- 1Shanghai Veterinary Research Institute, Chinese Academy of Agricultural Sciences, Shanghai, China
- 2College of Veterinary Medicine, Xinjiang Agricultural University, Urmuqi, China
- 3Tengzhou Animal Disease Prevention and Control Center of Shandong Province, Tengzhou, China
Actinobacillus pleuropneumoniae and Pasteurella multocida are two important bacterial pathogens in swine industry. In the present study, resistance profiles of nine commonly used antibiotics of A. pleuropneumoniae and P. multocida isolates of swine origin from different regions of China were investigated by determination of minimum inhibitory concentrations (MICs). In addition, genetic relationship of the florfenicol-resistant A. pleuropneumoniae and P. multocida isolates was determined by pulsed-field gel electrophoresis (PFGE). The genetic basis of florfenicol resistance in these isolates were explored by floR detection and whole genome sequencing. High resistance rates (>25%) of florfenicol, tetracycline and trimethoprim- sulfamethoxazole were observed for both bacteria. No ceftiofur- and tiamulin- resistant isolates were detected. Furthermore, all the 17 florfenicol-resistant isolates (nine for A. pleuropneumoniae and eight for P. multocida) were positive for floR gene. The presence of similar PFGE types in these isolates suggested that clonal expansion of some floR-producing strains occurred in the pig farms from same regions. WGS and PCR screening showed that three plasmids, named pFA11, pMAF5, and pMAF6, were the cargos of the floR genes in the 17 isolates. Plasmid pFA11 exhibited novel structure and carried several resistance genes, including floR, sul2, aacC2d, strA, strB, and blaROB − 1. Plasmids pMAF5 and pMAF6 were presented in A. pleuropneumoniae and P. multocida isolates from different regions, suggesting horizontal transfer of the two plasmids are important for the floR dissemination in these Pasteurellaceae pathogens. Further studies of florfenicol resistance and its transfer vectors in Pasteurellaceae bacteria of veterinary origin are warranted.
Introduction
Actinobacillus pleuropneumoniae and Pasteurella multocida, two members of the family Pasteurellaceae, are important pathogens and cause significant economic losses in swine industry worldwide (1). A. pleuropneumoniae is etiological agent of porcine contagious pleuropneumonia and could be divided into two biovars based on the NAD dependence (2). To date, a total of 19 serovars has been described, and of them, serovars 1, 5, 9, and 11 are highly virulent and frequently observed in the clinical cases of porcine pleuropneumonia (3). P. multocida is commensal bacterial inhabitant of the upper respiratory tract of pigs and associated with atrophic rhinitis and pneumonia in clinical diseased pigs (4). Besides pigs, P. multocida also causes different diseases in various animals, such as cattle, poultry, rabbit and pets, and even humans (5, 6). Using molecular typing methods, P. multocida isolates could be classified into five capsular serotypes (A, B, D, E, F) and eight LPS genotypes (L1–L8) (7). Of them, serotypes A and D are most commonly isolated in diseased pigs (8). Due to the diverse serotypes and geographical difference in their prevalence, there is presently no vaccine with satisfactory protection to control A. pleuropneumoniae and P. multocida infections (2). For this reason, antibiotic use is still the dominant strategy for the treatment and prevention of the infectious diseases caused by these two pathogenic bacteria. However, due to the massive and long-term antibiotic use in pig farm, resistance to commonly used antibiotics in A. pleuropneumoniae and P. multocida isolates has emerged and been increasingly reported all over the world (1, 9).
Florfenicol, one member of phenicol family, is exclusively approved for veterinary medicine. With the structural modification, florfenicol do not have the toxic side effect of chloramphenicol and could avoid the acetyltransferase-mediated drug resistance (10). In China, florfenicol was licensed in 1999 for treatment of bacterial infections in cattle, pig, chicken and fish (11). It is one of the most used antibiotics in food-animal industry and about 10,000 tons was used in China in 2013 (12). Florfenicol is a broad-spectrum antimicrobial drug that are widely used to treat respiratory diseases and other pathogenic bacterial infections in pig farm, including A. pleuropneumoniae and P. multocida infections. Florfenicol-resistant A. pleuropneumoniae and P. multocida isolates have been sporadically reported in different countries and, same to other Gram-negative bacteria, floR is the predominant determinant for resistance to florfenicol in those isolates (8, 9, 13, 14). In these isolates, floR was mainly carried by plasmids with various sizes or integrative conjugative elements (ICEs) (9, 15–21).
There are few data about the florfenicol resistance and its genetic basis in A. pleuropneumoniae and P. multocida isolates from swine in China. In the present study, we collected A. pleuropneumoniae and P. multocida isolates from pig farms in different regions of China, and characterized the genetic relationships of the florfenicol-resistant isolates and the structures of relevant floR-carrying plasmids.
