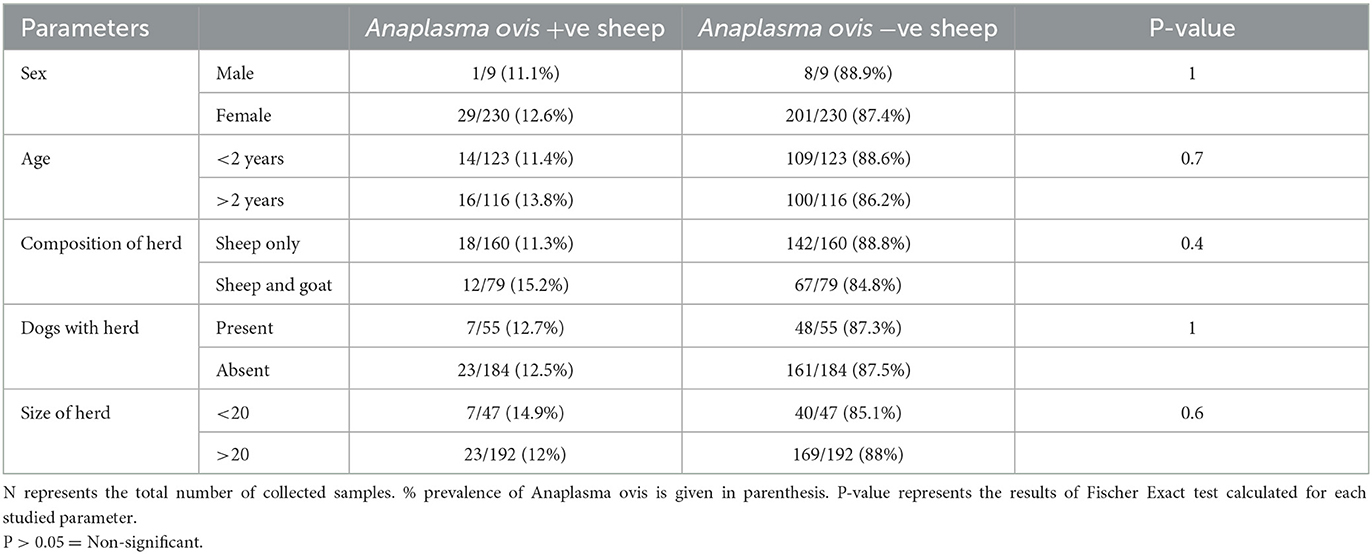
Table 2. Association of Anaplasma ovis prevalence with the studied epidemiological parameters describing sheep characters enrolled during the present study from District Dera Ghazi Khan.
Phylogenetic study and genetic diversity
During phylogenetic analysis, we have compared the partial msp4 (347 bp) gene sequences generated during this study (OP620757–OP620759) with those previously deposited GenBank from various parts of the World having sequence homology of 96% or more (Figure 1). Analysis revealed that all three A. ovis sequences generated during present study clustered in a single cluster as shown in Figure 1 along with those amplified from small ruminants in Kenya, China, Germany, Turkey, Portugal, Tunisia, and India (Figure 1).
