- 1College of Animal Science and Veterinary Medicine, Shandong Agricultural University, Tai'an, China
- 2Institute of Poultry Sciences, Shandong Academy of Agricultural Sciences, Jinan, China
Rabbit Viral Hemorrhagic Disease (RHD) is a highly contagious and fatal infection, resulting in considerable economic losses to the rabbit industry. Consequently, it is essential to develop a fast and accurate diagnostic method for RHDV GI.1. In this study, a rapid simple reverse transcriptase recombinase polymerase amplification (RTRPA) for RHDV GI.1 was successfully developed using specific primers to RHDV GI.1 VP60 gene. Results indicated that the entire amplification process could be achieved in an isothermal condition at 40°C for 30 minutes, with good specificity and no reaction to other common rabbit disease pathogens, and a high sensitivity of upto 0.1LD50 of RHDV GI.1. Then, RT-RPA method was used to detect 1144 clinical samples, and the positive rates were 0.95%, 1.29% and 2.50% in Zaozhuang, Linyi, and Liaocheng in Shandong Province, respectively (the Fisher's exact test, P = 0.413), suggesting that there is no significant difference in RHDV GI.1 infection among the different regions. In conclusion, this study established a RT-RPA assay which is suitable for quick detection and monitoring of RHDV GI.1, thus making it a viable option for epidemiological surveillance.
1. Introduction
Rabbit hemorrhagic disease (RHD) is an acute, septic, highly contagious and fatal disease caused by rabbit hemorrhagic virus (RHDV) infection (1). The disease first broke out in Wuxi, Jiangsu Province, China in 1984 and then spread to many regions around the world, which attracted the attention of scholars all over the world. The main clinical symptoms were neurological symptoms, respiratory symptoms, slow response and anorexia. The disease is characterized by rapid transmission and high mortality. Infected rabbit generally dies within 1–2 days after fever, and the mortality is as high as 70%-90%. The virus seriously damaged the ecological environment of rabbits, endangered rare hare species and affected the development of rabbit industry. In 2010, an additional RHDV, phylogenetically, and antigenically distinct from RHDV, emerged in Europe and was called RHDV2. A new RHDV isolate collected from infected rabbits at farms in the Sichuan province of China in April 2020, representing the first report of RHDV2 in China (2). According to the new genome classification, the genomes of classical RHDV are classified as GI.1, and the new emerged RHDV2 is now classified as GI.2. At present, RHD caused by RHDV GI.1 is highly epidemic in Chinese rabbit farms, even though some rabbits have been vaccinated with commercial vaccines, they still remain susceptible to the virus. Therefore, it is essential to create a rapid detection method for RHDV GI.1.
Since RHD can exist stably in the environment and transmit quickly, therefore, it is vital for RHDV GI.1 rapid detection in rabbit flocks. Various methods can be used to detect the virus, such as hemagglutination assay (HA), enzyme-linked immunosorbent assay (ELISA), reverse-transcription PCR (RT-PCR) (3) and quantitative real-time RT-PCR (qRT-PCR). However, these methods all have shortcomings such as time-consuming and low sensitivity. In addition, some methods are very demanding on instruments and equipment. Therefore, the development of a portable rapid test to monitor RHDV GI.1 infection is crucial in field diagnosis.
Recombinase polymerase amplification (RPA) is known as a novel nucleic acid detection technology that has several advantages compared with conventional PCR assay. RPA technology mainly relies on three enzymes: recombinase that can bind single stranded nucleic acid (oligonucleotide primer), single stranded DNA binding protein (SSB) and strand replacement DNA polymerase. Under isothermal conditions (generally constant at 37–39°C), the reaction process is very fast, amplification products usually can be detected within 20–40 min, then the amplified fragments can be observed by agarose gel electrophoresis (4). To our knowledge, there was no report on the RT-RPA method for the rapid detection of RHDV GI.1.