Materials and methods
Bacterial strains and susceptibility testing
A total of 19 A. pleuropneumoniae isolates and 31 P. multocida isolates were used in this study. These isolates were collected between September 2018 to December 2020 in routine clinical diagnosis in several pig farms from different provinces of China, including Jiangsu, Xinjiang, Shanghai and Hubei. Minimum inhibitory concentrations (MICs) were determined by broth microdilution method according to recommendation of the Clinical and Laboratory Standards Institute (CLSI) supplement VET08 (22). Nine commonly used antibiotics were tested, including florfenicol, ceftiofur, enrofloxacin, tetracycline, tiamulin, tilmicosin, gentamicin, ampicillin and trimethoprim-sulfamethoxazole. For A. pleuropneumoniae, clinical breakpoints were interpreted with CLSI-VET08 document for these antibiotics, except trimethoprim-sulfamethoxazole, for which the Epidemiological Cut-off value (ECOFF) value given by European Committee on Antimicrobial Susceptibility Testing (EUCAST) was temporarily used as resistance breakpoint (https://www.eucast.org/). In addition, CLSI has no clinical breakpoints of tiamulin, gentamicin, and trimethoprim-sulfamethoxazole for P. multocida and the ECOFF values from EUCAST were also used as resistance breakpoints for these antibiotics. Moreover, MIC50 and MIC90 were exhibited to reflect the MIC distributions. Escherichia coli ATCC 25922 and A. pleuropneumoniae ATCC 27090 were used as quality control strains.
Pulsed-field gel electrophoresis and floR detection
All the florfenicol-resistant isolates were screened for the presence of floR gene with specific primer sets (5′-CTGAACACGACGCCCGCTATG-3′ and 5′-CAGGACCGCTCCGCAAACAA-3′) with the amplification condition: 95°C for 5 min; followed by 30 cycles of 95°C for 30 s, 58°C for 30 s, and 72°C for 60 s; and extension at 72°C for 7 min. Sanger sequencing of the amplicons was performed to confirm the PCR results. To investigate the genetic relationship between the floR-positive isolates, ApaI-PFGE was performed as described previously with modified running parameter: 14°C, 6 V/cm, 2.16 s initial switching time, 54.2 s final switching time and a 18 h of total run (23, 24). The fingerprinting profiles were analyzed with BioNumerics v7.1 (Applied-Maths, Kortrijk, Belgium) to construct a dendrogram based on the Dice coefficient with 1.0% band-position tolerance and 1.5% optimization. Genetical relatedness between the floR-positive isolates were determined with the cut-off of 80% homology of the DNA restriction patterns (25, 26).
Whole genome sequencing and bioinformatic analysis
The genetically unrelated floR-positive three A. pleuropneumoniae and three P. multocida isolates were further subjected to WGS with the Illumina Hiseq 2000 platform. The raw data was assembled using the SPAdesv.3.13.0. Resfinder was used to analyze resistance genes and to find the floR-bearing contigs. Invert PCRs with the primers located at the both ends of the floR-bearing contigs were performed and the generated amplicons were subjected to Sanger sequencing, by which the complete sequences of the floR-carrying plasmids were obtained. Annotation was automatically generated using the RAST (https://rast.nmpdr.org/) (27). Structure comparisons with the plasmids deposited in Genbank were performed by BLAST (www.ncbi.nlm.nih.gov/BLAST) (28). Moreover, three specific primer sets were designated for the obtained three types of floR-carrying plasmids (P1: 5′-AGGTCGCCCTAAACTTCC-3′ and P2: 5′-TATCGCCTGCCATCCC-3′ for pFA11, P3: 5′-AATGGTTACAGGTGGAAGA-3′ and P4: 5′-GCACTGCTGCTGATGG-3′ for pMAF5, and P5: 5′-AGGGCGATTTATGATTGA-3′ and P6: 5′-TCGGCGAACTTTACGG-3′ for pMAF6). PCRs with the three primer sets were conducted to detect the plasmid types in the floR-producing isolates without WGS analysis. The amplicons were sequenced and compared with corresponding plasmids to confirm the PCR results.
Nucleotide sequence accession number
The complete sequences of the floR-carrying plasmids pFA11, pMAF5 and pMAF6 have been deposited in the GenBank database with the accession numbers CP100665, CP100664, CP100663.
