- 1School of Life Science and Engineering, Southwest University of Science and Technology, Mianyang, Sichuan, China
- 2College of Life Sciences, Yan'an University, Yan'An, Shaanxi, China
- 3Department of Biosciences, COMSATS University Islamabad, Islamabad, Pakistan
- 4Sichuan Key Laboratory of Conservation Biology on Endangered Wildlife, Chengdu Research Base of Giant Panda Breeding, Chengdu, Sichuan, China
Circular RNAs (circRNAs), as endogenous non-coding RNA with unique closed ring structure, is closely related to animal reproduction, and understanding the expression of circRNA in yak and cattleyak epididymal tissues is of great significance for understanding cattleyak sterility. Based on this, we screened and identified the differentially expressed circRNA in the epididymis of three yaks and two cattleyak. A total of 1,298 circRNAs were identified in the epididymis of yak and cattleyak, of which 137 differentially expressed (DE) circRNAs and the functions of some of them were elucidated in this research, as well as qPCR verification to 6 circRNAs from the 137 DE circRNAs. Gene Ontology (GO) enrichment analysis suggested that DE circRNAs were mainly related to metabolic process, development process, immune system process, reproductive process, reproduction, biological adhesion and growth. COG classification analysis showed that the DE circRNAs derived genes were mainly related to replication, recombination and repair. KEGG pathway analysis suggested that DE circRNAs were mainly involved in RNA degradation. In addition, we also screened Bta-mir-103, which is a circRNA binding miRNA related to sperm activity.
1. Introduction
A new group of circular RNAs (circRNAs) with covalently closed-loop structures have gained attention recently as novel molecules of gene regulation as other non-coding RNAs. CircRNAs were more stable than linear RNAs under RNAse R treatment because of the lack of free 5' and 3' ends (1). According to their sequence construction and sources from the genome, the so far discovered circRNAs can be mainly classified into three categories, such as exonic circRNAs, intronic circRNAs (ciRNAs) and exon-intron circRNAs (EIciRNAs). Exonic circRNAs are mainly found in the cytoplasm to act as miRNA sponges (2, 3). CiRNAs and EIciRNAs are speculated as efficient regulators in cis/trans for their parental genes in the nucleus (4, 5). Some circRNAs may also regulate gene expression by competitive alternative RNA splicing (6), or by acting as translation templates (7). Their main role is to modulate microRNAs (miRNAs) and protein availability working as sponges (8). This tethering activity inhibits miRNAs, protecting their mRNA targets from degradation. For this reason, circRNAs are part of the competitive endogenous RNA (ceRNA) network (ceRNET). Besides tethering activity, circRNAs also regulate transcription of their host genes (5) and splicing of their cognate mRNAs (9).
Cattleyak is a hybrid breed of male yak and female cattle. It shows obvious heterosis in meat and dairy quality and is also the main economic source of herdsmen in Plateau areas of China. However, the male offspring cattleyak is born through spermatogenesis dysfunction (10), which leads to their infertility. Our research team has studied the expression of miRNA (11) in yak epididymis and the expression of miRNA (12) and lncRNA (13) in cattleyak epididymis, and identified the proteins related to sperm maturation in yak and cattleyak epididymis by comparing iTRAQ (14) proteomics. Many scientists have carried out cellular and molecular level research to determine various factors causing the infertility. The cellular level research (15) shows that cattleyak infertility may be caused by meiotic chromosome pairing anomalies, which results in the stagnation of spermatogenesis in spermatogonial stem cells (SSCS) or primary spermatocyte phase. On the other hand, the molecular level (16), research shows that DNA hypermethylation and low expression of spermatogenesis related genes are the cause of male cattleyak infertility. During spermatogenesis, spermatozoa formed in the testis are carried to the epididymis, where post-testicular maturation of the sperm cells take place. Post-testicular maturation of spermatozoa mainly involves motility acquisition and the competence to undergo capacitation, which leads to fertilization of an egg (17, 18). The epididymis is a complex convoluted tube that connects efferent ducts to the vas deferens in the male reproductive tract (19, 20). The function of the epididymis is crucial to the maturation of normal sperm, as it is through this organ that sperms gain full activity for reproduction (21).
In the past couple of years, few studies have been conducted on circRNAs and have identified their critical role in sperm development (22, 23). CircRNAs have been identified in the testis of human (22) and cattle (23). However, none of such studies have been demonstrated in the epididymis. Therefore, epididymal based study may elucidate the causes of infertility in cattleyak and also enhance our understanding of molecular mechanisms underlying in this region. As it is mentioned earlier, circRNAs analysis on epididymal based study is yet to be explored. Therefore, we used RNA-seq to study the important role of circRNA in the regulation of gene expression in the epididymis of cattleyak and yak, so as to lay a foundation for yak and cattleyak breeding. In addition, circRNA: X:132347491|132372215 and 20:2680598|2714639 associated with spermatogenesis were successfully screened (24–27).
2. Materials and methods
2.1. Sample collection and treatment
Three adult male yak and three cattleyak testicles with 36 months of age, physique and health status were collected in the abattoir of Hongyuan County, Aba Prefecture, Sichuan Province. The epididymis was cleaned three times with Hanks solution and was separated from testis.
2.2. Total RNA isolation
Extraction of Total RNA from yak and cattleyak epididymis tissue was performed according to the instructions of Trizol reagent. The total RNA from yak and catleyak epididymis (caput, corpus and cauda) were extracted. The integrity of RNA was detected by 0.8 % agarose gel electrophoresis apparatus and the quality of RNA was detected by SpectraMax Quick Drop ultrafine spectrophotometer and Qubit 4.0. The extracted RNA was stored in an ultra-low temperature refrigerator (−80°C).
2.3. RNA sample detection
Nanodrop, Qubit 2.0, Agilent 2100 and electrophoresis were used to detect the purity, concentration and integrity of RNA samples. These instruments were used to determine whether there was genomic DNA contamination, so as to ensure the use of qualified samples for circRNAs sequencing.
2.4. Construction of RNA library
After qualified samples were detected, library construction was carried out and the main processes were as follows: rRNAs of samples were removed using Epicenter Ribo-ZerotM kit; fragmentation buffer was added to randomly break the rRNA- depleted RNA. The depleted RNA was cloned into the first cDNA strand using random hexamers. The second cDNA strand was synthesized by adding buffer, dATP, dUTP, dCTP, dGTP, RNAse H and DNA polymerase I and the cDNAs were purified using AMPure XP Beads. The purified double-stranded cDNA was repaired, A was added and sequenced. Then AMPure XP Beads were used for fragment size selection. Finally, the U chain was degraded and the cDNA library was enriched by PCR.
A qualified sequencing library is a necessary condition for sequencing. To ensure the quality of the library, the quality of the sequencing library is evaluated using the following two different perspectives: (1) The randomness of CircRNA fragmentation and the degradation of CircRNA is evaluated by checking the distribution of inserted fragments in genes; (2) The dispersion degree of the inserted fragment length is evaluated by the length distribution of the inserted fragment.
2.5. Library quality control
After library construction, the quality of the library was tested and machine sequencing was performed only after the test results meet the requirements. The detection methods proceeded: (1) Qubit2.0 was used for preliminary quantification; Agilent 2100 was used to detect the insert size of the library and the next experiment was carried out only after the insert size met expectations. (2) The effective concentration of the library was accurately quantified by q-PCR (effective concentration of library >2 nM) to complete library inspection.
The original sequence obtained by sequencing contains joint sequence or low-quality sequence. In order to ensure the accuracy of information analysis, it is necessary to conduct quality control on the original data to obtain high-quality sequences (i.e., Clean Reads). The quality control standard of the original sequence is: (1) Removed the read with connector; (2) Filtered and removed low quality data to ensure data quality; (3) Removed the reads containing N (unable to determine the base information) whose proportion is >5%. The high-quality read obtained after the above series of quality control are called Clean Data, which is also provided in FASTQ format.
2.6. Bioinformatics analysis
Raw data were filtered to remove joint sequences and low-quality reads to obtain high-quality clean data. The clean data was aligned against the specified reference genome to obtain the mapped data. Based on the mapped data, the quality assessment of sequencing library such as insert length test and randomness test were carried out. CIRI software (28) prediction circRNA, circRNA binding site analysis and CircRNA expression level analysis of different samples. RNAhybrid was used to predict the target gene of circRNA. The circRNA source genes (CSGs) were annotated with relevant functions, and the CSGs were analyzed with standardized biological annotation through GO database to compare the enrichment trend of circRNA source genes and all other genes. The CSGs were classified into lineal homologies using COG database. The function of CSGs was further interpreted by analyzing the KEGG pathway annotation of CSGs.
