- 1Fieldepi, Iowa State University College of Veterinary Medicine, Ames, IA, United States
- 2College of Veterinary Medicine, Michigan State University, Lansing, MI, United States
- 3Carthage Veterinary Service, Carthage, IL, United States
Introduction: The use of serum and family oral fluids for porcine reproductive and respiratory syndrome virus (PRRSV) surveillance in weaning-age pigs has been previously characterized. Characterizing more sample types similarly offers veterinarians and producers additional validated sample options for PRRSV surveillance in this subpopulation of pigs. Oral swab sampling is relatively easy and convenient; however, there is sparse information on how it compares to the reference sample type for PRRSV surveillance under field conditions. Therefore, this study's objective was to compare the PRRSV reverse-transcription real-time polymerase chain reaction (RT-rtPCR) test outcomes of oral swabs (OS) and sera samples obtained from weaning-age pig litters.
Method: At an eligible breeding herd, six hundred twenty-three weaning-age piglets from 51 litters were each sampled for serum and OS and tested for PRRSV RNA by RT-rtPCR.
Results and Discussion: PRRSV RT-rtPCR positivity rate was higher in serum samples (24 of 51 litters, 83 of 623 pigs, with a mean cycle threshold (Ct) value of RT-rtPCR-positive samples per litter ranging from 18.9 to 32.0) compared to OS samples (15 of 51 litters, 33 of 623 pigs, with a mean Ct of RT-rtPCR positive samples per litter ranging from 28.2 to 36.9); this highlights the importance of interpreting negative RT-rtPCR results from OS samples with caution. Every litter with a positive PRRSV RT-rtPCR OS had at least one viremic piglet, highlighting the authenticity of positive PRRSV RT-rtPCR tests using OS; in other words, there was no evidence of environmental PRRSV RNA being detected in OS. Cohen's kappa analysis (Ck = 0.638) indicated a substantial agreement between both sample types for identifying the true PRRSV status of weaning-age pigs.
1. Introduction
Porcine reproductive and respiratory syndrome virus (PRRSV) is an enveloped single-stranded positive-sense RNA virus belonging to the family Arteriviridae (1, 2). PRRSV is the etiological agent of Porcine Reproductive and Respiratory Syndrome (PRRS), an economically important swine disease costing the United States swine industry hundreds of millions of dollars in annual revenue losses (3). A typical PRRS outbreak in a sow herd is clinically characterized by respiratory difficulties in younger pigs, spikes in reproductive failures in sows, as well as accompanying increases in neonatal losses, pre-weaning mortality, and sow deaths (4–6).
Over the last three decades, the swine industry has significantly evolved in its effort to curb the menace of PRRSV; a variety of producer-driven PRRSV management programs have been instituted (7). A few factors that may have influenced the evolution of the mentioned programs include the growth and increased consolidation of swine enterprises (8), the emergence and reemergence of PRRSV variants (9, 10), improved understanding of the atypical ecology of PRRSV (11–13), improved diagnostic methods (14, 15) and tools (16–18), and the availability and adoption of technologies such as commercial vaccines (19) and air filtration systems (20).
PRRSV surveillance remains an important component of PRRSV management programs (18, 21, 22). Especially at low prevalence, effective PRRSV surveillance in breeding herds can be cost-prohibitive and usually requires a relatively large number of animals to be sampled. This challenge is a major reason aggregate samples (10, 21, 23, 24) have increasingly become the most predominant specimens submitted to US veterinary diagnostic laboratories for PRRSV molecular testing (10).
Weaning-age pigs (typically 2–3 weeks of age) are an epidemiologically important subpopulation for PRRSV control or elimination programs, not only because their PRRSV shedding status reflects the PRRSV status of a breeding herd (25), but they are also often translocated and are vehicles for swine disease pathogen spread.
Serum is the reference specimen to establish the PRRSV active circulation in pig populations (18, 25). With animal welfare concerns, sampling difficulty, needed expertise, and the cost of sampling materials associated with serum sampling, there is a need to evaluate alternative sampling strategies. One alternative is family oral fluids (FOFs) sampling, which has been shown to be a cost-efficient sample type for PRRSV surveillance, especially when PRRSV is at a low prevalence (26). Obtaining a FOF sample, however, requires voluntary interaction between litter mates and a sampling rope (23, 27).
There is therefore a need to evaluate other sampling options for PRRSV monitoring in weaning-age pigs, especially as swine practitioners are already submitting some of these sample types to veterinary diagnostic laboratories for testing of PRRSV and other swine pathogens (10, 28).
Thus, this study sought to compare PRRSV RNA RT-rtPCR detection rates in oral swabs to detection rates in serum samples from weaning-age pig litters.
2. Materials and methods
This study was approved by the Iowa State University Institutional Animal Care and Use Committee (IACUC-19-118).
2.1. Study design and eligibility criteria
This was a prospective field study conducted in a conveniently identified swine breeding herd that fit the eligibility criteria.
Serum samples and matched oral swabs were collected from all pigs within fifty-one litters. To be eligible, a breeding herd had to be PRRSV-positive unstable based on the American association of swine veterinarians' guidelines (18). Briefly, a PRRSV-positive unstable herd is a herd that tests RT-rtPCR positive for wildtype PRRSV on routine monitoring.
An eligible herd would also have diagnostic evidence of PRRSV circulation and would not have used PRRS virus vaccines during the study or in the previous 2 years.
