
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci., 15 September 2022
Sec. Veterinary Infectious Diseases
Volume 9 - 2022 | https://doi.org/10.3389/fvets.2022.987667
This article is part of the Research TopicEpidemic Status and Prevention of Swine Infectious DiseasesView all 11 articles
 Peixun Li1†
Peixun Li1† Yesheng Shen1†
Yesheng Shen1† Tailong Wang1
Tailong Wang1 Jing Li1
Jing Li1 Yan Li2
Yan Li2 Yiran Zhao1
Yiran Zhao1 Sidang Liu1
Sidang Liu1 Baoquan Li1*
Baoquan Li1* Mengda Liu3*
Mengda Liu3* Fanliang Meng1,4*
Fanliang Meng1,4*Since the rise of porcine reproductive and respiratory syndrome virus (PRRSV) in China, mutations have occurred regularly. In particular, the emergence of HP-PRRSV has significantly improved the pathogenicity of PRRSV. It has brought huge economic losses to the Chinese pig farming industry. To understand the current prevalence and evolution of PRRSV in Shandong Province, 1,344 samples suspected of having PRRSV were collected from local hog farms of different sizes. Genetic variation in the isolated PRRSV ORF5 gene was analyzed using the RT-PCR method. The results showed that the detection rate of PRRSV in the collected samples was 25.44%. The predominant strain of PRRSV in Shandong Province is still NADC30-like. However, it cannot be ignored that NADC34-like is also starting to become a prevalent strain. Mutations in ORF5 amino acids 13, 151 and neutralizing epitope (aa36-aa52) in some isolates can cause changes in virulence and ability to escape immunity. This study enriches the epidemiological data on PRRSV in Shandong Province, China. It provides an important reference for the development of new vaccines and for the prevention and control of PRRSV.
Porcine reproductive and respiratory syndrome (PRRS) is a highly contagious disease caused by the porcine reproductive and respiratory syndrome virus (PRRSV) (1). It is known as “blue ear disease” because it often leads to bluish purple ears in diseased pigs (2). In affected pigs, the disease primarily causes spontaneous abortion in late pregnancy, as well as stillbirths, mummified fetuses, or weak piglets. It also causes congenital dysplasia in piglets, respiratory distress, interstitial pneumonia and suppressed immune function. Moreover, it is often seen in mixed infections with other pathogens (3–5). PRRSV is currently one of the most serious pathogens posing a threat to global swine production. After beginning in Europe and the Americas in the 1990s, it spread across the globe (6, 7). The virus was first isolated in China in 1996 (5), and since then PRRSV has become widely prevalent there. A highly pathogenic strain of PRRSV emerged in China in 2006 and became the dominant epidemic strain (8–12). In 2012, in Henan Province, China, Zhou et al. (13) discovered for the first time a NADC30-like highly homologous strain with 131 aa discontinuous deletions in the nsp2 gene. In 2013–2015, NADC30-like strains were reported in many provinces in China (14). The high frequency of NADC30 recombination makes the prevention and control of PRRS extremely difficult (15–17). In 2017, the NADC34 strain was isolated for the first time in China (18). NADC34-like PRRSV is now mildly or moderately pathogenic to piglets (19–21). It primarily affects sows, often leading to severe spontaneous abortions among pregnant sows (19). NADC34-like PRRSV has become widespread in several provinces. The prevalence of NADC34 strains has been observed in 10 provinces and cities, including Heilongjiang, Liaoning, Jilin, Jiangsu, Henan, Hebei and Shandong (22–24). This makes it very necessary to monitor PRRSV and understand its prevalence in Shandong Province.
The genome of PRRSV is approximately 15 kb long, forms a cap structure at the 5′ end during mRNA processing, and has a Poly-A tail structure at the 3′ end (25). The structural proteins of this virus are GP2a, GP3, GP4, GP5, M, E, and N. Among these, GP5 and M proteins are the main envelope proteins of PRRSV (26). GP5 protein is the most variable protein in PRRSV. In addition, GP5 contains glycosylation sites that help recognize cell receptors and neutralize viruses. GP5 is also the main protein that promotes the production of neutralizing antibodies in the body. The rapid mutation and the high recombination frequency of PRRSV stimulate the prevalence of PRRS and exacerbate the difficulty of PRRS prevention and control (27). Therefore, the GP5 protein, as an extremely important PRRSV protein, has become an important indicator in the identification and analysis of PRRSV. To understand the latest epidemiological situation and epidemic strain types of PRRSV in 2020–2021, an experiment was conducted. In this experiment, clinical cases of suspected PRRSV infections were collected from swine farms of varying sizes in Shandong Province. The pathogens were detected by RT-PCR and were sequenced and analyzed for the GP5 protein gene.