2.7. qPCR validation of DE circRNAs
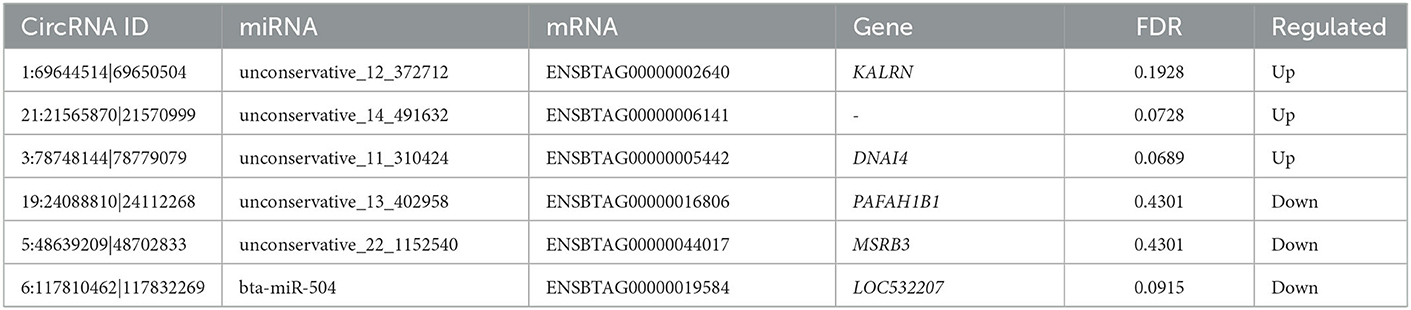
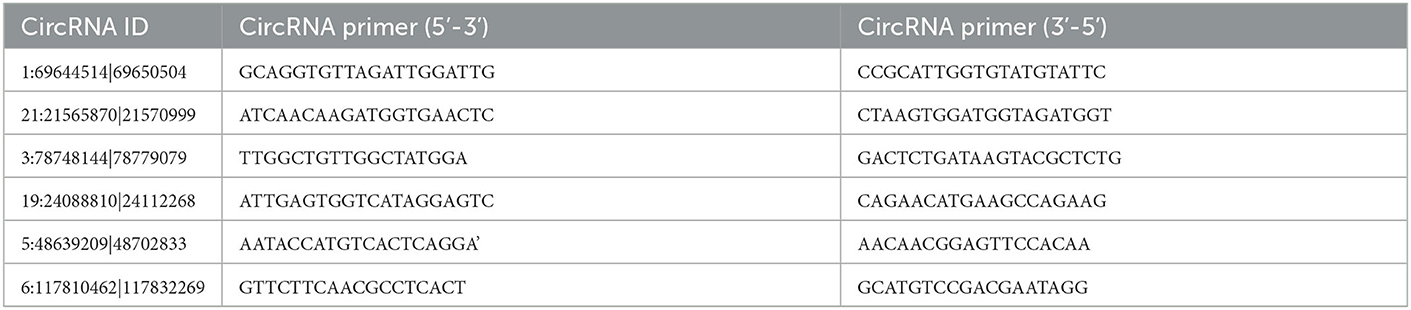
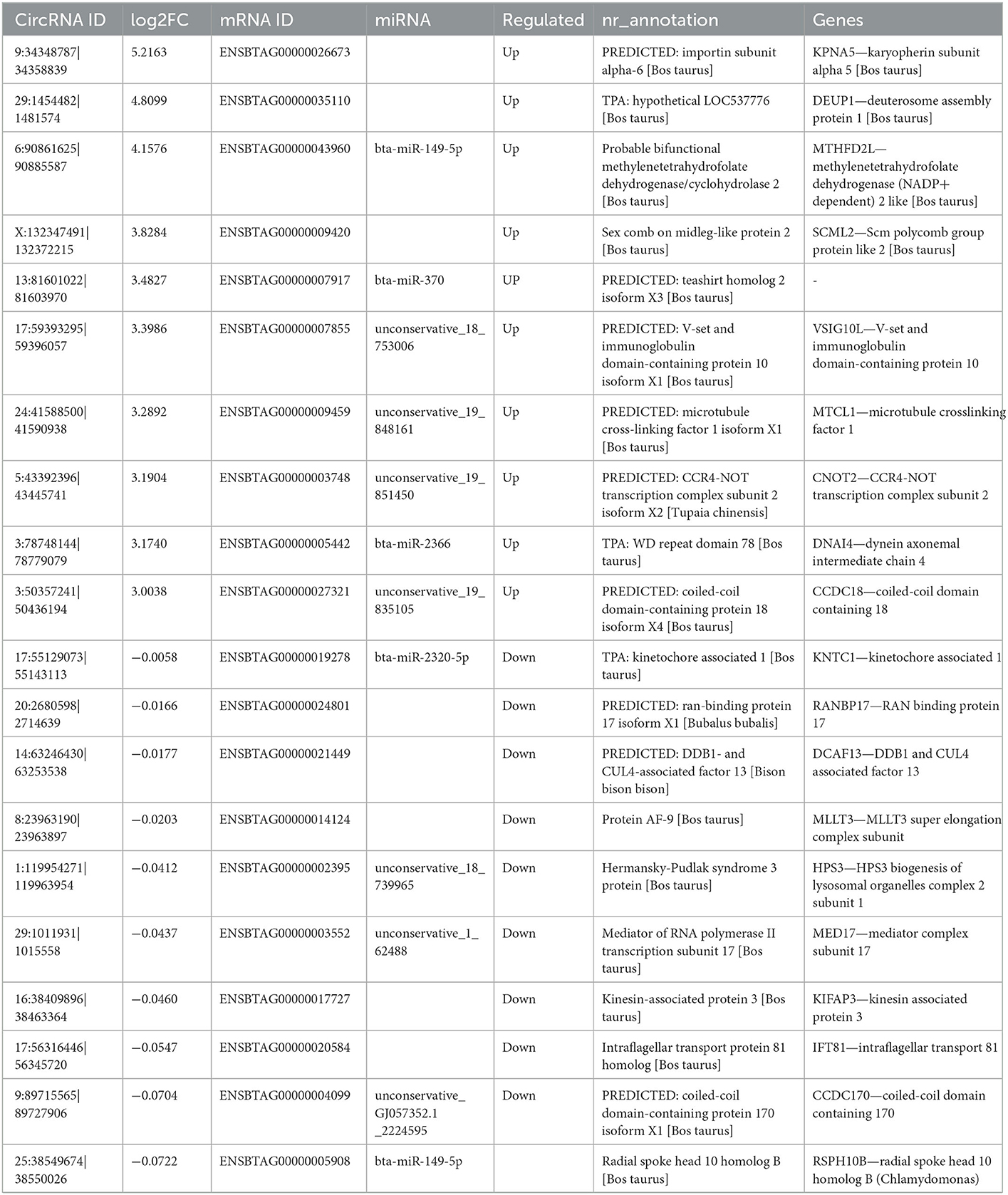
Based on the verified miRNA expression in our team's already published papers (29, 30), among the 137 DE circRNAs obtained in this study, six of them (see Table 1) were screened to test the differential expression of circRNA by qPCR, and the relationship between circRNA, miRNA and mRNA was verified. cDNA is obtained by reverse transcription from total RNA using SuperScript™ III reverse transcriptase (Invitrogen, Carlsbad, CA, USA). The SYBR Green assay was used in the CFX Connect™ real-time PCR detection system (Bio-Rad, Munich, Germany), and SYBR Premix Ex Taq (TaKaRa, Dalian, China) was used for qPCR validation using GAPDH as the internal reference gene. The qPCR reaction system was 95°C for 5 min, followed by 40 cycles of 95°C for 10 s, 60°C for 10 s and 72°C for 20 s. Each circRNA has two biological replicates. Data were processed using the 2−ΔΔCt method and statistical analysis was performed using Student's t-test in Simplot 11.2 software to compare the differences between yak and cattleyak epididymal samples at p<0.05. Primers for qPCR are listed in Table 2.
3. Results
3.1. Quality control of data
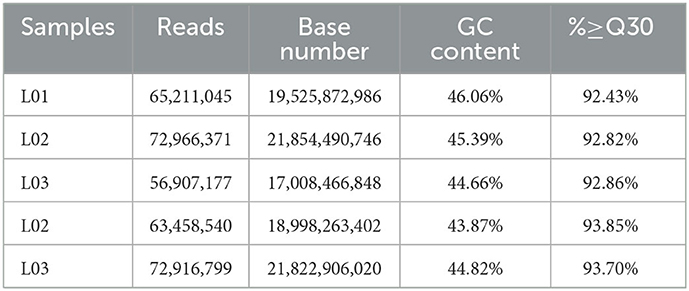
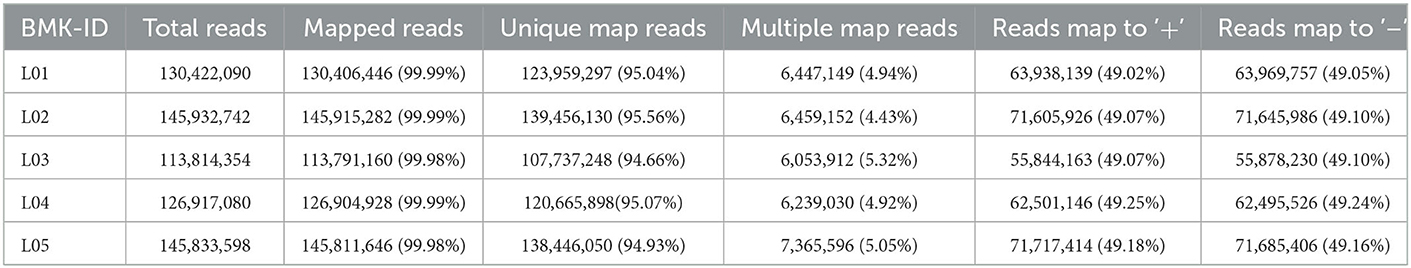
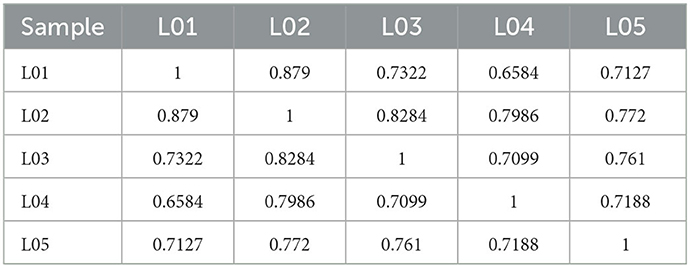
In this study, we have used six epididymal tissue samples of control (yak) and treatment (cattleyak) to construct cDNA libraries. Of the five tissue samples; three from yak (named L1, L2, and L3) and two from cattleyak (named L4 and L5) were completed, and then subjected to RNA-sequencing analysis. We obtained 99.21 Gb clean data from all five epididymal tissue samples. The clean data of each tissue sample reached 17.01Gb and the Q30 base percentage was not < 92.43% (Table 3). The efficiency of aligning reads with specified reference genome UMD3.1 of each tissue sample ranged from 99.98 to 99.99% (Table 4). The data utilization rate is normal and the selected reference genome can meet the needs of subsequent analysis. All data had good biological replicates and met the analytical requirements (Table 5).