2.2. Sample collection
Oral swabs were collected from each piglet in the study litters by rotating a polyester swab stick (Puritan, ME, USA) along the buccal mucosa between the back of the tongue and inner cheek of manually restrained pigs. Each swab was transferred to separate 2 ml microcentrifuge tubes (Fisher, MA, USA) containing 1 ml of Phosphate Buffered Saline (Gibco, MA, USA).
Serum samples were collected from each piglet in the study litters into 8.5 ml vacutainer tubes (Becton Dickinson, NJ, USA) via jugular venipuncture in manually restrained piglets.
All samples were matched by litter, i.e., identified by the sow identification number, stored at 4°C, and transported to an AALVD (American Association of Veterinary Laboratory Diagnosticians)-certified veterinary diagnostic laboratory for PRRSV RNA laboratory investigation by RT-rtPCR tests.
2.3. Sample size justification
The sow farm used for this study weaned an average of 238 litters a week. The sample size of 51 litters aimed at having an at least 95% confidence of selecting ≥1 PRRSV-positive litter (a litter having at least one PRRSV-viremic pig) assuming a 5% PRRSV-prevalence, sampling without replacement, and a perfect RT-rtPCR test (29, 30).
2.4. Statistical analysis
All statistical calculations and graphing were done on R statistical software (31).
2.4.1. Within litter prevalence and RT-rtPCR mean cycle threshold values
Within litter prevalence (WLP) by OS, and by serum for each litter were separately calculated as the proportion of piglets within a litter that tested RT-rtPCR positive for OS, and serum respectively.
A plot of OS WLP by serum WLP was made using the ggplot2 package (32) on the R statistical software (31).
The mean cycle threshold (Ct) value for RT-rtPCR PRRSV positive samples for both sample types for each sampled litter was calculated as:
Where Ctpos is the Ct for each RT-rtPCR positive sample i, and npos is the number of RT-rtPCR positive samples within that litter.
The effect of the serum WLP in the jth litter on the RT-rtPCR detection of PRRSV RNA in OS (POS) was characterized using a generalized linear model on the lme4 package (33) on R statistical software (31).
Where:
is the observed result of the Bernoulli trial (RT-rtPCR detection of PRRSV RNA in OS from at least one piglet, 1 or not, 0) in litter j,
α is the model's intercept,
β is the regression coefficient,
xj is the WLP in litter j, and
p is the estimated probability of detection.
2.4.2. Diagnostic performance assessment and Cohen's kappa analysis
Two by two contingency tables (Table 1) were constructed to compare litter-level PRRSV detection by sample type in each litter. When at least one piglet tested PRRSV RT-rtPCR positive by OS or serum, that litter was considered positive for that sample type. A litter was considered truly PRRSV-positive if serum from at least one piglet tested RT-rtPCR positive. Serum RT-rtPCR results, therefore, were considered as the reference in assessing the litter-level sensitivity and specificity of OS.
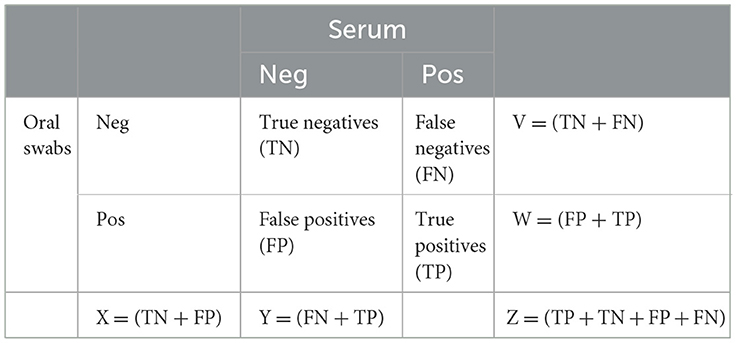
Table 1. 2 x 2 tables comparing PRRSV RT-rtPCR detection in oral swabs and serum in sampled litters.
Sensitivity and specificity were thereafter estimated using the formula:
Cohen's Kappa (Ck) analysis (34) was thereafter used to assess the litter-level agreement of PRRSV RT-rtPCR detection in OS and serum samples over chance, using the formula:
The scale (23) used to interpret the Ck is:
0 = no agreement
0 < Ck < 0.2 = slight agreement
0.21 ≤ Ck≥0.40 = fair agreement
0.41 ≤ Ck≥0.60 = moderate agreement
0.61 ≤ Ck≥ 0.80 = substantial agreement
0.81 ≤ Ck≥1.00 = almost perfect agreement
3. Results
A 5,300-head commercial breed-to-wean farm in the Midwestern United States was selected for this study based on eligibility criteria, and this study was conducted in the third quarter of 2021. At the time of the study, the herd was within 3 months of a PRRSV-2 outbreak with the PRRSV outbreak strain having an open reading frame 5 (ORF5) sequence restriction fragment length polymorphism (RFLP) pattern of 1-7-4.
Weaning-age pigs between 18 and 21 days of age belonging to fifty-one litters were conveniently selected, and both oral swabs and serum samples were collected. There was a total of 623 piglets across the 51 sampled litters.
3.1. Within-litter prevalence
Across all study litters, the within-litter prevalence ranged from 0.00 to 57.14% for oral swabs, and 0.00 to 100.00% for serum samples. There were more pigs testing RT-rtPCR positive by serum (83/623) than by OS (33/623).
The mean Ct of RT-rtPCR positive samples per litter ranged from 28.2 to 36.9 for OS samples, and 18.9 to 32.0 for serum samples. When the mean Ct of the PRRSV-positive serum samples for a litter was over 30 (n = 2), there was no OS from that litter testing PRRV-positive by RT-rtPCR. There was a positive linear relationship between the OS WLP and serum WLP (Figure 1) with a Pearson correlation coefficient value of 84.57%. Table 2 shows the actual WLP values by OS and serum and the mean Ct values of RT-rtPCR PRRSV-positive OS and serum for each litter (identified by sow number).
