- 1Plataforma de Investigación en Salud Animal, Instituto Nacional de Investigación Agropecuaria (INIA), Estación Experimental INIA La Estanzuela, Colonia, Uruguay
- 2Departamento de Desarrollo Biotecnológico, Instituto de Higiene, Facultad de Medicina, Universidad de la República, Montevideo, Uruguay
- 3Departamento de Bacteriología y Virología, Instituto de Higiene, Facultad de Medicina, Universidad de la República, Montevideo, Uruguay
Salmonella enterica is an important zoonotic pathogen that is frequently identified in dairy farming systems. An increase in antibiotic resistance has led to inadequate results of treatments, with impacts on animal and human health. Here, the phenotypic and genotypic susceptibility patterns of Salmonella isolates from dairy cattle and dairy farm environments were evaluated and compared. A collection of 75 S. enterica isolates were evaluated, and their phenotypic susceptibility was determined. For genotypic characterization, the whole genomes of the isolates were sequenced, and geno-serotypes, sequence types (STs) and core-genome-sequence types were determined using the EnteroBase pipeline. To characterize antibiotic resistance genes and gene mutations, tools from the Center for Genomic Epidemiology were used. Salmonella Dublin (SDu), S. Typhimurium (STy), S. Anatum (SAn), S. Newport (SNe), S. Agona (Sag), S. Montevideo (SMo) and IIIb 61:i:z53 were included in the collection. A single sequence type was detected per serovar. Phenotypic non-susceptibility to streptomycin and tetracycline was very frequent in the collection, and high non-susceptibility to ciprofloxacin was also observed. Multidrug resistance (MDR) was observed in 42 isolates (56.0%), with SAn and STy presenting higher MDR than the other serovars, showing non-susceptibility to up to 6 groups of antibiotics. Genomic analysis revealed the presence of 21 genes associated with antimicrobial resistance (AMR) in Salmonella isolates. More than 60% of the isolates carried some gene associated with resistance to aminoglycosides and tetracyclines. Only one gene associated with beta-lactam resistance was found, in seven isolates. Two different mutations were identified, parC_T57S and acrB_R717Q, which confer resistance to quinolones and azithromycin, respectively. The accuracy of predicting antimicrobial resistance phenotypes based on AMR genotypes was 83.7%. The genomic approach does not replace the phenotypic assay but offers valuable information for the survey of circulating antimicrobial resistance. This work represents one of the first studies evaluating phenotypic and genotypic AMR in Salmonella from dairy cattle in South America.
1. Introduction
Salmonella enterica subspecies enterica (from now S. enterica) is one of the main pathogens affecting humans and animals and can cause various symptoms and illnesses, ranging from self-limiting acute gastroenteritis to septicemia and death (1, 2). Humans can be affected by typhoidal and non-typhoidal Salmonella. Typhoidal serotypes include invasive Salmonella Typhi and Paratyphi, which cause enteric fever (3). On the other hand, non-typhoidal Salmonella can cause enteric or invasive salmonellosis and is a major cause of morbidity and mortality globally (4). Many Salmonella infections in humans are due to the consumption of contaminated food, including meat of bovine origin, since this is a potential reservoir of the pathogen (5). Salmonella is now considered to cause more than 1 million infections annually in the US and is among the leading causes of foodborne illness (6). Animals are affected by non-typhoid serotypes such as Abortusovis, Dublin, and Gallinarum, which are adapted to sheep, cattle and poultry hosts, respectively, as well as other ubiquitous serotypes such as S. Typhimurium and S. Enteritidis (7).
In cattle, non-typhoid serotypes are capable of causing systemic disease similar to typhoid fever in humans (8). Salmonellosis in these animals is characterized by a more frequent occurrence in intensive farming systems. The disease varies according to different factors, such as the serotype involved, and the age, physiological and immune status of the host (9). Salmonella Dublin is the serotype adapted to cattle and can cause septicemia and abortions. Salmonella Typhimurium is frequently identified in calving systems and, in addition to enteric disease, can cause invasive diseases (10, 11). Both serotypes can rapidly spread in a herd, causing mortality and presenting a challenge in disease management (9).
Antibiotics are one of the main tools for treating bacterial diseases. However, in recent decades, the increase in AMR has led to inadequate treatment results, driving the search for new therapeutic options (12). When treating salmonellosis, antibiotics are indicated when the risk of systemic infection is increased, as in immunocompromised persons (13). Ampicillin and trimethoprim-sulfamethoxazole are used for the treatment of bovine salmonellosis, and fluoroquinolones and β-lactams are used for human treatment (14). However, the effectiveness of these antibiotics has decreased, mainly due to antibiotic resistance, which is a trend that is increasing in this bacterial genus and others (15, 16).
Plasmids are circular or linear extrachromosomal genetic elements present in various organisms (17). Salmonella serovars cause infections in animals and humans and may carry plasmids conferring virulence and AMR with particular biological properties that are subsequently expressed in the host (18). Within plasmids, antibiotic resistance genes are generally located in transposons. In addition, integrons are involved in the recruitment and expression of antimicrobial resistance genes (19).
In bacteria of animal origin, AMR varies among regions of the world. In the USA, S. enterica of bovine origin commonly shows resistance to cephalosporins and quinolones (20), while in Canada, the most common AMR pattern is resistance to ampicillin, chloramphenicol, streptomycin, sulfamethoxazole and tetracycline (ACSSuT) (21). Additionally, in South America, the AMR Salmonella isolates obtained from calves differ among regions. In northeastern Brazil, the main type of resistance observed is to cefotaxime (22), whereas in southeastern Brazil, nalidixic acid and cefoxitin resistance is primarily observed to, with a high frequency of susceptibility among strains (23). In Uruguay, the main phenotypic resistance observed is to streptomycin, tetracyclines and ampicillin, with differences in susceptibility between serotypes, where Dublin is more susceptible to antibiotics than Typhimurium (24).
In recent years, the complete sequencing of microorganism genomes has made it possible to approach their study such that the information available about them from different approaches, such as clinical diagnosis, epidemiology and research, has increased considerably (25, 26). There are currently several bioinformatics tools for the analysis of complete genomes. For AMR investigation, there are specialized databases with information about resistance genes and mutations. These databases are generally free to access and are periodically updated (27). Although genome-wide analysis does not replace phenotypic methods of resistance analysis, there is a high degree of correlation between the two approaches, and genome-wide analysis is a useful complement for the identification of new mechanisms and relationships between isolates (28).
Studies characterizing bovine-derived Salmonella strains are scarce in our country. Hence, the objective of this work is to study the presence and distribution of resistance genes, plasmids and integrons using different bioinformatic evaluation systems in a diverse sample of Salmonella strains of bovine origin from Uruguay.
2. Materials and methods
2.1. Strain collection
The study was conducted with 75 strains of S. enterica obtained from 2016 to 2020 at the Plataforma de Investigación en Salud Animal—Instituto Nacional de Investigación Agropecuaria, La Estanzuela, Uruguay. The collection was represented by 75 isolates obtained from stool samples collected from 35 calves, 5 cows, and 1 heifer; 25 organ samples from autopsied animals; 5 samples from the dairy farm environment; 1 food sample; 1 udder swab sample from milking cows; 1 drinking water sample; and 1 bovine fetus autopsy sample. The fecal isolates came from 26 calves < 20 days old and 7 calves between 20 and 30 days old. There were also two isolates from calves aged 65 and 114 days. The cows from which Salmonella isolates were obtained from feces were between 2 and 4 years old, and the heifers were 1 year old. The calves had diarrhea in all cases, and the cows were pregnant. The calves from which samples of organs and fluids were obtained had ages of <10 days in 7 cases, between 11 and 20 days in 7 cases, between 21 and 35 days in 3 cases, between 45 and 60 days in 4 cases, and between 80 and 90 days in 3 cases (Supplementary Table 1).
2.2. Salmonella collection, maintenance and growth
For the isolation of Salmonella spp. from different samples collected, the procedures described by Casaux et al. (24) were used. The strains were conserved in 20% glycerol and stored at −80°C. For reuse, they were cultivated in trypticase soy agar at 37°C for 18–24 h.
2.3. Antimicrobial susceptibility testing
The antibiotic susceptibility test was performed using the Kirby Bauer disc diffusion method (29). The antibiotics evaluated were ampicillin (AMP, 10 μg), amoxicillin-clavulanic acid (AMC 20/10 μg), cefotaxime (CTX 30 μg), sulfamethoxazole/trimethoprim (SXT 23.75/1.25 μg), ciprofloxacin (CIP 5 μg), enrofloxacin (ENR 5 μg), chloramphenicol (C 30 μg), streptomycin (S 10 μg), gentamicin (CN 10 μg), tetracycline (TE 30 μg), nitrofurantoin (NF 300 μg), and azithromycin (AZT 15 μg). E. coli ATCC 25,922 and E. coli ATCC 35,218 were used for quality control. The results were interpreted according to Clinical and Laboratory Standards Institute (31), and to facilitate the description of the results, both resistant and intermediate results were classified as “not susceptible.” For enrofloxacin, the CLSI guidelines for bacteria isolated from animals were used (30) (Supplementary Table 1). In addition, multidrug-resistant isolates (MDR) were considered when the isolates showed non-susceptibility to 3 or more groups of antibiotics (32).
2.4. Whole-genome sequencing
The 75 isolates were sent to the sequencing facility of MicrobesNG (Birmingham, UK, http://www.microbesng.com) for whole-genome sequencing. Standard sequencing services were provided by the company. Genomic DNA libraries were prepared with the Nextera XT Library Prep Kit (Illumina, San Diego, USA) according to the manufacturer's protocol with the following modifications: input DNA was increased 2-fold, and the PCR elongation time was increased to 45 s. DNA quantification and library preparation were carried out on a Hamilton Microlab STAR automated liquid handling system (Hamilton Bonaduz AG, Switzerland). The pooled libraries were quantified using the Kapa Biosystems Library Quantification Kit for Illumina. Sequencing was performed with Illumina sequencers (HiSeq/NovaSeq) using a 250 bp paired-end protocol. Adapters were trimmed from the reads using Trimmomatic 0.30 with a sliding window quality cutoff of Q15 (33). De novo assembly was performed using SPAdes version 3.7 (34), and contigs were annotated using Prokka 1.11 (35).