3.2. Identification of circular RNAs (circRNAs)
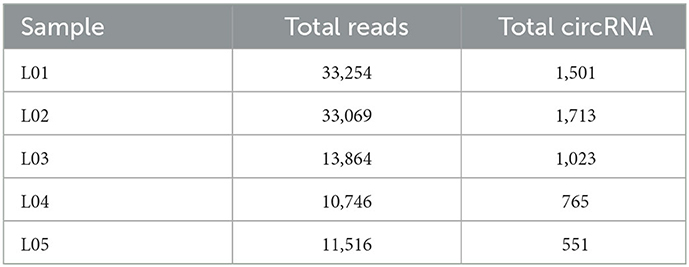
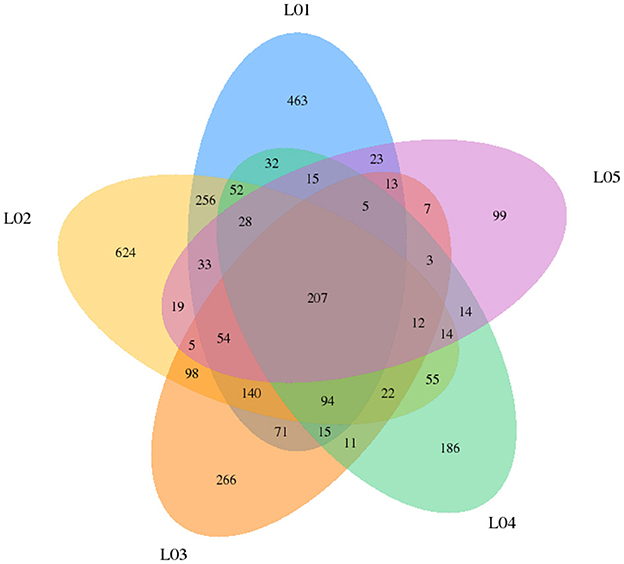
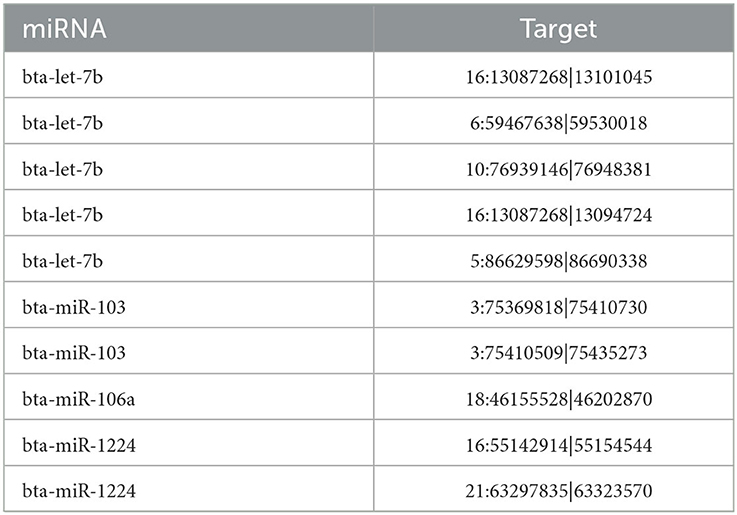
In yak verses cattleyak biological replicates (L01, L02, L03 vs. L04, L05), we identified a total of 1,298 candidate circRNAs by using CIRI and find-circ (8) software. 1,501, 1,713, and 1,023 circRNAs were found in three groups of yaks and 756 and 551 circRNAs were found in two groups of cattleyak (Table 6). 207 circRNAs were common among five replicates from the total identified circRNAs (Figure 1). Based on the position of circRNAs in the genome, these circRNAs were generally divided into three types: exonic, intronic and intergenic, from which most of the detected circRNAs in our study were originated from exonic regions and then intergenic regions and intronic regions respectively (Figure 2). Next, the distribution of circRNAs in different samples on different chromosomes was calculated and resulted that the mainly circRNAs were produced by chromosome 1 and chromosome 10 (Figure 3). The distribution statistics of circRNA length in each sample showed that those circRNAs who had ranged 200–2,200 bp in length were mainly originated from exons while those circRNAs ranged >2,600 were mainly originated from intergenic regions (Figure 4).
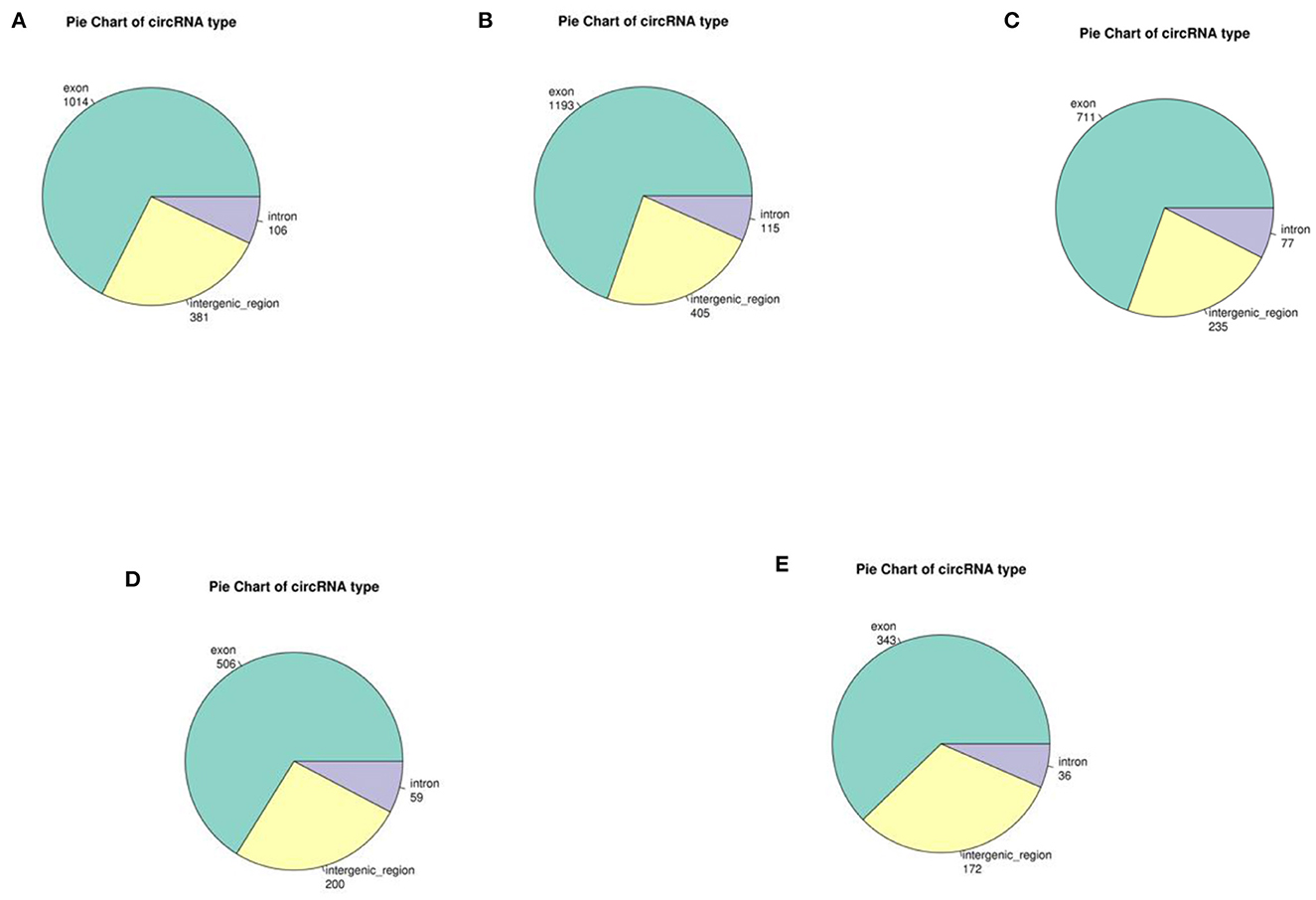
Figure 2. Distribution diagram of circRNA sources for each sample. Different colors represent different circRNA types, and numbers represent the proportion of CircRNA types in the total. (A–C) represent the source distribution of circRNA in three yak samples; (D, E) represent the source and distribution of circRNA in cattleyak samples.
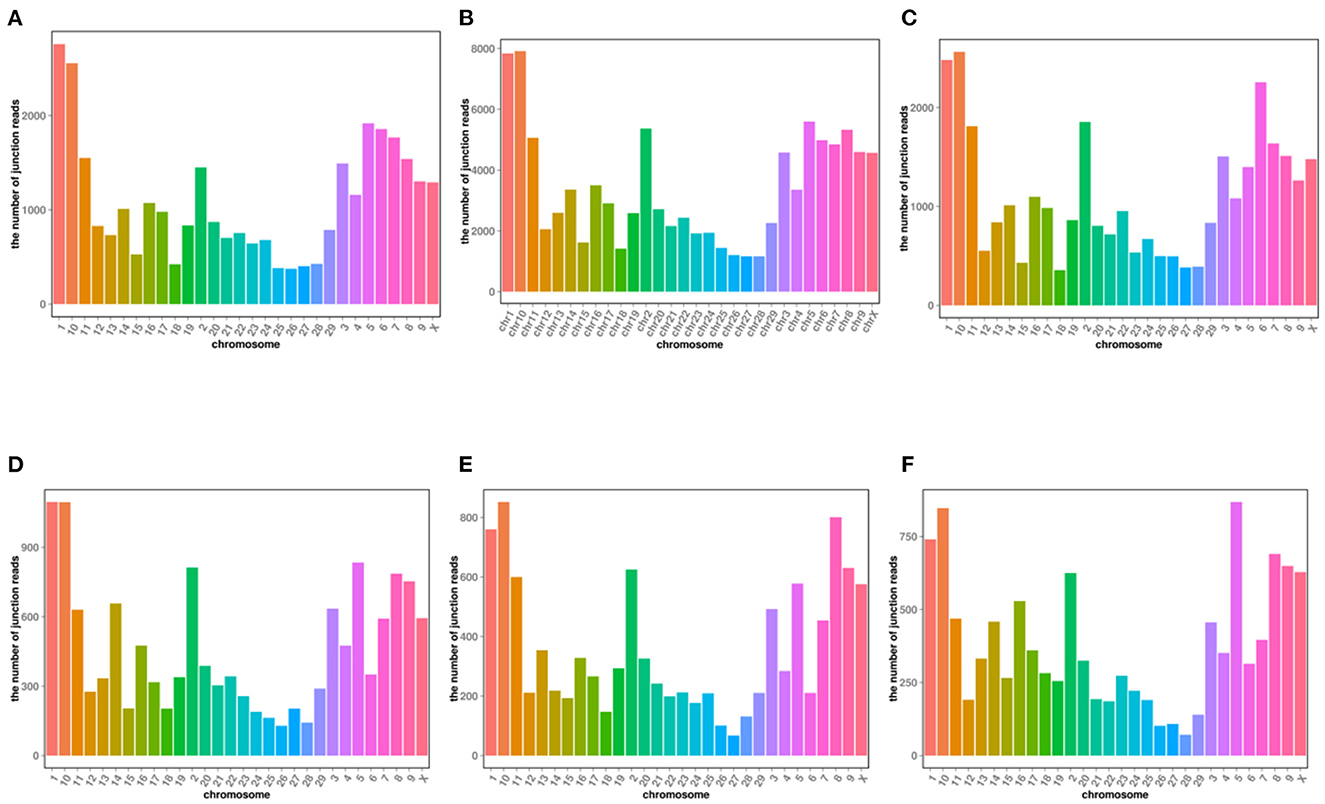
Figure 3. CircRNA chromosome reads distribution of each sample. (A) Distribution of RNA on all chromosomes; (B–D) distribution of circRNA on chromosomes in Yak samples; Distribution of circRNA on chromosomes in (E, F) cattleyak samples.