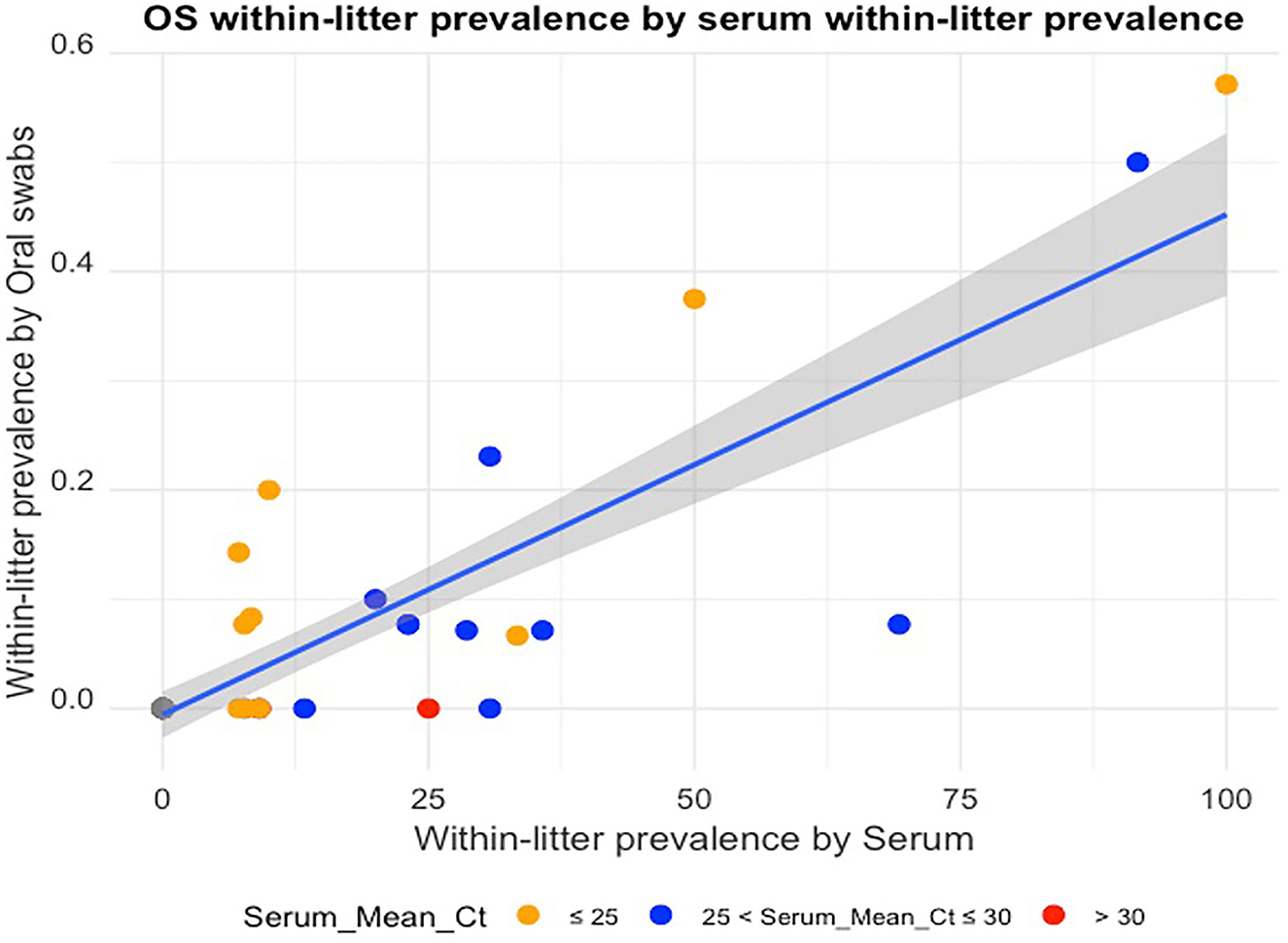
Figure 1. Oral swab within-litter prevalence plotted against serum within-litter prevalence, with colors indicating mean Ct ranges of the RT-rtPCR-positive serum samples for each litter.
