- Engineering and Technology Research Center of Traditional Chinese Veterinary Medicine of Gansu Province, Lanzhou Institute of Husbandry and Pharmaceutical Sciences, Chinese Academy of Agricultural Sciences, Lanzhou, China
Dampness heat diarrhea (DHD) is one of the most common syndromes of calf diarrhea. Its complex etiology and lack of objective diagnostic criteria bring great challenges to the diagnosis and treatment of this disease. This study aims to screen some prospective diagnostic biomarkers or therapeutic targets for calves with DHD by investigating the differential protein profiles of plasma between DHD calves and clinically healthy calves by mass spectrometry-based proteomic. A total of 120 DHD calves and 90 clinically healthy calves were divided into two groups randomly, 30 DHD calves and 30 clinically healthy calves in the test group, and 90 DHD calves and 60 clinically healthy calves in the validation group. In the test group, a total of 52 proteins were differentially expressed between calves with DHD and clinically healthy calves, 13 proteins were significantly increased and 39 proteins were significantly decreased. The differentially expressed proteins were associated with the intestinal immune network of IgA production, caffeine metabolism, purine metabolism, and PI3K signaling pathway. In the validation group, 13 proteins were selected from 52 differential expression proteins for parallel reaction monitoring validation to verify their associations with DHD calves. The targeted proteomic results showed that fibronectin precursor (FN1) and apolipoprotein C-IV precursor (APOC4) were significantly associated with DHD in calves, and they were downregulated in sick calves. In conclusion, the differential expression of plasma proteins was associated with DHD pathogenesis in calves, and the FN1 and APOC4 might be the potential clinical biomarkers for diagnosis of DHD in calves, and the intestinal immune network of IgA production, caffeine metabolism, purine metabolism, and PI3K signaling pathway are the candidate targets to treat DHD in calves. Our finding provides a reference for further investigating the pathogenesis, developing techniques of diagnosis, and screening treatment drugs for DHD in calves.
Introduction
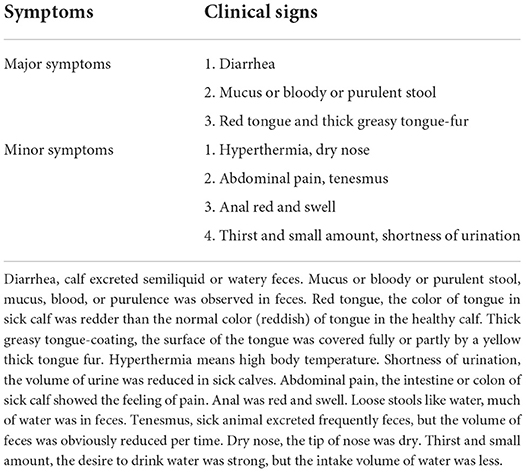
Diarrhea is one of the most common diseases worldwide in the cattle industry and can be caused by various complicated factors (1, 2). Especially, the morbidity and mortality of neonatal calf with diarrhea are high, which imposes a heavy economic burden on the cattle industry. According to the theory of traditional Chinese veterinary medicine (TCVM), diarrhea can be divided into dampness-heat diarrhea (DHD), dampness-cold diarrhea, spleen deficiency diarrhea, and kidney deficiency diarrhea (3–5). DHD is one of the most common syndromes in Chinese cattle herds, and its main clinical symptoms in calves are hyperthermia, sticky and loose stools with blood, mucus or purulence, red tongue, and thick greasy tongue-coating (3, 6). Although the management, feeding, nutrition, and equipment of cattle farms have been greatly improved, the DHD in calves is difficult to be effectively controlled in clinical practice due to the complicated etiology. Therefore, it is of great clinical significance to study the pathogenesis of calves with DHD from the whole perspective and to find suitable biomarkers for the diagnosis and targets for screening treatment drugs for this disease. However, there is a lack of systematic studies on the pathogenesis of DHD in calves, especially, since there are few of published evidence at the proteomics level. As an important method for screening biomarkers on various diseases, systems biology can help to find suitable and less invasive markers for diagnosis and targets for the treatment of calves with DHD.
With the rapid development of multi-omics analytical techniques, they have been widely used to explore the mechanism of various diseases, including gastrointestinal diseases (7, 8), cardiovascular diseases (9), and cancers (10). Few research on multi-omics techniques focused on calf diarrhea. Recently, metabolomics was used to study the mechanism of diarrhea in calves, and acetylcarnitine, indoxyl sulfate, and oxindole have been recognized to be potential biomarkers for calf diarrhea (11). Additionally, metabolites such as lecithin and choline have been found to affect the progression of DHD by interfering with the metabolism of arachidonic acid, linoleic acid, and glycerophospholipids in calves (12). Previous studies on the structure and function of fecal microflora of calves with DHD indicated that DHD had a significant effect on the intestinal microbial compositions in calves, including significant changes in Escherichia-Shigella, Bacteroides, and Fournierella in sick calves (12). These studies imply that systems biology makes it possible to screen potential pathological targets for calves with DHD.
At present, because there are few studies on the pathogenesis of calves with DHD, veterinarians can only confirm the diagnosis of this disease in calves by clinical manifestations. Biomarkers are objective measurements of biological processes, which can help diagnose DHD in calves more accurately and understand the mechanism of the disease. Therefore, systems biology is used to study calves' DHD, which might offer new practical information for the improvement of treatment strategies and diagnostics. However, there is no report about the systematic study on calves' DHD from the perspective of differential proteins. Blood samples in calves are easy to be obtained and have abundant proteins, which make them more suitable for clinical detection. The data-independent acquisition (DIA) is a high-throughput, high-sensitivity, and accurate approach for the identification of biomarkers in various diseases (13, 14).
This study aimed to identify potential biomarkers from plasma protein associated with DHD of calves. The experiment was conducted in two groups. In the test group, proteomic analysis of plasma was performed using DIA-mass spectrometry, the aim was to discover the proteins with significant changes in calves with DHD. In the validation group, parallel reaction monitoring (PRM) technology was used to validate the differential proteins for improving the reliability of these proteins as markers on calves with DHD. As a result, there were 52 proteins differentially expressed between calves with DHD and clinically healthy calves, 13 proteins were significantly increased and 39 proteins were significantly decreased. FN1 and APOC4 were significantly associated with DHD in calves. The differentially expressed proteins were related to the intestinal immune network of IgA production, caffeine metabolism, purine metabolism, and PI3K signaling pathway.
Materials and methods
Animal recruitment
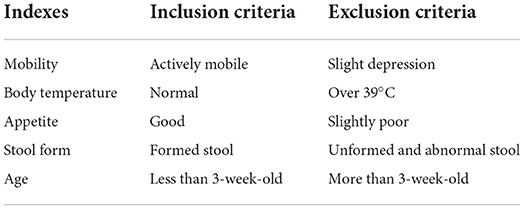
The trial was carried out on a commercial dairy farm in Hui Autonomous Prefecture of Changji, Xinjiang Uygur Autonomous Region, northwest of China. The breed of the calf is Holstein, and all enrolled calves were similar in genetic background and age (10- to 20-day old), and housed individually in a calf shed under the same conditions. The calves were fed colostrum at a dose of 4 kg/calf only on the first day of postpartum, and then fed the pasteurized normal milk at a dose of 6 kg/day, and the calf feed was fed from the third day of postpartum, and the feeding and drink tools were washed and disinfected two times daily. The diagnosis of calves with DHD followed the protocol in our previous study (12), which means that the DHD case was mainly characteristic with diarrhea as well as the symptom of red tongue coated with thick greasy tongue-fur or the symptom of feces with mucus, blood, or purulence. At the same time, the DHD case must show two out of total minor symptoms in Table 1. The inclusion and exclusion criteria of DHD cases are presented in Table 2. All of the clinically healthy calves showed normal body temperature without any abnormal clinical manifestations. The inclusion and exclusion criteria for the clinically health cases are presented in Table 3. The specific clinical syndrome differentiation was carried out by a senior expert and a Ph.D. student, their major is traditional Chinese veterinary medicine and clinical diagnosis. The screening protocol of DHD cases and clinically healthy calves were performed according to veterinary clinical examination such as the diagnosis protocol of DHD and the inclusion and exclusion criteria. The test calves were recruited from early of June to the end of July 2020. The enrolled calves were separated into a test group and a validation group. The test group consisted of 30 calves with DHD and 30 clinically healthy calves. The validation group consisted of 90 calves with DHD and 60 clinically healthy calves. The study protocols were approved by the guidelines of the Laboratory Animal Ethics Commission of the Lanzhou Institute of Husbandry and Pharmaceutical Sciences of CAAS (SYXK [Gan] 2019-0002).