2.5. Genoserotyping, MLST and core genome MLST
The genome sequences of the Salmonella collection were analyzed using the EnteroBase platform. The serotype was determined using the SISTR1 + SeqSero2 tools (36–38). The core-genome MLST V2 experimental scheme (cgMLST V2) was used to obtain the value of the sequence type (cgST), and the Achtman MLST scheme was used for the allelic profile and the sequence type (ST) (39) (Supplementary Table 1).
2.6. Antimicrobial resistance genes, mutations and plasmids
To identify AMR genes and to chromosomal point mutations, we used ResFinder and Point Finder 4.1 (40–42) from the Center for Genomic Epidemiology (CGE). For this analysis, we selected a threshold of 90% for the sequence ID and 80% for the minimum length coverage. Manual curation was performed in those cases where identification was not univocal and the gene sequence could contain a coding sequence disruption leading to a pseudogene. We also used the PlasmidFinder 2.1 (43) and IntFinder 1.0 (44) tools to identify plasmids and integrons. For PlasmidFinder, we used thresholds of 95 and 60% for identity and coverage, respectively, and for IntFinder, we used a threshold of 90% for both identity and coverage.
2.7. Correlation between phenotypic and genotypic resistance profiles
The phenotypic results of the antimicrobial susceptibility were used test as a reference and were correlated with the presence or absence of the resistance and/or mutation genes detected from the evaluation of the complete genome considering a resistant isolate. For each group of antibiotics, concordance was considered to exist for those results with a not susceptible phenotype for which at least one gene or mutation explained that finding, and concordance with phenotypic susceptibility was considered to exist when no corresponding gene or mutation was found. Sensitivity (Se), specificity (Sp), positive predictive values (PPVs), negative predictive values (NPVs), and accuracy were calculated according to Trevethan (45).
2.8. Phylogenetic analyses
The serovars SAn, SDu, Sty, and SNe were chosen to perform the phylogenetic analysis, as they were collected from more diverse sources and included more isolates for comparison. For each of these serovars, a phylogenetic tree was generated using different reference genomes: Anatum—USDA-ARS-USMARC-1728 (NCBI accession NZ_CP014664.1), Dublin—CT_02021853 (NCBI accession CP001144.1), Typhimurium—LT2 (NCBI accession NC_003197.2), and Newport 0007-33 (NCBI accession NZ_CP013685.1).
Snippy software (version 4.6.0) (Torsten Seemann. https://github.com/tseemann/snippy) was used to map the trimmed reads to the reference genome and obtain a whole-genome alignment. Snippy was run with the default parameters, and the snippy-core script was run to generate the whole-genome alignment file (.full.aln). This file was then used as the input for Gubbins software (version 3.1.6) to produce a new alignment, from which the regions of high polymorphism density (probable regions of recombination) were removed (46). Gubbins was used with the following options: –threads 8, –first_tree-biulder fasttree, –tree-builder raxmlng, and –first_model JC.
Whole-genome phylogenies were produced using RAxML-NG software (version 1.1.0) based on the filtered_polymorphic_sites files (obtained in the previous step). RAxML was used with the following options: –model GTR + G, –seed 3 and –bs-metric fbp (47). The branch support values were transferred to the best-scoring tree of each sample by the RAxML-NG pipeline. Bootstrapping converged after 50, 250, and 996 replicates for SDu, Sty, and SNe, respectively. For SAn, 1,000 replicates were performed, but convergence was not reached because of the very low divergence of the sequences.
Phylogenetic trees with annotations were generated using the online tool iTOL v5 (https://itol.embl.de/) (48). The final tree figures were edited with the software Inkscape 0.91 (https://inkscape.org/).
3. Results
3.1. Determination of serotypes by WGS
Seven serotypes were identified after analyzing the genomes of the 75 collected isolates. The greatest number of isolates (32) were classified as S. Typhimurium, 24 as S. Newport, 11 as S. Anatum, 6 as S. Dublin, 1 as S. Agona, 1 as S. Montevideo and 1 as IIIb 61:i:z53. Each serotype was represented by a single sequence type (ST). ST10, ST19, ST45, and ST64 were detected in all the isolates of S. Dublin, S. Typhimurium, S. Newport or S. Anatum, respectively. In addition, ST13 corresponded to S. Agona, ST138 corresponded to S. Montevideo and ST430 corresponded to the strain serotyped as IIIb 61:i:z53 were detected identified. Among a total of 31 S. Typhimurium genomes, 24 different cgMLSTs were distributed on 26 different farms. A farm with 24 isolates of S. Newport presented 9 different cgMLSTs. In S. Dublin, 6 different cgMLSTs were observed. In S. Anatum, 5 cgMLSTs were observed in 11 total genomes. cgMLSTs 261271, 260644, and 275391 were identified in IIIB 61:I:Z53, S. Agona and S. Montevideo, respectively (Supplementary Table 1).
3.2. Antimicrobial susceptibility testing
Most of the isolates showed non-susceptibility to quinolones, tetracyclines and aminoglycosides. Fifty-eight isolates were not susceptible to the second generation-quinolone ciprofloxacin (77.3%). In addition, in this group, 5 isolates (6.6%) were not susceptible to enrofloxacin. Fifty-eight isolates (67%) were not susceptible to tetracycline, 64 (85.3%) were not susceptible to streptomycin, and 7 were also not susceptible to gentamicin (9.3%).
Only nine isolates (12%) were not susceptible to beta-lactam ampicillin, and 4 (5.3%) were not susceptible to amoxicillin-clavulanic acid. Every isolate was susceptible only to cefotaxime, and only 3 (4%) of the isolates were not susceptible to trimethoprim/sulfamethoxazole. A similar situation was observed for chloramphenicol, to which only 1 (1.3%) of the strains was not susceptible. There were 15 (20%) isolates that were not susceptible to nitrofurantoin, and 7 (9.3%) were not susceptible to azithromycin.
Considering MDR, 42 isolates (56%) were MDR. Twenty-two isolates were not susceptible to three groups of antimicrobials (29.3%), 15 were not susceptible to 4 groups (20%), four were not susceptible to 5 groups (5.3%) and one was not susceptible to 6 groups of antimicrobials (1.3%). Three isolates of S. Anatum were MRD, as were 14 S. Newport isolates, 24 S. Typhimurium isolates, and the only IIIb 61:i:z53 isolate. The most frequent MDR phenotype was CIP-S-TE, observed in 15 isolates, followed by CIP-S-TE-NF, in 6 isolates, and S-TE-NF and CIP-S NF, in 2 isolates each (Supplementary Table 2).
3.3. Antibiotic resistance-related genes detected in Salmonella genomes
After using ResFinder, 21 genes that predict phenotypic resistance were detected. In addition to the presence of the aac(6')-Iaa gene in all 75 isolates, 50 (66.6%) presented the aph(3”)-Ib and aph(6)-Id genes. The aph(3”)-Ia, aadA1, aadA2, and aadA17 genes, which also confer resistance to aminoglycosides, were detected at a low frequency. Only three genes that confer resistance to tetracyclines were identified. The most abundant of these genes was tetA, found in 48 (64%) isolates, followed by tetB and tetM. Sulfonamide resistance genes were observed in 29 (38.6%) isolates, all of which presented the sul2 gene, and two of these isolates also presented the sul1 gene. The only identified gene for beta-lactam resistance was the blaTEM-1B gene, detected in 7 isolates, and the only gene for quinolone resistance was the qnrB19 gene, detected in 5 isolates. The floR and cmlA1, fosA, qac, dfrA, and lnu genes, which confer resistance to phenicols, fosfomycin, ammonium compounds, trimethoprim, and lincosamides, respectively, were detected in 4 or fewer isolates. According to serotype, the gene tetA was present in S. Typhimurium, S. Newport, S. Anatum and S. Agona. Only S. Typhimurium and S. Anatum presented the sul2 gene and the bla-TEM1 gene. Similarly, only S. Typhimurium and S. Dublin carried the qnrB19 gene. Two mutations were detected using the PointFinder database. The parC_T57S chromosomal mutation, conferring resistance to quinolones, was detected in 38 (50.6%) isolates, including all strains of S. Anatum, S. Newport, S. Montevideo, S. Agona and the IIIB 61:I:Z53 isolate. This mutation was not present in the S. Typhimurium and S. Dublin isolates. The other detected mutation, acrB_R717Q, which confers resistance to azithromycin, was detected in one strain of S. Dublin (Supplementary Tables 1, 2).