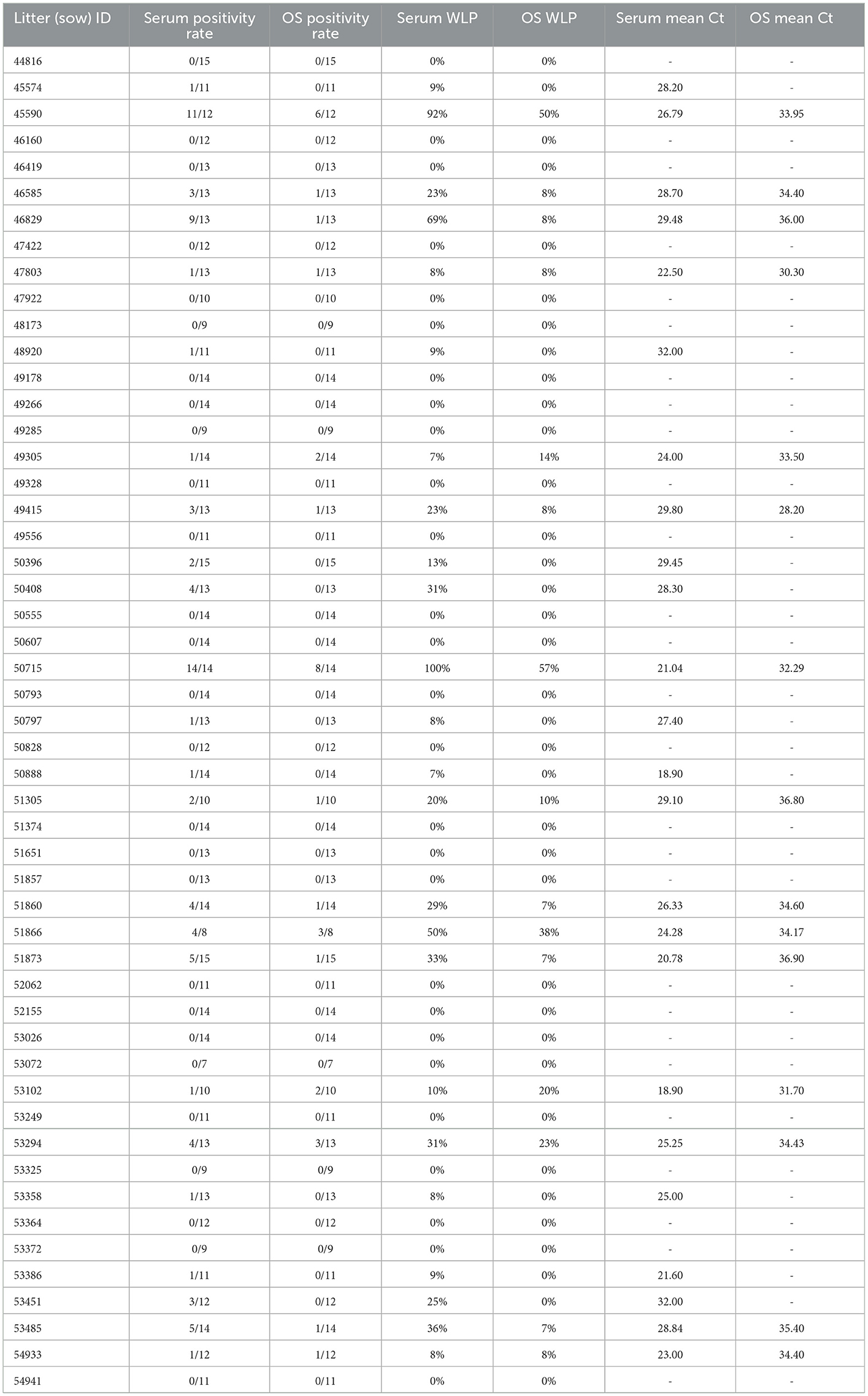
Table 2. Within-litter prevalence (WLP) and mean cycle threshold values for serum and oral swab (OS) sample types for each litter sampled.
From the regression output, the model's intercept (lower and upper 95% confidence intervals) was −2.82 (−4.420, −1.661) , and the regression coefficient (lower and upper 95% confidence intervals) was 0.16 (0.085, 0.268). Thus, the odds of detecting at least one PRRSV-positive piglet by RT-rtPCR in an OS sampled litter increased by e0.16 or 1.174 (or 17.4%) for every unit (1%) increase in WLP; the p−values for both coefficients were < 0.001. The probability of detecting at least one PRRSV-positive piglet by RT-rtPCR in OS rapidly increased as the within-littler prevalence increased (Figure 2).
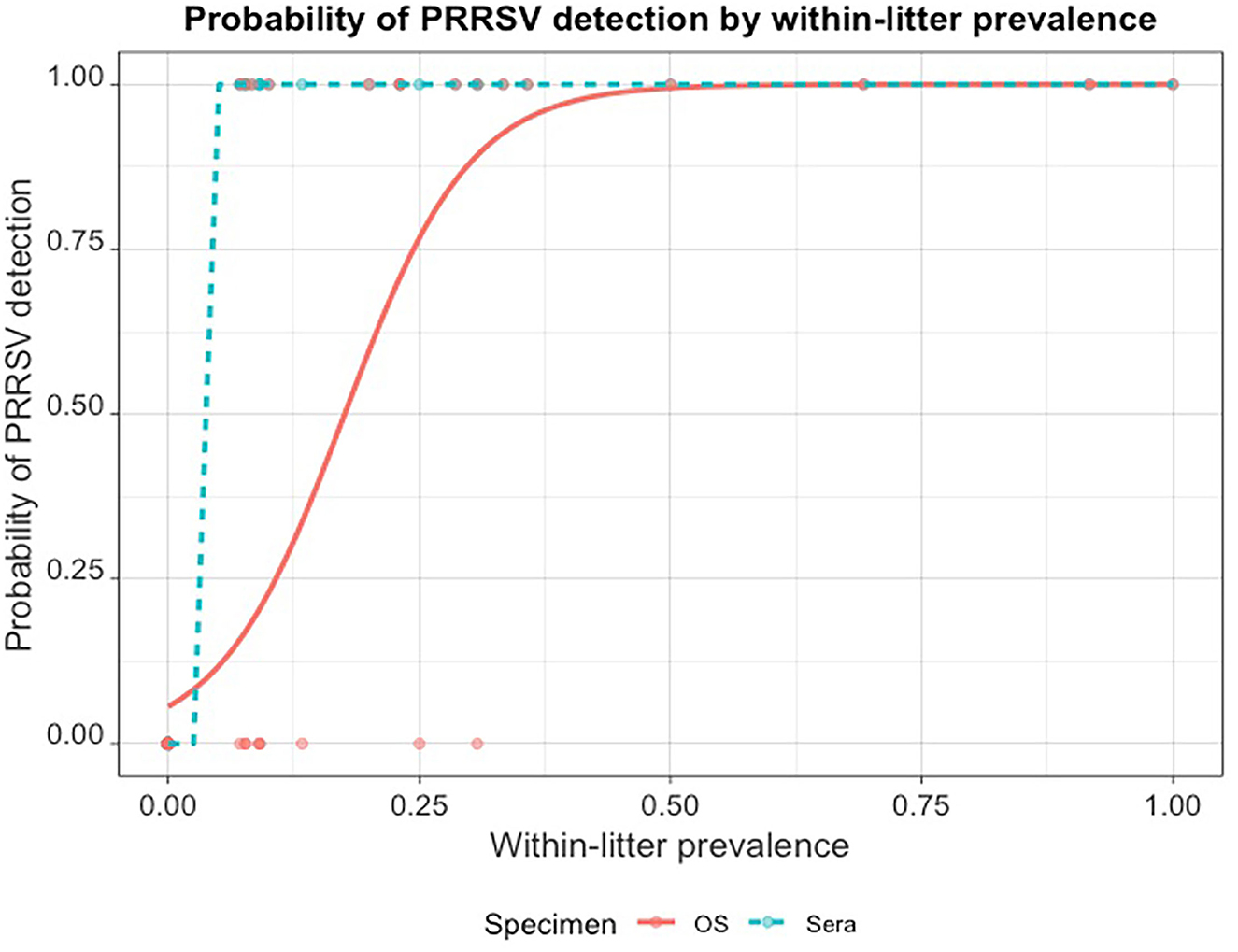
Figure 2. Probability of PRRSV RT-rtPCR detection in at least one pig when litters are fully sampled by oral swabs (OS) compared to within-litter prevalence established by serum testing.
3.2. Diagnostic performance assessment and Cohen's kappa analysis
A tabular comparison of the litter-level PRRSV RNA Rt-rtPCR detection using both sample types is shown in Table 3. Using equations (5) and (6), sensitivity and specificity of OS for fully sampled litters was 62.5% and 100%, respectively.
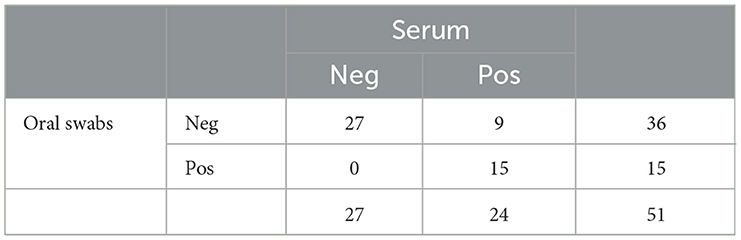
Table 3. Comparison of PRRSV RT-rtPCR detection in oral swabs and serum samples from litters of weaning age pigs.
Cohen's kappa analysis yielded a value of 0.638 (p−value < 0.001), indicating a substantial agreement between both sample types for identifying the true PRRSV-status of a fully sampled litter of weaning-age pigs.
4. Discussion
Oral swab sampling is a quick and non-invasive sampling technique that has been used in human populations for the detection/diagnosis of tuberculosis (35), Ebola (36), enteric viruses (37), streptococcosis (38), and more recently, Covid-19 (39–41). It has also been used extensively in animal disease surveillance for the detection of Influenza A virus in avian species (42), Bovine Viral Diarrhea virus (43), Foot and Mouth Disease virus (44), Peste des petits ruminants virus (45), Herpesvirus (46), and Nidoviruses (47), amongst others.
