
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci. , 28 September 2022
Sec. Zoological Medicine
Volume 9 - 2022 | https://doi.org/10.3389/fvets.2022.986332
 Fuzhong Wang1,2
Fuzhong Wang1,2 Lei Wang2
Lei Wang2 Haojie Ge1
Haojie Ge1 Xiaobo Wang1
Xiaobo Wang1 Yaxin Guo2
Yaxin Guo2 Zhengzhong Xu2
Zhengzhong Xu2 Shizhong Geng1
Shizhong Geng1 Xin'an Jiao2*
Xin'an Jiao2* Xiang Chen1*
Xiang Chen1*Salmonella enterica serovar Dublin (S. Dublin) is an important zoonotic pathogen with high invasiveness. In the prevention and control of the Salmonella epidemic, the live attenuated vaccine plays a very important role. To prevent and control the epidemic of S. Dublin in cattle farms, the development of more effective vaccines is necessary. In this study, we constructed two gene deletion mutants, Sdu189ΔspiC and Sdu189ΔspiCΔaroA, with the parental strain S. Dublin Sdu189. The immunogenicity and protective efficacy were evaluated in the mice model. First, both mutant strains were much less virulent than the parental strain, as determined by the 50% lethal dose (LD50) for specific pathogen-free (SPF) 6-week-old female BALB/c mice. Second, the specific IgG antibody level and the expression level of cytokine TNF-α, IFN-γ, IL-4, and IL-18 were increased significantly in the vaccinated mice compared to the control group. In addition, the deletion strains were cleared rapidly from organs of immunized mice within 14 d after immunization, while the parental strain could still be detected in the spleen and liver after 21 d of infection. Compared with the parental strain infected group, no obvious lesions were detected in the liver, spleen, and cecum of the deletion strain vaccinated groups of mice. Immunization with Sdu189ΔspiC and Sdu189ΔspiCΔaroA both provided 100% protection against subsequent challenges with the wild-type Sdu189 strain. These results demonstrated that these two deletion strains showed the potential as live attenuated vaccines against S. Dublin infection. The present study established a foundation for screening a suitable live attenuated Salmonella vaccine.
Salmonella enterica serovar Dublin (S. Dublin) is a serotype highly adapted to cattle, which causes enteritis and systemic diseases in bovine hosts (1). The clinical manifestations are watery or bloody diarrhea, accompanied by fever, depression, loss of appetite, dehydration, and bacteremia (2). It has also been reported that S. Dublin can cause respiratory diseases in calves of 10–30 d old, with an incidence rate of over 20% and a 75% mortality (3). S. Dublin is widely prevalent globally which causes enormous economic losses and severely constrains the development of the breeding industry (4). The transmission of S. Dublin to humans is usually associated with direct contact with infected animals or the consumption of contaminated raw meat, water, milk, and dairy products, which can lead to invasive infection and death in humans susceptible to diseases such as weakness and chronic infection (5). The consumption of beef and dried beef contaminated with S. Dublin has also been identified as the major cause of S. Dublin infection in humans (6). The traditional microbiological analysis found that S. Dublin has the highest isolated rate in raw milk and dairy products, meanwhile, S. Dublin in human blood and feces had the highest isolated rate (7).
To prevent and control the infection of this intestinal pathogen, multiple strategies need to be implemented, such as avoiding the introduction of pathogenic bacteria, controlling the transmission of pathogenic bacteria in livestock, vaccinating, and using antibiotics (8, 9). However, to avoid the introduction of pathogenic bacteria and control their transmission, farms need to spend huge human and material resources. Previous studies have also confirmed that the use of antibiotics can achieve specific therapeutic effects, which directly leads to the production of multiple drug-resistant strains and drug residues in meat (10). An American study reported that in the 8 years from 2005 to 2013 alone, the multi-drug resistance rate of S. Dublin increased to 55%, while the drug resistance rate of other serotypes was only 12% (11). Consequently, it will be increasingly difficult to treat S. Dublin infection with antibiotics, and vaccination will probably become the most important means to control S. Dublin infection in cattle. At present, three main types of vaccines are commercially available, including inactivated vaccine, live attenuated vaccine, and genetically engineered live attenuated vaccine (12). However, an inactivated vaccine has the disadvantages of low immunogenicity, weak protective immunity, short duration of effect, and requires multiple vaccinations (13). The drug resistance of live attenuated vaccines caused by the problem of preparation technology cannot be ignored. The genetically engineered live attenuated vaccine has shown sufficient advantages (14). In 1984, Smith and others developed a live vaccine SL1438 of the S. Dublin aro− gene deletion strain, which has been approved by the U.S. Department of Agriculture (USDA), but its clinical immune efficacy and protective efficacy were still insufficient (15, 16). The Salmonella vaccine used in the modern cattle industry decreased morbidity and mortality, and inhibit the bacterial excretion and persistent infection of S. Dublin. However, the use of these vaccines did not completely prevent the spread of S. Dublin in livestock (17). The ideal vaccine should be able to induce both humoral and cellular immune responses, with high safety and cross-protection against other serotypes of Salmonella (18). Therefore, it is necessary to develop new vaccines to prevent and control the infection and transmission of S. Dublin in cattle.
SpiC is an effector protein encoded by SPI-2, which is secreted by the Salmonella type III secretion system (T3SS) and injected into host cells (19, 20). It has been demonstrated that deleting spiC gene significantly reduces the virulence of Salmonella in mouse and chicken models (21). The spiC gene deletion strain of Salmonella Pullorum constructed by genetic engineering has been evaluated as a candidate vaccine for S. Pullorum (22, 23). In the breeding process, some nutrients can be added to improve the health of animals. Based on this, we consider whether we can adjust the state of vaccine strains in the body through feeding links, to achieve a better immune effect. The aroA gene is part of the shikimic acid pathway, which directly links glycolysis with the synthesis of aromatic amino acids (24). Mammals cannot synthesize aromatic amino acids by themselves, so it was difficult for S. Dublin to reproduce in mammalian tissues or survive in the environment without the aroA gene (25). aroA gene deletion is most commonly used as a metabolic mutation to reduce the virulence of Salmonella and other bacteria. Previous studies have confirmed that aroA-deficient S. Dublin strain is highly attenuated and considered to be a suitable carrier system (26). Therefore, aroA and spiC were selected as the target genes for the construction of candidate vaccines.
In this study, Sdu189ΔspiC and Sdu189ΔspiCΔaroA were constructed by employing the suicide plasmid pDM4. Bacterial virulence, host clearance, immune responses, pathological observation, and protective efficacy of these two deletion strains in mice model were analyzed to evaluate the potential of Sdu189ΔspiC and Sdu189ΔspiCΔaroA as live attenuated vaccines against S. Dublin infection.