Results and discussion
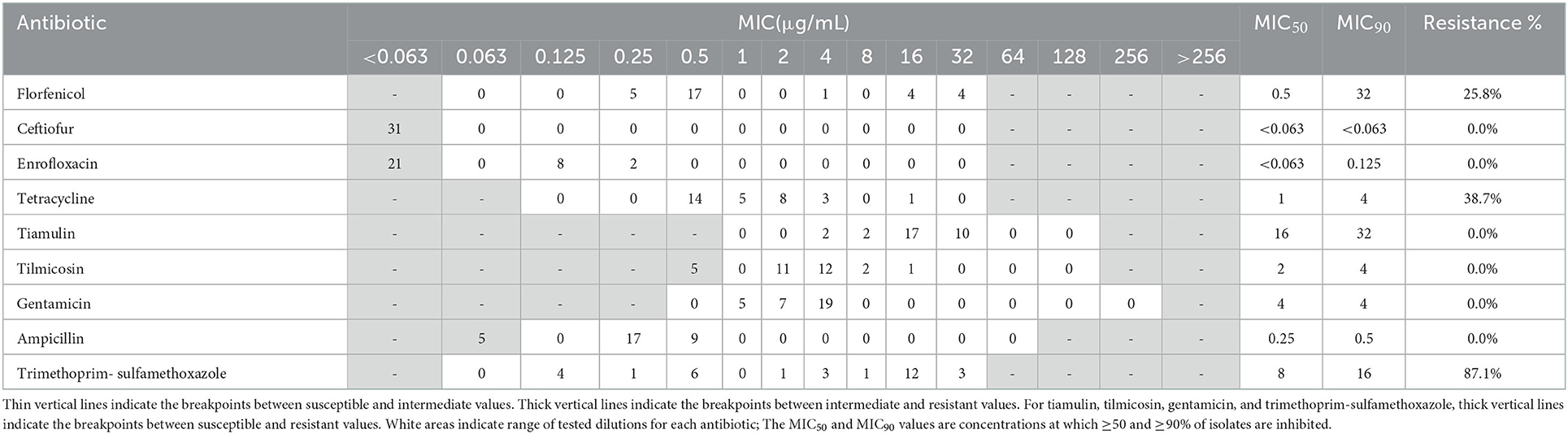
Resistance profiles of the 19 A. pleuropneumoniae and 31 P. multocida isolates were exhibited in Tables 1, 2. In A. pleuropneumoniae isolates, high resistance rates were observed for tetracycline and trimethoprim-sulfamethoxazole (89.5 and 68.4%, respectively), while florfenicol showed moderate degree of resistance (47.4%). Low frequencies of resistance were observed for ampicillin, enrofloxacin, tilmicosin and gentamicin (10.5, 10.5, 5.3, and 5.3%, respectively). No resistance of ceftiofur and tiamulin were detected for these A. pleuropneumoniae isolates. In P. multocida isolates, highest resistance rate was observed for trimethoprim-sulfamethoxazole (87.1%). Eight and twelve isolates showed resistance to florfenicol and tetracycline, corresponding to resistance rates 25.8 and 38.7%, respectively. For other test antibiotics, no resistance was detected.
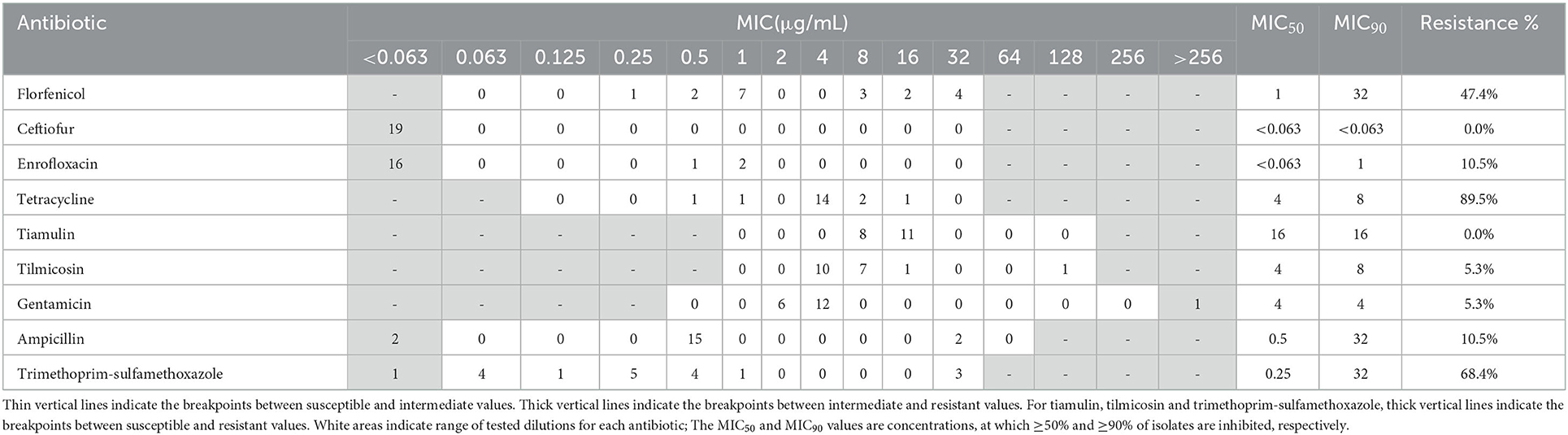
Table 1. MIC distribution and resistance profiles of nine antibiotics for 19 A. pleuropneumoniae isolates.