Oral swabs have also been used for PRRSV surveillance in swine herds, specifically, their use has been demonstrated in post-weaning age animals (48, 49).
Despite the epidemiological significance of the weaning age pig subpopulation in PRRSV control/elimination programs, serum samples and FOFs are the only two sample types validated specifically for this subpopulation (18, 24, 25). Given that there are potential constraints with the use of these tools, it is no surprise that swine practitioners opt for using other sample types, or use the validated tools conveniently rather than as recommended. Investing in a surveillance program using a sample type that has not been experimentally validated is not justifiable, as one cannot confidently interpret the laboratory results from testing these samples, coupled with the risks associated with a false negative test result.
This study evaluated the suitability of PRRSV RT-rtPCR testing on OS samples as a PRRSV surveillance tool in weaning age pig populations. From the findings of this study, RT-rtPCR tests on serum samples were consistently more likely to give a positive result than RT-rtPCR tests on OS samples for matched litters; this agrees with a previous study (49) that demonstrated that pigs could be viremic and not have detectable quantities of PRRSV in oral swab samples. In addition to this, all positive results from OS samples came from litters containing at least one viremic pig. This gives some credence to this sample type and supports that positive RT-rtPCR tests from OS samples are likely to come from only truly PRRSV-positive litters having at least one viremic pig.
Cycle threshold values from RT-rtPCR tests are a good proxy to estimating the amount of viral RNA present in a sample (50). The observed absence of detectable quantities of PRRSV RNA in oral swabs from a litter when the mean Ct values of RT-rtPCR tests on serum samples from the same litter was >30 (relatively low viremia), suggests that a negative PRRSV RT-rtPCR test on OS may reflect low to no viremia in a sampled pig or litter.