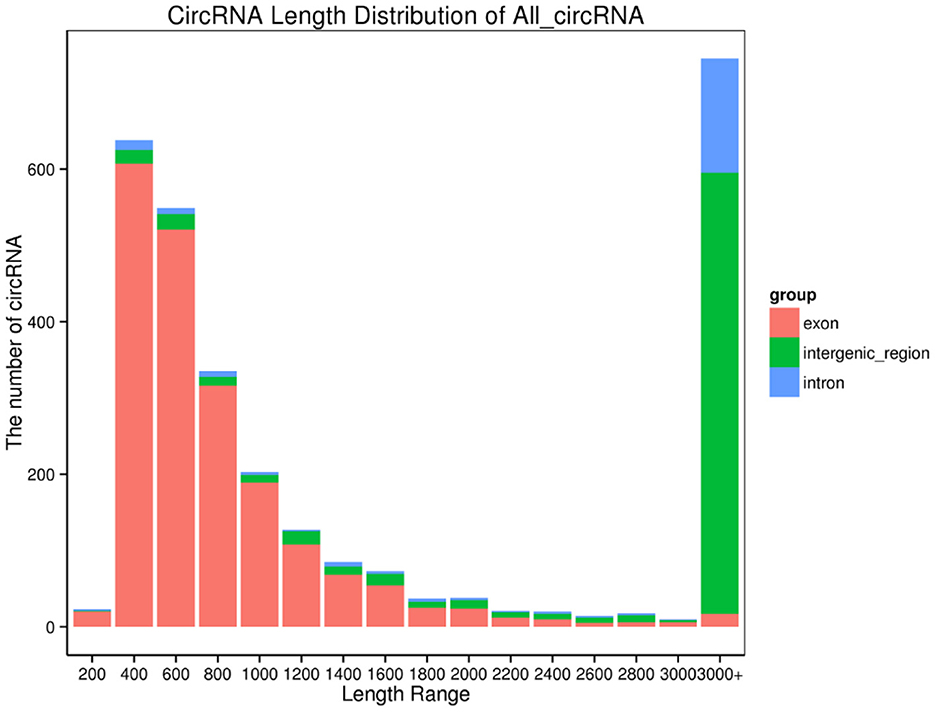
Figure 4. CircRNA length distribution. Abscissa, length interval; Ordinate, the number of circular RNAs in the length interval.
3.3. Differential expression profiling of circRNAs
In the differential expression circRNA detection process, we had set fold difference ≥2 and p-value < 0.05 as the screening criteria. The fold difference described the ratio of expression between two samples (groups), and a significant p-value was used as a screening key indicator of DE circRNAs. A total of 137 circRNAs were DE circRNA, of which 100 were up-regulated and the remaining 37 were down-regulated (see Tables 7, 8, Figure 5). Furthermore, hierarchical clustering analysis of DE circRNAs indicates that circRNA richness in yaks is higher than in cattleyak in the heat map (Figure 6).
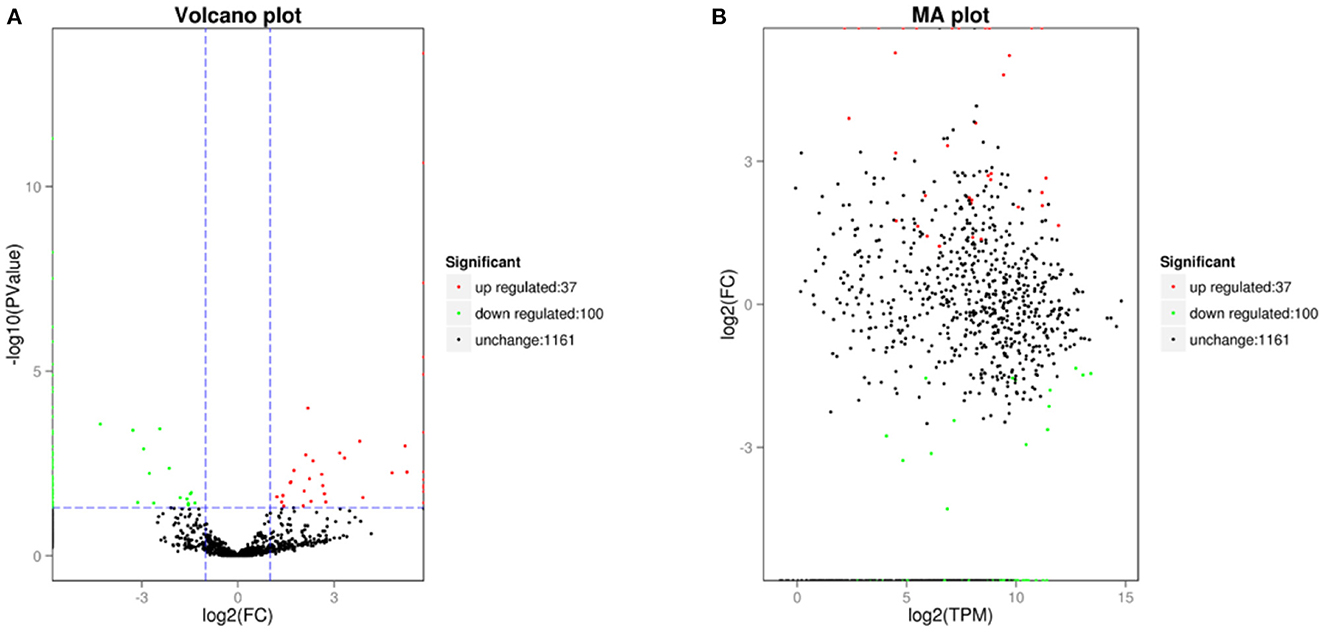
Figure 5. DE circRNAs plot. (A) is a volcano diagram of DE circRNA, that each point in the diagram represents a circRNA. (B) is a DE circRNA MA plot and each point in the diagram represents a gene.
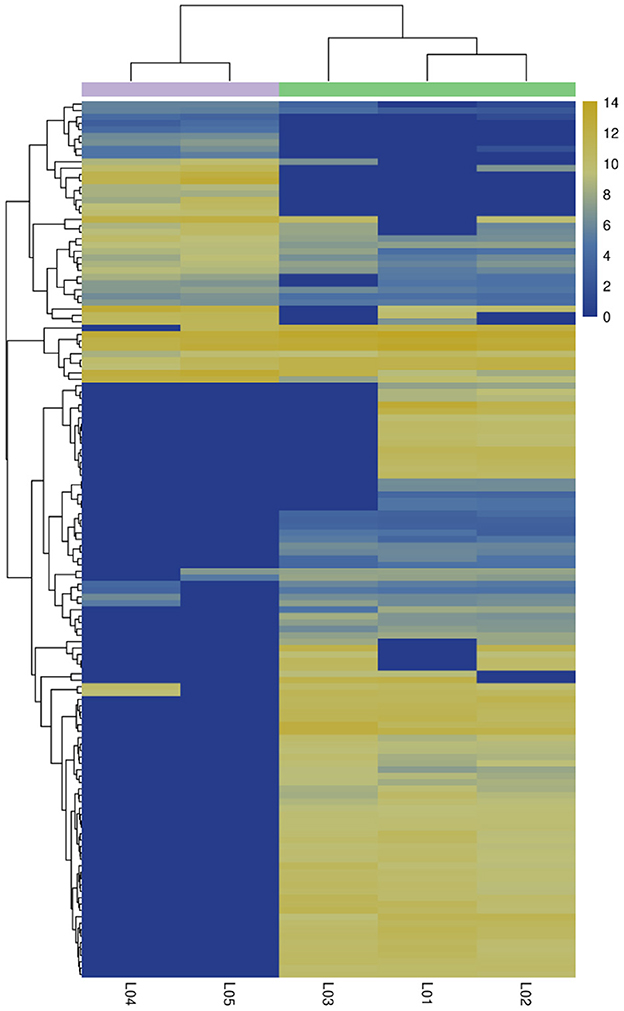
Figure 6. Correlation heat map. Different columns in the figure represent different samples, and different rows represent DE circRNAs. The color represents the expression level of circRNA in the sample (log2tpm + 1).
3.4. circRNA target gene prediction
circRNAs can regulate important biological processes by regulating the circRNA–miRNA–mRNA pathway. Therefore, we performed circRNA target gene prediction using RNAhybrid 2.1.2 software. Out of the total 137 DE circRNAs, 79 DE circRNAs were predicted that targeted 214 miRNAs. Since circRNA contains multiple miRNA binding sites, we used the method of miRNA target gene prediction to identify the circRNA bound to miRNA and clarify the function of this part of circRNA according to the functional annotation of miRNA target gene (Table 9).
3.5. GO annotation and KEGG enrichment analysis of circRNA host genes
The functional annotation of the source genes of circRNA was carried out to discover new circRNA for the yak and cattleyak to complement the original genome annotation information (Table 10). We conducted GO and KEGG pathway analysis on the genes producing the DE circRNAs. The GO annotation was arranged in three main categories, namely Cellular Component (CC), Molecular Function (MF), and Biological Process (BP). According to BP category, the DE circRNA host genes were involved in metabolic process, developmental process, immune system process, reproductive process, reproduction, biological adhesion, and growth; according to CC category, the DE circRNA host genes were involved in membrane-enclosed lumen, extracellular region and organelle; according to MF category, the DE circRNA host genes were involved in chemorepellent activity, eletron carrier activity and catalytic activity (Figure 7). Different gene products interact with each other to perform biological functions and the pathway annotations of host genes of circRNAs which could provide a better understanding of the functions of these genes. KEGG enrichment analysis showed that 18 pathways were related to circRNAs, mainly including RNA degradation and peroxisome (Figure 8). COG classification analysis of circRNA-derived genes showed that the genes were mainly related to replication, recombination and repair process (Figure 9).