The RHDV GI.1 genome is a single-stranded, positive-strand RNA which composed of 7,437 nucleotides (nt), including two open-reading frames (ORFs): positive sense ORF1 and ORF2. ORF1 encodes a large polyprotein which then hydrolyzed to some non-structural proteins and the capsid protein VP60. ORF2 encodes the minor structural protein VP10 (5–8). VP60 protein is not only the capsid protein of the virus, but also the only structural protein of the virus (9, 10). In this study, specific primers were designed based on RHDV GI.1VP60 gene, and the reaction conditions were optimized to establish the RT-RPA assay, which provide a suitable method for the diagnosis and detection of RHDV GI.1.
2. Materials and methods
2.1. Ethics statement
The animal experiments were approved by the Committee on the Ethics of Animal of Institute of Poultry Science, Shandong Academy of Agricultural Sciences (permit number: 2019005), according to the guidelines of the Review of Welfare and Ethics of Laboratory Animals authorized by the Shandong Municipality Administration Office of Laboratory Animals. The animal Experiments were conducted in the Biosafety Level 2 laboratory in Shandong Academy of Agricultural Sciences, in compliance with the ARRIVE guidelines. All applicable international, national, and/or institutional guidelines for the care and use of animals were followed. Rabbits were sacrificed by well-trained operators by inhaling CO2 (50–60%) to protect animal welfare using animal sacrifice device.
2.2. RT-RPA primer's design
To establish the RT-RPA assay, the primers were designed based on the VP60 nucleotide sequence of RHDV GI.1 LW/2015 strain (Genbank accession number: KY398843): Primer: F: 5'- TGTTATGGAGGGCAAAACCCGCACAGCGCCGCAA−3'; Primer R: 5'- CGGCTGTGAATGGGTTGTTCTGTGGAGAGTGTT -3'.
2.3. Optimization of reaction conditions of the RT-RPA assay
The RT-RPA assay was carried out by using TwistAmp Basic kit (TWISTDX Ltd., Babraham, UK) and in a 0.2 ml reaction tube, the total reaction volume is 50 mL, including rehydration buffer 29.5 μL, Primer F (10 μM) 2.4 μL, Primer R (10 μM) 2.4 μL, template 2.0 μL, ddH2O 11.2 μL, MgAc (280 mM) 2.5 μL. Then reaction tubes were incubated at 37°C for 30 min in a metal bath, and the reaction product was observed by electrophoresis in 1.2% agarose gel and purified by axyprep DNA gel extract Kit (AXYGEN Ltd., Silicon Valley, USA). The purified amplification product was verified by sequencing (Invitrogen Trading Ltd., Shanghai, China) and analyzed using the BLAST program in National Center for Biotechnology Information website. To achieve optimal amplification results, reaction temperature and concentrations of primers were optimized. Reaction was carried out 35 to 42°C for about 30 min according to TwistAmp Basic Kit. Then, different temperature (35, 37, 39, 40, 42°C) and various primer concentrations (0.12, 0.24, 0.48 μM) were also investigated in this study, respectively.
2.4. Specificity and sensitivity of the RT-RPA assay
The specificity of RT-RPA assay was assessed against other common infecting viruses in rabbit including rabbit rotavirus (Z3171 strain), rabbit coronavirus (J103 strain), rabbit astrovirus (Z721 strain), Salmonella (sm1378 strain), Klebsiella pneumoniae (kp1806 strain) and Pseudomonas Aeruginosa (pa2013 strain). In addition, the liver from SPF rabbits was used as negative control.
For sensitivity analysis, infected rabbit liver tissue was grinded with PBS buffer, diluted from 10−1 to 10−8, and RNAs were extracted from those samples. The extracted RNA was then analyzed with RT-RPA method to investigate the minimum detection dose. The RT-PCR was also performed using the same diluted samples.