Given that the reasons for evaluating alternative sample types include providing a convenient, animal-welfare friendly, and resource-savvy substitute to serum sampling, a logical next step would be to ascertain an appropriate sample size for OS samples that will provide the same probability of demonstrating disease freedom as serum samples for the same set of statistical assumptions. The design of the current study poses a challenge with determining sample size estimates as this was not the aim of the study; therefore, further studies may be needed in which samples will be matched by individual piglets, sourced from multiple herds, and include more sample-types for pairwise comparisons. Also, litters were conveniently selected and do not necessarily represent the status of other non-sampled litters. Therefore, the true PRRSV-prevalence in the herd could not be calculated with this dataset.
For the current study, when serum samples were first collected from the piglets, there was difficulty in obtaining oral swabs from those piglets as the buccal mucosa had often dried out, likely from vocalization. It is possible this affected the findings of the study, and the authors acknowledge this as a limitation; the design of future studies will be refined to minimize this occurrence.
In real world PRRSV surveillance scenarios for pathogen detection, sampling entire members of multiple litters using either sample type is cost-prohibitive and impractical; the purpose of sampling entire litter members was to facilitate an understanding of how the pressure of PRRSV infection within a litter influenced the probability of detecting at least one viremic pig by RT-rtPCR on OS samples. This was successfully characterized.
5. Conclusion
This study evaluated the use of OS under field conditions for PRRSV RNA detection by RT-rtPCR, as compared to serum samples. OS sampling offers a welfare-friendly, resource-savvy, easier, specific, but less sensitive alternative for PRRSV surveillance in weaning-age pig populations, negative RT-rtPCR results using this sample type should be interpreted with caution.
Further studies are needed to characterize the recommended sample size of OS needed to match serum sampling sensitivity for PRRSV surveillance.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Ethics statement
The animal study was reviewed and approved by Institutional Animal Care and Use Committee. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
DL, NV, and CL conceptualized the study. NV and CL carried out the sampling. OO and DL carried out the statistical analysis. NV, OO, CL, and DL discussed the output of all the analyses. CP and OO reassessed the findings of the study. OO drafted the first manuscript. All authors proofread, contributed to- and approved the final version of the manuscript.
Funding
This study was funded by the Veterinary Diagnostic and Production Animal Medicine, Iowa State University College of Veterinary Medicine, Ames, IA, United States.
Conflict of interest
CL was employed by Carthage Veterinary Services, Limited.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Dokland T. The structural biology of PRRSV. Virus Res. (2010) 154:86. doi: 10.1016/j.virusres.2010.07.029
2. Kappes MA, Faaberg KS. PRRSV structure, replication and recombination: origin of phenotype and genotype diversity. Virology. (2015) 479:475–86. doi: 10.1016/j.virol.2015.02.012
3. Holtkamp DJ, Kliebenstein JB, Neumann E, Zimmerman JJ, Rotto H, Yoder TK, et al. Assessment of the economic impact of porcine reproductive and respiratory syndrome virus on United States pork producers. J Swine Health Prod. (2013) 21:28. doi: 10.31274/ans_air-180814-28
4. Zimmerman JJ, Yoon KJ, Wills RW, Swenson SL. General overview of PRRSV: A perspective from the United States. In: Veterinary Microbiology. Elsevier (1997). p. 187–96. doi: 10.1016/S0378-1135(96)01330-2
5. Brockmeier SL, Halbur PG, Thacker EL. Porcine respiratory disease complex. In:Brigden KA, Guthmiller JM, , editors. Polymicrobial Diseases. doi: 10.1128/9781555817947.ch13
6. Halbur PG, Paul PS, Frey ML, Landgraf J, Eernisse K, Meng XJ, et al. Comparison of the pathogenicity of two US porcine reproductive and respiratory syndrome virus isolates with that of the Lelystad virus. Vet Pathol. (1995) 32:648–60. doi: 10.1177/030098589503200606
7. Corzo CA, Mondaca E, Wayne S, Torremorell M, Dee S, Davies P, et al. Control and elimination of porcine reproductive and respiratory syndrome virus. Virus Res. (2010) 154:185–92. doi: 10.1016/j.virusres.2010.08.016
8. USA: The world's third largest pork exporter and consumer—Pig Progress. Available online at: https://www.pigprogress.net/world-of-pigs/country-focus/usa-the-worlds-third-largest-pork-exporter-and-consumer/ (accessed January 2, 2023).
9. Balka G, Podgórska K, Brar MS, Bálint Á, Cadar D, Celer V, et al. Genetic diversity of PRRSV 1 in Central Eastern Europe in 1994–2014: origin and evolution of the virus in the region. Sci Rep. (2018) 8:7811. doi: 10.1038/s41598-018-26036-w
10. Trevisan G, Linhares LCM, Crim B, Dubey P, Schwartz KJ, Burrough ER, et al. Macroepidemiological aspects of porcine reproductive and respiratory syndrome virus detection by major United States veterinary diagnostic laboratories over time, age group, and specimen. PLoS ONE. (2019) 14:e0223544. doi: 10.1371/journal.pone.0223544
11. Linhares DCL, Cano JP, Torremorell M, Morrison RB. Comparison of time to PRRSv-stability and production losses between two exposure programs to control PRRSv in sow herds. Prev Vet Med. (2014) 116:111–9. doi: 10.1016/j.prevetmed.2014.05.010
12. Linhares DCL, Almeida M. PRRSv detection over time in different age groups in breeding herds attempting to produce PRRSv-negative piglets at weaning: part 2: suckling pig population (NPB 18-161). (2019). Available online at: https://www.pork.org/research/prrsv-detection-time-different-age-groups-breeding-herds-attempting-produce-prrsv-negative-piglets-weaning-part-2-suckling-pig-population/ (accessed December 28, 2020).