Florfenicol is an important antibiotic and extensively used in swine industry. Unlike other Gram-negative bacteria, such as Enterobacteriaceae, resistance to florfenicol in A. pleuropneumoniae and swine-origin P. multocida isolates has rarely been reported all over the world (1, 4, 29–31), except in Korea, where about 36% of A. pleuropneumoniae isolates collected from 2006 to 2013, and 16.3% swine-origin P. multocida isolates collected between 2008 and 2016 were resistant to florfenicol (13). In China, there is no epidemiological data about florfenicol resistance available for A. pleuropneumoniae. In 2009, a study concerning the antimicrobial resistance and virulence genes of swine-origin P. multocida isolates in China showed that no florfenicol-resistant strains were detected (4). In the present study, relatively high degree of florfenicol resistance were observed for A. pleuropneumoniae and swine-origin P. multocida isolates (9/19, 47.4% and 8/31, 25.8%, respectively). All of these strains were positive for floR gene. These data indicated that floR-mediated florfenicol resistance in P. multocida and A. pleuropneumoniae isolates have emerged and spread in pig farms of China in recent years. According to our survey, florfenicol is one of the most frequently applied antibiotics in the pig farms where the P. multocida and A. pleuropneumoniae strains used in this study were recovered. The high selection pressure due to extensive use of florfenicol in these farms may promote the dissemination of floR gene and associated resistant isolates. The genetic relationship of these floR-producing isolates was investigated by PFGE (Figure 1). The nine A. pleuropneumoniae isolates could be divided into three pulsotypes. Four strains from Shanghai showed highly similar band patterns. While another strain HD-5-Q-2 from Shanghai was located on a separated branch. The four isolates from Xinjiang exhibited ≥80% phylogenetic similarity. The eight P. multocida isolates showed highly various PFGE patterns. Six isolates from Shanghai exhibited three different types, while the isolates XY-5-Q-1 from Jiangsu and XN-PM-1 from Xinjiang were located on different branches. The same PFGE patterns observed in some isolates from same regions of China suggested that clonal expansion of floR-positive A. pleuropneumoniae and P. multocida isolates exists in these pig farms.
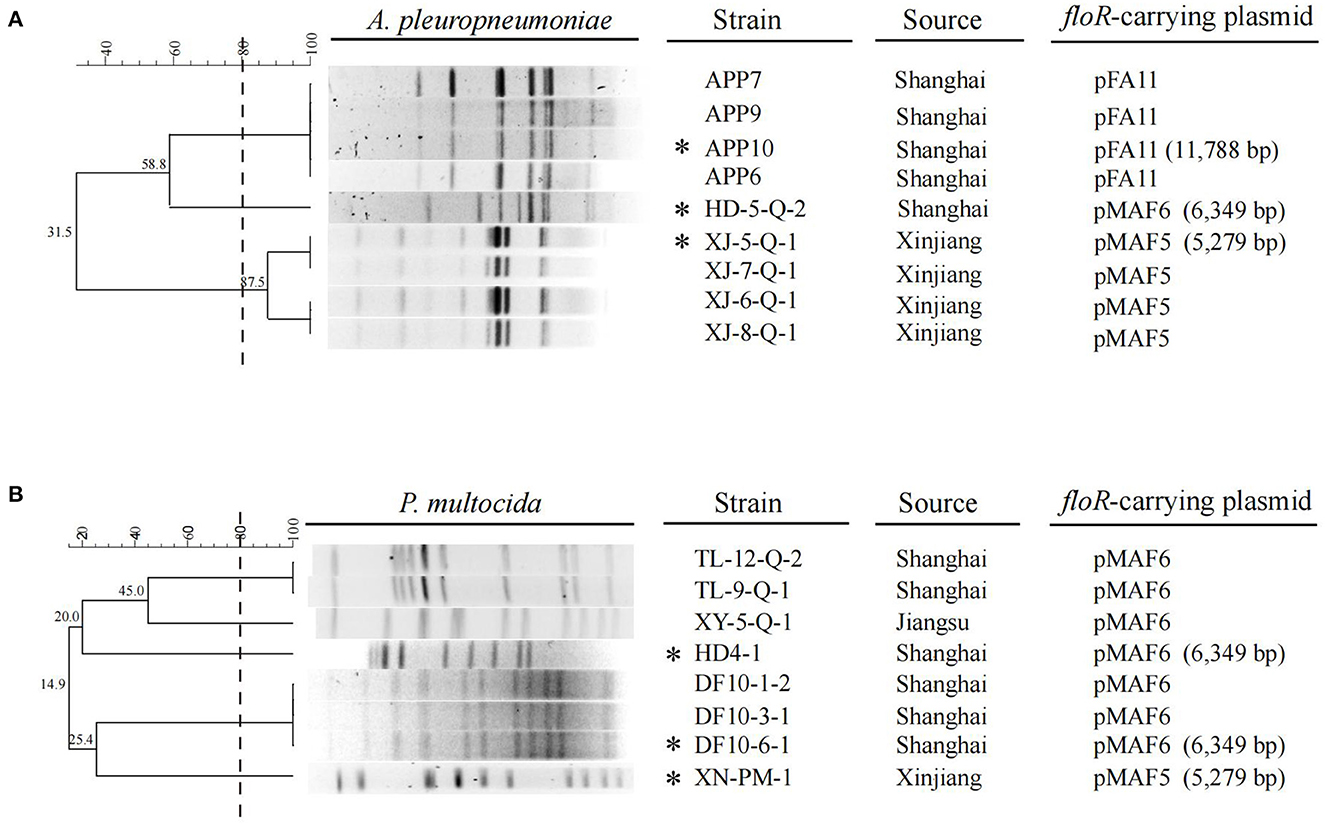
Figure 1. ApaI-PFGE patterns and dendrogram of the 9 floR-positive A. pleuropneumoniae isolates (A) and 8 floR-positive P. multocida isolates (B) in this study. The dashed line on the dendrogram indicates 80% similarity. The strain name, source and the floR-carrying plasmid corresponding to each isolate are also presented. The isolates marked with an asterisk are the ones characterized by WGS, while the floR-carrying plasmids in the isolates without asterisk are identified by specific PCRs.