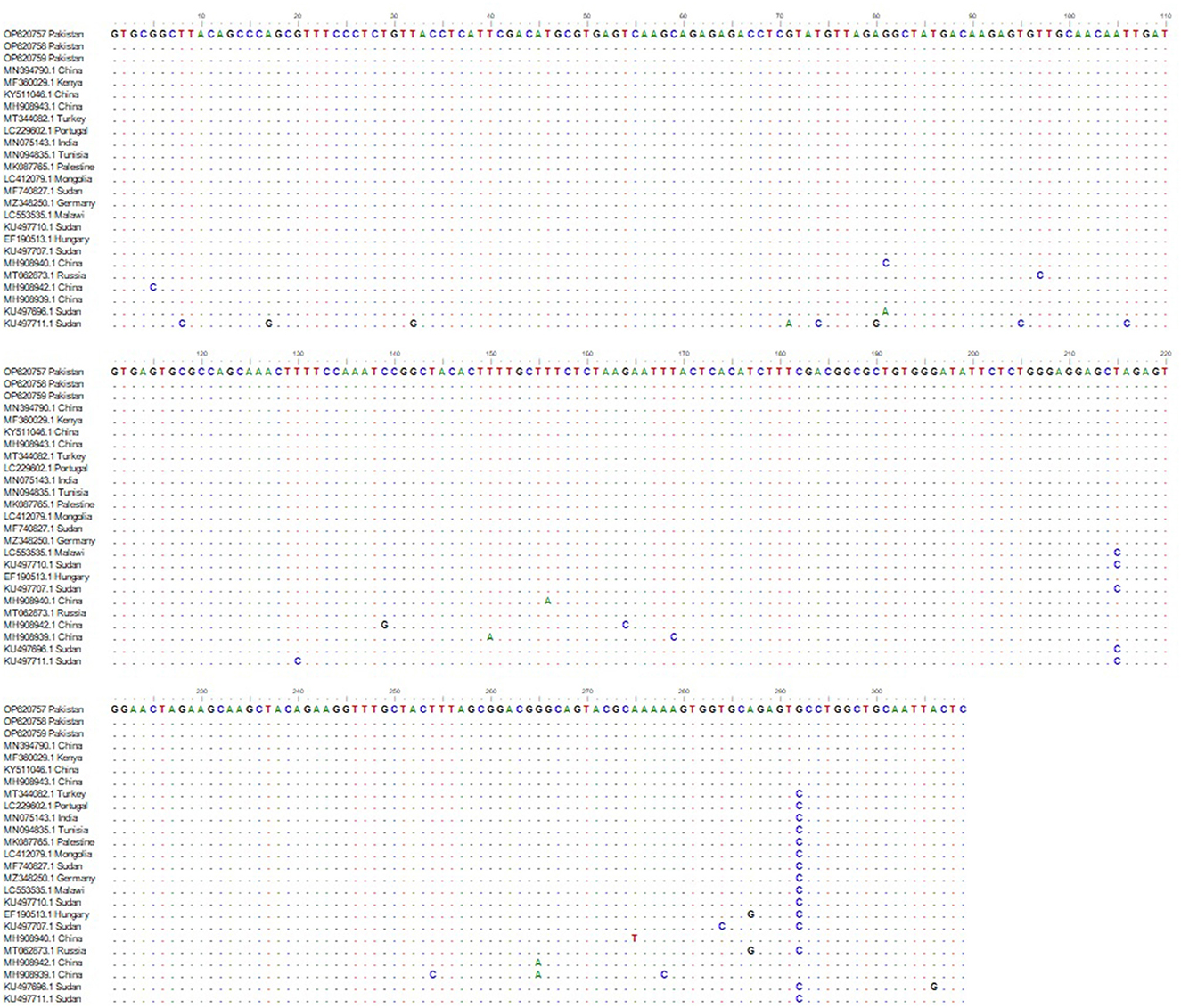
Figure 1. Phylogenetic analysis of Anaplasma ovis based on msp4 partial gene sequence. Sequences in the box were determined in this study. GeneBank accession numbers and country are shown for all sequences. Values at the nodes represent the number of occurrences of clades in 1,000 bootstrap replications of the taxa. The msp4 partial gene sequence of Anaplasma marginale from Mexico, Anaplasma centrale from Israel were and Anaplasma phagocytophilum from Italy were used as the outgroup. Maximum Likelihood method and Kimura 2-parameter model were used to infer the evolutionary history. The tree with the highest log likelihood (−1,168.91) is shown. Evolutionary analyses were conducted in MEGA11.
Alignment of msp4 partial sequences of msp4 gene from Pakistani sheep revealed a single genotype (Figure 2). All the three sequences generated during present study showed 100% genetic similarity with one another indicating that this msp4 sequence is highly conserved (Figure 2). While these sequences had 96–100% identity with the msp4 sequences submitted in GenBank from various countries (Figure 2). The phylogeny did not indicate a population structure that was based on geography. Thereof, the set of sequences generated in this study was taken as a single population for neutrality test. Calculated values for Fu and Li's D-test was −3.49615 with a statistical significance of P < 0.02 and the value for Tajima's D was −2.16609 with a statistical significance of P < 0.01.
