- 1Department of Pathobiology and Veterinary Science, University of Connecticut, Storrs, CT, United States
- 2College of Veterinary Medicine, Konkuk University, Seoul, Republic of Korea
- 3Department of Infectious Diseases and Microbiology, School of Veterinary Medicine, Mongolian University of Life Sciences, Ulaanbaatar, Mongolia
- 4Plum Island Animal Disease Center, Agriculture Research Service, US Department of Agriculture, Greenport, NY, United States
- 5KCAV Co., Ltd., Seoul, Republic of Korea
African swine fever (ASF) is a highly contagious and fatal disease affecting domestic and wild pigs caused by the African swine fever virus (ASFV). Since the first outbreak in China in August 2018, ASF has spread rapidly in Asia. and the first case in Mongolia was confirmed in January 2019. In this study, we report the first whole genome sequence of an ASFV (ASFV SS-3/Mongolia/2019) detected from a backyard pig in Mongolia in February 2019 using whole genome sequencing. We analyzed their phylogenetic relationship with other genotype II ASFVs from Eurasia. The ASFV SS-3/Mongolia/2019 belonged to genotype II (p72 and p54), serogroup 8 (CD2v), Tet-10a variant (pB602L), and IGRIII variant (intergenic region between the I73R/I329L genes). A total of five amino acid substitutions were observed in MGF 360-10L, MGF 505-4R, MGF 505-9R, NP419L, and I267L genes compared to the ASFV Georgia 2007/1 virus. ML phylogenetic analysis of the whole genome sequence showed that the virus shares a high nucleotide sequence identity with ASFVs recently identified in Eastern Europe and Asia and clustered with the ASFV/Zabaykali/WB5314/2020|Russia|2020 virus which was identified at the border between the Russian Federation and Mongolia in 2020. Our results suggest that trans boundary spread of ASF occurred through close geographic proximity.
Introduction
African swine fever (ASF) is caused by the African swine fever virus (ASFV), a virus within the genus Asfivirus of the family Asfarviridae. It is a highly contagious and high-mortality disease affecting domestic pigs and wild boars and a notifiable disease to the World Organization for Animal Health (WOAH) (1, 2). ASF was first described in Kenya in 1921; it subsequently re-emerged from Africa into Georgia and rapidly spread into Eastern Europe and Asia (1). In December 2007, ASF was reported in Russia, with subsequent outbreaks occurring between 2008 and 2009 and affecting domestic pigs and wild boars (1). In August 2018, the disease was reported in China; it spread rapidly across that country and Mongolia, and then to Vietnam, Cambodia, North Korea, Laos, the Philippines, Myanmar, South Korea, Timor-Leste, and Indonesia (2).
On 10 January 2019, Mongolia's State Central Veterinary Laboratory (SCVL) confirmed the first ASF outbreak in Bulgan province, Mongolia (3). After this first detection, 11 additional ASF outbreaks were recorded in Mongolia, involving seven provinces (4). A total of 105 farms and holdings were affected by the disease, resulting in the death or elimination of more than 3,000 exposed pigs (> 10 % of the total pig population in Mongolia) (4). While complete genome sequences of ASFV have been reported from several countries affected by ASF (5–12), a complete genome sequence of ASFV from Mongolia has not been published. However, several characterizations of the ASFV strain from the 2019 outbreak in Bulgan province, Mongolia have been reported in a previous study: partial p72, full p54, partial pB602L, and partial CD2v genes, as well as a 356-bp fragment between the I73R and I329L genes (2).
In this study, we report the first whole genome sequence of an ASFV detected in a backyard pig in Mongolia in February 2019 using next-generation sequencing (NGS). We also analyze its phylogenetic relationship with other genotype II ASFVs from Eurasia.