2.5. Clinical samples detection and epidemiological investigation and analysis
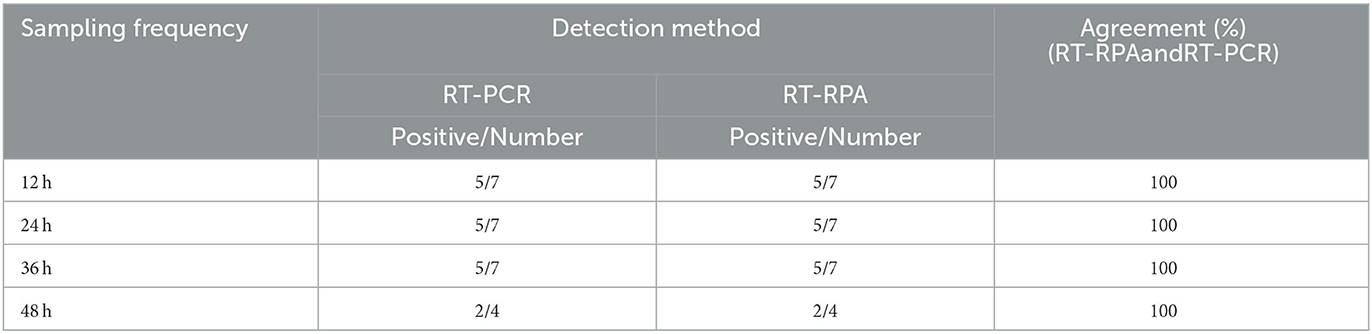
Seven 8-week-old SPF rabbits were purchased from Institute of animal husbandry and veterinary medicine, Shandong Academy of Agricultural Science. Five rabbits were inoculated intraperitoneally with 0.5 mL of the RHDV GI.1 NJ strain (Genbank: HM623309, provided by doctor Wang Fang of Jiangsu Academy of Agricultural Sciences) liver suspension (106 LD50) and two rabbits were utilized as negative control with Phosphate Buffered Saline (PBS) inoculation and two rabbits were used as negative control by inoculation of Phosphate Buffered Saline (PBS). Nasal swabs were collected from infected rabbits every 12 h, and liver tissues from dead rabbits and SPF rabbits were collected for detection by RT-RPA and RT-PCR. Shandong Province is a big rabbit breeding province in China, with the output ranking among the top in the country. Therefore, 1,144 nasal swabs were randomly collected from 56 rabbit farms in Liaocheng, Linyi and Zaozhuang counties of Shandong Province.
3. Results
3.1. Establishment of RT-RPA assay
The specificity of the amplification product was confirmed and visualized by agarose gel as 317 bp bands, which was consistent with the expected fragment size. So, the primer-set could be utilized in the later assays. Initially, we assessed a series of temperature during in cubation for 30 min. Amplification signals can be detected at 35, 37, 39, 40, 42°C, among which the amplification signal was in the highest at 40°C (Figure 1A). Amplification signals could be detected at 40°C various primer concentrations 0.12, 0.24, and 0.48 μM. However, there was no significant difference in the products yielded among the 0.12, 0.24, and 0.48 μM reactions (Figure 1B).
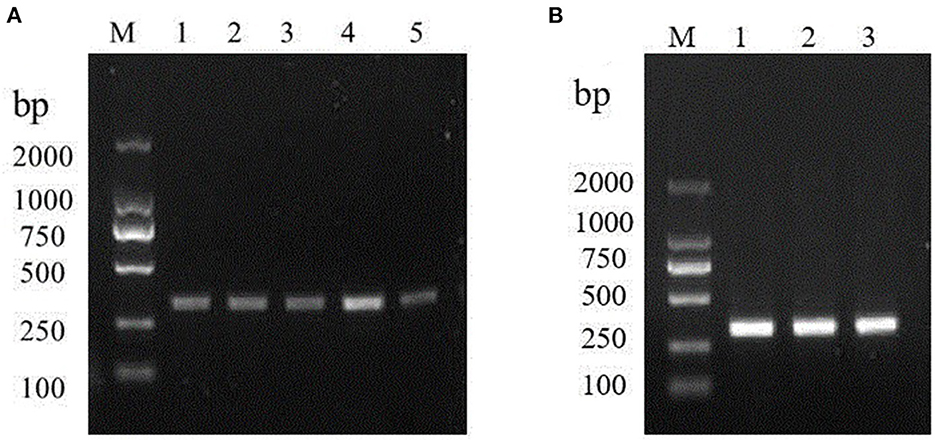
Figure 1. Optimizations of the RT-RPA assay components. (A) RT-RPA reaction temperature; M: DL2000 marker. Lanes 1–5: 35, 37, 39, 40, and 42°C, respectively. (B) Optimal primer concentration screening. M: DL2000 marker. Lanes 1-6: 0.12, 0.24, and 0.48 μM, respectively.