In this study, suspected cases of PRRS were collected from pig farms of different sizes in all cities of Shandong Province in 2020–2021. Blood and nasal cotton swabs were mainly collected from affected pigs, while lymph nodes and lung tissues were collected from dead pigs.
The lymph nodes and pulmonary tissues were collected aseptically excised and crushed in a suspension. The blood was mixed with the corresponding nasal cotton swab. All processed samples were centrifuged, and the supernatant was sucked out to extract the total RNA for the RT-PCR test. Samples containing the low-viral target bands with content were inoculated into Marc-145 cells and PAM cells for blind transmission in three generations. The virus fluid was collected for the RT-PCR test.
The viral solution was added to the RNA isolater, and the total RNA was extracted according to the instructions of RNA isolater Total RNA Extraction Reagent (Vazyme).
With the use of the HiScript III 1st Strand cDNA Synthesis Kit (Vazyme), the resulting total RNA was reverse transcribed into the cDNA in accordance with the instructions. PCR amplification was performed using the primers in Table 1. The results were observed through agarose gel electrophoresis under a gel system imager.
All samples with positive RT-PCR findings were sequenced for the ORF5 gene. The sequences related to PRRSV Sublineage 1.5, Sublineage 1.8, Lineage 3, Lineage 5, and Lineage 8 were downloaded from the NCBI database as reference strains. These were analyzed using MEGA X and MegAlign.
In this study, the full-length genome of SDHY-DZ037 was sequenced for recombination analysis. Alignment was screened using RDP4, implementing the RDP (28), GENECONV (29), Bootscan (30), Chimaera (31), SiScan (32), MaxChi (33), and 3Seq (34) algorithms. At least four of the above methods can identify a recombination event. In addition, if the breakpoint region of the recombination event is larger than 100 nt, the region can be regarded as a recombination region. To confirm these presumed recombinant events, we generated a series of phylogenetic trees for each sequence region identified during the analysis (35). For each region, evolutionary analysis of maximum likelihood was performed in MEGA X. To visualize the recombinant signal and inferred breakpoint locations, a similarity analysis between the presumptive recombinant sequences and the parental lineages was implemented in SIMPLOT v3.5.1 (36). The window size was set to 200 nt and the step size to 20 nt.
A total of 1,344 suspected PRRS-positive samples were collected from all cities in Shandong Province in 2020–2021. The number of samples in different regions is shown in Figure 1.

Figure 1. Map of sample collection and distribution in Shandong Province from 2020 to 2021 (the number represents the number of samples collected).
Total RNA was extracted from cultured virus fluid and reverse-transcribed into cDNA. PCR was performed using cDNA as the template. The results indicated that the number of PRRSV positive samples was 342, and the detection rate was 25.44%. PCR results from selected PRRSV-positive samples are presented in Figure 2.
Figure 2. PCR results of some PRRSV positive samples. M: 2,000 marker, +: positive control, –: negative control, 1–16: sample numbers.
In all, 76 strains were sequenced for the PRRSV ORF5 gene (Figure 3). Among them, 49 strains belonged to Sublineage 1.8 in the evolutionary genetic map of the ORF5 gene, representing 64.47% of isolates. Fifteen strains (19.74%) were classified as Sublineage 1.5. One strain (1.32%) was classified as Sublineage 5. Eleven strains (14.47%) were classified as Lineage 8.
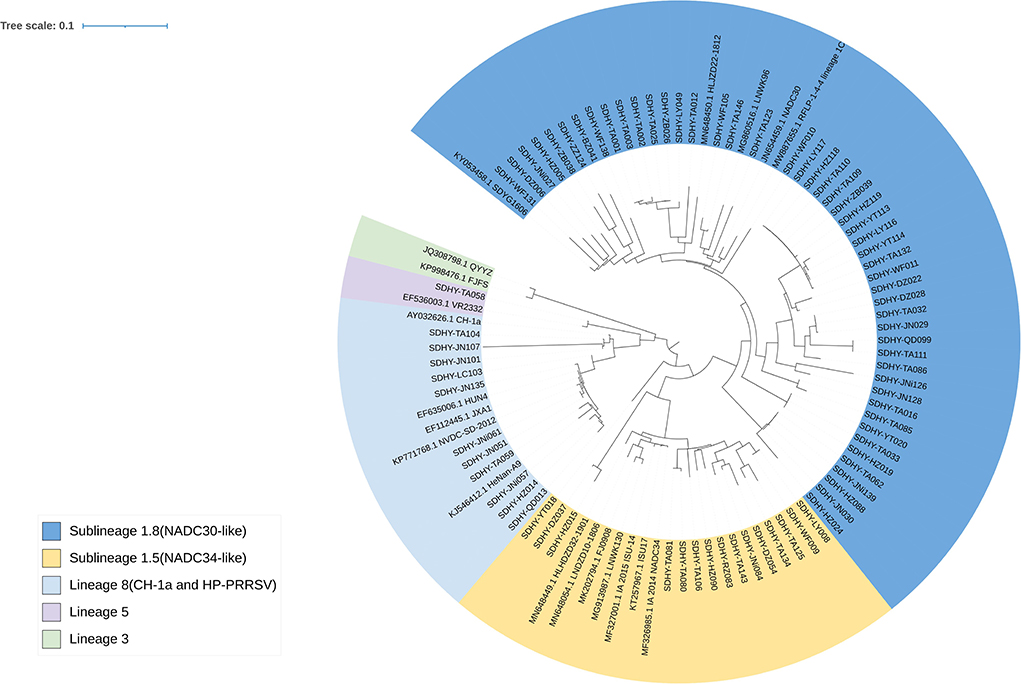
Figure 3. Phylogenetic tree based on the PRRSV ORF5 sequence. Evolutionary analysis of maximum likelihood performed in MEGA X. Multiple sequence alignments generated using Clustal W. ITOL was used to modify the genetic evolutionary tree, using different colors to distinguish different lineages and reference strains with GenBank sequence numbers.