3.4. Plasmid detection
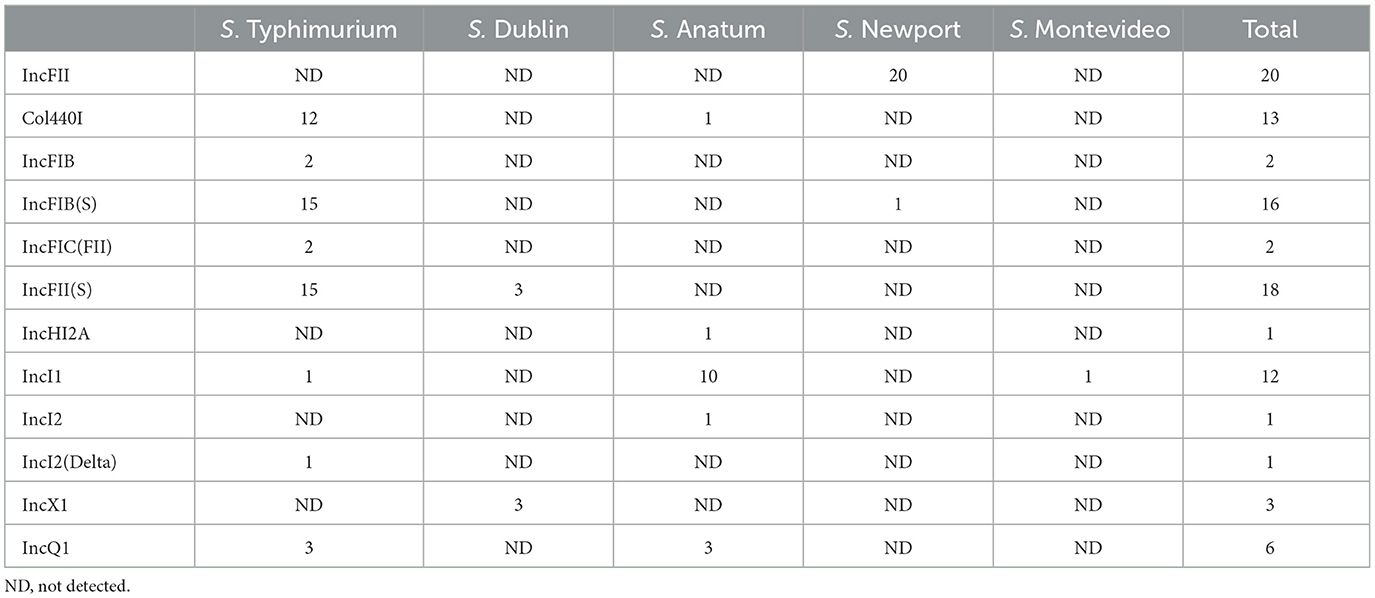
Twelve different incompatibility (Inc) groups were detected in 70 strains. Forty-nine strains showed one Inc group, 17 strains showed two, and 4 strains presented three. Three strains, S. Newport, S. Agona and IIIB 61:I:Z53, did not present any of these groups.
All but five isolates carried at least one Inc group. The most frequent of these groups were IncFII(pHN7A8), IncFII(S), IncFIB(S), Col440I and IncI1. All S. Typhimurium isolates showed Inc groups, and the most frequent combination was Col440/IncFIB(S).
Salmonella Dublin strains presented either IncX1 or IncFII(S). In S. Anatum, the combination of Col440I/IncHI2A/IncQ1 plasmids was present in 1 strain. Two strains presented 2 plasmids in the combinations IncI1/IncI2 and IncI1/IncQ1. Eight strains presented the IncI1 plasmid. A single strain of the S. Newport serotype presented the plasmids IncFII(pHN7A8) and IncFIB(S), and 20 strains presented the plasmid IncFII(pHN7A8). Salmonella serovar Montevideo had a single IncI1 plasmid (Table 1).
3.5. Plasmid localization of resistance genes and class 1 integrons
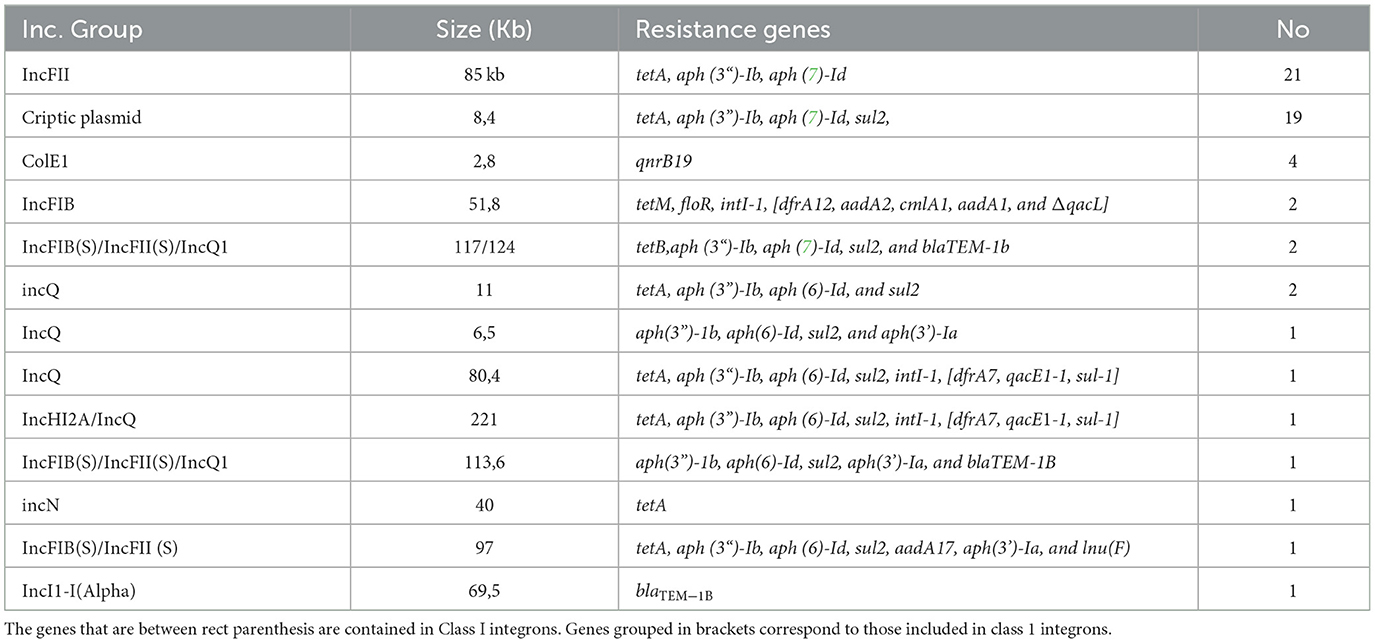
In 52/75 isolates, at least one plasmid incompatibility group and some antibiotic resistance genes were detected in the same contig (see the detailed results in Table 2).
The most frequently detected antibiotic resistance-related incompatibility group was IncFII, which was identified as an 85 kb contig in 21 isolates. This contig encoded the tetA, aph(3“)-Ib and aph(6)-Id genes, which confer resistance to tetracyclines and streptomycin. These 85 kb fragments were 98% identical to two plasmids available in the NCBI databases (accessions CP042245 and CP025329) from veterinary isolates in China.
Second, a small 8.4 kb non-conjugative cryptic plasmid was detected in 19 isolates that encoded sul2, aph(3”)-Ib, aph(6)-Id, and tetA, conferring resistance to sulfas, streptomycin, and tetracycline. This small plasmid without a defined incompatibility group is widely reported in databases such as CP023647 and CP049284.
A third group of plasmids that can be highlighted in the context of the accumulation of resistance genes was IncQ. Although only 8 isolates presented IncQ, these non-conjugative plasmids were highly heterogeneous in terms of the resistance genes involved and their sizes and mode of presentation. Thus, in four isolates, IncQ was found as the only incompatibility group of the contig, while in another four, it was cointegrated with other incompatibility groups, such as IncHI2a, or even with the virulence plasmid of Salmonella spp. IncFIB(S)/IncFII(S). All detected gene arrangements can be seen in Figure 1.
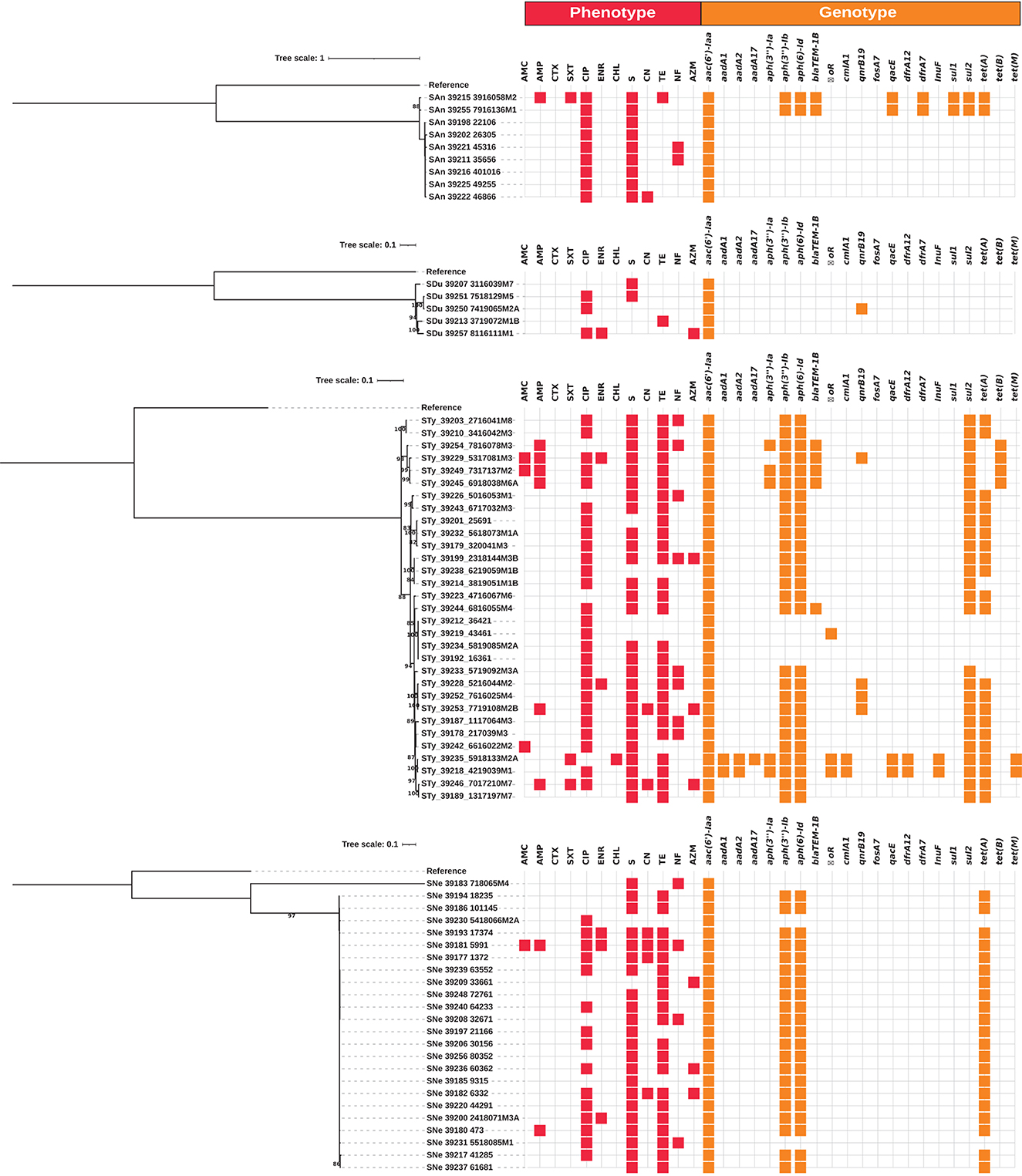
Figure 1. Whole-genome maximum likelihood phylogenetic trees (midpoint root) for the isolates from serovars Anatum (SAn), Dublin (SDu), Typhimurium (Sty) and Newport (SNe) in this study. Phenotypic antimicrobial resistance is indicated with red squares. Antibiotics: AMC, amoxicillin-clavulanic acid; AMP, ampicillin; CTX, cefotaxime; SXT, trimethoprim-sulfamethoxazole; CIP, ciprofloxacin; ENR, enrofloxacin; CHL, chloramphenicol; S, streptomycin; CN, gentamicin; TE, tetracycline; NF, nitrofurantoin; AZM, azithromycin. The presence of antimicrobial resistance genes is indicated with orange squares.