3.2. Specificity of the RT-RPA assay
To test the specificity of the RT-RPA assay, RHDV GI.1 and 6 other pathogens were detected by the assay. The results show that, only the RHDV GI.1 but no other pathogens used in this study was detected indicating that the primers showed a high specificity (Figure 2).
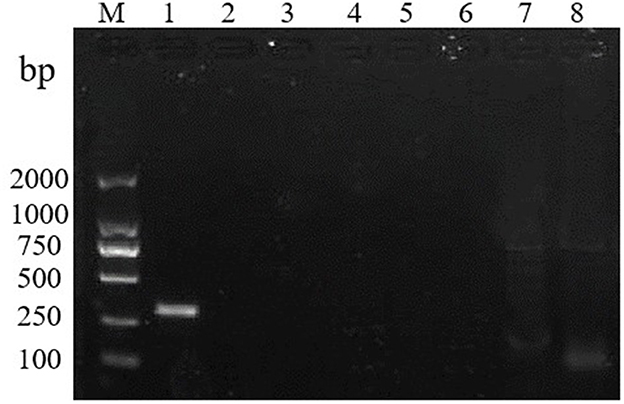
Figure 2. RT-RPA specificity test. M: DL2000 marker. Lanes 1–8: RHDV, rabbit rotavirus, rabbit coronavirus, rabbit astrovirus, Salmonella, Klebsiella pneumoniae, Pseudomonas Aeruginosa, and SPF rabbit liver.
3.3. Sensitivity of the RT-RPA assay
The RT-RPA assay was assessed for the detection sensitivity using different concentrations of the RHDV GI.1 RNA standards under the optimized reaction system. The RT-RPA assay showed high sensitivity, with the detection limit is 0.1LD50 of RHDV GI.1. The results show that RT-PCR assay and RT-RPA assay had the same detection limit 0.1LD50 of RHDV GI.1 (Figures 3A, B).
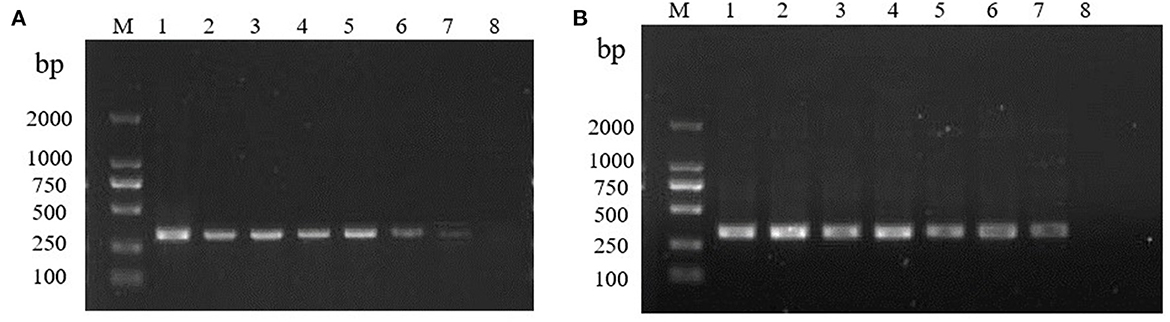
Figure 3. Sensitivity of the RT-RPA and RT-PCR. (A) Sensitivity of the RT-PCR. M: DL2000 marker. 10-fold gradient dilution of the RNA samples (10 −1 −10 −8, lanes 1–8). (B) Sensitivity of the RT-RPA. M: DL2000 marker. 10-fold gradient dilution of the RNA samples (10 −1 −10 −8, lanes 1–8).