The MegAlign module of DNASTAR Lasergene software was used to analyze the deduced amino acid mutation sites of 76 ORF5 genes in this study in comparison to some of the reference strains (Figure 4). A few typical mutations have been found. Amino acids 13 and 151 of ORF5-encoding GP5 protein were associated with virulence-related sites of the virus. Virulent strains of R13 and R151 are those that are often highly virulent (37–39). In this experiment, 13 strains of virus exhibited a Q13 → R13 mutation. Two strains were NADC30-like and the remaining 11 strains were CH-1a and HP-PRRSV-like.
Thirteen strains showed the K151 → R151 mutation, including two strains that were NADC30-like. One strain was NADC34-like and one strain was VR2332-like. The other nine strains were CH-1a and HP-PRRSV-like.
Mutations in the neutralizing epitope region at positions 36–52 of the amino acids encoded by ORF5 may cause the virus to escape the neutralizing effect induced by vaccine immunity. Reduce the protection effectiveness of vaccine immunity. In this study, multiple strains were isolated with mutations in the neutral epitope region. Twenty-three strains showed mutations in the neutral epitope region, of which 9 strains were NADC30-like, 2 strains were NADC34-like, 1 strain was VR2332-like, and the other 11 strains were CH-1a and HP-PRRSV-like.
Amino acid 137 (A137) of GP5 is unique to the VR2332, MLV, and RespPRRS/Repro vaccine strains and is considered to be a discriminating site between wild strains and vaccine strains (38, 40, 41). In this study, three isolates showed mutations from S137 to A137. Two of these were NADC34-like and one was VR2332-like.
In this study, we found that both SDHY-HZ015 and SDHY-DZ037 showed A137. A137 is generally considered a unique locus of the VR2332, MLV, and RespPRRS/Repro vaccine strains. We therefore conducted a genome-wide recombination analysis, using the HiScript III 1st Strand cDNA Synthesis Kit (Vazyme). The resultant total RNA was reverse transcribed into the cDNA, according to the instructions. PCR amplification was performed using the primers of Supplementary Table 1 (42). The results were observed through agarose gel electrophoresis under a gel system imager. We sent RT-PCR positive products for sequencing (BGI Genomics). Finally, an entire genome of SDHY-DZ037 was successfully isolated. Representative strains of each PRRSV strain were selected as reference. Recombination events and recombination breakpoints were confirmed through RDP4 software. The results are in Supplementary Table 2. Validation and presentation of results was done using SimPlot (Figure 5A). SDHY-DZ037 was a recombinant NADC30-like PRRSV and NADC34-like PRRSV. The primary parent strain was NADC30-like, and the secondary parent strain was NADC34-like. Four recombination events were identified by the RDP4 and SimPlot software. The results showed that: recombination event 1 occurred at nucleic acid 6515-10323 nt (Figure 5D); event 2 occurred at nucleic acid 10916-11773 nt (Figure 5E); event 3 occurred at nucleic acid 12543-12665 nt (Figure 5F); event 4 occurred at nucleic acid 14410-14591 nt (Figure 5G). The area of recombinant gene was shown in the NADC30 (GenBank: JN654459.1) genome (Figure 5B). A phylogenetic analysis was performed on the entire genome and each recombinant region. The genome-wide phylogenetic tree showed that SDHY-DZ037 belonged to Sublineage 1.8 (NADC30-like) (Figure 5C), and all the recombinant regions belonged to Sublineage 1.5 (NADC34-like).