Among the 75 isolates studied, only four carried class 1 integrons. Two highly related isolates of S. Typhimurium (39,218 and 39,235) presented the same integron on an IncFIB plasmid, with the arrangement intI-1, dfrA12, aadA2, cmlA1, aadA1, and qacL. Interestingly, isolate 39,235 but not 39,218 presented a second class 1 integron in the IncFIB(S)/IncFII(S) virulence plasmid encoding the aadA17 and lnu(F) genes.
On the other hand, two isolates of S. Anatum (39,215 and 39,255) carried the arrangement intI-1, dfrA7, qacEΔ-1, sul1 in a contig associated with the cointegrated plasmid IncHI2A/IncQ1 or IncQ1, respectively.
3.6. Correlation between phenotypic and genotypic resistance
We recorded 900 phenotypic antibiotic susceptibility results and 1,575 genotypic data from CGE on 75 S. enterica isolates. The phenotype matched the genotype in 709 results. A total of 191 differences between phenotype and genotype were observed, 65 of which were due to the presence of phenotypic non-susceptibility with the absence of analyzed resistance genes/mutations, and 126 were due to phenotypic sensitivity but with the presence of resistance genes/mutations, with a general accuracy between the two tests of 83.6% (range 48–100%).
The highest correlation between phenotypic and genotypic resistance was found for gentamicin (100%), chloramphenicol (97.3%), trimethoprim-sulfamethoxazole (96%), macrolides (92%), tetracyclines (85.3%) and streptomycin for aminoglycosides (85.3%). The greatest number of discrepancies between the susceptibility profile and genes conferring resistance were observed in the of quinolone group (35 isolates), followed by the nitrofurantoin group (15 isolates). Among these three groups of antibiotics, the observed differences were due to the presence of non-susceptibility and the absence of genes and mutations in 40 isolates and 36 isolates with phenotypic susceptibility but also genes or mutations. According to the reference method, the overall sensitivity was 85.4% (14.3–100%), the overall specificity was 82.7% (38.8–100%), the PPV was 71.1% (11.6–100%), and the NVP was 90% (2–100%) (Table 3).
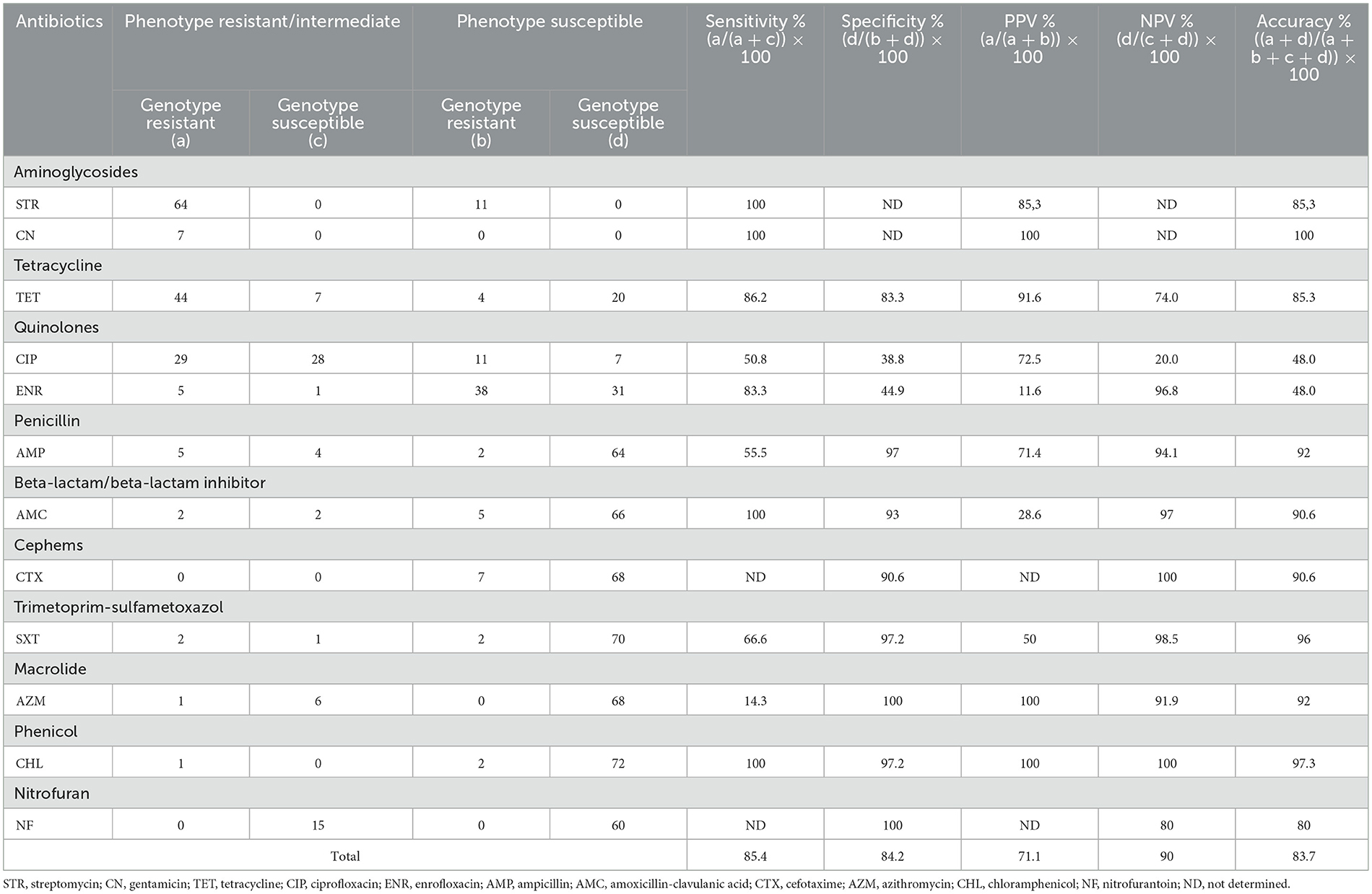
Table 3. Sensitivity and specificity of genotype predictions of resistant antimicrobial phenotypes for 75 Salmonella isolates.
3.7. Phylogenetic analysis
Whole-genome phylogenetic trees were produced for strains of the serovars SAn, SDu, STy and SNe, as they were the most frequent serovars present on more than one farm and/or included more isolates for comparison. Independent phylogenetic trees were produced for each serovar, as this approach could resolve in more detail the intraspecific differences among isolates, and the results are shown in Figure 1.
In broad terms, while the observed genotypes were roughly the same among a given serotype, greater phenotypic diversity was observed for AMR. Strains from the same farm were clustered together in their respective trees.
The STy strains were the most genetically diverse group, in accordance with the broader range of sources and AMR observed for this serovar, as described above, whereas the SAn isolates were less diverse.
4. Discussion
Salmonella enterica is a pathogen present on dairy farms in Uruguay. In this study, 8 serotypes were identified, and each showed a single sequence type, suggesting the presence of a single lineage of strains.
As observed in Canada (21), S. Typhimurium ST19 was the most frequent sequence type isolated from calves with diarrhea, being identified 26 different times on 16 different dairy farms. This contrasts with findings in the USA, where the most frequent serotype is S. Dublin (49). ST19 has been identified in other countries and is associated with gastrointestinal disease due to showing a greater immune response to the flagellar antigens of these strains (50, 51). On one dairy farm, we identified 24 S. Newport ST45 isolates, most of which were susceptible to antibiotics. This ST affects both humans and animals, including cattle, and has been reported to show an MDR phenotype (52–54). In agreement with other reports, S. Dublin ST10 was isolated from 5 different farms, and S. Anatum ST64 was isolated from 2 farms (55–60).