Sdu189 is a clinical strain obtained in 2017 from human anal swabs in Jiangsu Province, China. The spiC and aroA gene deletion mutant strains were constructed by using the suicide vector pDM4 based on homologous recombination, as previously described (27, 28). The open reading frame (ORF) of the targeted genes was completely deleted and confirmed by PCR analysis and sequencing. All of these strains were cultured in Luria-Bertani (LB) agar medium, LB broth, and Xylose Lysine Tergitol-4 (XLT4) agar at 37°C.
To evaluate the effect of deleted genes on the biological properties of S. Dublin mutants, biochemical tests were performed using the API 20E identification kit (BioMérieux, France) according to the manufacturer's protocol. The growth characteristics of Sdu189ΔspiC, Sdu189ΔspiCΔaroA, and Sdu189 were determined by measuring the optical density (OD600) of each strain cultured in 15 ml of LB broth at 37°C with shaking at 180 rpm. The OD600 was monitored every hour for 10 h as previously described.
Specific pathogen-free (SPF) 6-week-old female BALB/c mice were obtained from Beijing Weitonglihua Laboratory Animal Technology Co., Ltd. (Beijing, China). All mice were confirmed as free from Salmonella infection by both bacteriological examination and serum detection. Each group of mice was raised in separate rearing isolators and supplied with commercial feed and drinking water. The food and water for mice were tested to be Salmonella negative. All of the animal experiments and management procedures were undertaken with the permission of the Animal Welfare and Ethics Committees of Yangzhou University (IACUC license number: YZUDWLL-201811-001) and complied with the guidelines of the institutional administrative committee and the ethics committee for laboratory animals.
To evaluate the virulence of Sdu189ΔspiC and Sdu189ΔspiCΔaroA, 96 mice were divided into 16 groups (n = 6) randomly. Mice were administered 100 μl of Sdu189ΔspiC, Sdu189ΔspiCΔaroA (1 × 1010, 1 × 109, 1 × 108, 1 × 107, or 1 × 106 CFU/ml), or Sdu189 (1 × 107, 1 × 106, 1 × 105, 1 × 104, or 1 × 103 CFU/ml) in phosphate-buffered saline (PBS) by intramuscular injection. Control mice received 100 μl of PBS in the same way. The clinical symptoms and death of mice were recorded every day within 3 weeks after infection, and the LD50 was calculated by the Reed-Muench method (29).
A total of 63 6-week-old mice were randomly divided into three groups (n = 21). Each group was immunized intramuscularly with 1 × 105 CFU of Sdu189, Sdu189ΔspiC, or Sdu189ΔspiCΔaroA. Sections of the liver, spleen, ileum, and cecum were aseptically collected at 1, 3, 5, 7, 9, 14, and 21 d post-immunization. The organs were weighed and suspended in 1 ml of PBS for homogenization. The homogenates were diluted serially and subsequently inoculated on XLT4 agar plates for bacterial recovery at 37°C. After overnight cultivation, the bacterial number was counted and reported as log10 CFU/g.
The humoral immune response was evaluated by measuring specific IgG titers by indirect enzyme-linked immunosorbent assay (ELISA), using the heat-killed Sdu189 strain as the coating antigen. Mice blood samples were collected at 7, 14, and 21 d post-immunization, stored at 4°C for 4 h, and then centrifuged at 3,500 rpm for 10 min. Serum samples were diluted continuously as primary antibodies. Horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG (1:10,000 dilution, Sigma-Aldrich) was used as the secondary antibody. The HRP activity was determined using 3,30,5,50-tetramethylbenzidine (TMB, Sigma-Aldrich), and the OD450 value was determined with an ELISA reader (BioTek, USA) to detect the level of humoral immunity.
To evaluate the T-cell immune response and inflammatory response induced by the vaccine strains, the mRNA expression of cytokines IFN-γ, TNF-α, IL-4, and IL-18 was measured by qRT-PCR analysis with the Synthetic dye method (30, 31). GAPDH was used as an endogenous control gene. Gene-specific primers used in this analysis were listed in Table 1. Total RNA was extracted from spleen samples collected at 7, 14, and 21 d post-vaccination by using an RNA Plus Mini kit (Qiagen, Germany) according to the manufacturer's instructions. The RNA was reverse transcribed into cDNA by using PrimeScript RT Reagent Kit (TaKaRa) according to the manufacturer's instructions and then subjected to qRT-PCR analysis. The comparative threshold cycle (2–ΔΔC(T) method) was used to calculate relative concentrations. All qRT-PCR reactions were performed in triplicates and repeated three times.
To evaluate the protective efficacy of these two vaccine candidates, 36 6-week-old mice were divided into three groups (n = 12) randomly. Mice were immunized intramuscularly with 1 × 107 CFU of Sdu189ΔspiC or Sdu189ΔspiCΔaroA in 100 μl of PBS, and the control mice received 100 μl PBS. Two weeks post-infection, mice were immunized again with the same mutants using the same route and dose. Two weeks after the secondary immunization, each group of mice was challenged intramuscularly with 2 × 108 CFU of Sdu189. The number of surviving mice was recorded, and clinical symptoms were observed daily for 3 weeks. Samples of the liver, spleen, and cecum of each group were collected and fixed in 10% neutral-buffered formalin at 1, 3, 5, 7, 14, and 21 d post-immunization. The fixed samples were dehydrated, embedded in paraffin wax, sectioned to 5 μm, and stained with hematoxylin and eosin.
All data were described as mean ± standard error of the mean (SEM) unless otherwise specified and were analyzed using GraphPad Prism version 7.0. A p-value of <0.05 was considered statistically significant.
The API 20E identification kit was used to test the physiological and biochemical characteristics of deletion strains and parental strains. The results showed that the deletion of spiC and aroA genes markedly influenced bacterial metabolism (unpublished), and the knockout of the aroA gene in Sdu189 inhibited the ability to produce H2S. In addition, the growth curve of Sdu189ΔspiC was similar to the parental strain, reaching the logarithmic growth period at about 2 h, and gradually entering the stable period after 7 h (Figure 1). However, as we predicted, the growth curve of Sdu189ΔspiCΔaroA was different, reaching the logarithmic growth period at about 5 h, and gradually entering the stable period after 9 h.