3.4. Samples detection and epidemiological investigation using the RT-RPA assay
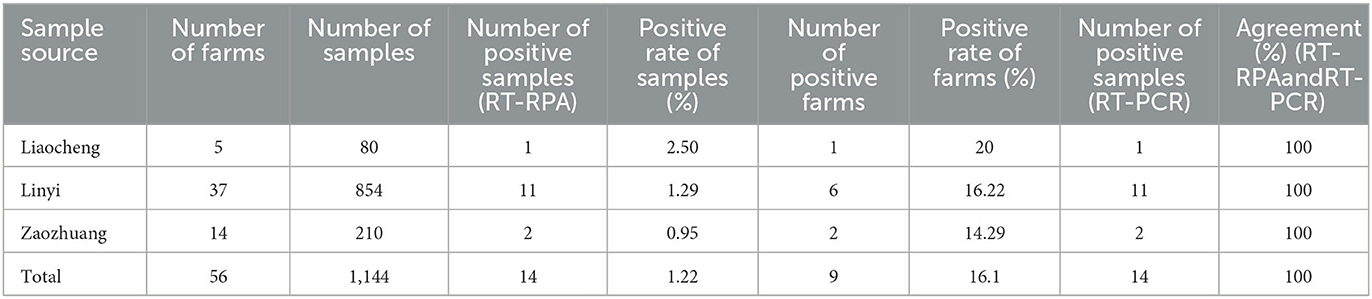
Nasal swabs were collected at 12, 24, and 36 h after infection, of which 3 rabbits died at 36 h and 2 rabbits died at 48 h. The results of RT-RPA and RT-PCR showed that all nasal swabs from five rabbits infected artificially with RHDV GI.1 were detected positive 12 h post infection, the liver of dead rabbits was positive, and the liver of SPF rabbits was negative (Table 1). Furthermore, among 1,144 nasal swab samples, 80 were from Liaocheng, 854 from Linyi, and 210 from Zaozhuang (Figure 4A). The results showed that the positive rate was 0.95, 1.29, 2.50% in Zaozhuang, Linyi, Liaocheng, respectively (Figure 4B). The average positive rate is 1.58% in three area, However, there was no significant difference in RHDV GI.1 infection in different regions [the Fisher's exact test, P > 0.05 (P = 0.413)]. The established RT-RPA method was consistent with one-step RT-PCR detection, with 1.22% positive rate (14/1,144) (Table 2). There were 16.1% (9/56) rabbit farms positive for RHDV GI.1. The detection rate of positive samples was 4.26% at the age of <3 months, 1.24% at the age of 4–8 months and 0.56% at the age of 9–12 months. Most of the positive samples were found in farm with the scale of about 500 rabbits. These data indicated that although RHDV GI.1 vaccine was immunized in selected rabbit farms, field RHDV GI.1 infection still existed in some rabbits.