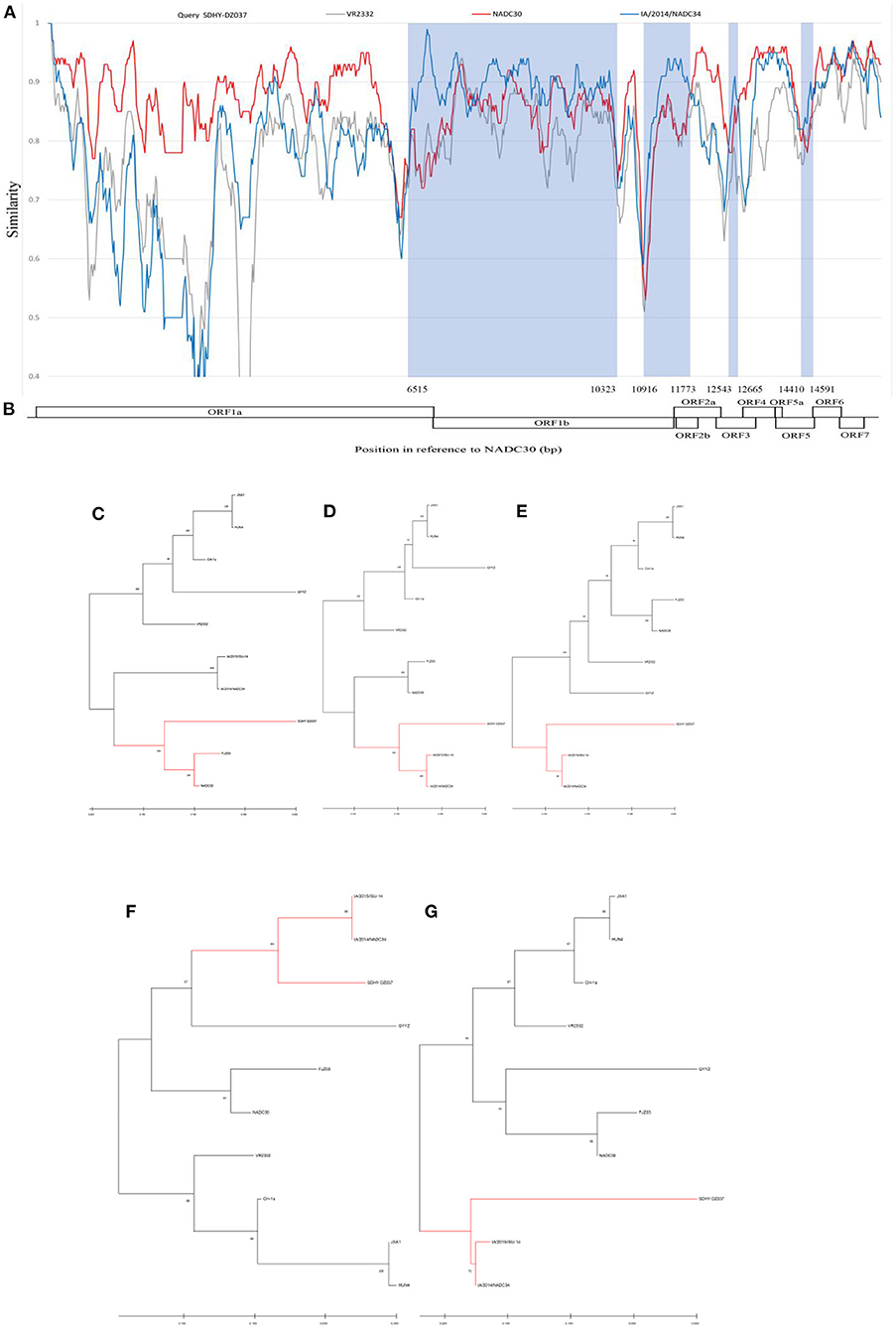
Figure 5. In this study, the PRRSV representative strains and SDHY-DZ037 were selected for genome-wide recombination analysis. Similarity maps were generated by the SimPlot v3.5 software (A). The gene region corresponding to the SDHY-DZ037 recombination was shown with NADC30 (GenBank: JN654459.1) (B). Phylogenetic tree based on full-length genome sequence (C). Phylogenetic tree of the nucleotide recombination region at positions 6,515–10,323 (D). Phylogenetic tree of the nucleotide recombination region at positions 10,916–11,773 (E). Phylogenetic tree of the nucleotide recombination region at positions 12,543–12,665 (F). Phylogenetic tree of the nucleotide recombination region at positions 14,410–14,591 (G). Evolutionary analysis of maximum likelihood performed in MEGA X. Multiple sequence alignments generated using Clustal W.