Plasma sample collection
Blood samples were drawn from the jugular vein and collected in tubes containing ethylene diamine tetraacetic acid (EDTA). Approximately 10 ml of venous blood was centrifuged at 5,000 g for 15 min at 4°C to obtain plasma. After all plasma samples of enrolled animals were collected, 30 plasma samples of DHD cases and 30 plasma samples of clinically healthy calves were divided into the test group, and 90 plasma samples of DHD cases and 60 plasma samples of clinically healthy calves were divided into the validation group. Before protein extraction, every 10 individual similar samples with equal volume were mixed to generate one pooled sample. Therefore, 3 DHD and 3 healthy pooled samples were, respectively, subjected to protein extraction for proteomic analysis in the test group, and 9 DHD and 6 healthy pooled samples were, respectively, subjected to protein extraction for proteomic analysis in the validation group. The plasma samples were immediately stored in liquid nitrogen until further protein extraction.
Protein sample preparation
After the pooled plasma samples were thawed at 4°C, which were transferred to a new centrifuge tube containing lysis buffer (2% SDS, 7 M urea, 1 mg/ml protease inhibitor cocktail) and homogenized by an ultrasonic homogenizer (PS-60AL, Leidebang Electronics Co., LTD, Shenzhen, China), proteins were isolated by centrifugation at 21,000 g for 15 min at 4°C (Eppendorf 5427R, Eppendorf, German) and their concentrations were measured using the BCA protein assay kit (Beyotime Biotechnology Co., LTD, Shanghai, China).
A total of 50 μg protein of each pooled sample was suspended in 50 μl deionized water and incubated at 55°C for 1 h after adding 1 μl 1 M dithiotreitol (DTT), then added 5 μl 20 mM iodoacetamide and alkylated at 37°C for 1 h in dark. Subsequently, 300 μl cold acetone was added to precipitate protein at −20°C overnight. The precipitate was washed twice with prechilled acetone and then resuspended in 50 mM ammonium bicarbonate. Finally, the protein was digested with sequence-grade modified trypsin (Promega, Madison, WI) according to a substrate/enzyme ratio of 50:1 (w/w) at 37°C for 16 h.
High-performance liquid chromatography-tandem mass spectrometry (HPLC-MS/MS) analysis
The pooled sample was re-dissolved in 30 μl solvent A (A: 0.1 % formic acid in water) and analyzed by online nanospray LC-MS/MS on an Orbitrap Fusion Lumos coupled to EASY-nLC 1200 system (Thermo Fisher Scientific, MA, USA). Three microliters plasma peptides were loaded onto the nano column (Acclaim PepMap C18, 75 μm × 25 cm) with a 120-min gradient, from 5 to 35% in solvent B (B: 0.1% formic acid in ACN). The column flow rate was maintained at 200 nl/min with a column temperature of 40°C. The electrospray voltage of 2 kV vs. the inlet of the mass spectrometer was used. The mass spectrometer was run under data independent acquisition mode and automatically switched between MS and MS/MS mode. Three replicates were performed for pooled samples in the protein identification. The specific parameters are in the Supplementary material.
Protein functional annotation and enrichment analysis
The biological significance and ontological functions of the differential proteins were analyzed using Gene Ontology (GO) annotation. The pathway of the proteins was annotated using the Kyoto Encyclopedia of Genes and Genomes (KEGG). Pathway-based analysis was used to further understand how different proteins coordinate each other to perform their biological functions. Significant ontological functions and pathways were examined within differentially expressed proteins with P < 0.05. We used the interaction relationships in the STRING protein interaction database (http://string-db.org) to analyze the differential protein interaction network and construct the protein interaction network diagram (15).
Database search and data analysis
Raw Data of DIA were processed and analyzed by Spectronaut X (Biognosys AG, Switzerland) with default settings to generate an initial target list. Spectronaut was set up to search the database of cattle along with the contaminant database assuming trypsin as the digestion enzyme. Carbamidomethyl was specified as the fixed modification for cysteine. Oxidation was specified as the variable modification for methionine. Comparisons between the DHD group and healthy group were performed using Student's t-test. Q-value cutoff on precursor and protein level was applied at 1%. Proteins with significant differences between groups must meet the absolute value of the fold change (FC) >1.5 times, and the Q value obtained by correcting the P-value should be <0.05. The R packages “ggpubr” and “ggthemes” were applied to construct a volcano plot of the different proteins between the DHD and clinically healthy calves.
Parallel reaction monitoring (PRM) and data analysis
To verify the reliability of the differential plasma proteins as diagnostic biomarkers for calf diarrhea, the expression levels of 13 proteins were further quantified by LC-PRM/MS analysis. These proteins were screened according to the unique peptides detected by mass spectrometry (>1), mass charge ratio, and the stability of retention. A total of 38 unique peptides were detected from these 13 differential proteins, which can be used for PRM. The PRM analysis was performed by Applied Protein Technology (Shanghai, China). The plasma proteins were isolated by centrifugation at 21,000 g for 15 min at 4°C, and their concentrations were measured using the BCA protein assay kit (Beyotime Biotechnology Co., LTD, Shanghai, China). Two hundred micrograms of protein of each sample was added to DTT at a final concentration of 100 mM. The samples were boiled for 15 min and cooled at room temperature. Then, each sample was mixed with 200 μl UA buffer (8 M urea, 150 mM Tris-HCl, pH 8.0) and centrifugated in an ultrafiltration tube at 14,000 g for 40 min. Discard the filtrate and add 100 μl IAA buffer [50 mM IAA (Bio-Rad, 163-2109) in UA] to shake at 150 g for 1 min. After incubation at room temperature for 30 min, each sample was centrifuged at 14,000 g for 20 min. Subsequently, each sample was washed with 100 μl UA buffer three times and with 100 μl NH4CO3 buffer (50 mM) for two times, and centrifuged at 14,000 g for 20 min. After adding 40 μl NH4CO3 buffer containing trypsin (1:50; Promega, 317107), the samples were shaken at 150 g for 1 min and incubated for 16 h at 37°C. Centrifugation at 14,000 g for 20 min and another 30 min following adding 40 μl NH4HCO3 buffer was used to collect the peptides. After desalting and lyophilization, the peptides were redissolved in buffer A (0.1% formic acid in water). The peptide content was estimated by UV light spectral density at 280 nm. Tryptic peptides were loaded onto a trap column (100 μm × 50 mm, 5 μm-C1) connected to a home-made tip column (75 μm × 200 mm, 3 μm-C18) for desalting before reversed-phase chromatography on an Easy nLC-1200 system (Thermo Fisher Scientific, MA, USA). One-hour liquid chromatography gradients with acetonitrile ranging from 5 to 35% in 45 min were used. PRM analysis was performed on a Q Exactive HF mass spectrometer (Thermo Scientific, USA). Methods optimized for collision energy, charge state, and retention times on the most significantly regulated peptides were generated experimentally using unique peptides of high intensity and confidence for each target protein. The working parameters of the mass spectrometer were shown in Supplementary material 1. The raw data were analyzed using Skyline (MacCoss Lab, University of Washington), where signal intensities for individual peptide sequences for each of the significantly altered proteins were quantified relative to each sample and normalized to a standard reference.