To further investigate the genetic basis of the floR transmission in A. pleuropneumoniae and swine-origin P. multocida isolates, three A. pleuropneumoniae and three P. multocida strains with different PFGE patterns were subjected to WGS and the complete floR-carrying plasmids in these isolates were obtained by PCR-based gap filling. Three types of floR-carrying plasmid, named pFA11, pMAF5, and pMAF6, were identified in the six isolates. Furthermore, specific PCRs corresponding to the three plasmids were performed to detect whether these plasmids were also present in other 11 floR-producing isolates. Taken together, plasmid pFA11 was detected in four A. pleuropneumoniae isolates, pMAF5 served as the carriers of floR gene in four A. pleuropneumoniae and one P. multocida isolates, and pMAF6 was present in one A. pleuropneumoniae and seven P. multocida isolates (Figure 1). These data demonstrated that the three plasmids mediated floR transmission in A. pleuropneumoniae and P. multocida isolates from swine in China.
Plasmid pFA11, identified in four A. pleuropneumoniae isolates from Shanghai, was 11,788 bp in size and consisted of 14 opening reading frames (ORFs). Besides floR, pFA11 also contained sulfonamide resistance gene sul2, aminoglycoside resistance genes aacC2d, strA, strB, and β-lactam resistance gene blaROB − 1. Comparison with sequences deposited in Genbank showed that pFA11 shared highly structural similarity with plasmids originated from other Pasteurellaceae bacteria such as pIMD50 from A. porcitonsillarum of swine origin in Switzerland (GenBank accession no. AJ830711) and pOV described in P. multocida isolated from Mexico (GenBank accession no. JX827416) (Figure 2) (32, 33). There are two main differences between pFA11 and the latter two plasmids. Firstly, compared to pFA11, additional genes associated with plasmid replication and mobilization were present within the other three plasmids, such as repA, repX, mobA, mobB and mobC. Secondly, plasmids pIMD50 and pOV shared highly similar resistance gene cluster. While, pFA11 harbored four additional resistance genes, including floR, aacC2d, and another copy of strA and strB. Since the antibiotics corresponding to these resistance genes are commonly used in pig farms, the multiple resistance gene cluster on pFA11 may facilitate the spread of this plasmid in the A. pleuropneumoniae isolates through the co-selection effect.

Figure 2. Schematic comparison of the floR-carrying plasmids pFA11, pMAF5, and pMAF6 with related plasmids in Genbank. Arrows represented the positions, orientations and relative sizes of the genes. The regions with ≥98% homology between these plasmids are marked by gray shading. The plasmid name, size, GenBank accession number, host bacteria and country of origin are given on the left of each plasmid map.
pMAF5 was 5,279 bp in length with simple structure, in which a plasmid replication gene rep, a resistance gene floR and four putative ORFs were present. Database searching showed that pMAF5 was highly similar to plasmid pHPSGC from G. parasuis HpsGC of sick pig in China (GenBank accession no. KX966395) (Figure 2). Structure comparison revealed that floR and its flanking region (lysR-floR-hp) showed high homology to the corresponding region of plasmid pCCK381 described in P. multocida isolate strain 381 from lung of a calf origin from UK (GenBank accession no. AJ871969) (18). The rep gene of pMAF5 was same to those of other resistance plasmids from different members of family Pasteurellaceae, such as pHPS1019 from G. parasuis lung6591 isolates in China (GenBank accession no. HQ622101), p780 originated from A. pleuropneumoniae MV780 in Brazil (GenBank accession no. MH457196) and pM3362MDR from A. pleuropneumoniae MID3362 originated from pig lung in UK (GenBank accession no. MH457202) (34). These plasmids possessed other resistance genes, such as tet (B), blaROB − 1, sul2 and strA, but not floR.