Salmonella Agona is a pathogen frequently detected in food worldwide (61). In this work, the representative ST13 was isolated from a mesenteric lymph node. The isolate of S. Montevideo belonged to ST138, and this microorganism has also been reported as a foodborne pathogen (62). This sequence type of bovine origin is implicated in foodborne illness and comes from a different clade from those isolated from human outbreaks. Isolate IIIB 61:I:Z53 belongs to sequence type ST430, which has only been reported in association with human infections in the US due to contact with small turtles and their environment (63, 64). Here, it was isolated from the feces of a calf suffering from an outbreak of neonatal diarrhea and calf mortality, so its implications for farm animals should be evaluated.
We observed differences in the susceptibility profiles between the different serotypes. Salmonella Typhimurium was the serotype with the greatest differences in susceptibility. This could be explained by the diversity of geographic locations represented by the isolates since it was the most numerous serotype. In this study, the most frequent non-susceptibility pattern was found for aminoglycosides, tetracyclines and quinolones, identified in 11 (34%) isolates, 81% of which were MDR. A similar characteristic was observed in Canada by Otto et al. (21). Great differences are found among the reports of resistance in S. Dublin (21), and in our work, the isolates presented susceptibility to all antibiotics except aminoglycosides. The most frequent non-susceptibility pattern observed in S. Newport isolates, obtained from a single farm, was similar to that of S. Typhimurium, and this serovar is usually reported as MDR (65, 66), unlike S. Anatum, which was not susceptible to only aminoglycosides and quinolones.
According to the evaluation of the obtained genomes, we observed that the most frequent resistance genes in all serotypes were those that confer resistance to aminoglycosides, tetracyclines and sulfonamides. Resistance to aminoglycosides mediated by the aac (6')-Iaa and aacC (6')-Iy genes is not considered by relevant platforms because studies indicate that they do not confer resistance and are ubiquitous in Salmonella (67). However, the aac(6')-Iaa gene was detected in 100% of the strains in our study. The aac(6')-Iaa gene has been previously detected in human Salmonella isolates from Uruguay (68). It confers resistance to tobramycin, amikacin and kanamycin, but this gene is less efficient in the acetylation of gentamicin, which may explain the low rate of detection of phenotypic resistance in our results (69). The aph(6)-Id, aph(3)-Ib and aph(3)-Ia genes were detected less frequently. These last three genes were recently reported in S. Dublin isolated from bovines in the USA (59).
The most frequent tetracycline resistance gene identified was the tetA gene, which was present in those strains that did not present the tetB gene; both of these genes expulsion pumps in gram-negative bacteria (70). In previous reports, it was observed that the tetA gene presented a greater dissemination capacity than the tetB gene on farms (71). This finding could be explained by the fact that these genes occur in mobile genetic elements that are frequently conjugative (72).
The sul2 gene was detected at the highest frequency, followed by sul1, both of which confer resistance to sulfonamides. This contrasts with findings in other animal sources, where sul1 is more prevalent (72, 73). This result was not confirmed phenotypically because we assayed trimethoprim and sulfamethoxazole antibiotics together. According to this combination, only 4 isolates carried sul1/sul2 and dfr genes that confer resistance to trimethoprim-sulfamethoxazole.
Plasmid-mediated quinolone resistance (PMQR) generates a low level of resistance to quinolones but promotes the selection of other resistance mechanisms, and their rapid spread and has been reported in different bacterial species of animal origin (74–77). In this study, the qnrB19 gene was detected in only 4 S. Typhimurium and 1 S. Dublin isolates. Similar isolates harboring this gene have been observed in South America (78–80). These results contrast with the published data from the region, where the most frequent variants are indicated to be qnrB2, qnrS1, and qnrE1 (80).
In this work, we only detected a mutation in parC at position 57, causing a Thr to Ser change in the topoisomerase that confers resistance to quinolones (80). Enrofloxacin is a representative antibiotic used in the veterinary industry, and due to its wide range of activity, it is frequently used for treating various clinical presentation (81, 82). The parCT57S mutation has been reported as the most frequent mutation in environmental samples and animal feces isolates in Brazil (80), and a single mutation in a topoisomerase IV or DNA gyrase gene confers high-level resistance to this group of drugs (75). In Uruguay, the parCT57S mutation and the presence of qnrB19 were detected in human S. Typhimurium isolates between 2011 and 2013 (68). Taken together, our findings suggest that these genes and mutations have existed for a long time.
In the USA, the detection of the blaTEM gene is very frequent (59, 83, 84); in contrast, only 7 strains carried this gene in this work, corresponding to <10% of the collection. Following the results obtained from Resfinder, the blaTEM-1 enzyme was only identified in 5 strains of S. Typhimurium and 2 strains of S. Anatum. Among these 7 isolates, only five showed reduced sensitivity to ampicillin and two to amoxicillin-clavulanic acid. Most of these blaTEM-1-carrying strains were isolated from neonatal calves.
We detected the aadA1 and aadA2 genes in two MDR S. Typhimurium isolates, and one of them even carried the aadA17 variant, which is frequently reported in England (85) but has not previously been reported in Salmonella isolates in South America. This class of genes is located within integrons (86), together with genes such as sul, qacEΔ1, drf, and cmlA that confer resistance to sulfonamides, antiseptics and disinfectants, trimethoprim and phenicol, respectively (73, 87–89). In our work, aadA17 was also found within a class 1 integron but was only accompanied by the lnu(F) gene encoded by the virulence plasmid of Salmonella spp. Inc FIB(S)/IncFII(S).
On the other hand, two isolates of S. Anatum with similar characteristics were detected on the same farm but came from two calves sampled months apart. These strains presented blaTEM, sul2, tetA, dfrA7, qacΔ1, and sul1, among which the last three genes are part of a class 1 integron.
The fosA gene is widely distributed among Enterobacteriaceae (90), and the fosA7 variant has been detected in plasmids in S. enterica from birds and in pigs and cattle in China (91, 92), conferring a high level of resistance to fosfomycin. In our study, this gene was detected in S. Agona and S. Montevideo isolated from mesenteric lymph nodes and feces, respectively. This gene was previously detected in Uruguay, but within E. coli STEC strains (93).
Interestingly, a single strain of S. Dublin had the acrB_R717Q mutation, which confers resistance to azithromycin, and was recently identified in Bangladesh in S. Typhi (94). Additionally, this mutation was recently reported in a study involving S. Dublin isolates of bovine origin by García-Soto et al. (95) in Germany.
In general, most plasmids were exclusive to a certain serotype. The plasmids shared by different serotypes were Col440I in S. Typhimurium and S. Anatum; IncFIB(S) in S. Typhimurium and S. Newport; IncI1 in S. Typhimurium, S. Anatum and Montevideo; and IncQ1 in S. Typhimurium and S. Anatum. This agrees with findings reported by Feng et al. (96), who indicated that serotypes generally do not share the same plasmids due to their mostly vertical inheritance. Previous studies reported that the most frequent plasmids detected in S. Dublin were IncX1, IncA/C2, and IncFII (59, 97). Although the number of S. Dublin strains evaluated in our study was very limited, the detected plasmids were similar, except for IncA/C2. While S. Typhimurium was the sole serovar harboring the plasmids IncA/C, IncI, Col, IncFII, IncFIB, and IncFIA (similar to the plasmids detected in our strains), there was a difference in the Newport serotype (in which the only detected plasmids shared with this work were the IncFII and IncFIB) and in Anatum and Agona (84). It is important to deepen the study of the plasmids present in the strains since they are used for epidemiological studies and the interpretation of the virulence and resistance observed in different regions.
Since 1960, the most commonly reported MDR phenotype in Salmonella has been resistance to ampicillin, chloramphenicol, streptomycin, sulfonamides, and tetracyclines (98). Most of the isolates were MDR, and 35.7% of these isolates showed the CIP-S-TE profile. Genotypically, we also observed MDR in 40% of the isolates, which carried resistance genes from 3 to 7 groups of antibiotics. Most of these MDR isolates were S. Anatum and S. Typhimurium.
Different publications have described notable correlations between resistance phenotypes and genotypes (83, 99). We detected a 100% correlation for gentamicin; above 90% for chloramphenicol, beta-lactams, trimethoprim-sulfamethoxazole and macrolides; and above 80% for tetracyclines, nitrofurans and streptomycin. Similar results were obtained by Campioni et al. (100) in Salmonella from different sources in Brazil.
In our work, although the estimated values of sensitivity, specificity, PPV and NPV were generally high, this was not observed in some specific drug groups (i.e., ciprofloxacin). With the strategy used in this work, we could not identify the presence of pseudogenes in the detected AMR genes that might account for part of this discrepancy. Other factors may be involved, such as low levels of expression or mutations in promoters or in Shine Dalgarno sequences. On the other hand, under the adopted strategy, we did not analyze the presence of efflux pumps since their effect depends on the level of expression. This aspect could explain certain levels of resistance that are not explained by our results.
Based on the above, phenotypic evaluation is still the most relevant approach when therapeutic decisions are being made.
This work represents one of the first studies in the region using the phenotypic resistance and WGS of S. enterica obtained from bovines as an analysis strategy. According to our results, S. Typhimurium was the serotype with the highest frequency and distribution, followed by S. Dublin, and all the serotypes detected were from a single sequence type. These serotypes were zoonotic, and we also found a high frequency of resistance to antibiotics used on farms and in human health. Taking these results into account, it is necessary to consider the implementation of strategies to limit the use of antibiotics that are considered critical and to mitigate the increase in resistance globally.