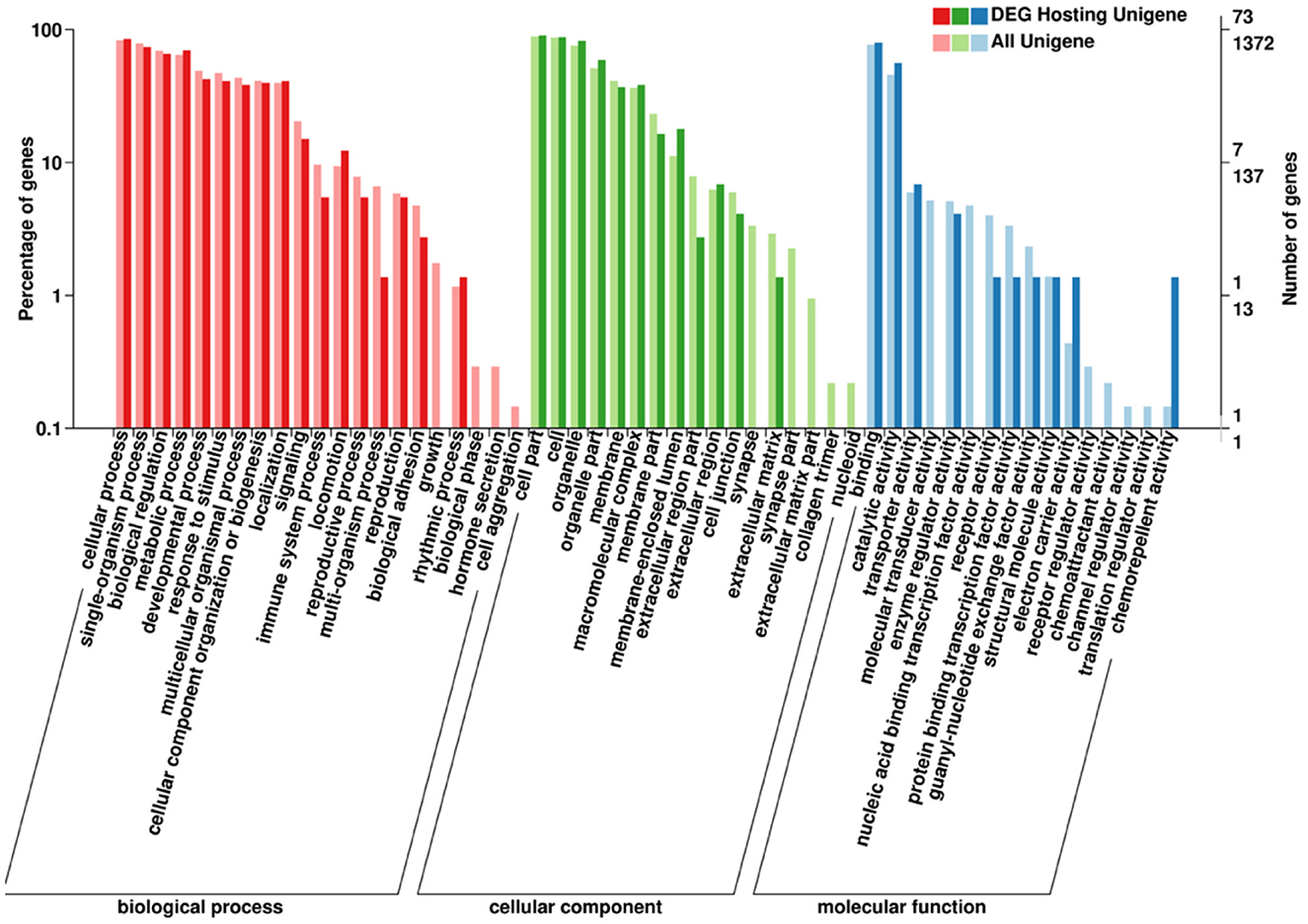
Figure 7. Annotated statistical diagram of circRNA-derived Gene Ontology Consortium (GO) secondary nodes. The abscissa is the go classification, the left side of the ordinate is the percentage of the number of genes, and the right side is the number of genes.
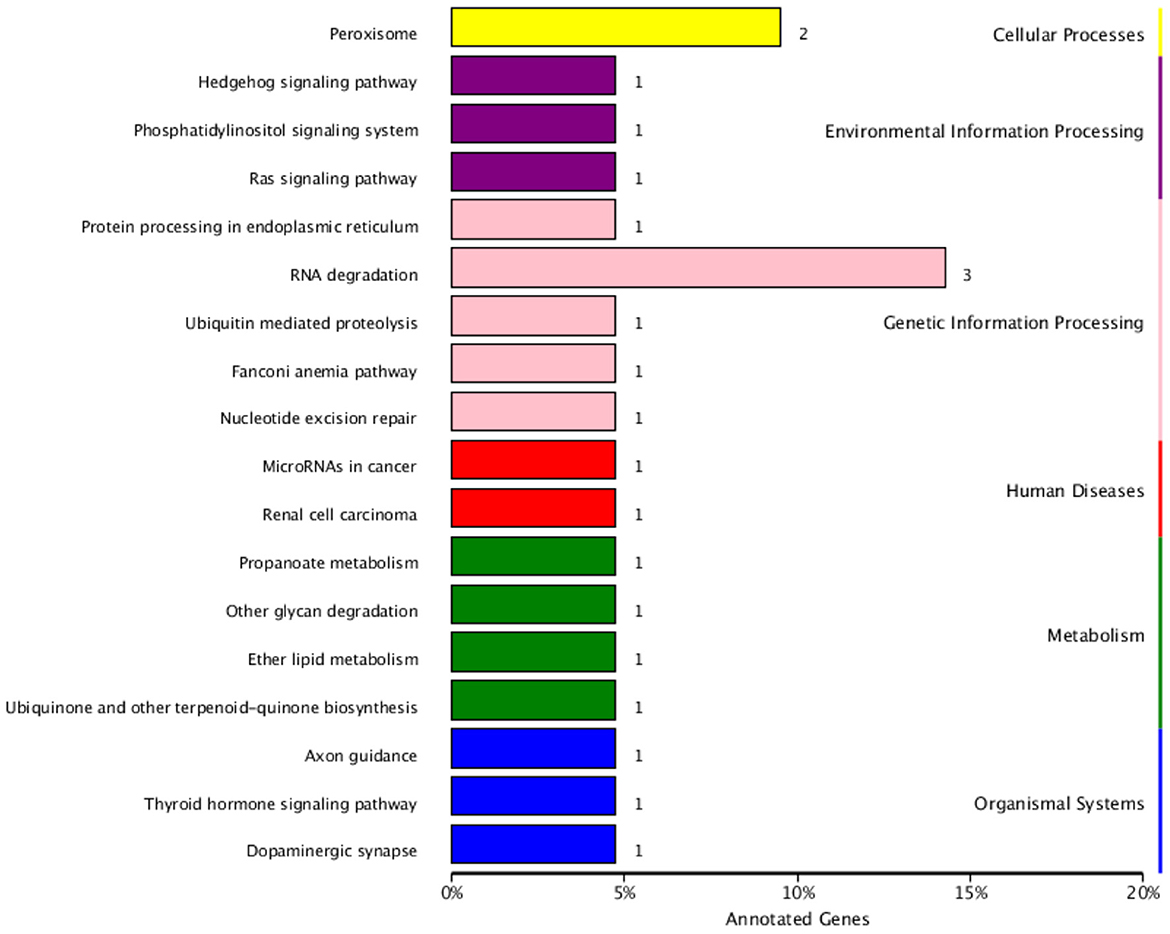
Figure 8. KEGG classification of circRNA-derived genes. The ordinate is the name of KEGG metabolic pathway, and the abscissa is the number of circRNA source genes annotated to this pathway and their proportion in the total number of genes annotated.
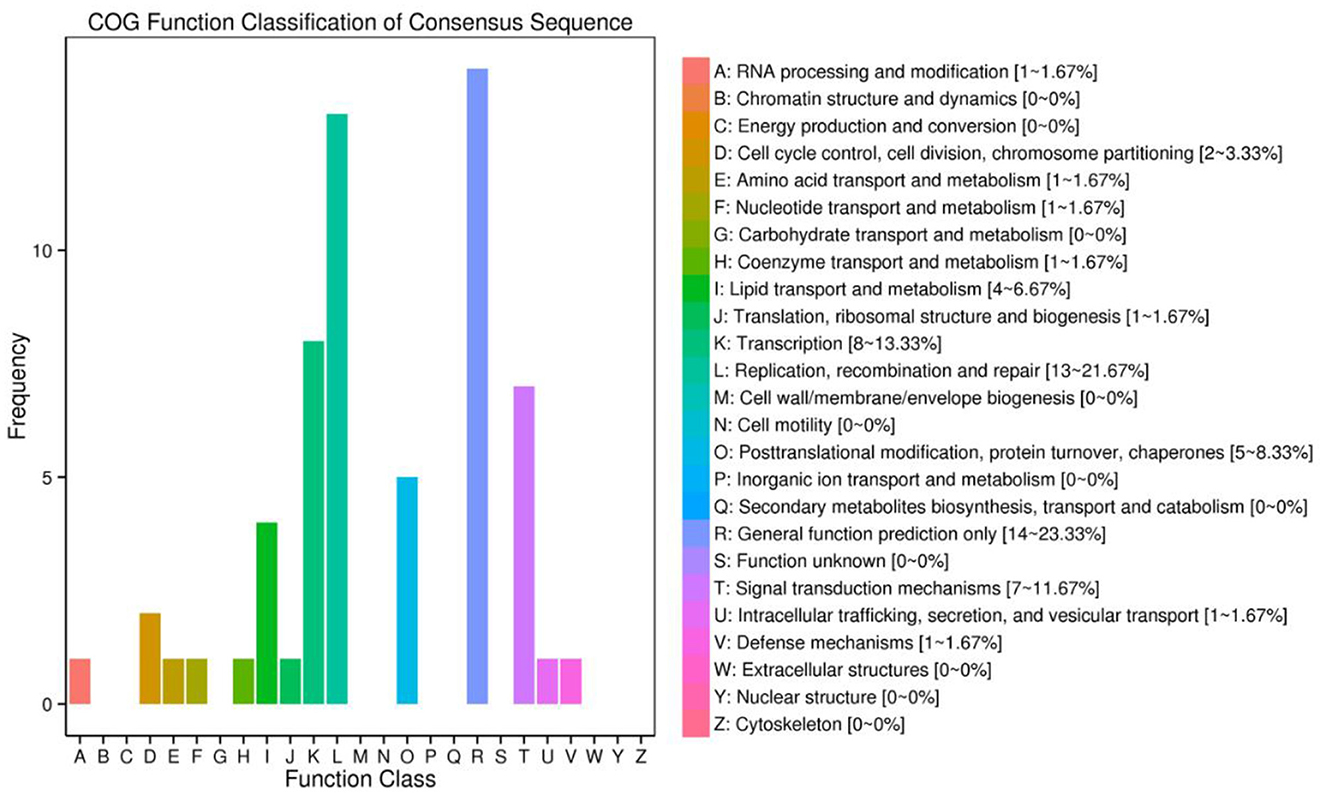
Figure 9. CircRNA origin gene Cluster of Orthologous Groups of proteins (COG) annotation classification statistical map. The abscissa is the classification content of COG, and the ordinate is the number of genes.