Plasmid pMAF6 was the most frequently observed floR-vector, which was detected in eight isolates from different regions and with various PFGE patterns. pMAF6 was 6, 349 bp in size and showed >99% nucleotide identity to previously reported floR-bearing plasmid pHPSF1 identified from G. parasuis ASB17W in China (GenBank accession no. KR262062) (35) (Figure 2). Compared to pHPSF1, pMAF6 had a 22 bp insertion at the end of the putative lysR gene, which generated an early stop codon and encoded a smaller putative LysR (101 amino acids for pMAF6 and 109 amino acids for pHPSF1). The same plasmid structure was also observed for plasmid p11745 in A. pleuropneumoniae APP11745 from Spain (GenBank accession no. DQ176855), which served as the cargo of tetracycline resistance gene tet (B) (36). Moreover, similar to plasmid pMAF5, the fragment lysR-floR-hp of pMAF6 was also highly similar to the corresponding region of plasmid pCCK381.
Conclusions
The genetic basis of the florfenicol resistance in A. pleuropneumoniae and P. multocida isolates of swine origin in China was analyzed in this study. Considering the importance of florfenicol in pig industry, the high prevalence of florfenicol resistance and floR gene observed in this study may significantly affect the control of A. pleuropneumoniae and P. multocida infections in swine respiratory diseases. Moreover, the prevalence of small plasmids with similar structure in various isolates of Pasteurellaceae bacteria from different regions revealed that this kind of plasmids are important cargos for the transmission of not only floR but also other resistance genes in porcine Pasteurellaceae pathogens. Continuous monitoring of florfenicol resistance and prevalence of floR gene and associated plasmids in Pasteurellaceae pathogens of veterinary origin are warranted, as this information is important for the control of infections caused by pathogenic species of Pasteurellaceae in food-producing animals.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Ethics statement
Ethical review and approval was not required for the animal study because the study was performed with resistant bacteria. Samples were taken by authorized veterinarians working in the farms. The researchers did not perform any animal studies.
Author contributions
BL and LX designed and supervised the study. WZ, YQ, KL, JW, ZL, QS, and ZM provided resources for this study. XY and QS laboratory analysis and statistical analysis. BL and XY wrote the manuscript. All authors contributed to this article and approved the submitted version.
Funding
This work was sponsored by Shanghai Agriculture Applied Technology Development Program, China (Grant No. X2022-02-08-00-12-F01100) and the National Natural Science Foundation of China (No. 32273056).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Michael GB, Bossé JT, Schwarz S. Antimicrobial resistance in pasteurellaceae of veterinary origin. Microbiol Spectr. (2018) 6:15–25. doi: 10.1128/microbiolspec.ARBA-0022-2017
2. Sassu EL, Bossé JT, Tobias TJ, Gottschalk M, Langford PR, Hennig-Pauka I. Update on Actinobacillus pleuropneumoniae-knowledge, gaps, and challenges. Transbound Emerg Dis. (2018) 65 Suppl 1:72–90. doi: 10.1111/tbed.12739
3. Stringer OW, Bossé JT, Lacouture S, Gottschalk M, Fodor L, Angen Ø, et al. Proposal of Actinobacillus pleuropneumoniae serovar 19, and reformulation of previous multiplex PCRs for capsule-specific typing of all known serovars. Vet Microbiol. (2021) 255:109021. doi: 10.1016/j.vetmic.2021.109021
4. Tang X, Zhao Z, Hu J, Wu B, Cai X, He Q, et al. Isolation, antimicrobial resistance, and virulence genes of Pasteurella multocida strains from swine in China. J Clin Microbiol. (2009) 47:951–8. doi: 10.1128/JCM.02029-08
5. Wilkie IW, Harper M, Boyce JD, Adler B. Pasteurella multocida: diseases and pathogenesis. Curr Top Microbiol Immunol. (2012) 361:1–22. doi: 10.1007/82_2012_216
6. Wilson BA, Ho M. Pasteurella multocida: from zoonosis to cellular microbiology. Clin Microbiol Rev. (2013) 26:631–55. doi: 10.1128/CMR.00024-13