Results
Identification of differentially expressed proteins in calves with diarrhea
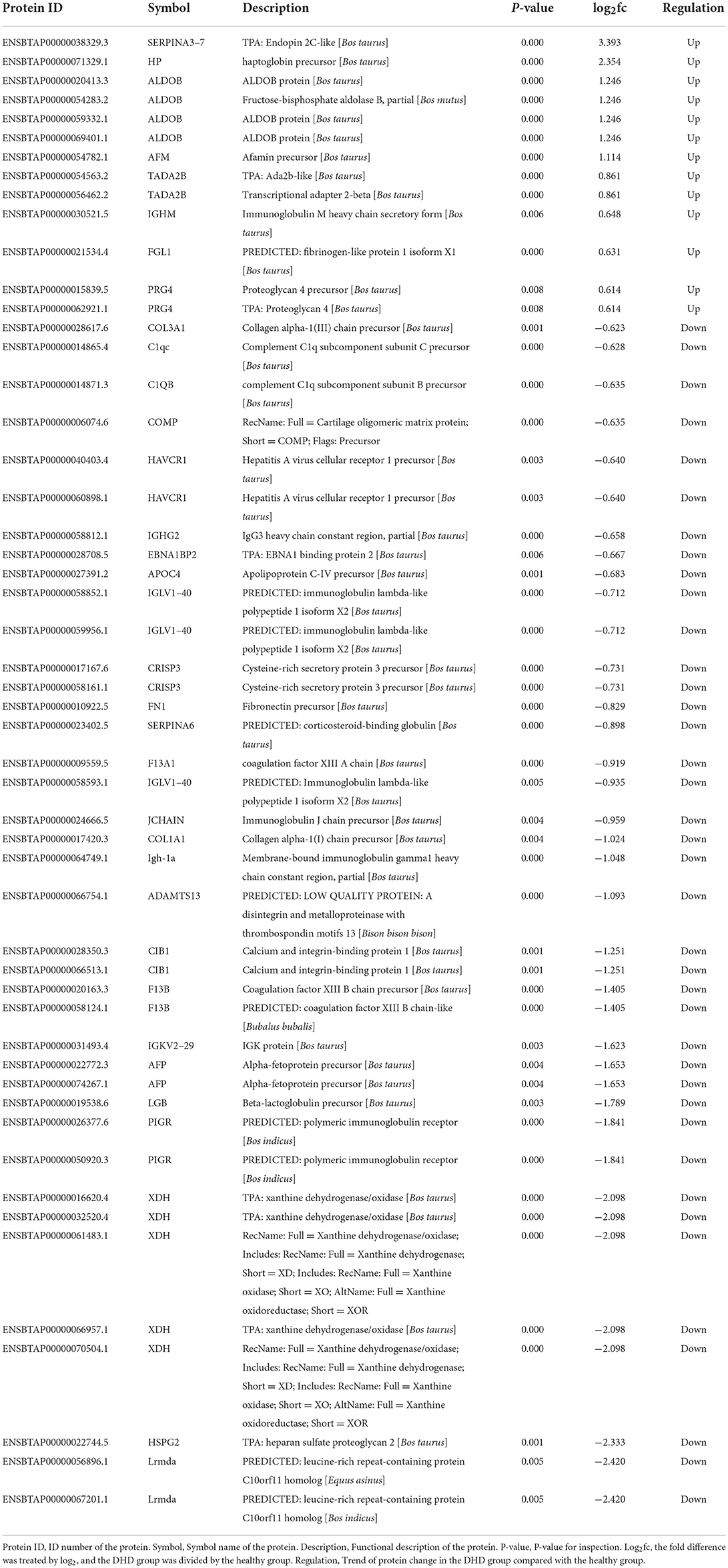
As shown in Figure 1A, the box plot analysis showed that the log10 protein intensity medians of all pooled samples were nearly at the same levels, which suggested that there was no bias toward samples. We used the local normalization method in Pulsar software to normalize the peak intensity of the overall sample spectra. After normalization, a total of 679 proteins were identified in the test group, in that 52 proteins were significantly different between healthy and DHD calves (|FC| > 1.5, P < 0.05; Table 4), and 13 proteins of them were upregulated and 39 proteins of them were downregulated in calves with DHD compared with clinically healthy calves (Figure 1B). The specific information of these significantly different proteins, such as gene names, fold change, and annotation information were listed in Table 2. Volcano plots revealed that haptoglobin precursor (HP) and endopin 2C-like (SERPINA3–7) were significantly increased (the right part of the axis), and xanthine dehydrogenase/oxidase (XDH), heparan sulfate proteoglycan 2 (HSPG2), leucine-rich repeat-containing protein C10orf11 homolog (Lrmda), and polymeric immunoglobulin receptor (PIGR) were significantly decreased (the left part of the axis; Figure 1C).
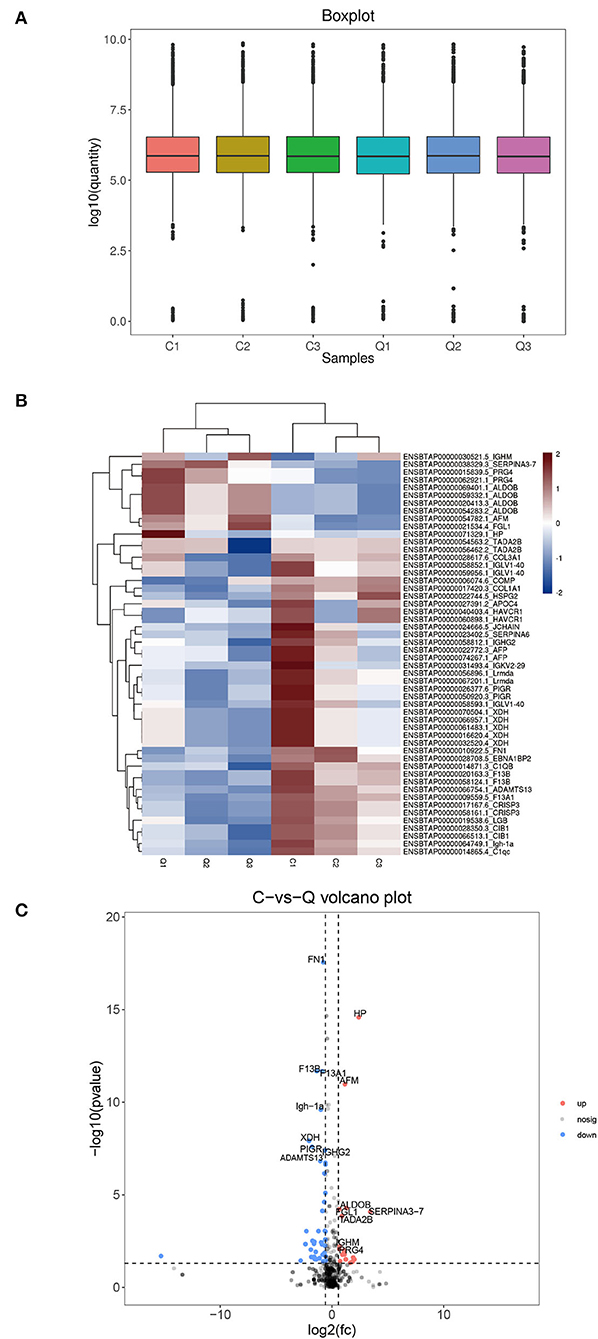
Figure 1. The differential plasma proteins in DHD calves and clinically healthy calves. C indicates clinically healthy calves and Q indicates DHD calves. (A) Normalized box diagram. The distribution of the normalized quantitative value of the peptide showed that the signal intensity of each group was basically the same. (B) Heatmap of the differentially expressed proteins. Plasma samples represent in the columns, and the differentially expressed proteins are delineated in rows. The color of each cell shows that red represents upregulation and blue represents downregulation. (C) Volcano plot of proteins with the most significantly different abundance levels in plasma samples of calves with DHD. Increased protein levels are presented on the positive X-axis and decreased levels are presented on the negative X-axis.