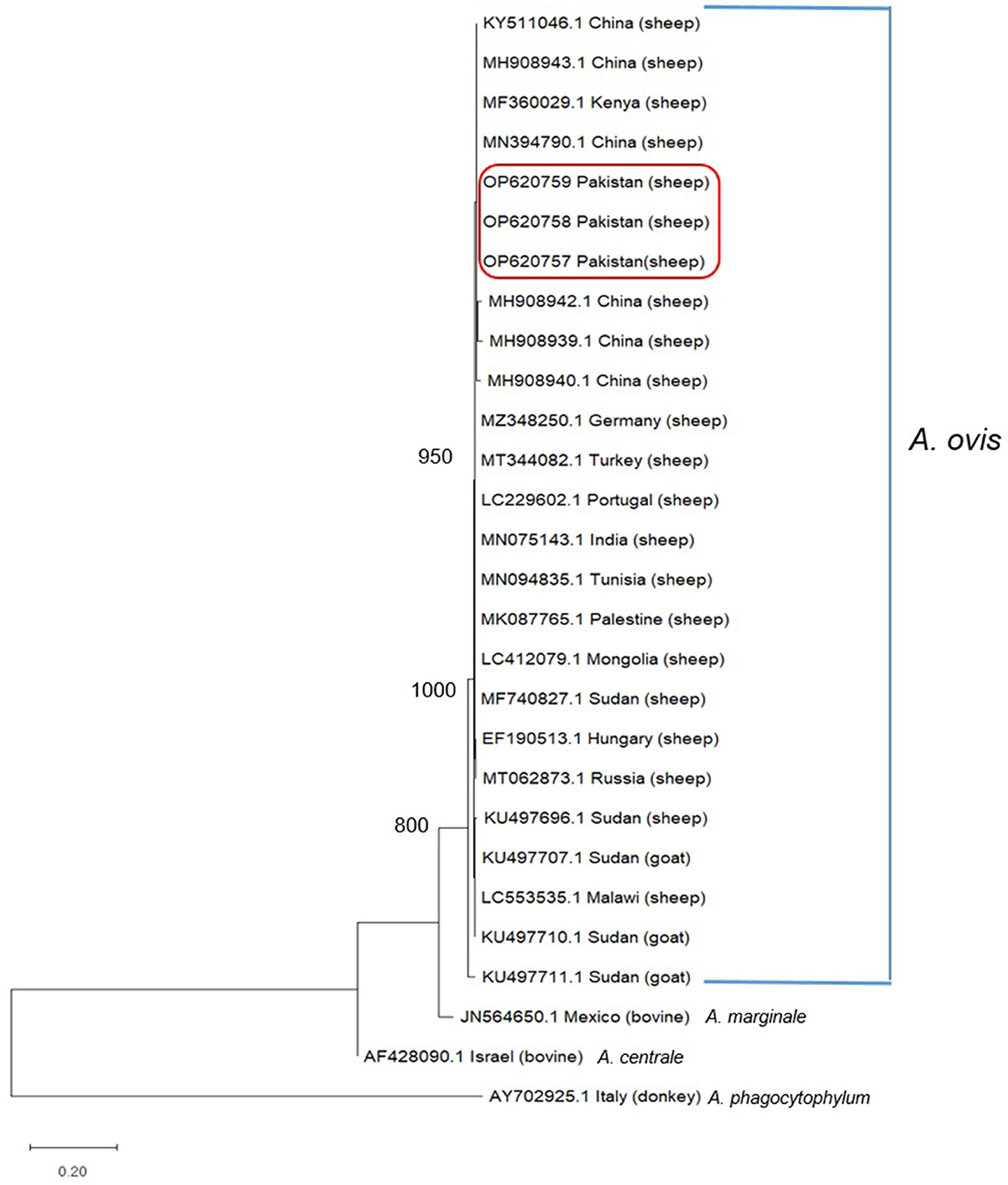
Figure 2. msp4 sequence alignment from Anaplasma ovis isolates amplified from Pakistani sheep and the sequences deposited in GenBank from various parts of world. Dashes indicate the conserved nucleotide positions. The positions with substitutions in DNA sequence of various Anaplasma spp. are represented by different colored nucleotides.