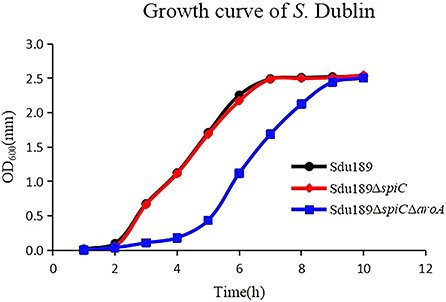
Figure 1. Growth curves of Sdu189, Sdu189ΔspiC, and Sdu189ΔspiCΔaroA. Bacteria were grown in LB broth at 37°C for 10 h with agitation, and the OD600 values of triplicate cultures in LB medium were determined in 1-h intervals.
The virulence of Sdu189, Sdu189ΔspiC, and Sdu189ΔspiCΔaroA was evaluated in 6-week-old BALB/c mice with intramuscular injection. As shown in Table 2, the LD50 of Sdu189ΔspiC and Sdu189ΔspiCΔaroA strains were 586-fold and 1,073-fold higher than that of parental strain Sdu189, respectively. In addition, there was no significant difference in the weight of decimals between the two groups compared with the PBS group (Figure 2).
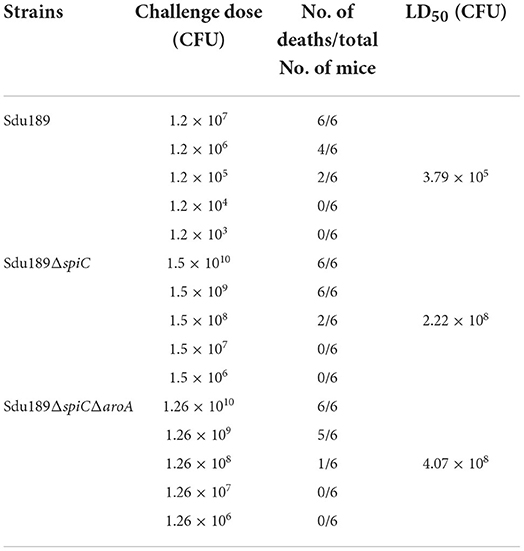
Table 2. The LD50 of S. Dublin Sdu189, Sdu189ΔspiC and Sdu189ΔspiCΔaroA, in 6-week-old mice after intramuscular immunization.
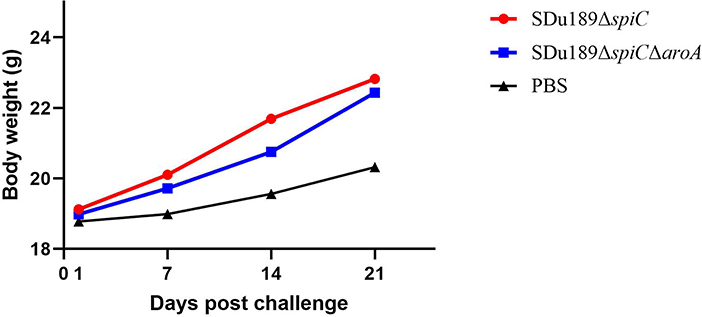
Figure 2. The body weight of mice after immunization. Groups of 6-week-old BALB/c mice were intramuscularly immunized with Sdu189ΔspiC and Sdu189ΔspiCΔaroA, and the control group received 100 μl PBS. The body weights of the experimental mice were determined at 1, 7, 14, and 21 days post-immunization. Data are presented as mean ± SEM.
The bacterial number was calculated in the liver, spleen, ileum, and cecum of immunized mice. All tissue samples from the negative control group were negative for S. Dublin. As shown in Figure 3, Sdu189, Sdu189ΔspiC, and Sdu189ΔspiCΔaroA colonization reached the highest level at 1 d post-immunization, thereafter it decreased gradually in the cecum. The bacterial colonization reached the highest level at 3 d post-immunization in the spleen and ileum, while the peak colonization of S. Dublin was found at 5 d post-immunization in the liver.
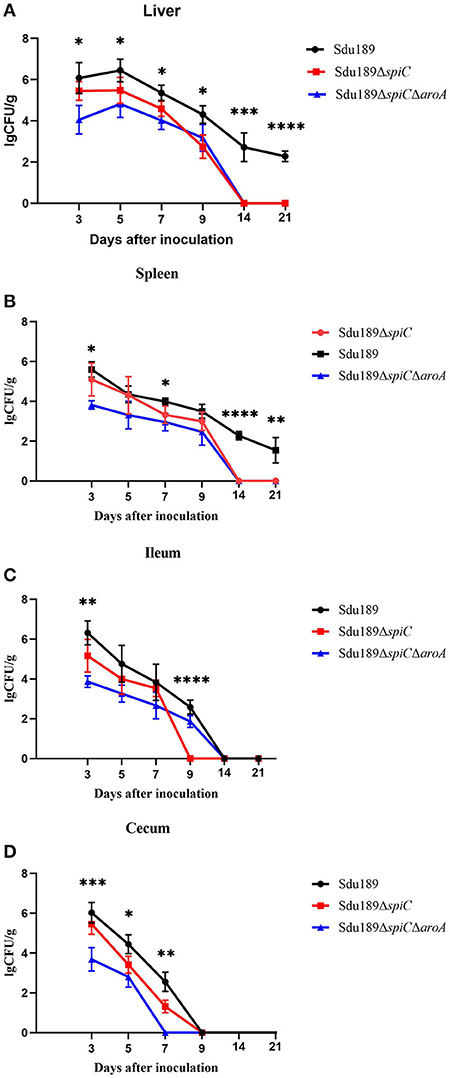
Figure 3. Bacterial colonization in tissues and organs of immunized mice. Mice were immunized with parental strain Sdu189, vaccine candidates Sdu189ΔspiC and Sdu189ΔspiCΔaroA. Bacterial colonization in the liver (A), spleen (B), ileum (C), and cecum (D) of the immunized mice. *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001 compared with the bacterial colonization number of control group mice by one-way ANOVA followed by Bonferroni's multiple comparison test. Data are presented as mean ± SEM of log10 CFU/g.
The viable counts in the liver, spleen, ileum, and cecum from the Sdu189ΔspiCΔaroA immunization group were lower than those from mice inoculated with Sdu189 at all tested time points. Overall, the bacterial clearance in Sdu189ΔspiC immunization group was also enhanced compared with the Sdu189 immunization group. None of S. Dublin was detected in the spleen and liver from mice immunized with Sdu189ΔspiC or Sdu189ΔspiCΔaroA at 14 d post-immunization. While, spleen and liver samples from mice immunized with Sdu189 were still positive at 21 d post-immunization (Figures 3A,B). In addition, all ileum samples from Sdu189ΔspiC immunization group were negative for bacterial recovery at 9 d post-immunization. And none of S. Dublin was detected in cecum samples from Sdu189ΔspiCΔaroA immunization group at 5 d post-immunization. These results indicated that the colonization ability of both Sdu189ΔspiC and Sdu189ΔspiCΔaroA was decreased compared with wild strain Sdu189.