Protein functional annotation and enrichment analysis
GO annotation of the significantly different proteins
Gene Ontology analysis was conducted to explore the ontological functions of 52 differential proteins. Totally, 21 proteins were annotated and classified as participating in certain biological processes, 19 proteins were annotated as related to some molecular function, and the cellular components of 22 proteins were noted. Biological process analysis showed that the majority of the significantly altered proteins were involved in biological regulation (10.36%), cellular process (10.36%), regulation of biological process (9.96%), and single-organism process (9.59%), and the proteins involved in these biological processes included FN1, PIGR, coagulation factor XIII A chain, and complement C1q subcomponent subunit C&B precursor (Figure 2A). Molecular functional analysis demonstrated that the proteins were associated with binding (61.90%), catalytic activity (16.67%), and structural molecule activity (7.14%; Figure 2C). The differential proteins associated with these GO terms include FN1, PIGR, alpha-fetoprotein precursor (AFM), and collagen alpha-1(I) chain precursor (COL1A1). Furthermore, cellular component analysis showed that these significantly altered proteins were located in the organelle (16.78%), extracellular region (14.77%), cell part (11.41%), and organelle part (7.38%; Figure 2B). We also used WoLFPSort to predict the more detailed subcellular localization of these proteins (Figure 2D). They were mainly located in extracellular space (48.08%) and cytoplasm (30.77%), accounting for more than 70% of all significantly different proteins. In addition, these proteins were found in the nucleus, anchored components of the plasma membrane, endomembrane system, and plasma membrane. To further determine which functional categories were closely associated with DHD calves, the significantly increased proteins and significantly decreased proteins were analyzed, respectively, by GO function annotation. For cellular components, the increased proteins were significantly enriched in organelles, whereas decreased proteins were enriched in the extracellular region. The major molecular functions of the increased proteins and the decreased proteins were both bindings. For the biological process, the increased proteins were mainly involved in biological regulation, while decreased proteins participated in the cellular process (Figures 3A,B).
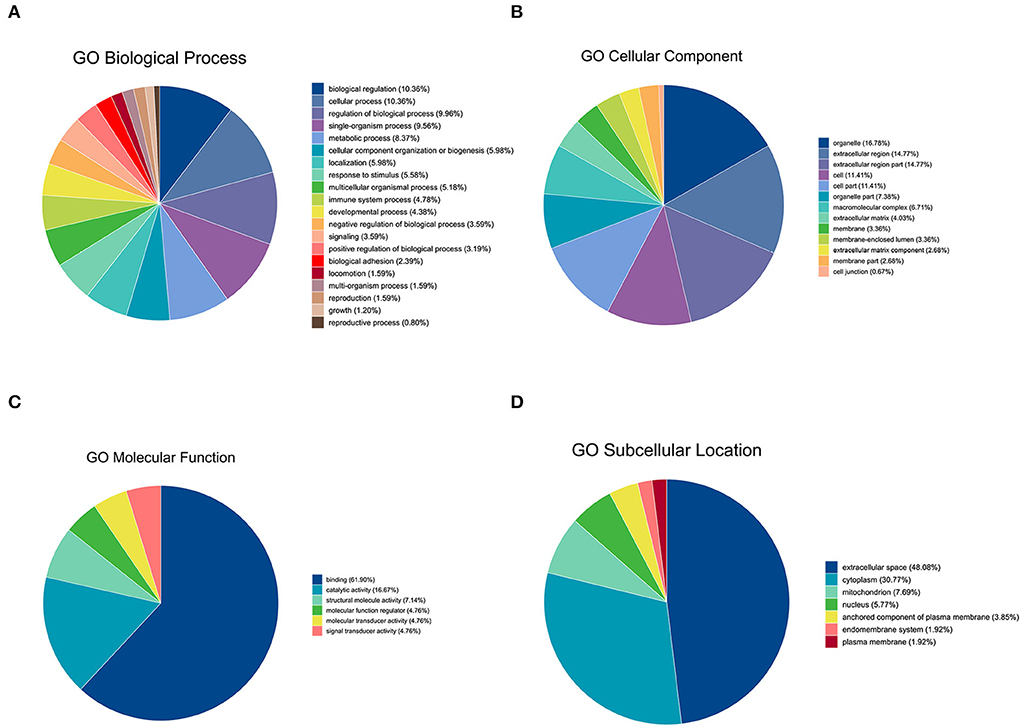
Figure 2. GO analysis of significantly differential proteins between DHD calves and clinically healthy calves. (A) Biological process-based analyses of significantly differential proteins. (B) Cellular component-based analyses of significantly differential proteins. (C) Molecular function-based analyses of significantly differential proteins. (D) Subcellular location-based analyses of significantly different expression proteins.
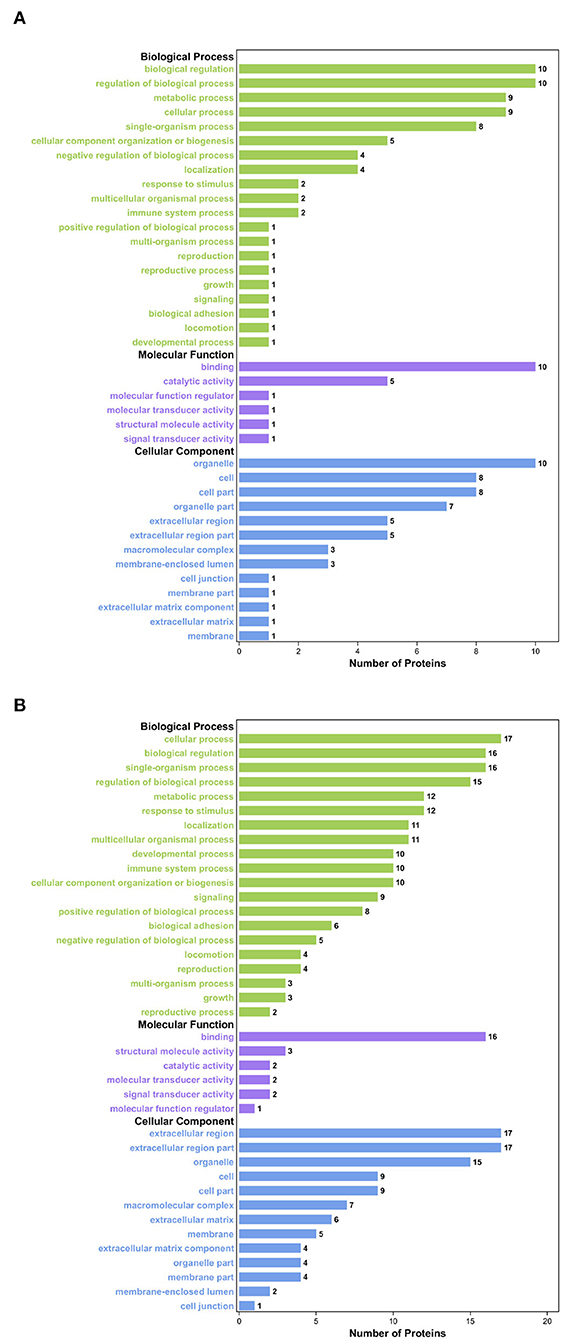
Figure 3. GO enrichment analysis of the significantly differential proteins between DHD calves and clinically healthy calves. (A) GO enrichment analysis on the ontology of molecular function, cellular component, and biological process of the significantly increased proteins. (B) GO enrichment analysis of the significantly decreased proteins.
KEGG annotation of the significantly different proteins
We used KEGG to annotate the information of the significantly different proteins at the level of biological pathways (16). As a result, 57 relevant KEGG signal pathways for 52 differential proteins were annotated, and these proteins were significantly enriched in 31 pathways (P < 0.05). The first 20 significantly enriched pathways were shown in Figure 4A, such as caffeine metabolism, intestinal immune network for IgA production, Fc epsilon RI signaling pathway, and NF-κB signaling pathway.