Since the emergence of PRRSV in China, PRRS has caused serious harm to the country's swine industry. The positive rate of PRRSV in Shandong Province was 9.58% in 2018–2019, and primarily Lineage 1 was predominant (43). To understand the prevalence of PRRSV in Shandong Province in 2020–2021, a total of 1,344 samples (mainly blood and nasal swabs from suspected sick pigs) were collected from pig farms of various sizes. The samples came from a wide array of sources, mainly Tai'an and the surrounding cities, and has covered pig farms of different scales in all cities of Shandong Province. A total of 342 samples were positive for PRRSV, giving a positive rate of 25.44%, showing that PRRSV is still one of the main pathogens threatening the swine industry in Shandong. Overall, 76 strains were isolated, of which 49 strains were NADC30-like, accounting for 64.47% of the isolated viruses. Fifteen strains were NADC34-like, representing 19.74% of the isolated viruses; 11 strains were CH-1a and HP-PRRSV-like, accounting for 14.47% of the isolated viruses; and 1 strain was VR2332-like and accounted for 1.32% of the isolated viruses. The NADC30-like strains remain the dominant strains in Shandong. This corresponds to the previous study (43).
The NADC34-like strain was first discovered in Liaoning in 2017 (18). To date, the prevalence of the NADC34-like strain has been reported in at least 10 provinces, including Heilongjiang, Liaoning, Henan, Hebei, Fujian, Jiangsu, Sichuan, Tianjin, and Shandong (24, 44). However, the prevalence of NADC34-like in Shandong Province is still unknown. We conducted an epidemiological survey of PRRSV in Shandong in 2020–2021. The study found that in 2020, NADC30-like strains accounted for 75.00% of PRRSV-positive samples and NADC34-like strains accounted for 15%. In 2021, NADC30-like strains accounted for 52.78% of PRRSV-positive samples and NADC34-like strains accounted for 25%. This indicates that the NADC34 strain is starting to show an epidemic trend in Shandong. One assumes that it could become the dominant strain in the years to come.
Analysis comparing the deduced amino acid loci of the ORF5 gene in the isolated strains and in some reference strains showed that the mutation Q13 → R13 was present in all viruses of Lineage 8, while the mutation K151 → R151 was absent in two viruses at amino acid position 151. SDHY-TA059 still had K at amino acid position 151, and SDHY-JN107 had the mutation K151 → Q151. It appears that some of the strong strains may have lost some of their strong virulence characteristics during the evolutionary process, thereby weakening their virulence (37). Two strains of Sublineage 1.8 showed mutations of Q13 → R13, as opposed to other strains. In addition, two strains of Sublineage 1.8, one strain of Lineage 5, and one strain of Sublineage 1.5 had mutations of K151 → R151. This indicated that certain mutations occur on specific sites during the continuous mutation of viruses. This may make it more virulent than other viruses of the same type. Mutations in the neutralizing epitope region may cause the virus to be insensitive to the neutralizing effect of vaccination and thus avoid it. In this study, 23 isolated strains showed mutations in the neutralizing region of the epitope, accounting for 30.26% of the isolates. These strains may be insensitive to vaccine immunization, which may be an important reason for the poor efficacy of the current PRRSV vaccine.
The S137 → A137 mutation occurred in three of the isolated strains, two of which were NADC34-like and one VR2332-like. It has been reported that amino acid 137 of GP5 was serine for wild strains and alanine for VR2332, MLV, and RespPRRS/Repro vaccine strains (38, 40, 41). Some scholars believe that A137 can be used to distinguish Lineage 5 and Lineage 1 (45). Among all the strains isolated in this study, the amino acid 137 of GP5 was mutated to alanine in three strains. The other strains were serine. The three mutated viruses are SDHY-HZ015, SDHY-DZ037, and SDHY-TA058. In this study, a genome-wide recombination of SDHY-DZ037 was analyzed by RDP4 and SimPlot software. The results showed that SDHY-DZ037 was found to be a recombinant strain of NADC30-like and NADC34-like strains but without recombinant VR2332. We speculated that perhaps amino acid position 137 of GP5 had mutated during the PRRSV epidemic. Therefore, perhaps A137 was no longer a specific amino acid site within VR2332, MLV, and RespPRRS/Repro vaccine strains, and A137 may no longer be suitable for distinguishing Lineage 1 and Lineage 5.
In summary, the NADC30-like strain remained the dominant epidemic strain of PRRSV in Shandong, China, in 2020–2021. It should be noted that NADC34-like strains have also been quite prevalent in this area. Some isolates had mutations in amino acids 13 and 151 of ORF5 and in the region of the neutralizing epitope (aa36–aa52), which may alter the virulence and increase the virus's ability to escape immunity.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s)can be found below: https://www.ncbi.nlm.nih.gov/genbank/, ON890168-ON890242 and OP168793.
The animal study was reviewed and approved by the Animal Ethics Committee of Shandong Agricultural University.