Subsequently, we analyzed the histopathological changes or lesions of the organs from mice infected with S. Dublin. The organs from the Sdu189 infected group of mice showed obvious splenic sinus expanding, the number of macrophages increasing, and infiltration of heterophilic granulocytes at 3 d post-challenge (Supplementary Figure S1). Furthermore, the spleen and liver from the mice of the Sdu189 infection group displayed severe focal necrosis, and massive macrophage infiltration at 7–21 d post-challenge (Supplementary Figure S2). No obvious histopathological changes were found in the liver, spleen, and cecum among mice originating from immunization groups. These data suggested that the vaccine candidate strains showed a great safety profile in the host.
To evaluate the humoral immune response induced by two deletion strains, the serum IgG level was measured by indirect ELISA. As compared to the control group, the serum S. Dublin antigen-specific IgG levels were significantly increased in Sdu189ΔspiC and Sdu189ΔspiCΔaroA immunization group at 14, 21, and 28 d post-immunization. The serum IgG levels of Sdu189ΔspiCΔaroA immunized group of mice were significantly higher than those of mice immunized with Sdu189ΔspiC (Figure 4A). These results demonstrated that both Sdu189ΔspiC and Sdu189ΔspiCΔaroA were able to strongly elicit humoral immune responses in mice, and the specific serum IgG level induced by Sdu189ΔspiCΔaroA was stronger. To determine the expression of cytokines mRNA in the spleen of immunized mice, qRT-PCR analysis was performed using GAPDH as the internal control. As shown in Figure 4, the mRNA levels of IFN-γ and IL-18 in the spleen of both Sdu189ΔspiC and Sdu189ΔspiCΔaroA immunized mice were strongly induced at 1 d post-immunization and remained high at 28 d post-immunization compared with that in the spleen of control mice. And the expression level of IFN-γ induced by Sdu189ΔspiCΔaroA was higher than Sdu189ΔspiC at 14, 21, and 28 d post-immunization. Sdu189ΔspiCΔaroA induced a remarkably higher level of IL-18 expression in mice spleen compared with Sdu189ΔspiC.
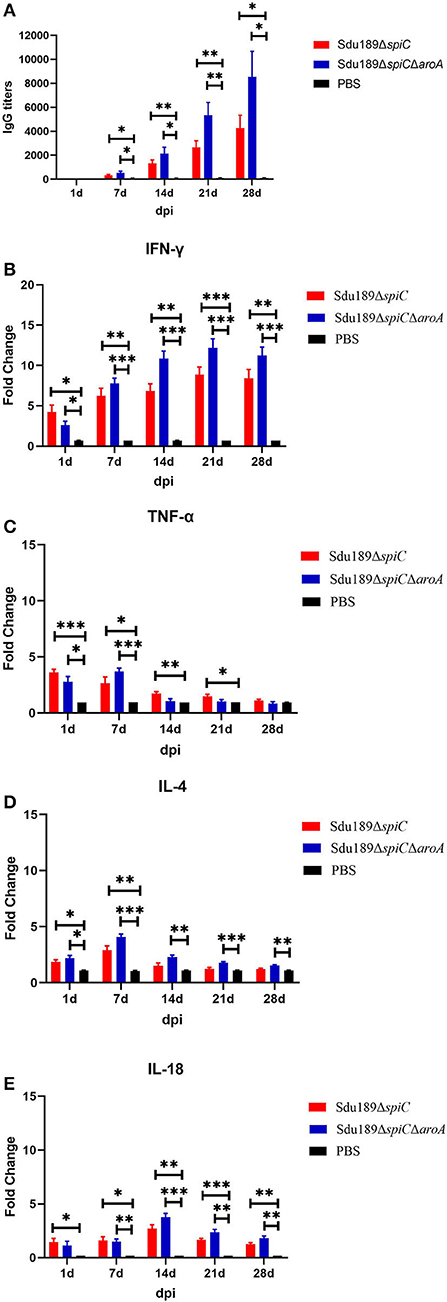
Figure 4. Humoral and cellular immune responses induced by vaccine candidates. The 6-week-old mice were immunized with 1×105 CFU of Sdu189ΔspiC or Sdu189ΔspiCΔaroA, and the serum S. Dublin-specific IgG antibody titer at 1, 7, 14, 21, and 28 d post-immunization were detected by Elisa (A). The total RNA was extracted from spleens of mice vaccinated by mutants at 1, 7, 14, 21, and 28 d post-immunization and subsequently subjected to the qRT-PCR analysis for detecting the expression levels of IFN-γ (B), TNF-α (C), IL-4 (D), and IL-18 (E). *p < 0.05, **p < 0.01, and ***p < 0.001 compared with control group by one-way ANOVA followed by Bonferroni's multiple comparison test. Data are presented as mean ± SEM.
The mRNA expression level of TNF-α as well as that of IL-4 was significantly upregulated in the spleen of mice at 1 d after immunization with Sdu189ΔspiC or Sdu189ΔspiCΔaroA, (Figure 4D) peaked at 7 d post-immunization and declined to basal level at 21 d post-immunization. These results suggested that Sdu189ΔspiC and Sdu189ΔspiCΔaroA tended to induce a Th2 immune response in mice at the early stage of immunization, while these two mutants tended to induce a Th1 immune response in mice after 7 d post-immunization. On balance, immunization with Sdu189ΔspiC or Sdu189ΔspiCΔaroA could induce strong cellular immune responses in the mice model.
The mice vaccinated intramuscularly with Sdu189ΔspiC or Sdu189ΔspiCΔaroA were challenged with the parental strain Sdu189 at 14 d post-secondary immunization (Figure 5). The percentage of surviving mice at 14 d post-challenge was shown in Figure 6. None of the immunized mice died in Sdu189ΔspiC and Sdu189ΔspiCΔaroA vaccinated groups. Whereas, all 12 mice in the control group died within 14 d post-challenge. The Sdu189ΔspiC and Sdu189ΔspiCΔaroA both conferred 100% protection against a lethal S. Dublin challenge in mice after second intramuscular injections. Whereas, 100% mortality was observed in the control group. Furthermore, vaccinated mice did not show clinical symptoms after the challenge. Compared with the non-immunized group, the increase in body weight of the mice immunized with the deletion strain was higher than that of the non-immunized group (Figure 2).