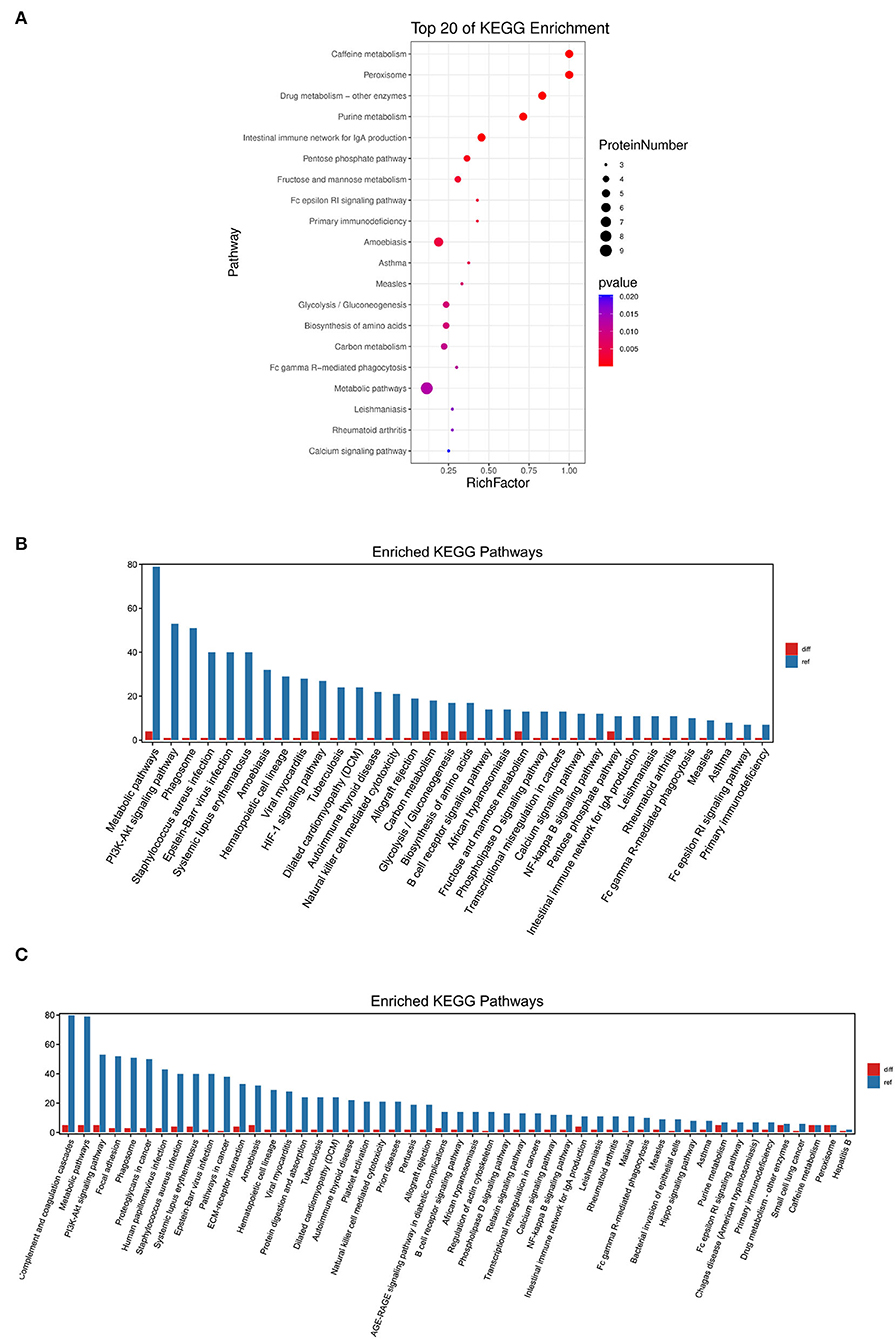
Figure 4. KEGG pathway enrichment analysis of the differentially expressed proteins. (A) The first 20 pathways with the lowest P-value were used for the diagram, with the ordinate as pathways and the abscissa as enrichment factors (The number of differential proteins in that pathway divided by the number of all proteins. The size indicated the number of different proteins, and the more red the color, the smaller the P-value). (B) KEGG enrichment analysis of the significantly increased proteins. (C) KEGG enrichment analysis of the significantly decreased proteins.
Further comparison of the KEGG pathway enrichment analysis revealed that the increased proteins in calves with DHD were mainly enriched in metabolic pathways, HIF-1 signaling pathway, and fructose and mannose metabolism (Figure 4B), and the decreased proteins were mainly enriched in caffeine metabolism, purine metabolism, and intestinal immune network for IgA production (Figure 4C).
Network Analysis
The protein-protein interaction (PPI) network consisted of 52 nodes and 113 edges with an average node degree of 8.96 (Figure 5). The expected number of edges for this analysis was 113. FN1, as an important node in the PPI network, was negatively correlated with fibrinogen-like protein 1 isoform X1 (FGL1) and endopin 2C-like (SERPINA3–7), and positively correlated with 15 proteins, such as xanthine dehydrogenase/oxidase (XDH), polymeric immunoglobulin receptor (PIGR), and complement C1q subcomponent subunit C precursor (C1qc). These data indicated that there was a partially biological connection among the differential proteins.
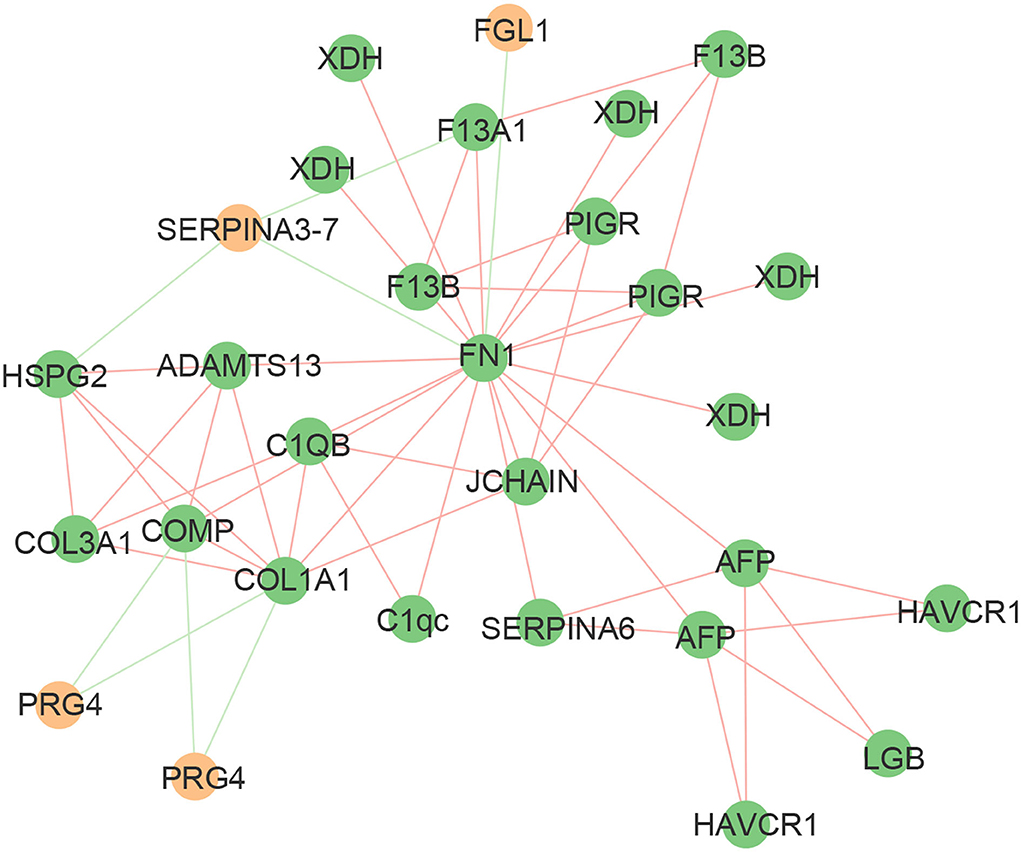
Figure 5. Mapping of significantly differential proteins onto a composite network based on predicted PPI. The edges of the network symbolize different interactions between proteins, based on the actions view of STRING v11.0. The green edges indicate a negative correlation and the red edges indicate a positive correlation. The green nodes indicate upregulated proteins and the yellow nodes indicate downregulated proteins.
PRM analysis of the differentially expressed plasma proteins
The results of the PRM pre-experiment showed that 13 of the 52 differential proteins in the test group were detected in the validation group. Thirteen differential proteins corresponding to 38 peptides were selected for PRM validation to verify whether they are associated with DHD. The results of the difference trend and t-test showed that fibronectin precursor and apolipoprotein C-IV precursor in plasma of calves with DHD were significantly lower than those in clinically healthy calves (P < 0.05), which were consistent with the results from the test group (Table 5). Therefore, the reliability of these two proteins as potential biomarkers for diagnosis of DHD was further validated.