7. Harper M, John M, Turni C, Edmunds M, St Michael F, Adler B, et al. Development of a rapid multiplex PCR assay to genotype Pasteurella multocida strains by use of the lipopolysaccharide outer core biosynthesis locus. J Clin Microbiol. (2015) 53:477–85. doi: 10.1128/JCM.02824-14
8. Kim J, Kim JW, Oh SI, So B, Kim WI, Kim HY. Characterisation of Pasteurella multocida isolates from pigs with pneumonia in Korea. BMC Vet Res. (2019) 15:119. doi: 10.1186/s12917-019-1861-5
9. Xu J, Jia H, Cui G, Tong H, Wei J, Shao D, et al. ICEAplChn1, a novel SXT/R391 integrative conjugative element. (ICE), carrying multiple antibiotic resistance genes in Actinobacillus pleuropneumoniae. Vet Microbiol. (2018) 220:18–23. doi: 10.1016/j.vetmic.2018.05.002
10. Jiang HX, Zeng ZL, Chen ZL, Liu JJ, Fung KF. Pharmacokinetics of florfenicol in pigs following intravenous, intramuscular or oral administration and the effects of feed intake on oral dosing. J Vet Pharmacol Ther. (2006) 29:153–6. doi: 10.1111/j.1365-2885.2006.00727.x
11. Li P, Gao M, Feng C, Yan T, Sheng Z, Shi W, et al. Molecular characterization of florfenicol and oxazolidinone resistance in Enterococcus isolates from animals in China. Front Microbiol. (2022) 13:811692. doi: 10.3389/fmicb.2022.811692
12. Zhang QQ, Ying GG, Pan CG, Liu YS, Zhao JL. Comprehensive evaluation of antibiotics emission and fate in the river basins of china: source analysis, multimedia modeling, and linkage to bacterial resistance. Environ Sci Technol. (2015) 49:6772–82. doi: 10.1021/acs.est.5b00729
13. Kim B, Hur J, Lee JY, Choi Y, Lee JH. Molecular serotyping and antimicrobial resistance profiles of Actinobacillus pleuropneumoniae isolated from pigs in South Korea. Vet Q. (2016) 36:137–44. doi: 10.1080/01652176.2016.1155241
14. Kucerova Z, Hradecka H, Nechvatalova K, Nedbalcova K. Antimicrobial susceptibility of Actinobacillus pleuropneumoniae isolates from clinical outbreaks of porcine respiratory diseases. Vet Microbiol. (2011) 150:203–6. doi: 10.1016/j.vetmic.2011.01.016
15. Bossé JT Li Y, Atherton TG, Walker S, Williamson SM, Rogers J, Chaudhuri RR, et al. Characterisation of a mobilisable plasmid conferring florfenicol and chloramphenicol resistance in Actinobacillus pleuropneumoniae. Vet Microbiol. (2015) 178:279–82. doi: 10.1016/j.vetmic.2015.05.020
16. da Silva GC, Rossi CC, Santana MF, Langford PR, Bossé JT, Bazzolli DMS. p518, a small floR plasmid from a South American isolate of Actinobacillus pleuropneumoniae. Vet Microbiol. (2017) 204:129–32. doi: 10.1016/j.vetmic.2017.04.019
17. Doublet B, Schwarz S, Kehrenberg C, Cloeckaert A. Florfenicol resistance gene floR is part of a novel transposon. Antimicrob Agents Chemother. (2005) 49:2106–8. doi: 10.1128/AAC.49.5.2106-2108.2005
18. Kehrenberg C, Schwarz S. Plasmid-borne florfenicol resistance in Pasteurella multocida. J Antimicrob Chemother. (2005) 55:773–5. doi: 10.1093/jac/dki102
19. Li Y, Li Y, Fernandez Crespo R, Leanse LG, Langford PR, Bossé JT. Characterization of the Actinobacillus pleuropneumoniae SXT-related integrative and conjugative element ICEApl2 and analysis of the encoded FloR protein: hydrophobic residues in transmembrane domains contribute dynamically to florfenicol and chloramphenicol efflux. J Antimicrob Chemother. (2018) 73:57–65. doi: 10.1093/jac/dkx342
20. Zhu D, Yuan D, Wang M, Jia R, Chen S, Liu M, et al. Emergence of a multidrug-resistant hypervirulent Pasteurella multocida ST342 strain with a floR-carrying plasmid. J Glob Antimicrob Resist. (2020) 20:348–50. doi: 10.1016/j.jgar.2019.09.012
21. Michael GB, Kadlec K, Sweeney MT, Brzuszkiewicz E, Liesegang H, Daniel R, et al. ICEPmu1, an integrative conjugative element. (ICE) of Pasteurella multocida: analysis of the regions that comprise 12 antimicrobial resistance genes. J Antimicrob Chemother. (2012) 67:84–90. doi: 10.1093/jac/dkr406
22. Clinical and Laboratory Standards Institute. (CLSI). Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated From Animals, 4th ed.; CLSI Supplement VET08; CLSI: Wayne, PA, USA (2018)