Data availability statement
The data presented in the study are deposited in the EnteroBase (https://enterobase.warwick.ac.uk) repository, accession numbers SAL_HB5975AA, SAL_HB5235AA, SAL_HB5236AA, SAL_HB5237AA, SAL_HB5238AA, SAL_HB5239AA, SAL_HB4194AA, SAL_HB5240AA, SAL_HB5241AA, SAL_HB5242AA, SAL_HB5243AA, SAL_HB5299AA, SAL_HB5300AA, SAL_HB5301AA, SAL_HB5302AA, SAL_IB4224AA, SAL_HB5471AA, SAL_HB5474AA, SAL_HB5476AA, SAL_HB5740AA, SAL_IB4223AA, SAL_HB5741AA, SAL_HB5742AA, SAL_HB5743AA, SAL_HB5744AA, SAL_HB5745AA, SAL_HB5746AA, SAL_HB5747AA, SAL_HB5748AA, SAL_HB5749AA, SAL_IB4193AA, SAL_HB5976AA, SAL_HB5977AA, SAL_HB5978AA, SAL_HB5979AA, SAL_HB5980AA, SAL_HB5981AA, SAL_HB5982AA, SAL_HB5983AA, SAL_HB5984AA, SAL_HB5985AA, SAL_HB5986AA, SAL_HB5987AA, SAL_HB5988AA, SAL_IB4194AA, SAL_HB5989AA, SAL_HB5990AA, SAL_HB5992AA, SAL_HB5993AA, SAL_HB5994AA, SAL_HB5995AA, SAL_HB5996AA, SAL_HB5997AA, SAL_HB6171AA, SAL_HB6172AA, SAL_HB6173AA, SAL_HB6179AA, SAL_HB6188AA, SAL_HB6194AA, SAL_HB6195AA, SAL_HB6201AA, SAL_HB6207AA, SAL_HB6208AA, SAL_HB6213AA, SAL_HB6217AA, SAL_HB6232AA, SAL_HB6235AA, SAL_HB6236AA, SAL_HB6238AA, SAL_HB6239AA, SAL_HB6240AA, SAL_HB6242AA, SAL_HB6244AA, SAL_HB6261AA, SAL_HB5975AA. Data has been available in the database since April 2021.
Author contributions
MC and MF conceptualized the study. MC performed the laboratory analysis and wrote the original draft. MC, BD'A, and RV performed data selection and data analysis. MC, BD'A, RV, and MF contributed to writing, proofreading, and editing. All authors contributed to the article and approved the submitted version.
Funding
This work was financed by the Instituto Nacional de Investigación Agropecuaria (INIA) through the PL-27 project and a postgraduate scholarship to MC. MC also received a grant to finish postgraduate studies from Comisión Asesora de Posgrado (CAP), Universidad dela República, Uruguay.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2023.1055432/full#supplementary-material
References
1. Coburn B, Grassl GA, Finlay BB. Salmonella, the host and disease: a brief review. Immunol Cell Biol. (2006) 85:112–8. doi: 10.1038/sj.icb.7100007
2. Kurtz JR, Goggins JA, McLachlan JB. Salmonella infection: Interplay between the bacteria and host immune system. Immunol Lett. (2017) 190:42–50. doi: 10.1016/j.imlet.2017.07.006
3. Bhan M, Bahl R, Bhatnagar S. Typhoid and paratyphoid fever. Lancet. (2005) 366:749–62. doi: 10.1016/S0140-6736(05)67181-4
4. Stanaway JD, Parisi A, Sarkar K, Blacker BF, Reiner RC, Hay SI, et al. The global burden of non-typhoidal Salmonella invasive disease: a systematic analysis for the Global Burden of Disease Study 2017. Lancet Infect Dis. (2019) 19:1312–24. doi: 10.1016/S1473-3099(19)30418-9
5. Carroll LM, Buehler AJ, Gaballa A, Siler JD, Cummings KJ, Cheng RA, et al. Monitoring the microevolution of S. enterica in healthy dairy cattle populations at the individual farm level using whole-genome sequencing. Front Microbiol. (2021) 12:763669. doi: 10.3389/fmicb.2021.763669
6. Centers for Disease Control Prevention (CDC). Salmonella infection from animals. Atlanta: Centers for Disease Control and Prevention (2022). Available online at: https://www.cdc.gov/salmonella/index (accessed April 21, 2022).
7. Wiedemann A, Virlogeux-Payant I, Chaussé AM, Schikora A, Velge P. Interactions of Salmonella with animals and plants. Front Microbiol. (2015) 21:791. doi: 10.3389/fmicb.2014.00791
8. Vohra P, Vrettou C, Hope JC, Hopkins J, Stevens MP. Nature and consequences of interactions between S. enterica serovar Dublin and host cells in cattle. Vet Res. (2019) 50:720. doi: 10.1186/s13567-019-0720-5
9. Mohler VL, Izzo MM, House JK. Salmonella in calves. Vet Clin North Am Food Anim Pract. (2009) 25:37–54. doi: 10.1016/j.cvfa.2008.10.009
10. Costa LF, Paixão TA, Tsolis RM, Bäumler AJ, Santos RL. Salmonellosis in cattle: advantages of being an experimental model. Vet Sci Res J. (2012) 93:1–6. doi: 10.1016/j.rvsc.2012.03.002
11. Casaux ML, Neto WS, Schild CO, Costa RA, Macías-Rioseco M, Caffarena RD, et al. Epidemiological and clinicopathological findings in 15 fatal outbreaks of salmonellosis in dairy calves and virulence genes in the causative S. enterica Typhimurium and Dublin strains. Braz J Microbiol. (2023) 48:1–16. doi: 10.1007/s42770-022-00898-9
12. Ventola CL. The antibiotic resistance crisis: part 1: causes and threats. P&T Peer-Rev J Form Manag. (2015) 40:277–83.
13. Medalla F, Gu W, Mahon BE, Judd M, Folster J, Griffin PM, et al. Estimated incidence of antimicrobial drug-resistant non-typhoidal Salmonella infections, United States, 2004–2012. Emerg Infect Dis. (2016) 23:29–37. doi: 10.3201/eid2301.160771
14. Constable PD, Hinchcliff KW, Done SH, Grünberg W. Alimentary tract diseases: non-ruminant. In: Constable PD, Hinchcliff KW, Done SH, Grünberg W, editors. Medicina Veterinaria. 11th ed. Filadelfia: WB Saunders (2017). p. 175–176.
15. Foley SL, Lynne AM. Food animal-associated Salmonella challenges: pathogenicity and antimicrobial resistance. J Anim Sci. (2008) 14:E173–87. doi: 10.2527/jas.2007-0447
16. Hur J, Jawale C, John Hwa Lee JH. Antimicrobial resistance of Salmonella isolated from food animals: a review. Food Res Int. (2012) 45:819–30. doi: 10.1016/j.foodres.2011.05.014
17. Shintani M, Sanchez ZK, Kimbara K. Genomics of microbial plasmids: classification and identification based on replication and transfer systems and host taxonomy. Front Microbiol. (2015) 31:242. doi: 10.3389/fmicb.2015.00242
18. Rychlik I, Gregorova D, Hradecka H. Distribution and function of plasmids in S. enterica Vet Microbiol. (2006) 112:1–10. doi: 10.1016/j.vetmic.2005.10.030
19. Carattoli A. Plasmid-mediated antimicrobial resistance in S. enterica. Curr Issues Mol Biol. (2003) 5:113–22. doi: 10.21775/cimb.005.113
20. Davidson KE, Byrne BA, Pires A, Magdesian KG, Pereira RV. Antimicrobial resistance trends in fecal Salmonella isolates from northern California dairy cattle admitted to a veterinary teaching hospital, 2002–2016. PLoS ONE. (2018) 13:928. doi: 10.1371/journal.pone.0199928
21. Otto SJG, Ponich KL, Cassis R, Goertz C, Peters D, Checkley SL, et al. Antimicrobial resistance of bovine S. enterica ssp. Enterica isolates from the Alberta agriculture and forestry disease investigation program (2006–2014). Can Vet J. (2018) 59:1195–201.
22. Vasconcelos AB, Andrade V, Moraes AC, Almeida EM, Silva D, França AC, et al. Occurrence and antimicrobial resistance profile of Salmonella spp. in calves from the Mesoregion Sertão of Alagoas. Acta Vet Brasil. (2020) 15:9363. doi: 10.21708/avb.2021.15.1.9363
23. Souto MS, Coura FM, Dorneles EM, Stynen AP, Alves TM, Santana JA, et al. Antimicrobial susceptibility and phylotyping profile of pathogenic Escherichia coli and S. enterica isolates from calves and pigs in Minas Gerais, Brazil. Trop Anim Health Prod. (2017) 49:13–23. doi: 10.1007/s11250-016-1152-0
24. Casaux ML, Caffarena RD, Schild CO, Giannitti F, Riet-Correa F, Fraga M, et al. Antibiotic resistance in S. enterica isolated from dairy calves in Uruguay. Brazil J Microbiol. (2019) 50:1139–44. doi: 10.1007/s42770-019-00151-w
25. van El C, Cornel M, Borry P, Hastings RJ, Fellmann F, Hodgson SV, et al. Whole-genome sequencing in health care. Eur J Hum Genet. (2013) 21:580. doi: 10.1038/ejhg.2013.46
26. Qin D. Next-generation sequencing and its clinical application. Cancer Biol Med. (2019) 16:4–10. doi: 10.20892/j.issn.2095-3941.2018.0055
27. Ellington MJ, Ekelund O, Aarestrup FM, Canton R, Doumith M, Giske M, et al. The role of whole genome sequencing in antimicrobial susceptibility testing of bacteria: report from the EUCAST Subcommittee. Clin Microbiol Infect. (2017) 23:2–22. doi: 10.1016/j.cmi.2016.11.012
28. Tyson GH, McDermott PF, Li C, Chen Y, Tadesse DA, Mukherjee S, et al. WGS accurately predicts antimicrobial resistance in E. coli. J Antimicrob Chemother. (2015) 70:2763–9. doi: 10.1093/jac/dkv186
29. Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing; 25th Informational Supplement, M100-S28. Wayne, PA: Clinical and Laboratory Standards Institute (2018).
30. Clinical and Laboratory Standards Institute. Performances standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals; Second Informational Supplement. In: CLSI Document VET01-S2, CLSI. Wayne, PA: Clinical and Laboratory Standards Institute (2013).
31. Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing. 31th ed. Wayne, PA: Clinical and Laboratory Standards Institute (2021).
32. Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. (2012) 18:268–81. doi: 10.1111/j.1469-0691.2011.03570.x
33. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illuminasequence data. Bioinformatics. (2014) 30:2114–20. doi: 10.1093/bioinformatics/btu170
34. Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. (2012) 19:455–77. doi: 10.1089/cmb.2012.0021
35. Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. (2014) 30:2068–9. doi: 10.1093/bioinformatics/btu153
36. Yoshida CE, Kruczkiewicz P, Laing CR, Lingohr EJ, Gannon VPJ, Nash JHE, et al. The Salmonella In Silico typing resource (SISTR): an open web-accessible tool for rapidly typing and subtyping draft Salmonella genome assemblies. PLoS ONE. (2016) 11:e0147101. doi: 10.1371/journal.pone.0147101
37. Zhang S, den Bakker HC, Li S, Chen J, Dinsmore BA, Lane C, et al. SeqSero2: rapid and improved Salmonella serotype determination using whole-genome sequencing data. Appl Environ Microbiol. (2019) 85:e01746–e01719. doi: 10.1128/AEM.01746-19
38. Robertson J, Yoshida C, Kruczkiewicz P, Nadon C, Nichani A, Taboada EN, et al. Comprehensive assessment of the quality of Salmonella whole genome sequence data available in public sequence databases using the Salmonella in silico Typing Resource (SISTR). Microb Genom. (2018) 4:e000151. doi: 10.1099/mgen.0.000151
39. Achtman M, Wain J, Weill FX, Nair S, Zhou Z, Sangal V, et al. Multilocus sequence typing as a replacement for serotyping in S. enterica. PLoS Pathog. (2012) 8:e10010. doi: 10.1371/journal.ppat.1002776
40. Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, et al. BLAST+: architecture and applications. BMC Bioinform. (2009) 10:421. doi: 10.1186/1471-2105-10-421
41. Bortolaia V, Kaas RF, Ruppe E, Roberts MC, Schwarz S, Cattoir V, et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J Antimicrob Chemother. (2020) 75:3491–500. doi: 10.1093/jac/dkaa345
42. Zankari E, Allesøe R, Joensen KG, Cavaco LM, Lund O, Aarestrup FM, et al. PointFinder: a novel web tool for WGS-based detection of antimicrobial resistance associated with chromosomal point mutations in bacterial pathogens. J Antimicrob Chemother. (2020) 72:2764–8. doi: 10.1093/jac/dkx217
43. Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O, Villa L, et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. (2014) 58:3895–903. doi: 10.1128/AAC.02412-14
44. Loaiza K. IntFinder Development Validation. (2021). Available online at: https://github.com/kalilamali/Integrons (accessed December, 2021).
45. Trevethan R. Sensitivity, specificity, and predictive values: foundations, pliabilities, and pitfalls in research and practice. Public Health Front. (2017) 5:307. doi: 10.3389/fpubh.2017.00307
46. Croucher NJ, Page AJ, Connor TR, Delaney AJ, Keane JA, Bentley SD, et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucl Acids Res. (2015) 18:e15. doi: 10.1093/nar/gku1196
47. Kozlov AM, Darriba D, Flouri T, Morel B, Stamatakis A. RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics. (2019) 35:4453–5. doi: 10.1093/bioinformatics/btz305
48. Letunic I, Bork P. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucl Acids Res. (2021) 49:W293–6. doi: 10.1093/nar/gkab301
49. Valenzuela JR, Sethi AK, Aulik NA, Poulsen KP. Antimicrobial resistance patterns of bovine S. enterica isolates submitted to the Wisconsin Veterinary Diagnostic Laboratory: 2006–2015. J Dairy Sci 100. (2017) 1319–30. doi: 10.3168/jds.2016-11419
50. Okoro CK, Kingsley RA, Connor TR, Harris SR, Parry CM, Al-Mashhadani S, et al. Intracontinental spread of human invasive Salmonella Typhimurium pathovariants in sub-Saharan Africa. Nat Genet. (2012) 44:1215–21. doi: 10.1038/ng.2423
51. Carden S, Okoro C, Dougan G, Monack D. Non-typhoidal Salmonella Typhimurium ST313 isolates that cause bacteremia in humans stimulate less inflammasome activation than ST19 isolates associated with gastroenteritis. Pathogens Dis. (2015) 73:ftu023. doi: 10.1093/femspd/ftu023
52. Alcaine SD, Soyer Y, Warnick LD, Su WL, Sukhnanand S, Richards J, et al. Multilocus sequence typing supports the hypothesis that cow- and human-associated Salmonella isolates represent distinct and overlapping populations. Appl Environ Microbiol. (2006) 72:7575–85. doi: 10.1128/AEM.01174-06
53. Sangal V, Harbottle H, Mazzoni CJ, Helmuth R, Guerra B, Didelot X, et al. Evolution and population structure of S. enterica serovar Newport. J Bacteriol. (2010) 192:6465–76. doi: 10.1128/JB.00969-10
54. Pan H, Zhou X, Chai W, Paudyal N, Li S, Zhou X, et al. Diversified sources for human infections by S. enterica serovar Newport Transbound. Emerg Dis. (2019) 66:1044–8. doi: 10.1111/tbed.13099
55. Ferrari RG, Rosario D, Cunha-Neto A, Mano SB, Figueiredo E, Conte-Junior CA. Worldwide epidemiology of Salmonella serovars in animal-based foods: a meta-analysis. Appl Environ Microbiol. (2019) 85:e00591–9. doi: 10.1128/AEM.00591-19
56. Paudyal N, Pan H, Elbediwi M, Zhou X, Peng X, Li X, et al. Characterization of Salmonella Dublin isolated from bovine and human hosts. BMC Microbiol. (2019) 19:226. doi: 10.1186/s12866-019-1598-0
57. Mangat CS, Bekal S, Avery BP, Côté G, Daignault D, Doualla-Bell F, et al. Canadian integrated program for antimicrobial resistance surveillance public health partnership. Genomic investigation of the emergence of invasive multidrug-resistant S. enterica Serovar Dublin in Humans and Animals in Canada. Antimicrob Agents Chemother. (2019) 63:e00108–19. doi: 10.1128/AAC.00108-19
58. Vilela FP, Dos Prazeres Rodrigues D, Costa RG, Casas M, Falcão JP, Campioni F. High similarity and high frequency of virulence genes among Salmonella Dublin strains isolated over a 33-year period in Brazil. Braz J Microbiol. (2020) 51:497–509. doi: 10.1007/s42770-019-00156-5
59. Srednik ME, Lantz K, Hicks JA, Morningstar-Shaw BR, Mackie TA, Schlater LK, et al. Antimicrobial resistance and genomic characterization of Salmonella Dublin isolates in cattle from the United States. PLoS ONE. (2021) 16:e0249617. doi: 10.1371/journal.pone.0249617
60. Levent G, Schlochtermeier A, Ives SE, Norman KN, Lawhon SD, Loneragan GH, et al. High-resolution genomic comparisons within S. enterica serotypes derived from beef feedlot cattle: parsing the roles of cattle source, pen, animal, sample type, and production period. Appl Environ Microbiol. (2021) 87:e00485–e00421. doi: 10.1128/AEM.00485-21
61. Dangel A, Berger A, Messelhäußer U, Konrad R, Hörmansdorfer S, Ackermann N, et al. Genetic diversity and delineation of Salmonella Agona outbreak strains by next generation sequencing, Bavaria, Germany, 1993 to 2018. EuroSurveillance. (2019) 24:1800303. doi: 10.2807/1560-7917.ES.2019.24.18.1800303
62. Bakker HC, Switt AI, Cummings CA, Hoelzer K, Degoricija L, Rodriguez-Rivera LD, et al. Whole-genome single nucleotide polymorphism-based approach to trace and identify outbreaks linked to a common S. enterica subsp enterica serovar Montevideo pulsed-field gel electrophoresis type. Appl Environ Microbiol. (2011) 77:8648–55. doi: 10.1128/AEM.06538-11
63. Lamas A, Miranda JM, Regal P, Vázquez B, Franco CM, Cepeda A. A comprehensive review of non-enterica subspecies of S. enterica. Microbiol Res. (2018) 206:60–73. doi: 10.1016/j.micres.2017.09.010
64. Centers for Disease Control Prevention (CDC). Cuatro brotes multiestatales de infecciones por Salmonella en seres humanos vinculadas a tortugas pequeñas. Atlanta: Centers for Disease Control and Prevention (2022). Available online at: https://www.cdc.gov/salmonella/small-turtles-10-15/index-esp.html (accessed January 17, 2022).
65. Cobbold RN, Rice DH, Davis MA, Besser TE, Hancock DD. Long-term persistence of multi-drug-resistant S. enterica serovar Newport in two dairy herds. J Am Vet Med Assoc. (2006) 228:585–91. doi: 10.2460/javma.228.4.585
66. Lopes VC, Wedel SD, Bender JB, Smith KE, Leano FT, Boxrud DJ, et al. Emergence of multidrug-resistant S. enterica serotype newport in Minnesota. Clin Infect Dis. (2006) 43:210–3. doi: 10.1086/505119
67. Feldgarden M, Brover V, Gonzalez-Escalona N, Frye JG, Haendiges J, Haft DH, et al. AMR FinderPlus and the reference gene catalog facilitate examination of the genomic links among antimicrobial resistance, stress response, and virulence. Sci Rep. (2021) 11:12728. doi: 10.1038/s41598-021-91456-0
68. Cordeiro NF, D'Alessandro B, Iriarte A, Pickard D, Yim L, Chabalgoity JA, et al. Draft genome sequences of two multidrug-resistant S. enterica serovar typhimurium clinical isolates from Uruguay. Microbiol Resour Announc. (2018) 7:e00917–8. doi: 10.1128/MRA.00917-18
69. Salipante SJ, Hall BG. Determining the limits of the evolutionary potential of an antibiotic resistance gene. Mol Biol Eur. (2003) 20:653–9. doi: 10.1093/molbev/msg074
70. Olowe OA, Idris OJ, Taiwo SS. Prevalence of tet genes mediating tetracycline resistance in E. coli clinical isolates in Osun State, Nigeria. Eur J Microbiol Immunol. (2013) 3:135–40. doi: 10.1556/EuJMI.3.2013.2.7
71. Skocková A, Cupáková Š, Karpíšková R, Janštová B. Detection of tetracycline resistance genes in E. coli from raw cow's milk. J Microbiol Biotechnol Food Sci. (2012) 1:777–84.