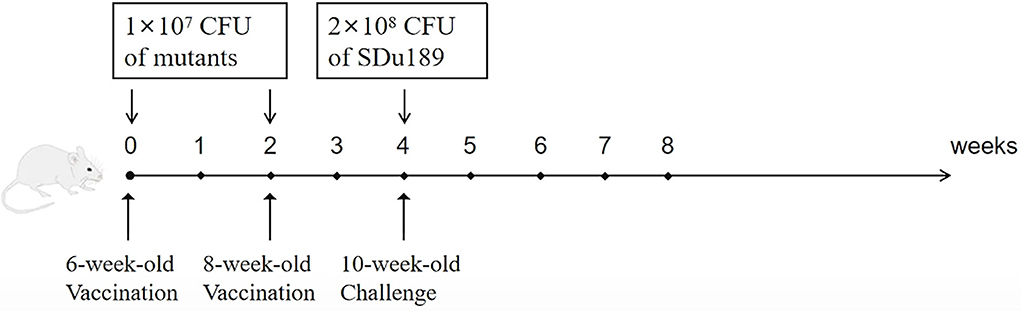
Figure 5. BALB/c mouse immunization protocol and protective efficacy of Sdu189 ΔspiC or Sdu189ΔspiCΔaroA. Mice immunized with different doses of Sdu189 ΔspiC, Sdu189ΔspiCΔaroA, or PBS were intramuscularly challenged at 2 weeks post-immunization with virulent S. Dublin strain Sdu189, and mortality was recorded.
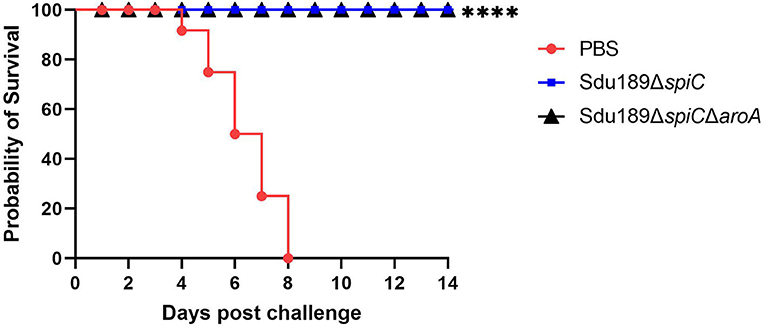
Figure 6. Protective efficiency of two vaccine candidates in balb/c mouse. * p < 0.05, ** p < 0.01, *** p < 0.001 and **** p < 0.0001 compared with the survival number of mice of control group mice by one-way ANOVA followed by Bonferroni's multiple comparison test.
S. Dublin is a strongly adapted serotype Salmonella in cattle, with high invasiveness and usually causes serious clinical symptoms and high mortality (32, 33). At present, the epidemiological investigation of S. Dublin has not been able to explore an effective and reliable control scheme. More research is needed to prevent the infection and transmission of S. Dublin in cattle farms. Vaccination is considered the best long-term way to control salmonellosis. Until now, two main types of vaccines have been developed, namely inactivated vaccine and live attenuated vaccine, for the control of S. Dublin in the cattle industry (34). Inactivated vaccines can induce the production of specific IgG antibodies to kill extracellular bacteria, but these antibodies are difficult to eliminate intracellular S. Dublin, which can be achieved by the live attenuated Salmonella vaccine (35, 36). Live attenuated Salmonella vaccine should be attenuated and safe to the host, with satisfied immunogenicity and great immune protection efficacy (37). The protective effect of the live attenuated vaccine was higher than that of an inactivated vaccine, the major reason was that the immune response induced by the live vaccine was stronger than inactivated vaccine (38, 39). In this study, spiC deletion strain and spiC/aroA double gene deletion strain were constructed based on the clinically isolated S. Dublin strain Sdu189. As we expected, the growth of Sdu189ΔspiCΔaroA was inhibited, and it would still reach a stable growth period similar to the parent plant in the later stage (Figure 1). It shows that when Sdu189ΔspiCΔaroA was used as a vaccine strain, some measures could be explored in the feeding link to regulate the state of bacteria. As shown in Table 2, the virulence of these two mutant strains was significantly attenuated compared with the parental strain, which meets the requirements of reducing bacterial virulence in the development of live attenuated vaccines. In addition, the double deletion mutant strains showed greater decreases in virulence, suggesting that Sdu189ΔspiCΔaroA was safer than Sdu189ΔspiC.
The primary importance for the host immunized with live vaccines is safety. Previous studies showed that abscess of the inoculation site and transient diarrhea may be induced by some live Salmonella vaccines (40). In contrast, the immunization with Sdu189ΔspiC or Sdu189ΔspiCΔaroA did not affect the body weight of mice, and no obvious clinical signs were found in immunized mice. A previous study showed that the immunization with a nuoG gene deletion Salmonella Gallinarum strain significantly reduce the mortality of chicken, whereas necrotic lesions were detected in the spleen and liver of immunized animals (41). Another live Salmonella Enteritidis vaccine was also reported to induce pathological changes in the tissues of the immunized chickens (42). Notably, no obvious pathological lesions were found in the spleen, liver, cecum, and ileum of mice immunized with Sdu189ΔspiC or Sdu189ΔspiCΔaroA, indicating that both single deletion and double deletion live vaccines strains were safe. In addition, vaccine strain persistence and clearance in the immunized host was also an important index to evaluate the safety of vaccines. Residual vaccine strains in immunized animals may result in the contamination of slaughterhouses and meat. A live Salmonella vaccine strain was able to preserve in the organs of immunized animals for a long time, which led to systemic disease (43). Chickens were still positive for Salmonella in cloacal swabs at 10 weeks post-immunization with another live Salmonella vaccine (44). While nine kinds of Salmonella vaccines were reported to be removed quickly from the spleen of immunized animals within 14 d after immunization (45). Similar results were found in this study, both Sdu189ΔspiC and Sdu189ΔspiCΔaroA can be cleared rapidly from organs of immunized mice with 14 d post-inoculation. It is undeniable that the clearance rate of vaccinated animals is an important aspect of vaccine efficacy evaluation. In addition, images of animal organs (including gross and histology) attacked by parental strains are also strong evidence for the protective efficacy of vaccines. As a preliminary inquiry, this study lacks the above evaluation data. It is necessary to use younger mice and calves for evaluation in the next evaluation to improve the value of the vaccine.