23. Chevallier B, Dugourd D, Tarasiuk K, Harel J, Gottschalk M, Kobisch M, et al. Chromosome sizes and phylogenetic relationships between serotypes of Actinobacillus pleuropneumoniae. FEMS Microbiol Lett. (1998) 160:209–16. doi: 10.1111/j.1574-6968.1998.tb12913.x
24. Kong LC, Wang Z, Wang YM, Dong WL, Jia BY, Gao D, et al. Antimicrobial susceptibility and molecular typing of Pasteurella multocida isolated from six provinces in China. Trop Anim Health Prod. (2019) 51:987–92. doi: 10.1007/s11250-018-1754-9
25. Speijer H, Savelkoul PH, Bonten MJ, Stobberingh EE, Tjhie JH. Application of different genotyping methods for Pseudomonas aeruginosa in a setting of endemicity in an intensive care unit. J Clin Microbiol. (1999) 37:3654–61. doi: 10.1128/JCM.37.11.3654-3661.1999
26. Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. (1995) 33:2233–9. doi: 10.1128/jcm.33.9.2233-2239.1995
27. Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics. (2008) 9:75. doi: 10.1186/1471-2164-9-75
28. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. (1990) 215:403–10. doi: 10.1016/S0022-2836(05)80360-2
29. Cid D, Fernández-Garayzábal JF, Pinto C, Domínguez L, Vela AI. Antimicrobial susceptibility of Pasteurella multocida isolated from sheep and pigs in Spain—Short communication. Acta Vet Hung. (2019) 67:489–98. doi: 10.1556/004.2019.048
30. Hayer SS, Rovira A, Olsen K, Johnson TJ, Vannucci F, Rendahl A, et al. Prevalence and time trend analysis of antimicrobial resistance in respiratory bacterial pathogens collected from diseased pigs in USA between 2006–2016. Res Vet Sci. (2020) 128:135–44. doi: 10.1016/j.rvsc.2019.11.010
31. Morioka A, Asai T, Nitta H, Yamamoto K, Ogikubo Y, Takahashi T, et al. Recent trends in antimicrobial susceptibility and the presence of the tetracycline resistance gene in Actinobacillus pleuropneumoniae isolates in Japan. J Vet Med Sci. (2008) 70:1261–4. doi: 10.1292/jvms.70.1261
32. López-Ochoa AJ, Sánchez-Alonso P, Vázquez-Cruz C, Horta-Valerdi G, Negrete-Abascal E, Vaca-Pacheco S, et al. Molecular and genetic characterization of the pOV plasmid from Pasteurella multocida and construction of an integration vector for Gallibacterium anatis. Plasmid. (2019) 103:45–52. doi: 10.1016/j.plasmid.2019.04.003
33. Matter D, Rossano A, Sieber S, Perreten V. Small multidrug resistance plasmids in Actinobacillus porcitonsillarum. Plasmid. (2008) 59:144–52. doi: 10.1016/j.plasmid.2007.11.003
34. Li Y, da Silva GC Li Y, Rossi CC, Fernandez Crespo R, Williamson SM, Langford PR, et al. Evidence of illegitimate recombination between two Pasteurellaceae plasmids resulting in a novel multi-resistance Replicon, pM3362MDR, in Actinobacillus pleuropneumoniae. Front Microbiol. (2018) 9:2489. doi: 10.3389/fmicb.2018.02489
35. Li B, Zhang Y, Wei J, Shao D, Liu K, Shi Y, et al. Characterization of a novel small plasmid carrying the florfenicol resistance gene floR in Haemophilus parasuis. J Antimicrob Chemother. (2015) 70:3159–61. doi: 10.1093/jac/dkv230
Keywords: pig, floR, PFGE, Pasteurellaceae, antibiotic resistance
Citation: Yao X, Song Q, Zhu W, Wei J, Shao D, Liu K, Li Z, Qiu Y, Ma Z, Xia L and Li B (2023) Characterization of small plasmids carrying florfenicol resistance gene floR in Actinobacillus pleuropneumoniae and Pasteurella multocida isolates from swine in China. Front. Vet. Sci. 10:1084491. doi: 10.3389/fvets.2023.1084491
Received: 01 November 2022; Accepted: 12 January 2023;
Published: 30 January 2023.
Edited by:
Moussa Sory Diarra, Agriculture and Agri-Food Canada (AAFC), CanadaReviewed by:
Ashenafi Feyisa Beyi, Iowa State University, United StatesKumaragurubaran Karthik, Tamil Nadu Veterinary and Animal Sciences University, India
Copyright © 2023 Yao, Song, Zhu, Wei, Shao, Liu, Li, Qiu, Ma, Xia and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lining Xia,  eGxuNzUwNTMwQDE2My5jb20=; Beibei Li,
eGxuNzUwNTMwQDE2My5jb20=; Beibei Li,  bGJiQHNodnJpLmFjLmNu
bGJiQHNodnJpLmFjLmNu
†These authors have contributed equally to this work
 Xiaohui Yao
Xiaohui Yao Qiangqiang Song1,2†
Qiangqiang Song1,2† Jianchao Wei
Jianchao Wei Donghua Shao
Donghua Shao Ke Liu
Ke Liu Zongjie Li
Zongjie Li Yafeng Qiu
Yafeng Qiu Zhiyong Ma
Zhiyong Ma Beibei Li
Beibei Li