3.6. Validation of DE circRNAs expression by qPCR
We performed qPCR validation on six DE circRNAs (Table 1), and although the folding rates differed, the overall trend was consistent, proving that our sequencing data was correct (Figure 10). At the same time, combined with previous articles (29, 30) on the expression of DE miRNA in yak and cattleyak epididymal tissues, the relationship chain between circRNA-miRNA-mRNA was confirmed.
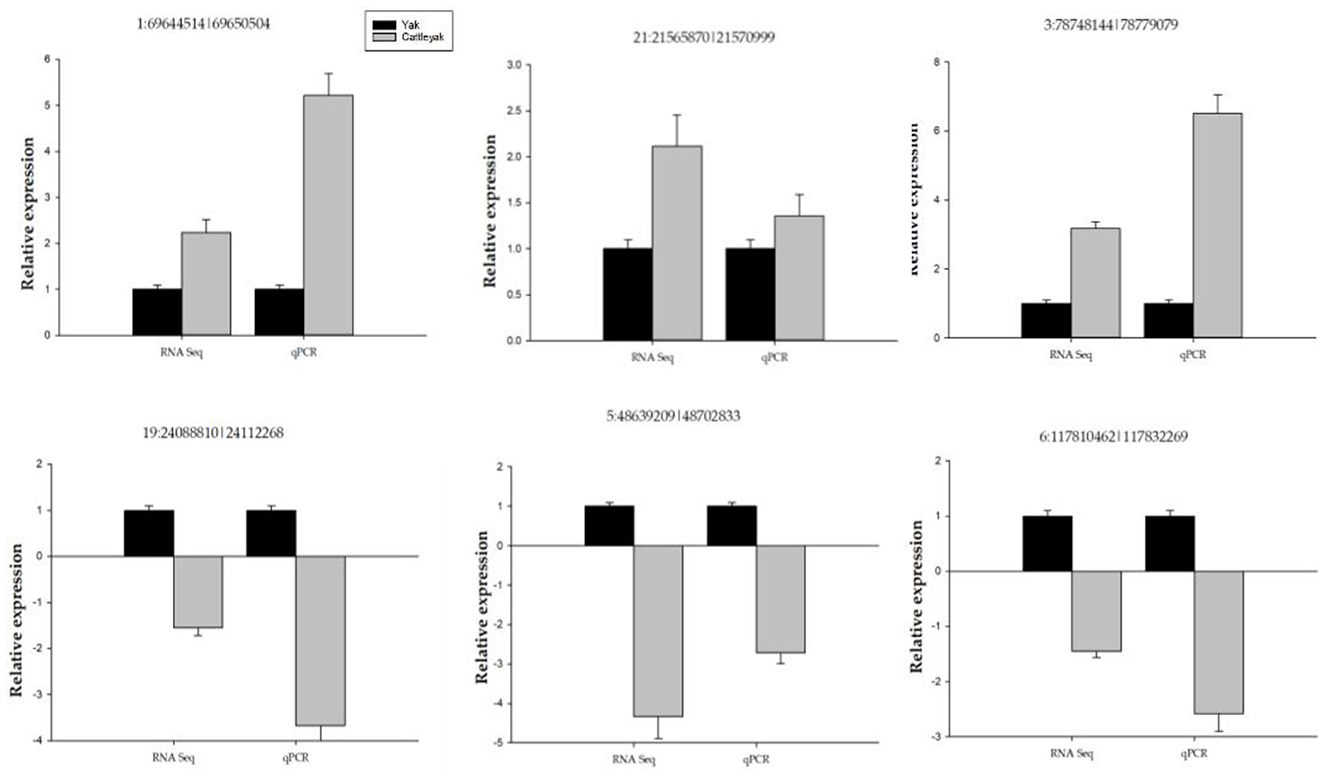
Figure 10. Shows qPCR validation of some selected DE circRNAs. The y-axis indicates the relative expression of six DE circRNAs between yaks and cattleyak.
4. Discussion
In previous studies, scientists have conducted a variety of studies on yak and cattleyak epididymal tissues (31), utilizing proteomics (15) and high-throughput sequencing techniques (29). These studies geared toward explaining the possible mechanism of cattleyak infertility in the epididymal region. CircRNAs are one of the emerging non-coding RNAs. Over the years, their physiological functions in various tissues of mammalian species have been studied. However, since its discovery in the 1980s, its exploration in mammalian epididymis is still lacking behind. Therefore, we intend to explore its potential role in the epididymis of yak and cattleyak for the first time. Another main factor of our motivation is to uncover the molecular mechanisms underlying the process of sperm maturation and cattleyak infertility.
In this study, we identified a total of 1,298 circRNAs between the epididymis of yak and cattleyak by employing the RNA-sequencing technique. Of the total 1,298 circRNAs, 137 circRNAs were differentially expressed and among them 100 were up-regulated while the remaining 37 were down-regulated in cattleyak. The circRNAs identified in the epididymal tissues of yak and cattleyak were less than that in testicular tissues, mammary gland tissues, adipose tissues and skeletal muscle tissues of other bovine animal species (23, 32–34). Furthermore, 1,753 CircRNAs were detected in the testis of newborn (1 week old) and adult (4 years old) Qinchuan yaks (23). There were significant differences in the expression of 4,248 circRNAs between neonatal and adult yak testis (23). Among these circRNAs, 2,225 were up-regulated, and the relative expression of circRNAs in chromosomes 1 and 10 was observed higher indicating that it was consistent with our results (23). A total of 7,203 circRNAs were found in the study of yak adipocyte differentiation on days 0, 2 and 12, of which 4,139 circRNAs were detected on day 0, 4,755 on day 2 and 5,505 on day 12, whereas 2,737 circRNAs were detected at all three time points (35). In the analysis of circRNA expression characteristics of testicular samples from 6-month-old bulls, a total of 17,013 circRNAs were identified, of which 681 circRNAs were differentially expressed. Of these DE circRNAs, 579 were up-regulated and 103 were down regulated in calf and bull testis (36). In consistent with previous reports, we identified that most of the circRNAs were derived from exonic regions, then intergenic and intronic regions, respectively.
Most of the DE circRNAs detected were from chromosomes 1 and 10. Related literatures showed that bovine chromosome 1 was related to the biological functions of α-1 crystal protein (CRYA1), γ -S crystal protein (CRYG8), superoxide dismutase I (SOD1) and uridine-monophosphates (UMPS) (24). In addition, a genetic map of DNA loci on Bovine Chromosome 1 occurred in comparison with that of other mammals during the evolutionary process (25). Chromosome 10 is associated with beef odor (25) and marbling of bovine muscle (26).
In DE circRNA genes, SCML2 (27) is a meiotic chromatin related protein encoded by the X chromosome. In mouse studies, SCML2 was found to be associated with phosphorylated H2AX. The loss of SCML2 in mice will lead to sperm production defects, resulting in a sharp decline in sperm production. SCML2 binds to the hypomethylation promoter rich in H3K4me2/3 in undifferentiated sperm to promote H3K27me3, thereby affecting sperm differentiation and the totipotency of oosperm (37). SCML2 was identified as a key regulator of heterochromatin tissue in spermatogenesis (38). RANBP17 (39) may play a role in the inactivation of sex chromosomes during the meristematic stage of spermatogenesis, as well as in the intragranular transport during spermatogenesis. PAFAH1B1 is a highly conserved protein, and studies in mice and other domesticated animals have found that PAFAH1B1 has an important relationship with spermatogenesis and embryonic development (40). MSRB3 plays an important role in ossification and fat deposition in cattle (41). KALRN is involved in synaptic regulation (42).
CircRNAs have demonstrated closer relationship with miRNAs and are known to be miRNA sponges. miRNAs are notably known as the regulators of genes at post-transcriptional level. Through circRNA-miRNA-mRNA pathway, the circRNAs regulate most important parameters of sperm maturation, such as circRNA_002581-miR-122-Slc1a5, and circRNA_007585-miR-326- UCP2 (43). CircRNAs function by binding to miRNA. Therefore, we used miRNA target gene prediction method to identify CircRNA bound to miRNA (Table 9). The results showed that bta-let-7b in the identification results were related to lipid metabolism and mitochondrial activity (44). In the study of vitrified bovine embryos, it was found that bta-let-7b was a potential target for cell pluripotency, cell programming, survival rate after cryopreservation, embryo quality, lipid metabolism and mitochondrial activity, which were related to miRNA targeting lipid metabolism and mitochondrial activity genes (44). In yak and cattleyak studies, downregulation of the bta-let-7 family, bta-miR-125, and bta-miR-23a may impair RA-induced sperm division (10). The target mRNA ENSBTAG000000022360 of circRNA 5:86629598|86690338 corresponds to the gene SOX5 associated with Polish Holstein-Friesian Bulls' semen quality (45), so there may be a 5:86629598|86690338 to bta-let-7b to SOX5 relationship chain in this study. Bta-mir-103 may be related to heat stress (46) and sperm motility (47) after cryopreservation in cattle and the expression of bta-mir-103 in yak and cattleyak testis is also different (10). Bta-mir-106a is widely expressed in yaks (48), and is related to TGFβ1/smad signaling pathway (49). Bta-mir-1224 is a fibroblast related miRNA that can inhibit the expression of COL11A2, FGFR3, ISLR2 and MYH3 genes in muscle tissue of Xinjiang brown cattle (50).
According to KEGG pathways analysis results (Figure 8), most of the DE circRNA came from peroxisome-related genes (51), and antioxygenation (52) and are closely related to the peroxisomal lipid metabolism. Peroxidase is also involved in regulating signal homeostasis of spermatogenesis (53), which may also be one of the reasons leading to the difference in reproduction between yak and cattleyak. From COG annotation analysis (Figure 9), it is speculated that circRNAs may be involved in signal transduction. The literatures also indicate that circRNAs have certain effects on signaling pathways, such as inhibition of Wnt/β-catenin signaling pathway by circRNA-ITCH (54) and regulation of circRNA-000911/ Mir-449a pathway in breast cancer (55). Furthermore, vesicle transport of circRNAs in vivo may indicate that these RNAs may also function as signaling molecules (24).