After Salmonella infection, the host humoral immunity and cellular immunity are indispensable in the process of eliminating pathogens. One field experiment showed that the low morbidity of salmonellosis in vaccinated flocks was largely due to the strong IgG antibody production (23). The S. Pullorum spiC mutant strain had been demonstrated to induce high levels of IgG antibody in chickens inoculated intramuscularly (46). Similarly, the S. Dublin-specific serum IgG levels in mice immunized with Sdu189ΔspiC or Sdu189ΔspiCΔaroA were significantly higher than those of the control mice (p < 0.01). The early humoral immune response induced by S. Dublin infection was usually not enough to effectively eliminate pathogens. The cellular immune responses also play an essential role in immune defense against S. Dublin infection (47). Intracellular Salmonella inhibits cell division and toxicity to escape the host defense mechanism and persists in the host cell. In the acute infection stage of S. Dublin, the host Th1 and Th2 immune responses were quickly induced to repress the replication of intracellular bacteria and subsequently remove these invading bacteria (48). However, in the second stage of persistent infection, the reduction of Th1 type response destroys the balance between Th1 and Th2. A new balance between host and persistent Salmonella infection has been established (11). Previous studies showed that elimination of primary Salmonella infection was associated with secretion of IFN-γ (49). High IFN-γ expression mediated by Th1 immune response was reported to be essential for clearance of Salmonella (50), which may explain why Sdu189ΔspiC and Sdu189ΔspiCΔaroA were cleared rapidly in organs of immunized mice. All of the mice immunized with these two deletion strains represented high expression levels of IFN-γ in the spleen at all tested time points after immunization, which was consistent with the findings that the live Salmonella vaccine was able to induce a Th1 immune response after immunization (51).
Immune protective efficacy is the most important indicator to evaluate the potential of attenuated strain as a live vaccine against pathogens infection. The live vaccines were known to have a higher protective effect than the killed vaccines. A previous study showed that a live strain only offers 80% protection in immunized animals against S. Enteritidis infection (52). While a live spiC and crp deletion mutant of S. Gallinarum vaccine strain provides 100% protection in chickens after challenge (53). Similar results were found in this study, Sdu189ΔspiC and Sdu189ΔspiCΔaroA both conferred 100% protection against a lethal S. Dublin challenge in mice, and no obvious lesions were found in all of the tested organs of immunized mice. These data indicated the potential of Sdu189ΔspiC and Sdu189ΔspiCΔaroA as effective vaccines against S. Dublin infection. We know that S. Dublin is more pathogenic to calves. The 6-week-old mice used in this experiment are not representative of calves. For further evaluation, animals younger than the 6-week-old mice need to be selected for immunization to make up for this deficiency.
In summary, the live attenuated S. Dublin strains Sdu189ΔspiC and Sdu189ΔspiCΔaroA were able to induce both strong humoral and cellular immune responses in mice, accompanied by high safety. These two vaccine strains were also conferred high protection against the lethal S. Dublin challenge in mice. Thus, the Sdu189ΔspiC and Sdu189ΔspiCΔaroA strains have the potential of being a safe, immunogenic, and effective vaccine against S. Dublin infection.
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author/s.
The animal study was reviewed and approved by Animal Welfare and Ethics Committees of Yangzhou University.
FW and LW contributed to the conception and design of the study. HG organized the database. FW and XW performed the statistical analysis. FW wrote the first draft of the manuscript. YG, ZX, SG, and XC wrote sections of the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
This work received financial support from the National Key Research and Development Program of China (2021YFD1800403), the Jiangsu Agricultural Science and Technology Independent Innovation Funds (CX(21)1004), the Science and Technology Program of Jiangsu (BE2021331), and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2022.986332/full#supplementary-material
1. Yim L, Sasias S, Martinez A, Betancor L, Estevez V, Scavone P, et al. Repression of flagella is a common trait in field isolates of Salmonella enterica serovar Dublin and is associated with invasive human infections. Infect Immun. (2014) 82:1465–76. doi: 10.1128/IAI.01336-13
2. Matthews TD, Schmieder R, Silva GGZ, Busch J, Cassman N, Dutilh BE, et al. Genomic comparison of the closely-related Salmonella enterica serovars enteritidis, dublin and gallinarum. PLoS ONE. (2015) 10:E0126883. doi: 10.1371/journal.pone.0126883
3. Uzzau S, Brown DJ, Wallis T, Rubino S, Leori G, Bernard S, et al. Host adapted serotypes of Salmonella enterica. Epidemiol Infect. (2000) 125:229–55. doi: 10.1017/S0950268899004379
4. Pezoa D, Blondel CJ, Silva CA, Yang HJ, Andrews-Polymenis H, Santiviago CA, et al. Only one of the two type VI secretion systems encoded in the Salmonella enterica serotype Dublin genome is involved in colonization of the avian and murine hosts. Vet Res. (2014) 45:2. doi: 10.1186/1297-9716-45-2
5. Funke S, Anker JC, Ethelberg S. Salmonella Dublin patients in Denmark and their distance to cattle farms. Infect Dis. (2017) 49:208–16. doi: 10.1080/23744235.2016.1249024
6. Eyler AB, M'ikanatha NM, Xiaoli L, Dudley EG. Whole-genome sequencing reveals resistome of highly drug-resistant retail meat and human Salmonella Dublin. Zoonoses Public Health. (2020) 67:251–62. doi: 10.1111/zph.12680
7. Ung A, Baidjoe AY, Van Cauteren D, Fawal N, Fabre L, Guerrisi C. Disentangling a complex nationwide Salmonella Dublin outbreak associated with raw-milk cheese consumption, France, 2015 to 2016. Euro Surveill. (2019) 24:1700703. doi: 10.2807/1560-7917.ES.2019.24.3.1700703
8. Jiang Z, Paudyal N, Xu Y, Deng T, Li F, Pan H, et al. Antibiotic resistance profiles of Salmonella recovered from finishing pigs and slaughter facilities in Henan, China. Front Microbiol. (2019) 10:1513. doi: 10.3389/fmicb.2019.01513
9. Paudyal N, Pan H, Liao X, Zhang X, Li X, Fang W, et al. A meta-analysis of major foodborne pathogens in Chinese food commodities between 2006 and 2016. Foodborne Pathog Dis. (2018) 15:187–97. doi: 10.1089/fpd.2017.2417
10. Pan H, Paudyal N, Li X, Fang W, Yue M. Multiple food-animal-borne route in transmission of antibiotic-resistant Salmonella Newport to humans. Front Microbiol. (2018) 9:23. doi: 10.3389/fmicb.2018.00023
11. Harvey RR, Friedman CR, Crim SM, Judd M, Barrett KA, Tolar B, et al. Epidemiology of Salmonella enterica serotype Dublin Infections among Humans, United States, 1968-2013. Emerg Infect Dis. (2017) 23:1493–501. doi: 10.3201/eid2309.170136
12. Smith BP, Reina-Guerra M, Stocker BA, Hoiseth SK, Johnson E. Aromaticdependent Salmonella Dublin as a parenteral modified live vaccine for calves. Am J Vet Res. (1984) 45:2231–4.