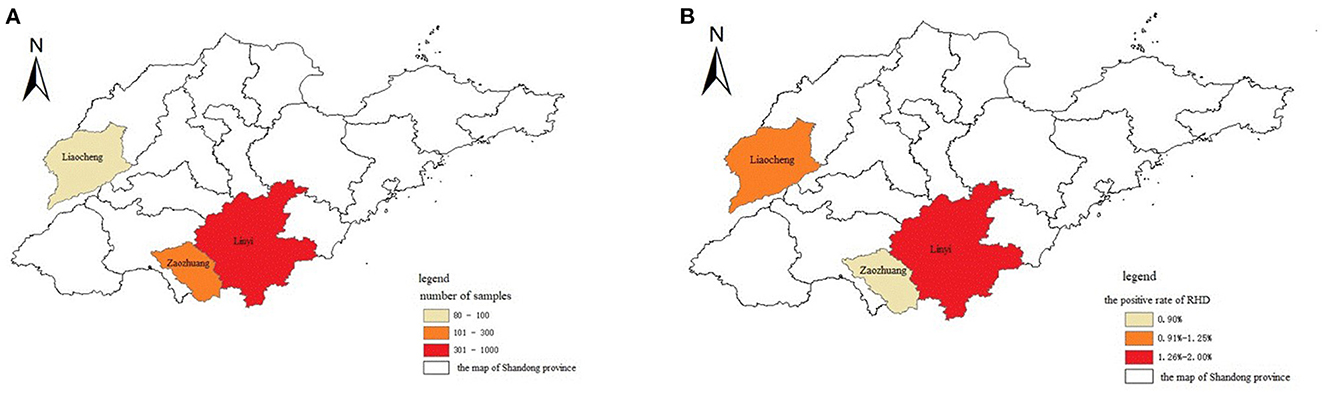
Figure 4. Geographic distribution and epidemic of RHDV. (A) The cities where the samples were collected were indicated in different colors in the map of Shandong province in China. (B) The cities affected with RHDV infection were indicated in different colors in the map of Shandong province in China.
4. Discussion
RHDV GI.1 generally infects adult rabbits and has the characteristics of rapid spread, highly contagious, short incubation period, and high rates of mortality and morbidity, which brings significant economic losses to the rabbit industry. Through the observation of five rabbits with acute death caused by artificial infection, it can be seen that the disease is a kind of systemic sepsis, there are serious material metabolism disorders in the body, many important parenchymal organs show progressive lesions and bleeding, and the defense mechanism of the body collapses, so the infected rabbits die quickly after infection. Although there is a commercial vaccine for RHDV GI.1, the rabbits still have infection after immunization Therefore, it is vital for establish a method for rapid detection of RHDV GI.1.
HA test was the first routine laboratory diagnostic method used for RHD detection. RHDV GI.1 only agglutinated with human red blood cells, but not agglutinate with rabbit or other mammalian red blood cells. However, it was difficult to obtain human O-type blood in most of laboratories because of the public health conditions (11). In addition, it was found that the emergence of no agglutinated “O” type red blood cell characteristic strain may lead to missed detection and wrong detection. Through electron microscopy (12), especially immune electron microscopy (IEM), virus particles and virus proteins can be accurately identified, but it was not suitable for clinical diagnosis since it required expensive equipment and well-trained technicians.
To date, molecular assays include conventional RT-PCR, real-time fluorescent quantitative RT-PCR, etc. have been widely used in most labs. However, these methods need special equipment, and require the experimenters to spend time exploring a stable reaction system, and the reaction time is too long. Compared with the previous detection techniques, RT-RPA method has the advantages of fast, sensitive, simple operation and short-time.
Nowadays, RPA method is widely used in the detection of a variety of different fields including human and veterinary pathogenic microorganism detection, food safety and agriculture (13–15). The method not only offset the faultiness of other methods, such as low detection rate, time-consuming, and high requirements for instruments, but also improves the detection efficiency, reduces the workload, and shortens the detection time. In addition, since all RT-RPA reagents were commercially available, it was conducive to the standardization of this assay.
However, RT-RPA detection methods still have some limitations. In the study, using the LW/2015 strain as a reference, we optimized the primer sequence by aligning it to the known RHDV GI.1 sequence in GenBank. The primer is relevantly conserved for RHDV GI.1 in theory. The above tests have demonstrated that RT-RPA is a viable method for detecting RHDV GI.1. In the future, we will consider the degenerate bases of the primer for the potential mutation of the VP60 gene of newly isolated RHDV GI.1.