Angiotensin converting enzyme (ACE) is a male sterility-related enzyme (56). Studies in male mice have found that ACE1 −/− male mice are infertile (57). ACE can be synthesized endogenously in the epididymis, independent of the testis (58). The activity of ace in epididymal epithelial cells depends on androgen activity (59). In the study of renin-angiotensin-aldosterone system (RAAS), it was found that the expression of ace was affected by circRNA. The interaction between hsa_ circ_ 0122153/hsa_ mir_ 483_ 3p affects the expression of Ang II, which can promote ACE and inhibit the expression of angiotensin converting enzyme 2 (ACE2) in neurons through p38 MAPK and erk1/2 signaling pathways. In addition, has_ mir_ 483_ 3 can also act directly on ACE (60). In our study, we found that the target miRNA (bta-let-7b) of the differentially expressed circRNA was related to fat synthesis, and related studies showed that ACE2 deficiency showed lipid accumulation and mitochondrial dysfunction in skeletal muscle (61). In subcutaneous fat, high-fat diet inhibited ACE2 mRNA (62). Besides, studies on the epididymal fat pad of normal mice showed that physiologically activated NKT cells express high levels of intracellular IFN-g (63). The epididymal fat pad is a warehouse of immune cells in the epididymis, especially under inflammatory conditions (64). To sum up, these findings imply that the circRNA target genes we predicted may be related to ACE, and the ACE in the epididymis of yak and cattleyak may affect their fertility by acting on the epididymis adipose tissue.
5. Conclusion
In conclusion, we identified DE circRNA in epididymis of yaks and cattleyak, and analyzed the source genes of DE circRNA by bioinformatics. So far, there are few reports about circRNA in epididymis of yaks and cattleyak. This study will lay a foundation for solving the infertility of cattleyak, and breeding of yaks and cattleyak in the future.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA883995.
Ethics statement
The animal study was reviewed and approved by Ethics Committee: Biological and Health Ethics Committee of Southwest University of Science and Technology affiliation: Southwest University of Science and Technology. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
CL: writing—original draft and conceptualization. YY: writing—review and editing. CP: data curation. MA: methodology. KS: visualization. PW: investigation. MP: supervision. KL: software. YW: methodology and supervision. WZ: funding acquisition. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by Qinghai Provincial Central Government Guidance Local Science and Technology Development Fund Project (2023ZY026), Sichuan Science and Technology Program (2021YFH0101), the National Natural Science Foundation of China (31601946), and Longshan Academic Talent Research Supporting Program of SWUST (17LZX671 and 18lzx661).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Suzuki H, Zuo Y, Wang J, Zhang MQ, Malhotra A, Mayeda A. Characterization of RNAse R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic Acids Res. (2006) 34:e63. doi: 10.1093/nar/gkl151
2. Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature. (2013) 495:384–8. doi: 10.1038/nature11993
3. Liu Q, Zhang X, Hu X, Dai L, Fu X, Zhang J, et al. Circular RNA related to the chondrocyte ECM regulates MMP13 expression by functioning as a MiR-136 'sponge' in human cartilage degradation. Sci Rep. (2016) 6:22572. doi: 10.1038/srep22572
4. Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, et al. Circular intronic long noncoding RNAs. Mol Cell. (2013) 51:792–806. doi: 10.1016/j.molcel.2013.08.017
5. Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. (2015) 22:256–64. doi: 10.1038/nsmb.2959
6. Burd CE, Jeck WR, Liu Y, Sanoff HK, Wang Z, Sharpless NE. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet. (2010) 6:e1001233. doi: 10.1371/journal.pgen.1001233
7. Abe N, Matsumoto K, Nishihara M, Nakano Y, Shibata A, Maruyama H, et al. Rolling circle translation of circular RNA in living human cells. Sci Rep. (2015) 5:16435. doi: 10.1038/srep16435
8. Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. (2013) 495:333–8. doi: 10.1038/nature11928
9. Conn VM, Hugouvieux V, Nayak A, Conos SA, Capovilla G, Cildir G, et al. A circRNA from SEPALLATA3 regulates splicing of its cognate mRNA through R-loop formation. Nat Plants. (2017) 3:17053. doi: 10.1038/nplants.2017.53
10. Xu C, Shah MA, Mipam T, Wu S, Yi C, Luo H, et al. Bovid microRNAs involved in the process of spermatogonia differentiation into spermatocytes. Int J Biol Sci. (2020) 16:239–50. doi: 10.7150/ijbs.38232
11. Zhao W, Quansah E, Yuan M, Li P, Yi C, Cai X, et al. Next-generation sequencing analysis reveals segmental patterns of microRNA expression in yak epididymis. Reprod Fertil Dev. (2020) 32:1067–83. doi: 10.1071/RD20113
12. Zhao W, Hussain Solangi T, Wu Y, Yang X, Xu C, Wang H, et al. Comparative RNA-seq analysis of region-specific miRNA expression in the epididymis of cattleyak. Reprod Domest Anim. (2021) 56:555–76. doi: 10.1111/rda.13893
13. Zhao W, Ahmed S, Ahmed S, Yangliu Y, Wang H, Cai X. Analysis of long non-coding RNAs in epididymis of cattleyak associated with male infertility. Theriogenology. (2021) 160:61–71. doi: 10.1016/j.theriogenology.2020.10.033
14. Zhao W, Ahmed S, Liu J, Ahmed S, Quansah E, Solangi TH, et al. Comparative iTRAQ proteomics identified proteins associated with sperm maturation between yak and cattleyak epididymis. BMC Vet Res. (2021) 17:255. doi: 10.1186/s12917-021-02907-9
15. Fedyk S, Krasińska M. Spermatogenesis in backcross generations of European bison and domestic cattle hybrids. Acta Theriol. (1980) 25:201–12. AT.arch.80-15. doi: 10.4098/AT.arch.80-15
16. Niayale R, Cui Y, Adzitey F. Male hybrid sterility in the cattle-yak and other bovines: a review. Biol Reprod. (2021) 104:495–507. doi: 10.1093/biolre/ioaa207
17. Turner TT. On the epididymis and its role in the development of the fertile ejaculate. J Androl. (1995) 16:292–8.
18. Visconti PE, Bailey JL, Moore GD, Pan D, Olds-Clarke P, Kopf GS. Capacitation of mouse spermatozoa. I Correlation between the capacitation state and protein tyrosine phosphorylation. Development. (1995) 121:1129–37. doi: 10.1242/dev.121.4.1129
19. Robaire B, Hinton BT, Orgebin-Crist MC. The Epididymis[M]//Knobil and Neill's Physiology of Reproduction. https://www.google.com/search?biw=1536&bih=670&q=Cambridge,+Massachusetts&stick=H4sIAAAAAAAAAOPgE-LUz9U3MM8rzi5X4gAxDQszzLW0spOt9POL0hPzMqsSSzLz81A4VhmpiSmFpYlFJalFxYtYJZwTc5OKMlPSU3UUfBOLixOTM0qLU0tKinewMu5iZ-JgAAAm2NNbZgAAAA&sa=X&ved=2ahUKEwjt5P6lo9j8AhX6F1kFHWJwAYoQmxMoAHoECFwQAg Cambridge, MA: Academic Press (2006), 1071–148.