Writing–original draft preparation was done by PL and YS. Writing–review and editing was done by BL, ML, and FM. Software was done by PL and YL. Investigation was done by JL, YZ, and TW. Funding acquisition was done by SL. All authors have read and agreed to the published version of the manuscript.
This study was funded by the Shandong Provincial Agricultural Major Application Technology Innovation Project (Establishment and Demonstration of Healthy Pig Breeding Technology Integration and Product Supply Chain Traceability System).
Author FM was employed by Huayun (Shandong) Inspection and Quarantine Service Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2022.987667/full#supplementary-material
1. Albina E. Epidemiology of porcine reproductive and respiratory syndrome (PRRS): an overview. Vet Microbiol. (1997) 55:309–16. doi: 10.1016/S0378-1135(96)01322-3
2. Guo A, Wu G, Gong W, Luo X, Zheng H, Jia H, et al. Outbreaks of highly pathogenic porcine reproductive and respiratory syndrome in Jiangxi province, China. Ir Vet J. (2012) 65:1–4. doi: 10.1186/2046-0481-65-14
3. Wang G, Yu Y, Cai X, Zhou EM, Zimmerman JJ. Effects of PRRSV infection on the porcine thymus. Trends Microbiol. (2020) 28:212–23. doi: 10.1016/j.tim.2019.10.009
4. Zimmerman JJ, Dee SA, Holtkamp DJ, Murtaugh MP, Stadejek T, Stevenson GW, et al. Porcine reproductive and respiratory syndrome viruses (porcine arteriviruses). In: Diseases of Swine, 11th edn (Hoboken, NJ: John Wiley) (2019). p. 685–708.
5. Guo B Q, Chen Z S, Liu W X, Cui Y Z. Isolation and identification of porcine reproductive and respiratory syndrome (PRRS) virus from aborted fetuses suspected of PRRS. Chin J Anim Poult Infect Dis. (1996) 2:117–24.
6. Stadejek T, Stankevicius A, Murtaugh MP, Oleksiewicz MB. Molecular evolution of PRRSV in Europe: current state of play. Vet Microbiol. (2013) 165:21–8. doi: 10.1016/j.vetmic.2013.02.029
7. Canelli E, Catella A, Borghetti P, Ferrari L, Ogno G, De Angelis E, et al. Phenotypic characterization of a highly pathogenic Italian porcine reproductive and respiratory syndrome virus (PRRSV) type 1 subtype 1 isolate in experimentally infected pigs. Vet Microbiol. (2017) 210:124–33. doi: 10.1016/j.vetmic.2017.09.002
8. Guo B, Lager KM, Henningson JN, Miller LC, Schlink SN, Kappes MA, et al. Experimental infection of United States swine with a Chinese highly pathogenic strain of porcine reproductive and respiratory syndrome virus. Virology. (2013) 435:372–84. doi: 10.1016/j.virol.2012.09.013
9. Li Y, Wang X, Bo K, Wang X, Tang B, Yang B, et al. Emergence of a highly pathogenic porcine reproductive and respiratory syndrome virus in the Mid-Eastern region of China. Vet J. (2007) 174:577–84. doi: 10.1016/j.tvjl.2007.07.032
10. Tian K, Yu X, Zhao T, Feng Y, Cao Z, Wang C, et al. Emergence of fatal PRRSV variants: unparalleled outbreaks of atypical PRRS in China and molecular dissection of the unique hallmark. PLoS ONE. (2007) 2:e526. doi: 10.1371/journal.pone.0000526
11. Tong GZ, Zhou YJ, Hao XF, Tian ZJ, An TQ, Qiu HJ. Highly pathogenic porcine reproductive and respiratory syndrome, China. Emerg Infect Dis. (2007) 13:1434–6. doi: 10.3201/eid1309.070399
12. Wu J, Li J, Tian F, Ren S, Yu M, Chen J, et al. Genetic variation and pathogenicity of highly virulent porcine reproductive and respiratory syndrome virus emerging in China. Arch Virol. (2009) 154:1589–97. doi: 10.1007/s00705-009-0478-6
13. Zhou F, Chang HT, Zhao J, Chen L, Wang XW, Liu HY, et al. Identification and molecular epidemiology of porcine reproductive and respiratory syndrome virus prevailing in Henan province from 2012 to 2013. Chin J Vet Sci. (2014) 9:1398–404. doi: 10.16303/j.cnki.1005-4545.2014.09.002
14. Li C, Zhuang J, Wang J, Han L, Sun Z, Xiao Y, et al. Outbreak Investigation of NADC30-Like PRRSV in South-East China. Transbound Emerg Dis. (2016) 63:474–9. doi: 10.1111/tbed.12530