72. Pavelquesi S, Oliveira F, Rodrigues A, de Souza Silva A, Orsi CMDC, da Silva I. Presence of tetracycline and sulfonamide resistance genes in Salmonella spp.: literature review. Antibiotics. (2021) 10:1314. doi: 10.3390/antibiotics10111314
73. Antunes P, Machado J, Sousa JC, Peixe L. Dissemination of sulfonamide resistance genes (sul1, sul2, and sul3) in Portuguese S. enterica strains and relation with integrons. Antimicrobial Agents Chemother. (2005) 49:836–9. doi: 10.1128/AAC.49.2.836-839.2005
74. Drlica K. The mutant selection window and antimicrobial resistance. J Antimicrob Chemother. (2003) 52:11–17. doi: 10.1093/jac/dkg269
75. Correia S, Poeta P, Hébraud M, Capelo JL, Igrejas G. Mechanisms of quinolone action and resistance: where do we stand? J Med Microbiol. (2017) 66:551–9. doi: 10.1099/jmm.0.000475
76. Li P, Liu D, Zhang X, Tuo H, Lei C, Xie X, et al. Characterization of plasmid-mediated quinolone resistance in gram-negative bacterial strains from animals and humans in China. Microb Drug Resist. (2019) 25:1050–6. doi: 10.1089/mdr.2018.0405
77. Ayobola ED, Oscar WO, Ejovwokoghene EF. Plasmid-mediated quinolone resistance genes transfer among enteric bacteria isolated from human and animal sources. AIMS Microbiol. (2021) 7:200–15. doi: 10.3934/microbiol.2021013
78. Moreno-Switt AI, Pezoa D, Sepúlveda V, González I, Rivera D, Retamal P, et al. Transduction as a potential dissemination mechanism of a clonal qnrB19-carrying plasmid isolated from Salmonella of multiple serotypes and isolation sources. Front Microbiol. (2019) 10:2503. doi: 10.3389/fmicb.2019.02503
79. Coppola N, Freire B, Umpiérrez A, Cordeiro NF, Ávila P, Trenchi G, et al. Transferable resistance to highest priority critically important antibiotics for human health in E. coli strains obtained from livestock feces in Uruguay. Front Vet Sci. (2020) 7:588919. doi: 10.3389/fvets.2020.588919
80. Rodrigues GL, Panzenhagen P, Ferrari RG, Dos Santos A, Paschoalin V, Conte-Junior CA, et al. Frequency of antimicrobial resistance genes in Salmonella from Brazil by in silico whole-genome sequencing analysis: an overview of the last four decades. Front Microbiol. (2020) 11:1864. doi: 10.3389/fmicb.2020.01864
81. Otero JL, Mestorino ON, Errecalde JO. Enrofloxacin: a fluorquinolone of exclusive use in veterinary. Part I: chemical characteristics, mechanism of action, antimicrobial activity and bacterial resistance. Analecta Vet. (2001) 21:1.
82. Trouchon T, Lefebvre S. A review of enrofloxacin for veterinary use. Open J Vet Med. (2016) 6:40–58. doi: 10.4236/ojvm.2016.62006
83. McDermott PF, Tyson GH, Kabera C, Chen Y, Li C, Folster JP, et al. Whole-genome sequencing for detecting an timicrobial resistance in non-typhoidal Salmonella. Antimicrob Agents Chemother. (2016) 60:5515–20. doi: 10.1128/AAC.01030-16
84. Zhao S, Li C, Hsu CH, Tyson GH, Strain E, Tyson GH, et al. Comparative genomic analysis of 450 strains of S. enterica isolated from diseased animals. Genes. (2020) 11:1025. doi: 10.3390/genes11091025
85. Neuert S, Nair S, Day MR, Doumith M, Ashton PM, Mellor KC, et al. Prediction of phenotypic antimicrobial resistance profiles from whole genome sequences of non-typhoidal S. enterica. Front Microbiol. (2018) 5:92. doi: 10.3389/fmicb.2018.00592
86. Mazel D. Integrons: agents of bacterial evolution. Nat Rev Microbiol. (2006) 4:608–20. doi: 10.1038/nrmicro1462
87. Paulsen IT, Littlejohn TG, Rådström P, Sundström L, Sköld O, Swedberg G, et al. The 3' conserved segment of integrons contains a gene associated with multidrug resistance to antiseptics and disinfectants. Antimicrob Agents Chemother. (1993) 37:761–8. doi: 10.1128/AAC.37.4.761
88. Schwarz S, Kehrenberg C, Doublet B, Cloeckaert A. Molecular basis of bacterial resistance to chloramphenicol and florfenicol. FEMS Microbiol Rev. (2004) 28:519–42. doi: 10.1016/j.femsre.2004.04.001
89. Deng Y, Bao X, Ji L, Chen L, Liu J, Miao J, et al. Resistance integrons: class 1, 2, and 3 integrons. Ann Clin Microbiol. (2015) 14:45. doi: 10.1186/s12941-015-0100-6
90. Ito R, Mustapha MM, Tomich AD, Callaghan JD, McElheny CL, Mettus RT, et al. Widespread fosfomycin resistance in gram-negative bacteria attributable to the chromosomal fosA gene. MBio. (2017) 8:e00749–e00717. doi: 10.1128/mBio.00749-17
91. Rehman MA, Yin X, Persaud-Lachhman MG, Diarra MS. First detection of a fosfomycin resistance gene, fosA7, in S. enterica serovar heidelberg isolated from broiler chickens. Antimicrob Agents Chemother. (2017) 61:e00410–7. doi: 10.1128/AAC.00410-17
92. Wang J, Wang Y, Wang ZY, Wu H, Mei CY, Shen PC, et al. Chromosomally located fosA7 in Salmonella isolates from China. Front Microbiol. (2021) 12:781306. doi: 10.3389/fmicb.2021.781306
93. Mota MI, Vázquez S, Cornejo C, D'Alessandro B, Braga V, Caetano A, et al. Does Shiga toxin-producing E. coli and listeria monocytogenes contribute significantly to the burden of antimicrobial resistance in Uruguay? Front. Vet Sci. (2020) 7:583930. doi: 10.3389/fvets.2020.583930
94. Hooda Y, Sajib MSI, Rahman H, Luby SP, Bondy-Denomy J, Santosham M, et al. Molecular mechanism of azithromycin resistance among typhoidal Salmonella strains in Bangladesh identified through passive pediatric surveillance. PLoS Negl Trop Dis. (2019) 13:e0007868. doi: 10.1371/journal.pntd.0007868
95. García-Soto S, Tomaso H, Linde J, Methner U. Epidemiological analysis of S. enterica subsp enterica serovar dublin in german cattle herds using whole-genome sequencing. Microbiol Spectr. (2021) 9:e0033221. doi: 10.1128/Spectrum.00332-21
96. Feng Y, Liu J, Li YG, Cao FL, Johnston RN, Zhou J, et al. Inheritance of the Salmonella virulence plasmids: mostly vertical and rarely horizontal. Infect Genet E12. (2012) 1058–63. doi: 10.1016/j.meegid.2012.03.004
97. Hsu CH, Li C, Hoffmann M, McDermott P, Abbott J, et al. Comparative genomic analysis of virulence, antimicrobial resistance, and plasmid profiles of Salmonella Dublin isolated from sick cattle, retail beef, and humans in the United States. Microb Drug Resist. (2019) 25:1238–49. doi: 10.1089/mdr.2019.0045
98. Economou V, Gousia P. Agriculture and food animals as a source of antimicrobial-resistant bacteria. Infect Drug Resist. (2015) 8:49–61. doi: 10.2147/IDR.S55778
99. Sodini SM, Kemper KE, Wray NR, Trzaskowski M. Comparison of genotypic and phenotypic correlations: Cheverud's conjecture in humans. Genetics. (2018) 209:941–8. doi: 10.1534/genetics.117.300630
Keywords: bovine salmonellosis, antimicrobial resistance, calves, WGS, genotyping
Citation: Casaux ML, D'Alessandro B, Vignoli R and Fraga M (2023) Phenotypic and genotypic survey of antibiotic resistance in Salmonella enterica isolates from dairy farms in Uruguay. Front. Vet. Sci. 10:1055432. doi: 10.3389/fvets.2023.1055432
Received: 27 September 2022; Accepted: 15 February 2023;
Published: 09 March 2023.
Edited by:
Julio Alvarez, VISAVET Health Surveillance Centre (UCM), SpainReviewed by:
Abraham Loera Muro, Centro de Investigación Biológica del Noroeste (CIBNOR), MexicoRuixi Chen, Massachusetts Institute of Technology, United States
Takele Beyene Tufa, Addis Ababa University, Ethiopia
Copyright © 2023 Casaux, D'Alessandro, Vignoli and Fraga. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: María Laura Casaux, bGNhc2F1eEBpbmlhLm9yZy51eQ==; Martín Fraga, bWZyYWdhQGluaWEub3JnLnV5
 María Laura Casaux
María Laura Casaux Bruno D'Alessandro2
Bruno D'Alessandro2 Rafael Vignoli
Rafael Vignoli Martín Fraga
Martín Fraga