For this RT-RPA assay, the amplified fragments detected by agarose gel electrophoresis can be observed within 30 min. The optimal reaction conditions were determined through optimizing the concentration of the primers and reaction temperature, and the optimal reaction temperature was determined to be 40°C. Then, specificity and sensitivity tests were performed compared with normal RT-PCR assay. The results showed that the method had good specificity and had no cross-reaction with other pathogen which are often susceptible rabbits, such as E. coli, Pasteurella, Salmonella, Bordetella and SPF rabbit liver. The sensitivity of the RT-RPA method and normal RT-PCR method was the same, with the detect limit is 0.1LD50 of RHDV GI.1.
The most effective preventive measure for RHDV GI.1 is vaccination. Despite the vaccination, the disease is still sporadic and epidemic. To understand the prevalence of RHDV GI.1 in Shandong Province, 1,144 samples were detected with the RT-RPA method established in this study. The results showed that the established RT-RPA method was consistent with RT-PCR one-step detection, with 14 RHDV GI.1 positive samples were detected, the positive detection rate was 1.22%, which had good clinical applicability. And the veterinary epidemiological investigation and monitoring sampling calculator showed that 23 samples were collected in farm with the scale of about 500 rabbits. To provide basis for local RHD etiology monitoring and RHD prevention and control, so as to better ensure the healthy and sustainable development of rabbit industry.
In conclusion, an RT-RPA assay was established for RHDV GI.1 detection in this study. The method has the advantages of good stability, strong specificity and high sensitivity, which had a good application prospect and provided a technique tool for clinical diagnoses and epidemic disease monitoring of RHDV GI.1 in the future.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
Animal experiments were approved by the Committee on the Ethics of Animal of Institute of Poultry Science, Shandong Academy of Agricultural Sciences (permit number: 2019005), according to the guidelines of the Review of Welfare and Ethics of Laboratory Animals authorized by the Shandong Municipality Administration Office of Laboratory Animals.
Author contributions
Conceived and designed the study: BH and LZZ. Performed the study: LZZ, QYZ, YTi, and YTa. The data analysis and manuscript revision: LZZ, YXW, and BH. Wrote the manuscript: LZZ. All authors have read and approved the final manuscript.
Funding
This research was supported by Shandong Modern Agricultural Technology and Industry System, Grant Number SDAIT-21-06.
Acknowledgments
The authors acknowledge Institute of Animal Husbandry and Veterinary Medicine, Shandong Academy of Agricultural Science for providing the rabbits for this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Blancou J. Preface of viral haemorrhagic disease of rabbits and the European brown bare syndrome. Sci Tech Rev. (1991) 24:1–10. doi: 10.20506/rst.10.2.558
2. Hu B, Wei H, Fan Z, Song Y, Chen M, Qiu R, et al. Emergence of rabbit haemorrhagic disease virus 2 in China in 2020. Vet Med Sci. (2021) 7:236–9. doi: 10.1002/vms3.332
3. Le Gall-Reculé G, Zwingelstein F, Portejoie Y, Le Gall G. Immunocapture-RT-PCR assay for detection and molecular epidemiology studies of rabbit haemorrhagic disease and european brown hare syndrome viruses. J Virol Methods. (2001) 97:49–57. doi: 10.1016/S0166-0934(01)00336-6