2. Materials and methods
Spleen tissue from a backyard pig carcass found at a dumping ground near Ulaanbaatar was collected during passive surveillance on 14 February 2019, and it was confirmed to be ASFV-positive using quantitative reverse transcription real-time PCR (RT-qPCR) assay (13, 14). DNA was extracted from spleen tissue from the ASF-positive backyard pig carcass using a DNeasy Blood and Tissue kit (Qiagen, Valencia, CA) in accordance with the manufacturer's instructions and eluted in distilled water. DNA concentrations were determined using a Qubit BR dsDNA assay kit (Invitrogen, Carlsbad, CA). DNA samples were diluted to 0.2 ng/μl and libraries were prepared using the Illumina Nextera XT DNA Library Prep Kit (Illumina, San Diego, CA). The concentration of sample libraries was determined using the Qubit dsDNA HS assay kit, and libraries were diluted to a 2 nM concentration and combined in equal volumes to form the pooled library. Subsequently, 600 μl of the 10 pM libraries were submitted for pair-end sequencing using the MiSeq reagent kit v2 (500 cycles) (Illumina, San Diego, CA).
The raw reads were adapter-trimmed for known Illumina adapters and quality-trimmed (Q >20 and minimum length >50) with Bbduk (https://sourceforge.net/projects/bbmap). Trimmed reads were mapped to the reference sequence, the ASFV Georgia 2007/1 (GenBank acc. FR682468.2), using minimap2 (15) with the default settings in Geneious Prime 10 Software (https://www.geneious.com/), and the consensus genome sequences (hereafter referred to as ASFV SS-3/Mongolia/2019) were called using Geneious Prime 10 with default parameter settings. To demonstrate the phylogenetic organization of the ASFV, ASFV genotype I 56/Ca/1978 (GenBank acc. MN270969) and all available full-length genome sequences (>150,000 bp) of genotype II ASFVs (n = 70) were downloaded from the NCBI GenBank database. The online multiple alignment server MAFFT, version 7 (https://mafft.cbrc.jp/alignment/software/), was used for sequence alignment of whole genome sequences with the default settings (16). Maximum likelihood (ML) phylogenies were constructed using RAxML-HPC v.8 using the general time-reversible (GTR) nucleotide substitution model with gamma distribution and with 1,000 rapid bootstrap replicates (17). Phylogenetic trees were rooted to the ASFV genotype I 56/Ca/1978 as an outgroup and converted to cladogram form for better visualization of the genetic relationships.
3. Results and discussion
From 14,542,882 total raw reads, 111,352 reads were mapped to the reference ASF sequence. The length of the assembled genome was 190,591 bp, with a mean coverage depth of 162.3 reads (minimum: 43; maximum: 2,638). The molecular characteristics of ASFV SS-3/Mongolia/2019 were consistent with previous findings on ASFVs from the 2019 outbreaks in Mongolia (2), specifically genotype II (p72 and p54), serogroup 8 (CD2v), Tet-10a (pB602L), IGRIII variant (intergenic region between the I73R/I329L genes). It was found to have a high degree of nucleotide identity (99.98%) with ASFV Georgia 2007/1(data not shown). A total of five amino acid substitutions were observed in genes MGF 360-10L, MGF 505-4R, MGF 505-9R, NP419L, and I267L relative to ASFV Georgia 2007/1 (Supplementary Table 1). MGF360 and MGF505 gene products have been reported to be related to suppression of a type I interferon response, as analyzed using a swine cDNA microarray (18). NP419L encodes DNA ligase involved in DNA replication, repair, nucleotide metabolism, transcription, and other enzymatic activities or host defense evasion (19). Finally, I267L is an important virulence factor that operates by impairing innate immune responses mediated by the RNA Pol-III-RIG-I axis (20).
ML phylogenetic analysis of the whole genome sequence suggested that the virus belongs to the Georgia-07-like genotype II ASF virus (Figure 1). It has a high degree of nucleotide identity (>97%) with ASFVs recently detected in Eastern Europe and Asia (data not shown). It was found to cluster with the ASFV/Zabaykali/WB5314/2020|Russia|2020 virus, which was identified at the border between Russia and Mongolia in 2020 (Figure 1) (5), suggesting that transboundary spread of ASF occurred through close geographic proximity. In late 2020, ASF-related mass mortality of wild boar was observed in MinJiin Khangai mountain, Mungunmorit soum of Tuv province, and Yeroo soum of Selenge province, which are remote areas of Mongolia bordering with Russia. These areas are far from domestic animal farms. We assume that the virus might have been disseminated by the movement of wild boar carriers between Russia and Mongolia.