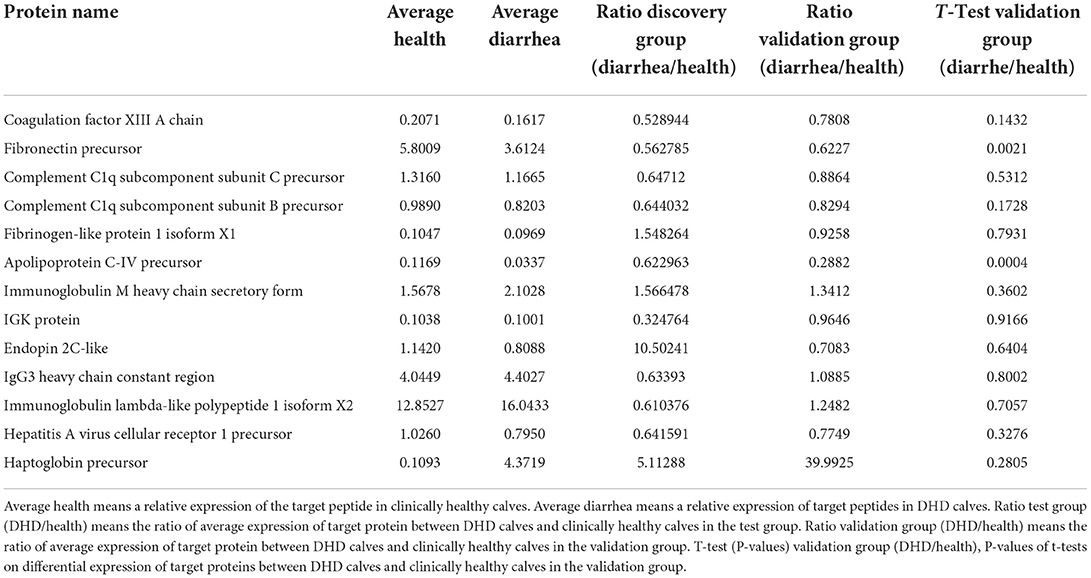
Table 5. Differential proteins identified in plasma samples of DHD calves and clinically healthy calves by PRM.
Discussion
Calf diarrhea is a growing concern worldwide because it can cause serious economic losses. DHD is a common type of diarrhea in calves, but the exact pathogenesis has not been determined yet, so the current treatment strategy cannot satisfactorily control the occurrence of DHD in calves. Therefore, it is necessary to further study the pathogenesis of DHD in calves. Proteomics is an effective technique to help characterize diseases by identifying subtle changes in protein abundance between the sick and healthy states of the body (17). To date, although the proteomic has been widely used in the study of many diseases including diarrhea, little information is available about the protein profiles of DHD in the calf. In the present study, proteomics was used to identify significantly differential proteins in calves with DHD, and their possible related functions were analyzed to elucidate the pathogenesis of DHD in calves at the proteomic level.
Intestinal tissue from animals cannot easily be sampled in general, especially in calves, the individual economic value is high. In contrast, the plasma is an ideal sample for studying the pathogenesis of calves with DHD because of its relative chemical and physical stability, abundant protein content, and relatively reliable collection methods in large animals. In this study, 52 proteins were detected to be significantly altered in calves with DHD. KEGG annotated that these proteins are involved in caffeine metabolism, Fc epsilon RI signal pathway, purine metabolism, intestinal immune network for IgA production, NF-κB signal pathway, and PI3K/Akt signal pathway. Caffeine metabolism and purine metabolism are important metabolic processes in the body, and their metabolic disorder will lead to the occurrence of many diseases, such as rheumatic immune diseases and obesity (18, 19). Caffeine was verified to be closely related to inhibiting diarrhea, the reason is related to its function of sterilization and anti-inflammatory effect (20, 21). Purines are natural substances found in virtually all foods, and they can be metabolized into uric acid in the body, which has antioxidant properties (22). Besides, purines as chemical messengers could transmit throughout tissues and species, and cross-linked with other transmitter networks to coordinate numerous aspects of cell behavior such as proliferation, migration, apoptosis, and other physiological processes of organisms (18). It has also been shown that regulation of purine metabolism can effectively alleviate colitis (23). Fc epsilon RI signal pathway plays a central role in the IgE-mediated allergic response and mast cell inflammation, which could be activated in some diseases with increased inflammation such as digestive diseases and heart diseases (24, 25). The significant increase in IGHM associated with this pathway in calves with DHD may indicate an enhanced inflammatory response in sick calves. PI3K/Akt signal pathway participates in the pathogenesis of diarrhea-related diseases by regulating various inflammatory factors (26). The NF-κB transcription factor is a typical pro-inflammatory signaling factor, and the activation of NF-κB can promote the occurrence and development of downstream inflammatory reactions (27). Abnormal regulation of these pathways has been found in several diseases presenting with diarrhea (28, 29).
Most of the significantly differential proteins (39/52) in this study were decreased in calves with DHD, including fibronectin, coagulation factor XIII B chain, and coagulation factor XIII A chain. Fibronectin is a glycoprotein that is involved in various biochemical processes, such as wound healing, cell adhesion, and blood coagulation (30, 31). Fibronectin decreased in calves with DHD, which may be responsible for the increased mucosal permeability (32). Adhesion is a crucial step in bacterial infection. Fibronectin was found to be associated with adhesion in many intestinal pathogens (33). Previous studies have considered fibronectin as a biomarker for some diseases, such as sepsis, where low fibronectin levels seem to be a marker of poor prognosis, and meningitis and asthma, where fibronectin levels are significantly increased (34, 35). But it is the first time that fibronectin could be a biomarker for calf diarrhea. Coagulation factor XIII is a coagulation factor with many cellular functions, which is involved in the wound healing process, proangiogenic function, and monocyte/macrophage functions (36). It has been reported that the levels and activity of coagulation factor XIII antigen were reduced in inflammatory bowel disease (37). Plasma coagulation factor XIII could promote cross-linking of the extracellular matrix components fibrin and fibronectin in response to tissue injury (38). The cross-linking of coagulation factor XIII A to fibronectin could promote the wound healing process (39). In this study, the reduction of these proteins may slow down the healing process of intestinal injury in calves with DHD. Heparan sulfate proteoglycans, complex molecules in cell membrane and extracellular matrix, play vital roles in tumorigenesis due to its mediating cell adhesion, differentiation, migration, and signal transduction. It also has been considered as an important target for the treatment and diagnosis of colorectal cancer (40). Xanthine dehydrogenase (XDH) and Xanthine oxidase (XO) are two interconvertible forms of Xanthine oxidoreductase (XOR). XO activity could be inhibited in some diseases that can lead to intestinal injury (41). Metabolism of intestinal tissue is usually very active, even short periods of hypoxia or ischemia may cause oxidative damage to the intestinal mucosa, which is due to an increase in the amount or activity of XO. It has been demonstrated that XDH is converted to XO when intestinal mucosa was oxidative damaged (42). Therefore, this may also be one of the reasons for the significant decrease in XDH in calves with DHD. In addition, alpha-fetoprotein (AFP) (43), IGK protein (44), and collagen alpha-1 (I) chain precursor (COL1A1) (45) were also used as biomarkers for some intestinal diseases.