4. Wang JC, Wang JF, Li RW, Shi RH, Liu LB, Yuan WZ, et al. Evaluation of an incubation instrument-free reverse transcription recombinase polymerase amplification assay for rapid and point-of-need detection of canine distemper virus. J Virol Methods. (2018) 260:56–61. doi: 10.1016/j.jviromet.2018.07.007
5. Gould AR, Kattenbelt JA, Lenghaus C, Morrissy C, Chamberlain T, Collins BJ, et al. The complete nucleotide sequence of rabbit haemorrhagic disease virus (Czech strain V351): use of the polymerase chain reaction to detect replication in Australian vertebrates and analysis of viral population sequence variation. Virus Res. (1997) 47:7–17. doi: 10.1016/S0168-1702(96)01399-8
6. Abrantes J, van der Loo W, Le Pendu J, Esteves PJ. Rabbit haemorrhagic disease (RHD) and rabbit haemorrhagic disease virus (RHDV): a review. Vet Res. (2012) 43:12. doi: 10.1186/1297-9716-43-12
7. Wirblich C, Thiel HJ, Meyers G. Genetic map of the calicivirus rabbit hemorrhagic disease virus as deduced from in vitro translation studies. J Virol. (1996) 70:7974–83. doi: 10.1128/jvi.70.11.7974-7983.1996
8. Wang BB, Zhe MJ, Chen ZY, Li CF, Meng CC, Zhang MT, et al. Construction and applications of rabbit hemorrhagic disease virus replicon. PLoS ONE. (2013) 8:e60316. doi: 10.1371/journal.pone.0060316
9. Esteves PJ, Abrantes J, Carneiro M, Müller A, Thompson G, van der Loo W. Detection of positive selection in the major capsid protein VP60 of the rabbit haemorrhagic disease virus (RHDV). Virus Res. (2008) 137:253–6. doi: 10.1016/j.virusres.2008.07.025
10. Fouchet D, Le Pendu J, Guitton JS, Guiserix M, Marchandeau S, Pontier D. Evolution of microparasites in spatially and genetically structured host populations: the example of RHDV infecting rabbits. J Theor Biol. (2009) 257:212–27. doi: 10.1016/j.jtbi.2008.11.021
11. Ruvoën-Clouet N, Ganière JP, André-Fontaine G, Blanchard D, Le Pendu J. Binding of rabbit hemorrhagic disease virus to antigens of the ABH histo-blood group family. J Virol. (2000) 74:11950–4. doi: 10.1128/JVI.74.24.11950-11954.2000
12. Capucci L, Fallacara F, Grazioli S, Lavazza A, Brocchi E. A further step in the evolution of rabbit hemorrhagic disease virus: the appearance of the first consistent antigenic variant. Virus Res. (1998) 58:115–26. doi: 10.1016/S0168-1702(98)00106-3
13. Abd El Wahed A, El-Deeb A, El-Tholoth M, Abd El Kader H, Ahmed A, Hassan S, et al. A portable reverse transcription recombinase polymerase amplification assay for rapid detection of foot-and-mouth disease virus. PLoS ONE. (2013) 8:1–10. doi: 10.1371/journal.pone.0071642
14. James A, Macdonald J. Recombinase polymerase amplification: emergence as a critical molecular technology for rapid, low-resource diagnostics. Expert Rev Mol Diagn. (2015) 15:1475–89. doi: 10.1586/14737159.2015.1090877
Keywords: rabbit hemorrhagic disease virus, GI.1 genotype, isothermal, recombinase polymerase amplification, rapid detection method, epidemiological surveillance
Citation: Zhang L, Zhao Q, Tian Y, Tang Y, Wang Y and Huang B (2023) A novel reverse transcription recombinase polymerase amplification assay for rapid detection of GI.1 genotype of rabbit hemorrhagic disease virus. Front. Vet. Sci. 10:1056601. doi: 10.3389/fvets.2023.1056601
Received: 29 September 2022; Accepted: 21 February 2023;
Published: 10 March 2023.
Edited by:
Guillermo Roberto Risatti, University of Connecticut, United StatesReviewed by:
Amaresh Das, United States Department of Agriculture (USDA), United StatesJie Zhu, Shanghai Veterinary Research Institute (CAAS), China
Copyright © 2023 Zhang, Zhao, Tian, Tang, Wang and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bing Huang, aGJpbmRAMTYzLmNvbQ==
 Lianzhi Zhang
Lianzhi Zhang Qiaoya Zhao2
Qiaoya Zhao2 Yi Tang
Yi Tang Yixin Wang
Yixin Wang Bing Huang
Bing Huang