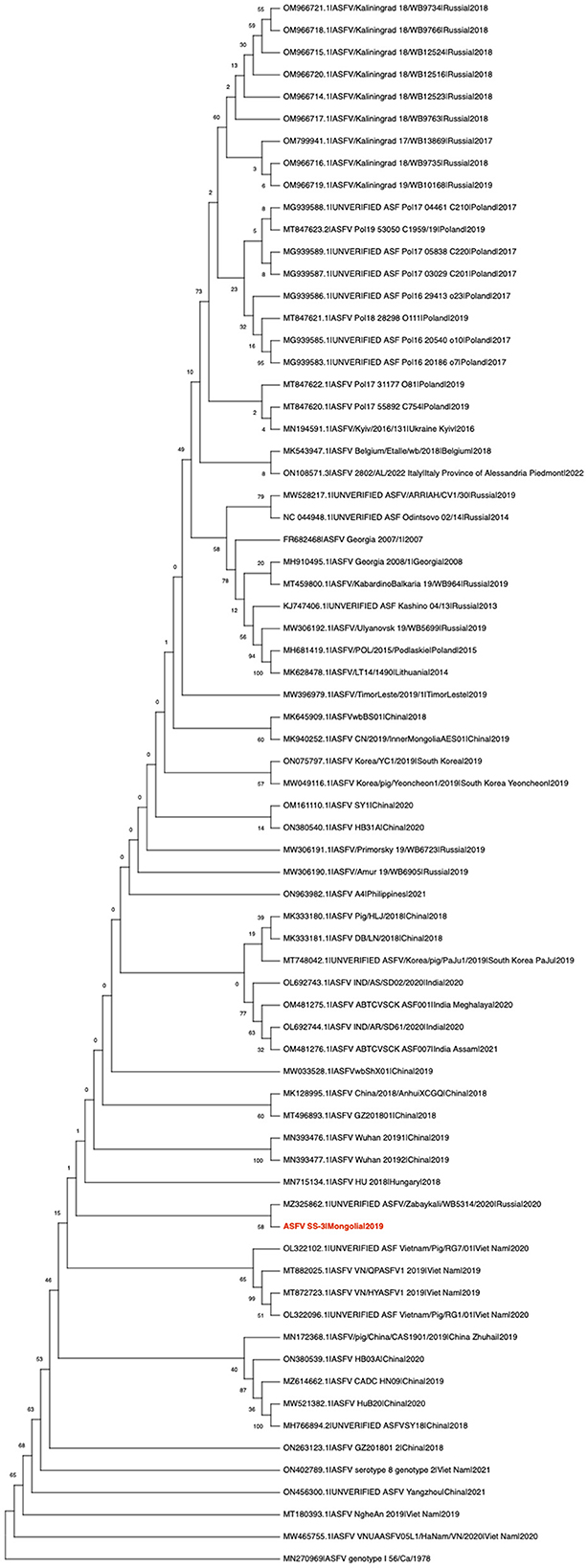
Figure 1. Maximum likelihood analysis of 70 complete coding sequences of ASFV, including the ASFV SS-3|Mongolia|2019 sequenced in this study (in red). The phylogeny was rooted to the ASFV genotype I 56/Ca/1978 as an outgroup and converted to cladogram form for better visualization of the genetic relationships. Numerical values represent 1,000 bootstrap replicate values expressed as percentages.
Data availability statement
The data presented in the study are deposited in GenBank under the accession number OP787478 and Bioproject accession number PRJNA924538.