The significantly increased proteins of plasma in calves with DHD included haptoglobin precursor, afamin precursor, and ALDOB protein. Haptoglobin, an acute phase response protein secreted by the liver, has the activities of antioxidant, immunomodulatory, antibacterial, and anti-inflammatory (46, 47). The aberrant glycosylation of haptoglobin is associated with many diseases, especially cancer (48). In addition, as a biomarker of diabetic cardiovascular disease and steroid-resistant nephrotic syndrome, haptoglobin can also be used to evaluate and diagnose diarrhea in calves due to its significantly increased expression in calves with DHD (49–51). The significantly increased haptoglobin in the plasma of calves with DHD may be associated with an acute inflammatory response caused by diarrhea. Afamin is a vitamin E-binding glycoprotein expressed mainly in the liver and is associated with many metabolic diseases, such as type 2 diabetes, metabolic syndrome, and obesity (52–54). Afamin is also considered to be a marker of increased hepatic lipid content, and fatty liver has been found in damp-heat diarrhea in rats (52, 55). Although this protein has not been reported in diarrhea-related diseases, it is one of the biomarkers for the detection of gastric cancer (56), and it plays an important role in the anti-apoptotic cellular processes related to oxidative stress, which may be the reason for the significantly increased afamin precursor in the plasma of calves with DHD (57). ALDOB played an important role in glycolysis and was annotated by metabolic pathways, carbon metabolism, glycolysis/gluconeogenesis, and HIF-1 signaling pathway by KEGG pathway analysis. ALDOB is mainly used for the diagnosis of liver diseases and is used as a biomarker for acute liver injury (58). ALDOB has also been poorly studied in relation to diarrhea, but the plasma glucose and lactate concentrations could change in calves with DHD (59), which could account for the significant changes in ALDOB protein. In addition, proteoglycan 4 is a protein that is critical for the virulence factor binding to the cell surface, so it plays an important role in colitis (60).
In the biomarker validation group, the reliability of FN1 and APOC4 as biomarkers for a calf with DHD was verified by targeted quantification. FN1 is related to tissue repair and host defense, and it is also annotated by KEGG into the PI3K-Akt signaling pathway, focal adhesion, and bacterial invasion of epithelial cells. Apolipoprotein could be synthesized in the intestine and reduced inflammatory responses (61). A significant reduction of Apolipoprotein has also been reported in some cases of diarrhea, which may be related to its important role in lipid metabolism in intestinal diseases (62, 63). Therefore, these two proteins may be potential biomarkers for diagnosis and targets for therapy on DHD in calves. In addition, although there were no significant differences in coagulation factor XIII A chain, IGK protein, and haptoglobin precursor in the invalidation group, their changing trends were consistent with those in the test group. These proteins may be closely associated with the pathogenesis of DHD in calves.
This is a preliminary study that indicates the feasibility of plasma proteins as a potential diagnostic biomarker approach for calves with DHD. Since our experiments were conducted on one farm, there are significant limitations in the results and conclusion. In our follow-up experiments, the number of clinical samples and farms should be increased, and more testing methods, such as enzyme-linked immunosorbent assay (ELISA) and western blot, are needed to verify these potential biomarkers for the clinical diagnosis and reliable targets for therapy DHD in calves. In addition, a more in-depth analysis of plasma proteomics in combination with other omics can provide a more complete and accurate understanding of calves with DHD.
Conclusion
Dampness heat diarrhea disturbed the composition of plasma proteins in calves. The changes in plasma protein levels, especially fibronectin precursor, haptoglobin precursor, coagulation factor XIII A chain, and apolipoprotein C-IV precursor, might affect the progression of DHD in calves by interfering with complement and coagulation cascades, PI3K-Akt signaling pathway, and focal adhesion. The reliability of fibronectin precursor and apolipoprotein C-IV precursor as biomarkers of calf with DHD was verified using PRM analysis. The findings provide a new insight for further exploring the mechanism of DHD in calves and evaluating the clinical meaning of fibronectin precursor and apolipoprotein C-IV precursor for diagnosis or treatment of the disease.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: http://www.proteomexchange.org/, PXD027099.
Ethics statement
The animal study was reviewed and approved by Laboratory Animal Ethics Commission of the Lanzhou Institute of Husbandry and Pharmaceutical Sciences of CAAS.
Author contributions
JL and KaiZ conceived and designed the work. JL coordinated technical support and funding. ZY wrote the manuscript. ZY, KanZ, KaiZ, GW, LW, JZ, ZQ, and ZG performed the experiments and collected the samples. JL and KaiZ reviewed the manuscript. All authors have read and approved the published version of the manuscript.
Funding
This work was financially supported by the Earmarked Fund for CARS-36, the National Key Research and Development Program of China (2017YFE0114400 and 2018YFD0502403), and Special Fund of Chinese Central Government for Basic Scientific Research Operations in Commonwealth Research Institutes (1610032021002).
Acknowledgments
Sincerely thanks to Chaoying Luo and other colleagues in the lab for their help in the clinical trial. Many thanks to the China Agriculture Research System of MOF and MARA and the National Key Research and Development Program of China for their support. We are also grateful to Guangzhou Genedenovo Biotechnology Co., Ltd., and Shanghai Applied Protein Technology Co., Ltd., for assisting in bioinformatics analysis.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2022.986329/full#supplementary-material
References
1. Caffarena RD, Casaux ML, Schild CO, Fraga M, Castells M, Colina R, et al. Causes of neonatal calf diarrhea and mortality in pasture-based dairy herds in Uruguay: a farm-matched case-control study. Braz J Microbiol. (2021) 52:977–88. doi: 10.1007/s42770-021-00440-3
2. Meganck V, Hoflack G, Opsomer G. Advances in prevention and therapy of neonatal dairy calf diarrhoea: a systematical review with emphasis on colostrum management and fluid therapy. Acta Vet Scand. (2014) 56:75. doi: 10.1186/s13028-014-0075-x
3. Hua YL, Ma Q, Zhang XS, Jia YQ, Peng XT, Yao WL, et al. Pulsatilla decoction can treat the dampness-heat diarrhea rat model by regulating glycerinphospholipid metabolism based lipidomics approach. Front Pharmacol. (2020) 11:197. doi: 10.3389/fphar.2020.00197
4. Xiong R, Li W, Li Y, Zheng K, Zhang T, Gao M, et al. Er Shen Wan extract reduces diarrhea and regulates AQP 4 and NHE 3 in a rat model of spleen-kidney Yang deficiency-induced diarrhea. Biomed Pharmacother. (2018) 98:834–46. doi: 10.1016/j.biopha.2018.01.023
5. Pan X, Xu K, Xiong R, Chen ZM, Hu CJ, Feng J, et al. Therapeutic effect of Ershen Pill () extract on Pi (Spleen)-Shen (Kidney) Yang deficiency-induced diarrhea in rat model. Chin J Integr Med. (2019) 25:536ntegr Medect 1007/s11655-017-2766-4
6. Chen XL, Wen Y, Wu ZC, Zhang BP, Hou ZK, Xiao JL, et al. Development of a traditional chinese medicine syndrome-specific scale for ulcerative colitis: the large intestine dampness-heat syndrome questionnaire. Evid Based Complement Alternat Med. (2018) 2018:4039019. doi: 10.1155/2018/4039019