13. Barrow PA. Salmonella infections: immune and non-immune protection with vaccines. Avian Pathol. (2007) 36:1–13. doi: 10.1080/03079450601113167
14. Jiao H, Yang H, Zhao D, Chen J, Zhang Q, Liang J, et al. Design and immune characterization of a novel Neisseria gonorrhoeae DNA vaccine using bacterial ghosts as vector and adjuvant. Vaccine. (2018) 36:4532–9. doi: 10.1016/j.vaccine.2018.06.006
15. Hoiseth SK, Stocker BA. Aromatic-dependent Salmonella Typhimurium are non-virulent and effective as live vaccines. Nature. (1981) 291:238–9. doi: 10.1038/291238a0
16. Smith BP, Reina-Guerra M, Stocker BA, Hoiseth SK, Johnson E. Aromatic-dependent Salmonella Dublin as a parenteral modified live vaccine for calves. Am J Vet Res. (1984) 45:2231–5.
17. Selim SA, Cullor JS, Smith BP, Blanchard P, Farver TB, Hoffman R, et al. The effect of Escherichia coli J5 and modified live Salmonella Dublin vaccines in artificially reared neonatal calves. Vaccine. (1995) 13:381–90. doi: 10.1016/0264-410X(95)98262-9
18. Jung B, Park S, Kim E, Yoon H, Hahn TW. Salmonella Typhimurium lacking phoBR as a live vaccine candidate against poultry infection. Vet Microbiol. (2022) 266:109342. doi: 10.1016/j.vetmic.2022.109342
19. Wang Y, Liu G, Zhang J, Gu D, Hu M, Zhang Y, et al. WbaP is required for swarm motility and intramacrophage multiplication of Salmonella Enteritidis spiC mutant by glucose use ability. Microbiol Res. (2021) 245:126686. doi: 10.1016/j.micres.2020.126686
20. Bao H, Wang S, Zhao JH, Liu SL. Salmonella secretion systems: Differential roles in pathogen-host interactions. Microbiol Res. (2020) 241:126591. doi: 10.1016/j.micres.2020.126591
21. Zhang JF, Shang K, Wei B, Lee YJ, Park JY, Jang HK, et al. Evaluation of safety and protective efficacy of a waaJ and spiC double deletion Korean epidemic strain of Salmonella enterica serovar Gallinarum. Front Vet Sci. (2021) 8:756123. doi: 10.3389/fvets.2021.756123
22. Wang Y, Huang C, Tang J, Liu G, Hu M, Kang X, et al. Salmonella Pullorum spiC mutant is a desirable LASV candidate with proper virulence, high immune protection and easy-to-use oral administration. Vaccine. (2021) 39:1383–91. doi: 10.1016/j.vaccine.2021.01.059
23. Geng S, Jiao X, Barrow P, Pan Z, Chen X. Virulence determinants of Salmonella Gallinarum biovar Pullorum identified by PCR signature-tagged mutagenesis and the spiC mutant as a candidate live attenuated vaccine. Vet Microbiol. (2014) 168:388–94. doi: 10.1016/j.vetmic.2013.11.024
24. Bentley R. The shikimate pathway—a metabolic tree with many branches. Crit Rev Biochem Mol Biol. (1990) 25:307–84. doi: 10.3109/10409239009090615
25. McKelvie ND, Khan SA, Karavolos MH, Bulmer DM, Lee JJ, DeMarco R, et al. Genetic detoxification of an aroA Salmonella enterica serovar Typhimurium vaccine strain does not compromise protection against virulent Salmonella and enhances the immune responses towards a protective malarial antigen. FEMS Immunol Med Microbiol. (2008) 52:237–46. doi: 10.1111/j.1574-695X.2007.00368.x
26. Lindberg AA, Segall T, Weintraub A, Stocker BA. Antibody response and protection against challenge in mice vaccinated intraperitoneally with a live aroA O4-O9 hybrid Salmonella Dublin strain. Infect Immun. (1993) 61:1211–21. doi: 10.1128/iai.61.4.1211-1221.1993
27. Ge H, Zhang K, Gu D, Chen X, Wang X, Li G et al. The rfbN gene of Salmonella Typhimurium mediates phage adsorption by modulating biosynthesis of lipopolysaccharide. Microbiol Res. (2021) 250:126803. doi: 10.1016/j.micres.2021.126803
28. Zhang Y, Liu H, Gu D, Lu X, Zhou X, Xia X. Transcriptomic analysis of PhoR reveals its role in regulation of swarming motility and T3SS expression in Vibrio parahaemolyticus. Microbiol Res. (2020) 235:126448. doi: 10.1016/j.micres.2020.126448
29. Guo R, Jiao Y, Li Z, Zhu S, Fei X, Geng S, et al. Safety, protective immunity, and diva capability of a rough mutant Salmonella Pullorum vaccine candidate in broilers. Front Microbiol. (2017) 8:547. doi: 10.3389/fmicb.2017.00547
30. Vinod N, Noh HB, Oh S, Ji S, Park HJ, Lee KS, et al. A Salmonella Typhimurium ghost vaccine induces cytokine expression in vitro and immune responses in vivo and protects rats against homologous and heterologous challenges. PLoS ONE. (2017) 12:e0185488. doi: 10.1371/journal.pone.0185488
31. Li N, Ansari AR, Sun Z, Huang H, Cui L, Hu Y, et al. Toll like receptor 4 signaling pathway participated in Salmonella lipopolysaccharide-induced spleen injury in young chicks. Microb Pathog. (2017) 112:288–94. doi: 10.1016/j.micpath.2017.10.004
32. Qu F, Cui EB, Fan ZP, Zhang WJ, Bao CM, Chen SM, et al. First report of liver abscess caused by Salmonella enterica serovar Dublin. J Clin Microbiol. (2013) 51:3140–2. doi: 10.1128/JCM.01034-13
33. Mohammed M, Le Hello S, Leekitcharoenphon P, Hendriksen R. The invasome of Salmonella Dublin as revealed by whole genome sequencing. BMC Infect Dis. (2017) 17:544. doi: 10.1186/s12879-017-2628-x
34. Mohler VL, Heithoff DM, Mahan MJ, Walker KH, Hornitzky MA, Shum LW, et al. Cross-protective immunity in calves conferred by a DNA adenine methylase deficient Salmonella enterica serovar Typhimurium vaccine. Vaccine. (2006) 26:1751–8. doi: 10.1016/j.vaccine.2008.01.018