Discussion
Ovine anaplasmosis is among the most common TBDs reported in sheep from worldwide especially in tropical and sub-tropical regions (17). Since its identification in 1912, Anaplasma ovis has been reported from Asia, Europe, Africa, and the United States (8). Usually infection caused by Anaplasma ovis is not severe but cases with severe pathology in small ruminants have been documented from Northern United States and from Africa (18). In Haibei County of western China, during 2008, infection due to A. ovis resulted in 17% mortality and 40–50% sheep morbidity (19). Due to increased tick infestation, the incidence of ovine and caprine anaplasmosis is rising worldwide (20). Data regarding the prevalence of A. ovis in sheep and goats of Pakistan is very limited and only one previous report is available in literature to date.
In present study, we have reported that 12.5% of collected blood samples from sheep were infected with Anaplasma ovis. The only study from Pakistan is reported by Niaz et al. (21) in which sheep were enrolled from northern areas of Pakistan and they have used molecular tool (PCR) for the detection of Anaplasma spp. and they had found that 21.7% of enrolled sheep were infected with A. spp. and DNA sequencing of the amplified PCR products confirmed the presence of A. ovis in enrolled sheep. The prevalence of A. ovis has been reported from various parts of the world. The prevalence of A. ovis was reported to be 70.1% in sheep of Tunisa (22), 69% in sheep of Mogolia (23), 54.5% in sheep of Qinghai, China (24), 34.2% in sheep of central and Western Kenya (25), 29.7% in sheep of Turkey (26), 20.8% in sheep of Iran (27), 10% in sheep of West Iran (28), 5.7% in sheep of South Western China (29), and 2.6% in sheep of North East China (30). The variation in the prevalence of Anaplasma ovis between different studies is due to difference in the geographical and climatic conditions of sampling sites, age, gender, immunity of the host animal, tick density in a specific area, and also depends upon type of farm management technique that was followed during a specific study (31).
Phylogenetic and sequence analysis of msp4 partial sequence amplified during this study revealed that this gene sequence highly conserved (Figures 1, 2). Phylogenetic analysis revealed that our amplified DNA sequences are placed in stable monophyletic with 100% homology with msp4 partial sequences from China, Kenya, Germany, Turkey, Portugal, Tunisia, and India (Figure 1). However, our isolates are relatively distant from the strains isolated from small ruminants in Sudan, Hungry and Russia. The genetic variations between msp4 sequences of A. ovis that we have generated and those deposited in GenBank are probably due to the difference of geographic conditions of the areas from the bacterium samples were detected as not only the tick diversity and density varies with the geographic and climatic conditions but also the pathogenicity of A. ovis strains is also affected (32). Negative values of Tajima's D and Fu's F were obtained during present study that indicates an excess of rare variation, deviations from neutrality and a recent expansion of the population. This is consistent due to the origin of the sequences used. However, there number of DNA sequences that were used in the analysis was limited and use of large number of DNA sequences are recommended in future studies for a thorough evolutionary analysis. Further, we are not excluding that other msp4 variants may be circulating in the small ruminants of Pakistan.
In present study, all the epidemiological factors (age, sex, breed, size of herd, dogs with herd, and composition of herd studied) were not found associated with the prevalence of A. ovis. Contrary to our result, it is reported that eve are more susceptible to A. ovis infection as compared to ram. This is because of the fact that eve faces more hormonal fluctuations due to their reproductive cycles that makes them more susceptible to infections (22, 28). It is also reported that adult sheep were more affected than lambs. This is probably due to the fact that adults are more exposed to the environment (for grazing and for marketing) and hence they have higher chances to encounter vectors rather than lambs that are mostly kept at farms (22, 27).