15. Zhou L, Wang Z, Ding Y, Ge X, Guo X, Yang H. NADC30-like strain of porcine reproductive and respiratory syndrome virus, China. Emerg Infect Dis. (2015) 21:2256–7. doi: 10.3201/eid2112.150360
16. Guo Z, Chen XX Li X, Qiao S, Deng R, Zhang G. Prevalence and genetic characteristics of porcine reproductive and respiratory syndrome virus in central China during 2016-2017: NADC30-like PRRSVs are predominant. Microb Pathog. (2019) 135:103657. doi: 10.1016/j.micpath.2019.103657
17. Zhou L, Kang R, Yu J, Xie B, Chen C, Li X, et al. Genetic characterization and pathogenicity of a novel recombined porcine reproductive and respiratory syndrome virus 2 among Nadc30-Like, Jxa1-Like, and Mlv-like strains. Viruses. (2018) 10:551. doi: 10.3390/v10100551
18. Zhang HL, Zhang WL, Xiang LR, Leng CL, Tian ZJ, Tang YD, et al. Emergence of novel porcine reproductive and respiratory syndrome viruses (ORF5 RFLP 1-7-4 viruses) in China. Vet Microbiol. (2018) 222:105–8. doi: 10.1016/j.vetmic.2018.06.017
19. Song S, Xu H, Zhao J, Leng C, Xiang L, Li C, et al. Pathogenicity of NADC34-like PRRSV HLJDZD32-1901 isolated in China. Vet Microbiol. (2020) 246:108727. doi: 10.1016/j.vetmic.2020.108727
20. Xie CZ, Ha Z, Zhang H, Zhang Y, Xie YB, Zhang H, et al. Pathogenicity of porcine reproductive and respiratory syndrome virus (ORF5 RFLP 1-7-4 viruses) in China [published online ahead of print, 2020 Mar 18]. Transbound Emerg Dis. (2020) 10.1111/tbed.13549. doi: 10.1111/tbed.13549
21. Li C, Gong B, Sun Q, Xu H, Zhao J, Xiang L, et al. First Detection of NADC34-like PRRSV as a Main Epidemic Strain on a Large Farm in China. Pathogens. (2021) 11:32. doi: 10.3390/pathogens11010032
22. Xu H, Li C, Li W, Zhao J, Gong B, Sun Q, et al. Novel characteristics of Chinese NADC34-like PRRSV during 2020-2021 [published online ahead of print, 2022 Feb 19]. Transbound Emerg Dis. (2022) 10.1111/tbed.14485. doi: 10.1111/tbed.14485
23. Zhao HZ, Wang FX, Han XY, Guo H, Liu CY, Hou LN, et al. Recent advances in the study of NADC34-like porcine reproductive and respiratory syndrome virus in China. Front Microbiol. (2022) 13:950402. doi: 10.3389/fmicb.2022.950402
24. Yuan L, Zhu Z, Fan J, Liu P, Li Y, Li Q, et al. High pathogenicity of a Chinese NADC34-like PRRSV on Pigs [published online ahead of print, 2022 Jun 29]. Microbiol Spectr. (2022) e0154122. doi: 10.1128/spectrum.01541-22
25. Snijder EJ, Meulenberg JJ. The molecular biology of arteriviruses. J Gen Virol. (1998) 79:961–79. doi: 10.1099/0022-1317-79-5-961
26. Dokland T. The structural biology of PRRSV. Virus Res. (2010) 154:86–97. doi: 10.1016/j.virusres.2010.07.029
27. Jiang Y, Li G, Yu L, Li L, Zhang Y, Zhou Y, et al. Genetic diversity of Porcine Reproductive and Respiratory Syndrome Virus (PRRSV) From 1996 to 2017 in China. Front Microbiol. (2020) 11:618. doi: 10.3389/fmicb.2020.00618
28. Martin D, Rybicki E. RDP detection of recombination amongst aligned sequences. Bioinformatics. (2000) 16:562–3. doi: 10.1093/bioinformatics/16.6.562
29. Padidam M, Sawyer S, Fauquet CM. Possible emergence of new geminiviruses by frequent recombination. Virology. (1999) 265:218–25. doi: 10.1006/viro.1999.0056
30. Martin DP, Posada D, Crandall KA, Williamson C. A modified bootscan algorithm for automated identification of recombinant sequences and recombination breakpoints. AIDS Res Hum Retroviruses. (2005) 21:98–102. doi: 10.1089/aid.2005.21.98
31. Posada D, Crandall KA. Evaluation of methods for detecting recombination from DNA sequences: computer simulations. Proc Natl Acad Sci USA. (2001) 98:13757–62. doi: 10.1073/pnas.241370698
32. Gibbs MJ, Armstrong JS, Gibbs AJ. Sister-scanning: a Monte Carlo procedure for assessing signals in recombinant sequences. Bioinformatics. (2000) 16:573–82. doi: 10.1093/bioinformatics/16.7.573