13. de Almeida MN, Corzo CA, Zimmerman JJ, Linhares DCL. Longitudinal piglet sampling in commercial sow farms highlights the challenge of PRRSV detection. Porcine Health Manag. (2021) 7:1–10. doi: 10.1186/s40813-021-00210-5
14. Lalonde C, Provost C, Gagnon CA. Whole-genome sequencing of porcine reproductive and respiratory syndrome virus from field clinical samples improves the genomic surveillance of the virus. J Clin Microbiol. (2020) 58:20. doi: 10.1128/JCM.00097-20
15. Risser J, Ackerman M, Evelsizer R, Wu S, Kwon B, Hammer JM. Porcine reproductive and respiratory syndrome virus genetic variability a management and diagnostic dilemma. Virol J. (2021) 18:1–12. doi: 10.1186/s12985-021-01675-0
16. Lopez WA. Porcine reproductive and respiratory syndrome monitoring in breeding herds using processing fluids. J Swine Health Prod. (2018) 26:146–50.
17. Almeida MN, Zhang M, Zimmerman JJ, Holtkamp DJ, Linhares DCL. Finding PRRSV in sow herds: family oral fluids vs. serum samples from due-to-wean pigs. Prev Vet Med. (2021) 193:105397. doi: 10.1016/j.prevetmed.2021.105397
18. Holtkamp DJ, Torremorell M, Corzo CA, L Linhares DC, Almeida MN, Yeske P, et al. Proposed modifications to porcine reproductive and respiratory syndrome virus herd classification. J Swine Health Prod. (2021) 29:261–70.
19. Olanratmanee EO, Thanawongnuwech R, Kunavongkrit A, Tummaruk P. Reproductive performance of sows with and without PRRS modified live virus vaccination in PRRS-virus-seropositive herds. Trop Anim Health Prod. (2014) 46:1001–7. doi: 10.1007/s11250-014-0606-5
20. Dee S, Batista L, Deen J, Pijoan C. Evaluation of an air-filtration system for preventing aerosol transmission of porcine reproductive and respiratory syndrome virus. Can J Vet Res. (2005) 69:293–8.
21. Perez AM, Linhares DCL, Arruda AG, Van Der Waal K, Machado G, Vilalta C, et al. Individual or common good? Voluntary data sharing to inform disease surveillance systems in food animals. Front Vet Sci. (2019) 6:194. doi: 10.3389/fvets.2019.00194
22. Rovira A, Reicks D, Muñoz-Zanzi C. Evaluation of surveillance protocols for detecting porcine reproductive and respiratory syndrome virus infection in boar studs by simulation modeling. J Vet Diag Invest. (2007) 19:492–501. doi: 10.1177/104063870701900506
23. Osemeke OH, de Freitas Costa E, Almeida MN, Trevisan G, Ghosh AP, Silva GS, et al. Effect of pooling family oral fluids on the probability of PRRSV RNA detection by RT-rtPCR. Prev Vet Med. (2022) 206:105701. doi: 10.1016/j.prevetmed.2022.105701
24. Nunes de, Almeida M. Improved Porcine Reproductive and Respiratory Syndrome Virus Surveillance in Swine Breeding Herds ProQuest Dissertations and Theses. Iowa: State University (2020).
25. Holtkamp DJ, Polson DD, Torremorell M, Morrison B, Classen DM, Becton L, et al. Terminology for classifying swine herds by porcine reproductive and respiratory syndrome virus status. J Swine Health Prod. (2011) 19:44–56.
26. Almeida MN, Rotto H, Schneider P, Robb C, Zimmerman JJ, Holtkamp DJ, et al. Collecting oral fluid samples from due-to-wean litters. Prev Vet Med. (2020) 174:4810. doi: 10.1016/j.prevetmed.2019.104810
27. Boulbria G, Normand V, Leblanc-Maridor M, Belloc C, Berton P, Bouchet F, et al. Feasibility of pooled oral fluid collection from pre-weaning piglets using cotton ropes. Vet Anim Sci. (2020) 9:100099. doi: 10.1016/j.vas.2020.100099
28. Osemeke O, Donovan T, Dion K, Holtkamp D, Linhares D. Characterization of changes in productivity parameters as breeding herds transitioned through the 2021 PRRSV breeding herd classification system. J Swine Health Prod. (2022) 30:145–8. doi: 10.54846/jshap/1269
29. Cannon RM, Roe RT. Livestock Disease Surveys. A Field Manual for Veterinarians. Bureau of Rural Science, Department of Primary Industry. Canberra: Australian Government Pub. Service (1982). 14–17 p.
30. Fosgate GT. Practical sample size calculations for surveillance and diagnostic investigations. J Vet Diag Invest. (2009) 21:3–14. doi: 10.1177/104063870902100102
31. R Core Team. R: A Language and Environment for Statistical Computing. Vienna. R Foundation for Statistical Computing Vienna (2019).
32. Wickham H. CRAN - Package ggplot2. (2020). Available online at: https://cran.r-project.org/web/packages/ggplot2/index.html (accessed July 12, 2022).