Conclusions
In conclusion, we are reporting a moderate prevalence of A. ovis in sheep blood samples that were collected from Dera Ghazi Khan in Punjab (Pakistan). None of the enrolled sheep breed was specifically susceptible to A. ovis. Data generated in this study will pave the way for the prophylactic detection and control of ovine anaplasmosis in Pakistan. We recommend that similar and large scale studies must be conducted in all those areas of Pakistan that are unexplored for the incidence and prevalence of A. ovis. This will significantly help in control of tis bacterium and will improve the output of livestock sector in Pakistan.
Data availability statement
The data presented in the study are deposited in the GenBank, accession numbers OP620757-59.
Ethics statement
The animal study was reviewed and approved by Ethical Committee of Institute of Pure and Applied Biology, Bahauddin Zakariya University Multan (Pakistan). Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
FI and AdK designed and supervised the study. MN, MA, and ATar collected blood samples from sheep and noted epidemiological data. MN, MS, AsK, ATaq, and SA extracted DNA from blood samples and carried out PCR assays. IA-E, AS, and AA performed or assisted with the statistical analysis, sequence alignment, and phylogenetic study. MN, AS, AA, and FI wrote the text and edited and finalized the article. All authors approved the final version of the article.
Funding
The authors extend their appreciation to the Deputyship for Research & Innovation, Ministry of Education in Saudi Arabia for funding this research work through the project no. (IFKSURG-2-913).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Tanveer M, Farooq M, Amjad M, Asif M, Kashif M, Latif M, et al. Molecular prevalence, associated risk factors and phylogeny of Anaplasma marginale, Theileria ovis and T. lestoquardi in sheep from Pakistan. Comp Immunol Microbiol Infect Dis. (2022) 86:101822. doi: 10.1016/j.cimid.2022.101822
2. Abid K, Bukhari S, Asif M, Sattar A, Arshad M, Aktas M, et al. Molecular detection and prevalence of Theileria ovis and Anaplasma marginale in sheep blood samples collected from district Layyah in Punjab Pakistan. Trop Anim Health Prod. (2021) 53:439. doi: 10.1007/s11250-021-02870-5
3. Government of Pakistan. Pakistan Economic Survey. Islamabad: Ministry of Finance. (2021). p. 1–5.
4. Karim S, Budachetri K, Mukherjee N, Williams J, Kausar A, Hassan MJ, et al. A study of ticks and tick-borne livestock pathogens in Pakistan. PLoS Negl Trop Dis. (2017) 11:e0005681. doi: 10.1371/journal.pntd.0005681
5. Miranda EA, Han SW, Cho YK, Choi KS, Chae JS. Co-Infection with Anaplasma species and novel genetic variants detected in cattle and goats in the republic of Korea. Pathol. (2021) 10:28. doi: 10.3390/pathogens10010028
6. Li H, Zheng YC, Ma L, Jia N, Jiang BG, Jiang RR, et al. Human infection with a novel tick-borne Anaplasma species in China: A surveillance study. Lan Infect Dis. (2015) 15:663–70. doi: 10.1016/S1473-3099(15)70051-4
7. Rymaszewska A, Grenda S. Bacteria of the genus Anaplasma, characteristics of Anaplasma and their vectors. Rev Vet Med. (2008) 53:573–84. doi: 10.17221/1861-VETMED
8. Renneker S, Abdo J, Salih DE, Karagenc T, Bilgic H. Can Anaplasma ovis in small ruminants be neglected any longer? Transbound Emerg Dis. (2013) 60:105–12. doi: 10.1111/tbed.12149