Ethics statement
Ethical review and approval was not required for the study on animals in accordance with the local legislation and institutional requirements.
Author contributions
J-YH: data analysis and manuscript writing. D-HL: study design, data analysis, and manuscript editing. E-OT-O: data collection and manuscript editing. DG, MB, and GR: study design and manuscript editing. S-SN and C-SS: data analysis, study design, and manuscript editing. All authors contributed to the article and approved the submitted version.
Funding
J-YH and S-SN are supported by the Korea Institute of Planning and Evaluation for Technology in Food, Agriculture, and Forestry (IPET) through the Animal Disease Management Technology Development Program, funded by the Ministry of Agriculture, Food, and Rural Affairs (MAFRA) (122016-02).
Conflict of interest
C-SS was employed by KCAV Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2023.1094052/full#supplementary-material
References
1. Mazur-Panasiuk N, Wozniakowski G, Niemczuk K. The first complete genomic sequences of african swine fever virus isolated in Poland. Sci Rep. (2019) 9:4556. doi: 10.1038/s41598-018-36823-0
2. Ankhanbaatar U, Sainnokhoi T, Khanui B, Ulziibat G, Jargalsaikhan T, Purevtseren D, et al. African Swine Fever Virus Genotype Ii in Mongolia, 2019. Transbound Emerg Dis. (2021) 68:2787–94. doi: 10.1111/tbed.14095
3. Heilmann M, Lkhagvasuren A, Adyasuren T, Khishgee B, Bold B, Ankhanbaatar U, et al. African Swine Fever in Mongolia: Course of the Epidemic and Applied Control Measures. Vet Sci. (2020) 7:24. doi: 10.3390/vetsci7010024
4. Food and Agriculture Organization of the United Nations. South Asia Animal Disease Outbreaks and News 61 ed. Rome, Italy: Food and Agriculture Organization of the United Nations (2020).
5. Mazloum A, Igolkin AS, Shotin AR, Zinyakov NG, Vlasova NN, Aronova EV, et al. [Analysis of the Whole-Genome Sequence of an Asf Virus (Asfarviridae: Asfivirus: African Swine Fever Virus) Isolated from a Wild Boar (Sus Scrofa) at the Border between Russian Federation and Mongolia]. Vopr Virusol. (2022) 67:153–64. doi: 10.36233/0507-4088-104
6. Mileto P, da Conceicao F, Stevens V, Cummins D, Certoma A, Neave MJ, et al. Complete genome sequence of african swine fever virus isolated from a domestic pig in timor-leste, 2019. Microbiol Resour Announc. (2021) 10:e0026321. doi: 10.1128/MRA.00263-21
7. Hien ND, Nguyen LT, Hoang LT, Bich NN, Quyen TM, Isoda N, et al. First Report of a Complete Genome Sequence of a Variant African Swine Fever Virus in the Mekong Delta, Vietnam. Pathogens. (2022) 11:797. doi: 10.3390/pathogens11070797
8. Granberg F, Torresi C, Oggiano A, Malmberg M, Iscaro C, De Mia GM, et al. Complete Genome Sequence of an African Swine Fever Virus Isolate from Sardinia, Italy. Genome Announc. (2016) 4:e01220–16. doi: 10.1128/genomeA.01220-16
9. Hakizimana JN, Ntirandekura JB, Yona C, Nyabongo L, Kamwendo G, Chulu JLC, et al. Complete Genome Analysis of African Swine Fever Virus Responsible for Outbreaks in Domestic Pigs in 2018 in Burundi and 2019 in Malawi. Trop Anim Health Prod. (2021) 53:438. doi: 10.1007/s11250-021-02877-y