35. Selke M, Meens J, Springer S, Frank R, Gerlach GF. Immunization of pigs to prevent disease in humans: Construction and protective efficacy of a Salmonella enterica serovar Typhimurium live negative-marker vaccine. Infect Immun. (2007) 75:2476–83. doi: 10.1128/IAI.01908-06
36. Mizuno T, Ploeg R, Trott D. A new concept to stimulate mucosal as well as systemic immunity by parenteral vaccination as applied to the development of a live attenuated Salmonella serovar Dublin vaccine. Vet Res. (2007) 38:773–94. doi: 10.1051/vetres:2007031
37. Groves PJ, Sharpe SM, Muir WI, Pavic A, Cox JM. Live and inactivated vaccine regimens against caecal Salmonella Typhimurium colonisation in laying hens. Aust Vet J. (2016) 94:387–93. doi: 10.1111/avj.12490
38. Kong Q, Yang J, Liu Q, Alamuri P, Roland KL, Curtiss R. Effect of deletion of genes involved in lipopolysaccharide core and o-antigen synthesis on virulence and immunogenicity of Salmonella enterica serovar Typhimurium. Infect Immun. (2011) 79:4227–39. doi: 10.1128/IAI.05398-11
39. Nagy G, Palkovics T, Otto A, Kusch H, Kocsis B, Dobrindt U, et al. “Gently rough”: the vaccine potential of a Salmonella enterica regulatory lipopolysaccharide mutant. J Infect Dis. (2008) 198:1699–706. doi: 10.1086/593069
40. Zhang-Barber L, Turner AK, Dougan G, Barrow PA. Protection of chickens against experimental fowl typhoid using a nuoG mutant of Salmonella serotype Gallinarum. Vaccine. (1998) 16:899–903. doi: 10.1016/S0264-410X(97)00300-9
41. Nandre R, Matsuda K, Lee JH. Efficacy for a new live attenuated Salmonella Enteritidis vaccine candidate to reduce internal egg contamination. Zoonoses Public Health. (2014) 61:55–63. doi: 10.1111/zph.12042
42. Silva EN, Snoeyenbos GH, Weinack OM, Smyser CF. Studies on the use of 9R strain of Salmonella Gallinarum as a vaccine in chickens. Avian Dis. (1981) 25:38–52. doi: 10.2307/1589825
43. Varmuzova K, Faldynova M, Elsheimer-Matulova M, Sebkova A, Polansky O, Havlickova H, et al. Immune protection of chickens conferred by a vaccine consisting of attenuated strains of Salmonella Enteritidis, Typhimurium and Infantis. Vet Res. (2016) 47:94. doi: 10.1186/s13567-016-0371-8
44. Pei Y, Parreira VR, Roland KL, Curtiss R III, Prescott JF. Assessment of attenuated Salmonella vaccine strains in controlling experimental Salmonella Typhimurium infection in chickens. Can J Vet Res. (2014) 78:23–30.
45. Betancor L, Schelotto F, Fernandez M, Pereira M, Rial A, Chabalgoity JA. An attenuated Salmonella Enteritidis strain derivative of the main genotype circulating in Uruguay is an effective vaccine for chickens. Vet Microbiol. (2005) 107:81–9. doi: 10.1016/j.vetmic.2005.01.004
46. Chausse AM, Grepinet O, Bottreau E, Robert V, Hennequet-Antier C, Lalmanach AC, et al. Susceptibility to Salmonella carrier-state: a possible Th2 response in susceptible chicks. Vet Immunol Immunopathol. (2014) 159:16–28. doi: 10.1016/j.vetimm.2014.03.001
47. Moore KW, de Waal Malefyt R, Coffman RL, O'Garra A. Interleukin-10 and the interleukin-10 receptor. Annu Rev Immunol. (2001) 19:683–765. doi: 10.1146/annurev.immunol.19.1.683
48. Gal-Mora O. Persistent infection and long-term carriage of typhoidal and nontyphoidal Salmonella. Clin Microbiol Rev. (2019) 32:e00088–18. doi: 10.1128/CMR.00088-18
49. Withanage GS, Wigley P, Kaiser P, Mastroeni P, Brooks H, Powers C, et al. Cytokine and chemokine responses associated with clearance of a primary Salmonella enterica serovar Typhimurium infection in the chicken and in protective immunity to rechallenge. Infect Immun. (2005) 73:5173–82. doi: 10.1128/IAI.73.8.5173-5182.2005
50. Chappell L, Kaiser P, Barrow P, Jones MA, Johnston C, Wigley P. The immunobiology of avian systemic salmonellosis. Vet Immunol Immunopathol. (2009) 128:53–9. doi: 10.1016/j.vetimm.2008.10.295
51. Kamble NM, Jawale CV, Lee JH. Interaction of a live attenuated Salmonella Gallinarum vaccine candidate with chicken bone marrow-derived dendritic cells. Avian Pathol. (2016) 45:235–43. doi: 10.1080/03079457.2016.1144919
52. Si W, Wang X, Liu H, Yu S, Li Z, Chen L, Zhang W, Liu S. Physiology, pathogenicity and immunogenicity of live, attenuated Salmonella enterica serovar Enteritidis mutants in chicks. Microb Pathog. (2015) 83–4:6–11. doi: 10.1016/j.micpath.2015.03.018
Keywords: Salmonella enterica serovar Dublin, spiC, aroA, live attenuated vaccine, immune protection
Citation: Wang F, Wang L, Ge H, Wang X, Guo Y, Xu Z, Geng S, Jiao X and Chen X (2022) Safety of the Salmonella enterica serotype Dublin strain Sdu189-derived live attenuated vaccine—A pilot study. Front. Vet. Sci. 9:986332. doi: 10.3389/fvets.2022.986332
Received: 05 July 2022; Accepted: 30 August 2022;
Published: 28 September 2022.
Edited by:
Min Yue, Zhejiang University, ChinaReviewed by:
Takashi Taguchi, Western University of Health Sciences, United StatesCopyright © 2022 Wang, Wang, Ge, Wang, Guo, Xu, Geng, Jiao and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiang Chen, Y2hlbnhpYW5nQHl6dS5lZHUuY24=; Xin'an Jiao, amlhb0B5enUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.