9. Kocan KM, De la Fuente J, Guglielmone AA, Melendez RD. Antigens and alternatives for control of Anaplasma marginale infection in cattle. Clin Microbiol Rev. (2003) 16:698–712. doi: 10.1128/CMR.16.4.698-712.2003
10. Kocan KM, Busby AT, Allison RW, Breshears MA, Coburn L, Galindo RC, et al. Sheep experimentally infected with a human isolate of Anaplasma phagocytophilum serve as a host for infection of Ixodes scapularis ticks. Tick Tick Born Dis. (2012) 3:147–53. doi: 10.1016/j.ttbdis.2012.01.004
11. Grimberg J, Nawoschik S, Belluscio L, McKee R, Turck A, Eisenberg A, et al. simple and efficient non-organic procedure for the isolation of genomic DNA from blood. Nucl Acid Res. (1989) 17:8390. doi: 10.1093/nar/17.20.8390
12. Torina A, Agnone A, Blanda V, Alongi A, D'Agostino R, Caracappa S, et al. Development and validation of two PCR tests for the detection of and differentiation between Anaplasma ovis and Anaplasma marginale. Tick Tick Born Dis. (2012) 3:283–7. doi: 10.1016/j.ttbdis.2012.10.033
13. Tamura, K, Stecher G, Kumar S. MEGA 11: Molecular evolutionary genetics analysis version 11. Mol Biol Evol. (2021) 2021:msab120. doi: 10.1093/molbev/msab120
14. Kimura M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. (1980) 16:111–20. doi: 10.1007/BF01731581
15. Hall TA. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acid Symp Ser. (1999) 41:95–8.
16. Rozas J, Ferrer-Mata A, Sánchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, et al. DnaSP 6: DNA sequence polymorphism analysis of large datasets. Mol Biol Evol. (2017) 34:3299–302. doi: 10.1093/molbev/msx248
17. Ros-García A, Barandika JF, Garcia-Perez AL, Juste RA, Hurtado A. Assesment of exposure to piroplasms in sheep grazing in communal mountain pastures by using a multiplex DNA bead-based suspension array. Parasitol Vect. (2013) 6:277. doi: 10.1186/1756-3305-6-277
18. Tibbitts T, Goff W, Foreyt W, Stiller D. Susceptibility of two rocky mountains bighorn sheep to experimental infection with Anaplasma ovis. J Wildl Dis. (1992) 28:125–9. doi: 10.7589/0090-3558-28.1.125
19. Cui Y, Yan Y, Wang X, Cao S, Zhang Y, Jian F, et al. First molecular evidence of mixed infections of Anaplasma spp. in dogs in Henan, China. Tick Tick Born Dis. (2017) 8:283–9. doi: 10.1016/j.ttbdis.2016.12.001
20. Chochlakis D, Ioannou I, Sharif L, Kokkini S, Hristophi N, Dimitriou T, et al. Prevalence of Anaplasma spp. in goats and sheep in Cyprus. Vect Born Zoonot Dis. (2009) 9:457–63. doi: 10.1089/vbz.2008.0019
21. Niaz S, Rahman ZU, Ali I, Cossio-Boyugar R, Amaro-Estrada I, Alanazi AD, et al. Molecular prevalence, characterization and associated risk factors of Anaplasma spp. and Theileria spp in small ruminants in Northern Pakistan. Parasitol. (2021) 28:3. doi: 10.1051/parasite/2020075
22. Belkahia H, Said MB, El Hamdi S, Yahiaoui M, Gharbi M, Daaloul-Jedidi M, et al. First molecular identification and genetic characterization of Anaplasma ovis in sheep from Tunisia Small. Rumin Res. (2014) 121:404–10. doi: 10.1016/j.smallrumres.2014.07.009
23. Enkhtaivan B, Narantsatsrala S, Davaasurena B, Otgonsurena D, Amgalanbaatara T, Uuganbayara E, et al. Molecular detection of Anaplasma ovis in small ruminants and ixodid ticks from Mongolia. Parasitol Int. (2019) 69:47–53. doi: 10.1016/j.parint.2018.11.004