10. Koltsov A, Tulman ER, Namsrayn S, Kutish GF, Koltsova G. Complete genome sequence of virulent genotype I African swine fever virus strain K49 from the democratic Republic of the Congo, Isolated from a Domestic Pig (Sus Scrofa Domesticus). Arch Virol. (2022) 167:2377–80. doi: 10.1007/s00705-022-05543-2
11. Kovalenko G, Ducluzeau AL, Ishchenko L, Sushko M, Sapachova M, Rudova N, et al. Complete genome sequence of a virulent african swine fever virus from a domestic pig in Ukraine. Microbiol Resour Announc. (2019) 8:e00883–19. doi: 10.1128/MRA.00883-19
12. Senthilkumar D, Rajukumar K, Venkatesh G, Singh F, Tosh C, Kombiah S, et al. Complete genome analysis of african swine fever virus isolated from domestic pigs during the first asf outbreaks in India. Transbound Emerg Dis. (2022) 69:e2020–e7. doi: 10.1111/tbed.14536
13. Fernandez-Pinero J, Gallardo C, Elizalde M, Robles A, Gomez C, Bishop R, et al. Molecular diagnosis of African swine fever by a new real-time pcr using universal probe library. Transbound Emerg Dis. (2013) 60:48–58. doi: 10.1111/j.1865-1682.2012.01317.x
14. King DP, Reid SM, Hutchings GH, Grierson SS, Wilkinson PJ, Dixon LK, et al. Development of a Taqman Pcr assay with internal amplification control for the detection of african swine fever virus. J Virol Methods. (2003) 107:53–61. doi: 10.1016/S0166-0934(02)00189-1
15. Li H. Minimap2: Pairwise Alignment for Nucleotide Sequences. Bioinformatics. (2018) 34:3094–100. doi: 10.1093/bioinformatics/bty191
16. Katoh K, Rozewicki J, Yamada KD. Mafft online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. (2019) 20:1160–6. doi: 10.1093/bib/bbx108
17. Stamatakis A. Raxml Version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. (2014) 30:1312–3. doi: 10.1093/bioinformatics/btu033
18. Afonso CL, Piccone ME, Zaffuto KM, Neilan J, Kutish GF, Lu Z, et al. African swine fever virus multigene family 360 and 530 genes affect host interferon response. J Virol. (2004) 78:1858–64. doi: 10.1128/JVI.78.4.1858-1864.2004
19. Dixon LK, Chapman DA, Netherton CL, Upton C. African Swine Fever Virus Replication and Genomics. Virus Res. (2013) 173:3–14. doi: 10.1016/j.virusres.2012.10.020
Keywords: African swine fever (ASF), whole genome sequencing (WGS), Mongolia, backyard pig, phylogenetic analysis
Citation: Hyeon J-Y, Tseren-Ochir E-O, Lee D-H, Nahm S-S, Gladue DP, Borca MV, Song C-S and Risatti GR (2023) Whole genome sequencing and phylogenetic analysis of African swine fever virus detected in a backyard pig in Mongolia, 2019. Front. Vet. Sci. 10:1094052. doi: 10.3389/fvets.2023.1094052
Received: 09 November 2022; Accepted: 23 January 2023;
Published: 20 February 2023.
Edited by:
Iryna Goraichuk, Agricultural Research Service (USDA), United StatesReviewed by:
Oliver Lung, Canadian Food Inspection Agency (CFIA), CanadaChristopher Lewis Netherton, The Pirbright Institute, United Kingdom
Denis V. Kolbasov, Federal Research Center of Virology and Microbiology, Russia
Copyright © 2023 Hyeon, Tseren-Ochir, Lee, Nahm, Gladue, Borca, Song and Risatti. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dong-Hun Lee,  ZG9uZ2h1bmxlZUBrb25rdWsuYWMua3I=
ZG9uZ2h1bmxlZUBrb25rdWsuYWMua3I=
 Ji-Yeon Hyeon
Ji-Yeon Hyeon Erdene-Ochir Tseren-Ochir
Erdene-Ochir Tseren-Ochir Dong-Hun Lee
Dong-Hun Lee Sang-Soep Nahm
Sang-Soep Nahm Douglas P. Gladue4
Douglas P. Gladue4 Manuel V. Borca
Manuel V. Borca Guillermo R. Risatti
Guillermo R. Risatti