33. Bates D, Mächler M, Bolker B, Walker S. Fitting linear mixed-effects models using lme4. J Stat Softw. (2014) 67:1. doi: 10.18637/jss.v067.i01
34. McHugh ML. Interrater reliability: the kappa statistic. Biochem Med. (2012) 22:276. doi: 10.11613/BM.2012.031
35. Luabeya AK, Wood RC, Shenje J, Filander E, Ontong C, Mabwe S, et al. Non-invasive detection of tuberculosis by oral swab analysis. J Clin Microbiol. (2019) 57:18. doi: 10.1128/JCM.01847-18
36. Spengler JR, Chakrabarti AK, Coleman-McCray JD, Martin BE, Nichol ST, Spiropoulou CF, et al. Utility of oral swab sampling for Ebola virus detection in guinea pig model. Emerg Infect Dis. (2015) 21:1816–9. doi: 10.3201/eid2110.150840
37. Zhuo R, Parsons BD, Lee BE, Drews SJ, Chui L, Louie M, et al. Identification of enteric viruses in oral swabs from children with acute gastroenteritis. J Mol Diagn. (2018) 20:56–62. doi: 10.1016/j.jmoldx.2017.09.003
38. Adler L, Parizade M, Koren G, Yehoshua I. Oral cavity swabbing for diagnosis of group a Streptococcus: a prospective study. BMC Fam Pract. (2020) 21:6. doi: 10.1186/s12875-020-01129-6
39. Labbé AC, Benoit P, Gobeille Paré S, Coutlée F, Lévesque S, Bestman-Smith J, et al. Comparison of saliva with oral and nasopharyngeal swabs for SARS-CoV-2 detection on various commercial and laboratory-developed assays. J Med Virol. (2021) 93:5333–8. doi: 10.1002/jmv.27026
40. Ku CW, Shivani D, Kwan JQ, Loy SL, Erwin C, Ko KK, et al. Validation of self-collected buccal swab and saliva as a diagnostic tool for COVID-19. Int J Infect Dis. (2021) 104:255–61. doi: 10.1016/j.ijid.2020.12.080
41. Mane A, Jain S, Jain A, Pereira M, Sirsat A, Pathak G, et al. Diagnostic performance of oral swab specimen for SARS-CoV-2 detection with rapid point-of-care lateral flow antigen test. Sci Rep. (2022) 12:8. doi: 10.1038/s41598-022-11284-8
42. Arnold ME, Slomka MJ, Coward VJ, Mahmood S, Raleigh PJ, Brown IH. Evaluation of the pooling of swabs for real-time PCR detection of low titre shedding of low pathogenicity avian influenza in turkeys. Epidemiol Infect. (2013) 141:1286–97. doi: 10.1017/S0950268812001811
43. Tajima M, Ohsaki T, Okazawa M, Yasutomi I. Availability of oral swab sample for the detection of bovine viral diarrhea virus (BVDV) gene from the cattle persistently infected with BVDV. Jap J Vet Res. (2008) 56:3–8.
44. Stenfeldt C, Lohse L, Belsham GJ. The comparative utility of oral swabs and probang samples for detection of foot-and-mouth disease virus infection in cattle and pigs. Vet Microbiol. (2013) 162:330–7. doi: 10.1016/j.vetmic.2012.09.008
45. Kuiek AM, Ooi PT, Yong CK, Ng CF. Comparison of serum and oral fluid antibody responses after vaccination with a modified live (MLV) porcine reproductive and respiratory syndrome virus (PPRSV) vaccine in PRRS endemic farms. Trop Anim Health Prod. (2015) 47:1337–42. doi: 10.1007/s11250-015-0868-6
46. Agrba VZ, Lapin BA, Timanovskaya V V, Dzhachvliany MC, Kokosha L V, Chuvirov GN, et al. Isolation of lymphotropic baboon herpesvirus (HVP) from oral swabs of hamadryas baboons of the Sukhumi monkey colony. Exp Pathol. (1980) 18:269–74. doi: 10.1016/S0014-4908(80)80031-7
47. Marschang RE, Kolesnik E. Detection of nidoviruses in live pythons and boas. Tierarztl Prax Ausg K Kleintiere Heimtiere. (2016) 45:22–6. doi: 10.15654/TPK-151067
48. Reicks DL, Muñoz-Zanzi C, Rossow K. Sampling of adult boars during early infection with porcine reproductive and respiratory syndrome virus for testing by polymerase chain reaction using a new blood collection technique (blood-swab method). J Swine Health Prod. (2006) 14:258–64.
49. Prickett J, Simer R, Christopher-Hennings J, Yoon KJ, Evans RB, Zimmerman JJ. Detection of porcine reproductive and respiratory syndrome virus infection in porcine oral fluid samples: a longitudinal study under experimental conditions. J Vet Diag Invest. (2008) 20:156–63. doi: 10.1177/104063870802000203
Keywords: PRRSV, surveillance, sampling, serum, swabs, weaning-age
Citation: Osemeke OH, VanKley N, LeFevre C, Peterson C and Linhares DCL (2023) Evaluating oral swab samples for PRRSV surveillance in weaning-age pigs under field conditions. Front. Vet. Sci. 10:1072682. doi: 10.3389/fvets.2023.1072682
Received: 17 October 2022; Accepted: 16 January 2023;
Published: 03 February 2023.
Edited by:
Jose L. Gonzales, Wageningen University and Research, NetherlandsReviewed by:
Eleftherios Meletis, University of Thessaly, GreeceLars Erik Larsen, University of Copenhagen, Denmark
Copyright © 2023 Osemeke, VanKley, LeFevre, Peterson and Linhares. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daniel C. L. Linhares,  bGluaGFyZXNAaWFzdGF0ZS5lZHU=
bGluaGFyZXNAaWFzdGF0ZS5lZHU=
 Onyekachukwu Henry Osemeke
Onyekachukwu Henry Osemeke Nathan VanKley
Nathan VanKley Claire LeFevre
Claire LeFevre Christina Peterson
Christina Peterson Daniel C. L. Linhares
Daniel C. L. Linhares