20. Dacheux JL, Dacheux F, Druart X. Epididymal protein markers and fertility. Anim Reprod Sci. (2016) 169:76–87. doi: 10.1016/j.anireprosci.2016.02.034
21. Leir SH, Browne JA, Eggener SE, Harris A. Characterization of primary cultures of adult human epididymis epithelial cells. Fertil Steril. (2015) 103:647–54.e1. doi: 10.1016/j.fertnstert.2014.11.022
22. Dong WW Li HM, Qing XR, Huang DH Li HG. Identification and characterization of human testis derived circular RNAs and their existence in seminal plasma. Sci Rep. (2016) 6:39080. doi: 10.1038/srep39080
23. Gao Y, Wu M, Fan Y, Li S, Lai Z, Huang Y, et al. Identification and characterization of circular RNAs in Qinchuan cattle testis. R Soc Open Sci. (2018) 5:180413. doi: 10.1098/rsos.180413
24. Chioccarelli T, Pierantoni R, Manfrevola F, Porreca V, Fasano S, Chianese R, Cobellis G. Histone post-translational modifications and CircRNAs in mouse and human spermatozoa: potential epigenetic marks to assess human sperm quality. J Clin Med. (2020) 9:640. doi: 10.3390/jcm9030640
25. Barendse W, Armitage SM, Ryan AM, Moore SS, Clayton D, Georges M, et al. genetic map of DNA loci on bovine chromosome 1. Genomics. (1993) 18:602–8. doi: 10.1016/S0888-7543(05)80362-3
26. Casas E, Shackelford SD, Keele JW, Koohmaraie M, Smith TP, Stone RT. Detection of quantitative trait loci for growth and carcass composition in cattle. J Anim Sci. (2003) 81:2976–83. doi: 10.2527/2003.81122976x
27. Luo M, Zhou J, Leu NA, Abreu CM, Wang J, Anguera MC, et al. Polycomb protein SCML2 associates with USP7 and counteracts histone H2A ubiquitination in the XY chromatin during male meiosis. PLoS Genet. (2015) 11:e1004954. doi: 10.1371/journal.pgen.1004954
28. Gao Y, Wang J, Zhao F, CIRI. an efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol. (2015) 16:1. doi: 10.1186/s13059-014-0571-3
29. Wang C, Hussain Solangi T, Wang H, Yang L, Adjei M, Ahmed S, et al. High-throughput sequencing reveals differential expression of miRNAs in yak and cattleyak epididymis. Reprod Domest Anim. (2022) 57:125–40. doi: 10.1111/rda.13973
30. Lang X, Adjei M, Wang C, Chen X, Li C, Wang P, et al. RNA-Seq reveals the functional specificity of epididymal caput, corpus, and cauda genes of cattleyak. Anim Sci J. (2022) 93:e13732. doi: 10.1111/asj.13732
31. Wu C, Wang C, Zhai B, Zhao Y, Zhao Z, Yuan Z, et al. Study of microRNA expression profile in different regions of ram epididymis. Reprod Domest Anim. (2021) 56:1209–19. doi: 10.1111/rda.13978
32. Zhang C, Wu H, Wang Y, Zhu S, Liu J, Fang X, et al. Circular RNA of cattle casein genes are highly expressed in bovine mammary gland. J Dairy Sci. (2016) 99:4750–60. doi: 10.3168/jds.2015-10381
33. Huang J, Zhao J, Zheng Q, Wang S, Wei X, Li F, et al. Characterization of circular RNAs in Chinese Buffalo (Bubalusbubalis) adipose tissue: a focus on circular RNAs involved in fat deposition. Animals (Basel). (2019) 9:403. doi: 10.3390/ani9070403
34. Wei X, Li H, Yang J, Hao D, Dong D, Huang Y, et al. Circular RNA profiling reveals an abundant circLMO7 that regulates myoblasts differentiation and survival by sponging miR-378a-3p. Cell Death Dis. (2017) 8:e3153. doi: 10.1038/cddis.2017.541
35. Zhang Y, Guo X, Pei J, Chu M, Ding X, Wu X, et al. CircRNA expression profile during yak adipocyte differentiation and screen potential circRNAs for adipocyte differentiation. Genes (Basel). (2020) 11:414. doi: 10.3390/genes11040414
36. Khan IM, Liu H, Zhuang J, Khan NM, Zhang D, Chen J, et al. Circular RNA expression and regulation profiling in testicular tissues of immature and mature Wandong cattle (Bos taurus). Front Genet. (2021) 12:685541. doi: 10.3389/fgene.2021.685541
37. Maezawa S, Hasegawa K, Yukawa M, Kubo N, Sakashita A, Alavattam KG, et al. Polycomb protein SCML2 facilitates H3K27me3 to establish bivalent domains in the male germline. Proc Natl Acad Sci U S A. (2018) 115:4957–62. doi: 10.1073/pnas.1804512115
38. Maezawa S, Hasegawa K, Alavattam KG, Funakoshi M, Sato T, Barski A, et al. SCML2 promotes heterochromatin organization in late spermatogenesis. J Cell Sci. (2018) 131:jcs217125. doi: 10.1242/jcs.217125
39. Bao J, Wu Q, Song R, Jie Z, Zheng H, Xu C, et al. RANBP17 is localized to the XY body of spermatocytes and interacts with SPEM1 on the manchette of elongating spermatids. Mol Cell Endocrinol. (2011) 333:134–42. doi: 10.1016/j.mce.2010.12.021
40. Yao GD, Shi SL, Song WY, Jin HX, Peng ZF, Yang HY, et al. Role of PAFAH1B1 in human spermatogenesis, fertilization and early embryonic development. Reprod Biomed Online. (2015) 31:613–24. doi: 10.1016/j.rbmo.2015.07.010
41. Wu M, Li S, Zhang G, Fan Y, Gao Y, Huang Y, et al. Exploring insertions and deletions (indels) of MSRB3 gene and their association with growth traits in four Chinese indigenous cattle breeds. Arch Anim Breed. (2019) 62:465–75. doi: 10.5194/aab-62-465-2019
42. Parnell E, Shapiro LP, Voorn RA, Forrest MP, Jalloul HA, Loizzo DD, et al. KALRN: A central regulator of synaptic function and synaptopathies. Gene. (2021) 768:145306. doi: 10.1016/j.gene.2020.145306
43. Jin X, Feng CY, Xiang Z, Chen YP Li YM. CircRNA expression pattern and circRNA-miRNA-mRNA network in the pathogenesis of nonalcoholic steatohepatitis. Oncotarget. (2016) 7:66455–67. doi: 10.18632/oncotarget.12186
44. Saber Y H A, Ibrahim S, Mahmoud K G M, et al. Changes in the relative abundance of miR-205, miR-26a-5p, let-7b and their target genes in vitrified bovine embryos after phenazineethosulfate supplementation. Adv Anim Vet Sci. (2021) 9:2157–67. doi: 10.17582/journal.aavs/2021/9.12.2157.2167
45. Hering DM, Kamiński S. Association between SOX5 genotypes and semen quality in polish holstein-friesian bulls. Pol J Vet Sci. (2016) 19:651–3. doi: 10.1515/pjvs-2016-0082
46. Kumar M. Noyonika, Aggarwal A, Kaul G. Novel and known miRNAs in zebu (Tharparkar) and crossbred (Karan-Fries) cattle under heat stress. Funct Integr Genomics. (2021) 21:405–19. doi: 10.1007/s10142-021-00785-w
47. Shangguan A, Zhou H, Sun W, Ding R, Li X, Liu J, et al. Cryopreservation induces alterations of mirna and mrna fragment profiles of bull sperm. Front Genet. (2020) 11:419. doi: 10.3389/fgene.2020.00419
48. Wang YH, Xu YF, Niu JQ, Wang H, Suolang S, Qiangba Y, et al. Preliminary studies on tissue expression profiles of Smad 4 mRNA and its miRNAs in yak. Acta Veterinaria et Zootechnica Sinica. (2017) 48:1882–91. doi: 10.11843/j.issn.0366-6964.2017.10.011
49. Xu Y, Niu J, Xi G, Niu X, Wang Y, Guo M, et al. TGF-β1 resulting in differential microRNA expression in bovine granulosa cells. Gene. (2018) 663:88–100. doi: 10.1016/j.gene.2018.04.036
50. Na L. Molecular mechanism of miRNA and mRNA regulation of lipid forming traits in Xinjiang brown cattle and Kazakh beef. Lanzhou, China: Gansu Agricultural University (2019).
51. Reddy JK, Mannaerts GP. Peroxisomal lipid metabolism. Annu Rev Nutr. (1994) 14:343–70. doi: 10.1146/annurev.nu.14.070194.002015
52. Walbrecq G, Wang B, Becker S, Hannotiau A, Fransen M, Knoops B. Antioxidant cytoprotection by peroxisomal peroxiredoxin-5. Free Radic Biol Med. (2015) 84:215–26. doi: 10.1016/j.freeradbiomed.2015.02.032
53. Dastig S, Nenicu A, Otte DM, Zimmer A, Seitz J, Baumgart-Vogt E, et al. Germ cells of male mice express genes for peroxisomal metabolic pathways implicated in the regulation of spermatogenesis and the protection against oxidative stress. Histochem Cell Biol. (2011) 136:413–25. doi: 10.1007/s00418-011-0832-0
54. Yang B, Zhao J, Huo T, Zhang M, Wu X. Effects of CircRNA-ITCH on proliferation and apoptosis of hepatocellular carcinoma cells through inhibiting Wnt/β-catenin signaling pathway. J BUON. (2020) 25:1368–74.
55. Wang H, Xiao Y, Wu L, Ma D. Comprehensive circular RNA profiling reveals the regulatory role of the circRNA-000911/miR-449a pathway in breast carcinogenesis. Int J Oncol. (2018) 52:743–54. doi: 10.3892/ijo.2018.4265
56. Pan PP, Zhan QT, Le F, Zheng YM, Jin F. Angiotensin-converting enzymes play a dominant role in fertility. Int J Mol Sci. (2013) 14:21071–86. doi: 10.3390/ijms141021071
57. Kessler SP, Rowe TM, Gomos JB, Kessler PM, Sen GC. Physiological non-equivalence of the two isoforms of angiotensin-converting enzyme. J Biol Chem. (2000) 275:26259–64. doi: 10.1074/jbc.M004006200
58. Seike H, Yokoyama M, Iwata H, Ochi K, Takeuchi M, Takada Y, Hiwada K, Kokubu T. Production of angiotensin-converting enzyme in rat epididymis. Andrologia. (1985) 17:406–10. doi: 10.1111/j.1439-0272.1985.tb01030.x
59. Wong PY, Uchendu CN. The role of angiotensin-converting enzyme in the rat epididymis. J Endocrinol. (1990) 125:457–65. doi: 10.1677/joe.0.1250457
60. He X, Tao Z, Zhang Z, He W, Xie Y, Zhang L. The potential role of RAAS-related hsa_circ_0122153 and hsa_circ_0025088 in essential hypertension. Clin Exp Hypertens. (2021) 43:715–22. doi: 10.1080/10641963.2021.1945077
61. Cao X, Lu XM, Tuo X, Liu JY, Zhang YC, Song LN, et al. Angiotensin-converting enzyme 2 regulates endoplasmic reticulum stress and mitochondrial function to preserve skeletal muscle lipid metabolism. Lipids Health Dis. (2019) 18:207. doi: 10.1186/s12944-019-1145-x
62. Kai H, Kai M, Niiyama H, Okina N, Sasaki M, Maeda T, et al. Overexpression of angiotensin-converting enzyme 2 by renin-angiotensin system inhibitors. Truth or myth? A systematic review of animal studies. Hypertens Res. (2021) 44:955–68. doi: 10.1038/s41440-021-00641-1
63. Caspar-Bauguil S, Cousin B, Galinier A, Segafredo C, Nibbelink M, André M, et al. Adipose tissues as an ancestral immune organ: site-specific change in obesity. FEBS Lett. (2005) 579:3487–92. doi: 10.1016/j.febslet.2005.05.031
Keywords: cattle, cattleyak, DE circRNA, epididymis, transcriptome sequencing
Citation: Li C, Yan Y, Pan C, Adjei M, Shahzad K, Wang P, Pan M, Li K, Wang Y and Zhao W (2023) Identification and analysis of differentially expressed (DE) circRNA in epididymis of yak and cattleyak. Front. Vet. Sci. 10:1040419. doi: 10.3389/fvets.2023.1040419
Received: 13 September 2022; Accepted: 18 January 2023;
Published: 07 February 2023.
Edited by:
Mohammad Reza Bakhtiarizadeh, University of Tehran, IranReviewed by:
Zeying Wang, Shenyang Agricultural University, ChinaSuneel Kumar Onteru, National Dairy Research Institute (ICAR), India
Copyright © 2023 Li, Yan, Pan, Adjei, Shahzad, Wang, Pan, Li, Wang and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ye Wang,  eWV3YW5nZmFpdGhAZ21haWwuY29t; Wangsheng Zhao,
eWV3YW5nZmFpdGhAZ21haWwuY29t; Wangsheng Zhao,  d2FuZ3NoZW5nemhhbzAxQDE2My5jb20=
d2FuZ3NoZW5nemhhbzAxQDE2My5jb20=
†These authors have contributed equally to this work
 Chunhai Li
Chunhai Li Yan Yan2†
Yan Yan2† Khuram Shahzad
Khuram Shahzad Wangsheng Zhao
Wangsheng Zhao