33. Smith JM. Analyzing the mosaic structure of genes. J Mol Evol. (1992) 34:126–9. doi: 10.1007/BF00182389
34. Lam HM, Ratmann O, Boni MF. Improved algorithmic complexity for the 3SEQ recombination detection algorithm. Mol Biol Evol. (2018) 35:247–51. doi: 10.1093/molbev/msx263
35. Boni MF, de Jong MD, van Doorn HR, Holmes EC. Guidelines for identifying homologous recombination events in influenza A virus. PLoS ONE. (2010) 5:e10434. doi: 10.1371/journal.pone.0010434
36. Lole KS, Bollinger RC, Paranjape RS, Gadkari D, Kulkarni SS, Novak NG, et al. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol. (1999) 73:152–60. doi: 10.1128/JVI.73.1.152-160.1999
37. Fang K, Liu S, Li X, Chen H, Qian P. Epidemiological and genetic characteristics of porcine reproductive and respiratory syndrome virus in South China between 2017 and 2021. Front Vet Sci. (2022) 9:853044. doi: 10.3389/fvets.2022.853044
38. Yin B, Qi S, Sha W, Qin H, Liu L, Yun J, Zhu J, Li G, Sun D. Molecular characterization of the Nsp2 and ORF5 (ORF5a) genes of PRRSV strains in nine provinces of china during 2016-2018. Front Vet Sci. (2021) 8:605832. doi: 10.3389/fvets.2021.605832
39. Do DT, Park C, Choi K, Jeong J, Nguyen TT, Le DT, et al. Nucleotide sequence analysis of Vietnamese highly pathogenic porcine reproductive and respiratory syndrome virus from 2013 to 2014 based on the NSP2 and ORF5 coding regions. Arch Virol. (2016) 161:669–75. doi: 10.1007/s00705-015-2699-1
40. Wesley RD, Mengeling WL, Lager KM, Clouser DF, Landgraf JG, Frey ML. Differentiation of a porcine reproductive and respiratory syndrome virus vaccine strain from North American field strains by restriction fragment length polymorphism analysis of ORF 5. J Vet Diagn Invest. (1998) 10:140–4. doi: 10.1177/104063879801000204
41. Zhou L, Chen S, Zhang J, Zeng J, Guo X, Ge X, et al. Molecular variation analysis of porcine reproductive and respiratory syndrome virus in China. Virus Res. (2009) 145:97–105. doi: 10.1016/j.virusres.2009.06.014
42. Zhang HL. Characteristic Analysis of Various Subgenotype PRRSV and Construction of DIVA Vaccine Strain. (Doctoral Dissertation, Chinese Academy of Agricultural Sciences). Available online at: https://kns.cnki.net/KCMS/detail/detail.aspx?dbname=CDFDLAST2022&filename=1019108143.nh (accessed August 15, 2019).
43. Xue RX, Sun SF Li YG, Wang ML, Wang GS Li YJ, et al. Diversity of porcine reproductive and respiratory syndrome virus in Shandong, China. Acta Virol. (2021) 65:303–6. doi: 10.4149/av_2021_305
44. Sun YF, Liu Y, Yang J, Li WZ, Yu XX, Wang SY, Li LA, Yu H. Recombination between NADC34-like and QYYZ-like strain of porcine reproductive and respiratory syndrome virus with high pathogenicity for piglets in China [published online ahead of print, 2022 Feb 4]. Transbound Emerg Dis. (2022) 10.1111/tbed.14471. doi: 10.1111/tbed.14471
Keywords: PRRSV, GP5, epidemiology, genetic evolutionary analysis, prevention and control
Citation: Li P, Shen Y, Wang T, Li J, Li Y, Zhao Y, Liu S, Li B, Liu M and Meng F (2022) Epidemiological survey of PRRS and genetic variation analysis of the ORF5 gene in Shandong Province, 2020–2021. Front. Vet. Sci. 9:987667. doi: 10.3389/fvets.2022.987667
Received: 06 July 2022; Accepted: 25 August 2022;
Published: 15 September 2022.
Edited by:
Lian-Feng LI, Harbin Veterinary Research Institute (CAAS), ChinaReviewed by:
Hongliang Zhang, Harbin Veterinary Research Institute (CAAS), ChinaCopyright © 2022 Li, Shen, Wang, Li, Li, Zhao, Liu, Li, Liu and Meng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Baoquan Li, bGlicUBzZGF1LmVkdS5jbg==; Fanliang Meng, MTg3NTQ4NzU5MjFAMTYzLmNvbQ==; Mengda Liu, bGl1bWVuZ2RhQGNhaGVjLmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.