24. Zhang X, Liu Z, Yang J, Chen Z, Guan G, Ren Q, et al. Multiplex PCR for diagnosis of Theileria uilenbergi, Theileria luwenshuni, and Theileria ovis in small ruminants. Parasitol Res. (2014) 113:527–31. doi: 10.1007/s00436-013-3684-9
25. Ringo AE, Aboge GO, Adjou Moumouni PF, Hun Lee S, Jirapattharasate C, Liu M, et al. Molecular detection and genetic characterisation of pathogenic Theileria, Anaplasma and Ehrlichia species among apparently healthy sheep in central and western Kenya. Onder J Vet Res. (2019) 86:1630. doi: 10.4102/ojvr.v86i1.1630
26. Zhou M, Cao S, Sevinc F, Sevinc M, Ceylan O, Ekici S, et al. Molecular detection and genetic characterization of Babesia, Theileria and Anaplasma amongst apparently healthy sheep and goats in the central region of Turkey. Tick Tick Born Dis. (2017) 8:246–52. doi: 10.1016/j.ttbdis.2016.11.006
27. Yousefi A, Rahbari S, Shayan P, Sadeghi-dehkordi Z, Bahonar A. Molecular detection of Anaplasma marginale and Anaplasma ovis in sheep and goat in west highland pasture of Iran. Asia Pac J Trop Biomed. (2017) 7:455–9. doi: 10.1016/j.apjtb.2017.01.017
28. Mohammadian B, Noaman V, Emami SJ. Molecular survey on prevalence and risk factors of Anaplasma spp. infection in cattle and sheep in West of Iran. Trop Anim Health Prod. (2021) 53:266. doi: 10.1007/s11250-021-02707-1
29. Yang J, Hana R, Niua Q, Liua Z, Guana G, Liua G, et al. Occurrence of four Anaplasma species with veterinary and public health significance in sheep, Northwestern China. Tick Tick Born Dis. (2017) 9:82–5. doi: 10.1016/j.ttbdis.2017.10.005
30. Shi Y, Yang J, Guan G, Liu Z, Luo J, Song M. Molecular investigation of Anaplasma species in sheep from Heilongjiang Province, northeast China identified four Anaplasma species and a novel genotype of Anaplasma capra. Parasitol Int. (2020) 76:102072. doi: 10.1016/j.parint.2020.102072
31. Rar V, Golovljova I. Anaplasma, Ehrlichia, and “Candidatus neoehrlichia” bacteria: Pathogenicity, biodiversity, and molecular genetic characteristics, a review. Infect Genet Evol. (2011) 11:1842–61. doi: 10.1016/j.meegid.2011.09.019
Keywords: Anaplasma ovis, sheep, msp4 gene, Pakistan, phylogeny
Citation: Naeem M, Amaro-Estrada I, Taqadus A, Swelum AA, Alqhtani AH, Asif M, Sajid M, Khan AU, Tariq A, Anjum S, Khan A and Iqbal F (2023) Molecular prevalence and associated risk factors of Anaplasma ovis in Pakistani sheep. Front. Vet. Sci. 10:1096418. doi: 10.3389/fvets.2023.1096418
Received: 12 November 2022; Accepted: 13 March 2023;
Published: 29 March 2023.
Edited by:
Khalid Mehmood, Islamia University of Bahawalpur, PakistanReviewed by:
Muhammad Ehsan, Islamia University of Bahawalpur, PakistanMehmet Fatih Aydin, Karamanoǧlu Mehmetbey University, Türkiye
Copyright © 2023 Naeem, Amaro-Estrada, Taqadus, Swelum, Alqhtani, Asif, Sajid, Khan, Tariq, Anjum, Khan and Iqbal. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Furhan Iqbal, ZnVyaGFuLmlxYmFsQGJ6dS5lZHUucGs=; Adil Khan, em9vbG9neWF3a3VtQGdtYWlsLmNvbQ==
†These authors share first authorship
 Muhammad Naeem1†
Muhammad Naeem1† Itzel Amaro-Estrada
Itzel Amaro-Estrada Ayman A. Swelum
Ayman A. Swelum Abdulmohsen H. Alqhtani
Abdulmohsen H. Alqhtani Adil Khan
Adil Khan Furhan Iqbal
Furhan Iqbal