7. Chai Y-N, Qin J, Li Y-L, Tong Y-L, Liu G-H, Wang X-R, et al. TMT proteomics analysis of intestinal tissue from patients of irritable bowel syndrome with diarrhea: implications for multiple nutrient ingestion abnormality. J Proteom. (2021) 231:103995. doi: 10.1016/j.jprot.2020.103995
8. Nirujogi RS, Muthusamy B, Kim M-S, Sathe GJ, Lakshmi PTV, Kovbasnjuk ON, et al. Secretome analysis of diarrhea-inducing strains of Escherichia coli. Proteomics. (2017) 17:10.1002. doi: 10.1002/pmic.201600299
9. Barrachina MN, Calderon-Cruz B, Fernandez-Rocca L, Garcia A. Application of extracellular vesicles proteomics to cardiovascular disease: guidelines, data analysis, and future perspectives. Proteomics. (2019) 19:e1800247. doi: 10.1002/pmic.201800247
10. Tang M, Zeng L, Zeng Z, Liu J, Yuan J, Wu D, et al. Proteomics study of colorectal cancer and adenomatous polyps identifies TFR1, SAHH, and HV307 as potential biomarkers for screening. J Proteomics. (2021) 243:104246. doi: 10.1016/j.jprot.2021.104246
11. Huang M-Z, Cui D-A, Wu X-H, Hui W, Yan Z-T, Ding X-Z, et al. Serum metabolomics revealed the differential metabolic pathway in calves with severe clinical diarrhea symptoms. Animals. (2020) 10:769. doi: 10.3390/ani10050769
12. Yan Z, Zhang K, Zhang K, Wang G, Wang L, Zhang J, et al. Integrated 16S rDNA gene sequencing and untargeted metabolomics analyses to investigate the gut microbial composition and plasma metabolic phenotype in calves with dampness-heat diarrhea. Front Vet Sci. (2022) 9:703051. doi: 10.3389/fvets.2022.703051
13. Ma L, Muscat J, Sinha R, Sun D, Xiu G. Proteomics of exhaled breath condensate in lung cancer and controls using data-independent acquisition (DIA): a pilot study. J Breath Res. (2020) 15:026002. doi: 10.1088/1752-7163/abd07e
14. Cang S, Liu R, Jin W, Tang Q, Li W, Mu K, et al. Integrated DIA proteomics and lipidomics analysis on non-small cell lung cancer patients with TCM syndromes. Chin Med. (2021) 16:126. doi: 10.1186/s13020-021-00535-x
15. Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, et al. STRING v10: protein-protein interaction networks integrated over the tree of life. Nucleic Acids Res. (2015) 43:D447–52. doi: 10.1093/nar/gku1003
16. Okuda S, Yamada T, Hamajima M, Itoh M, Katayama T, Bork P, et al. KEGG atlas mapping for global analysis of metabolic pathways. Nucleic Acids Res. (2008) 36:W423–6. doi: 10.1093/nar/gkn282
17. Sobsey CA, Ibrahim S, Richard VR, Gaspar V, Mitsa G, Lacasse V, et al. Targeted and untargeted proteomics approaches in biomarker development. Proteomics. (2020) 20:e1900029. doi: 10.1002/pmic.201900029
18. Huang Z, Xie N, Illes P, Virgilio FDi, Ulrich H, Semyanov A, et al. From purines to purinergic signalling: molecular functions and human diseases. Signal Transduct Target Ther. (2021) 6:162. doi: 10.1038/s41392-021-00553-z
19. Jing N, Liu X, Jin M, Yang X, Hu X, Li C, et al. Fubrick tea attenuates high-fat diet induced fat deposition and metabolic disorder by regulating gut microbiota and caffeine metabolism. Food Funct. (2020) 11:6971–86. doi: 10.1039/D0FO01282C
20. Mo L, Zeng Z, Li Y, Li D, Yan CY, Xiao S, et al. Animal study of the anti-diarrhea effect and microbial diversity of dark tea produced by the Yao population of Guangxi. Food Funct. (2019) 10:1999–2009. doi: 10.1039/C9FO00110G
21. Gökcen BB, Sanlier N. Coffee consumption and disease correlations. Crit Rev Food Sci Nutr. (2019) 59:336–48. doi: 10.1080/10408398.2017.1369391
22. Kaneko K, Aoyagi Y, Fukuuchi T, Inazawa K, Yamaoka N. Total purine and purine base content of common foodstuffs for facilitating nutritional therapy for gout and hyperuricemia. Biol Pharm Bull. (2014) 37:709–21. doi: 10.1248/bpb.b13-00967
23. Wu J, Wei Z, Cheng P, Qian C, Xu F, Yang Y, et al. Rhein modulates host purine metabolism in intestine through gut microbiota and ameliorates experimental colitis. Theranostics. (2020) 10:10665–79. doi: 10.7150/thno.43528
24. Frydas S, Varvara G, Murmura G, Saggini A, Caraffa A, Antinolfi P, et al. Impact of capsaicin on mast cell inflammation. Int J Immunopathol Pharmacol. (2013) 26:597–600. doi: 10.1177/039463201302600303
25. Wang Y, Ding Y, Li C, Gao J, Wang X, An H. Alpha-linolenic acid inhibits IgE-mediated anaphylaxis by inhibiting Lyn kinase and suppressing mast cell activation. Int Immunopharmacol. (2022) 103:108449. doi: 10.1016/j.intimp.2021.108449
26. Wu XF, Xu R, Ouyang ZJ, Qian C, Shen Y, Wu XD, et al. Beauvericin ameliorates experimental colitis by inhibiting activated T cells via downregulation of the PI3K/Akt signaling pathway. PLoS ONE. (2013) 8:e83013. doi: 10.1371/journal.pone.0083013
27. Saleh HA, Yousef MH, Abdelnaser A. The anti-inflammatory properties of phytochemicals and their effects on epigenetic mechanisms involved in TLR4/NF-κB-mediated inflammation. Front Immunol. (2021) 12:606069. doi: 10.3389/fimmu.2021.606069
28. Yan Z, Zhang K, Zhang K, Wang G, Wang L, Zhang J, et al. Huang Bai Jian Pi decoction alleviates diarrhea and represses inflammatory injury via PI3K/Akt/NF-κB pathway: in vivo and in vitro studies. J Ethnopharmacol. (2022) 115212. doi: 10.1016/j.jep.2022.115212
29. Kong N, Wu Y, Meng Q, Wang Z, Zuo Y, Pan X, et al. Suppression of virulent porcine epidemic diarrhea virus proliferation by the PI3K/Akt/GSK-3α/β Pathway. PLoS ONE. (2016) 11:e0161508. doi: 10.1371/journal.pone.0161508
30. Zhou Y, Cao G, Cai H, Huang H, Zhu X. The effect and clinical significance of FN1 expression on biological functions of gastric cancer cells. Cell Mol Biol. (2020) 66:191–8. doi: 10.14715/cmb/2020.66.5.32
31. Evans BA, Kamyszek RW, Piegore M, Stoner K, Fuller M, Welsby IJ, et al. Global sources of cryoprecipitate demonstrate variability in coagulant factor levels and functional hemostasis. Blood Coagul Fibrinolysis. (2021) 32:87–102. doi: 10.1097/MBC.0000000000000982
32. Camilleri M, Carlson P, Acosta A, Busciglio I, Nair AA, Gibbons SJ, et al. RNA sequencing shows transcriptomic changes in rectosigmoid mucosa in patients with irritable bowel syndrome-diarrhea: a pilot case-control study. Am J Physiol Gastrointest Liver Physiol. (2014) 306:C1089–98. doi: 10.1152/ajpgi.00068.2014
33. Guo G, Qin S, Kong X, Wang Z, Shen Y, Huo X, et al. Identification of novel fibronectin-binding proteins by 2D-far Western blot in atypical enteropathogenic Escherichia coli serotype O55:H7. Microb Pathog. (2021) 150:104682. doi: 10.1016/j.micpath.2020.104682
34. Shyu L-Y, Hu M-E, Chou C-H, Chen K-M, Chiu P-S. Fibronectin changes in eosinophilic meningitis with blood-CSF barrier disruption. Exp Parasitol. (2015) 151:73–79. doi: 10.1016/j.exppara.2015.02.002
35. Bazan-Socha S, Kuczia P, Potaczek DP, Mastalerz L, Cybulska A, Zareba L, et al. Increased blood levels of cellular fibronectin in asthma: relation to the asthma severity, inflammation, and prothrombotic blood alterations. Respir Med. (2018) 141:64–71. doi: 10.1016/j.rmed.2018.06.023
36. Muszbek L, Bereczky Z, Bagoly Z, Komaromi I, Katona E. Factor XIII: a coagulation factor with multiple plasmatic and cellular functions. Physiol Rev. (2011) 91:931–72. doi: 10.1152/physrev.00016.2010
37. Soendergaard C, Kvist PH, Seidelin JB, Nielsen OH. Tissue-regenerating functions of coagulation factor XIII. J Thromb Haemost. (2013) 11:806–16. doi: 10.1111/jth.12169
38. Tsujimoto I, Moriya K, Sakai K, Dickneite G, Sakai T. Critical role of factor XIII in the initial stages of carbon tetrachloride-induced adult liver remodeling. Am J Pathol. (2011) 179:3011–9. doi: 10.1016/j.ajpath.2011.08.037
39. Richardson VR, Cordell P, Standeven KF, Carter AM. Substrates of factor XIII-A: roles in thrombosis and wound healing. Clin Sci. (2013) 124:123–37. doi: 10.1042/CS20120233
40. Vicente CM, Silva DAA, Sartorio PV, Silva TD, Saad SS, Nader HB, et al. Heparan sulfate proteoglycans in human colorectal cancer. Anal Cell Pathol. (2018) 2018:8389595. doi: 10.1155/2018/8389595
41. Khan R, Khan AQ, Qamar W, Lateef A, Ali F, Rehman MU, et al. Chrysin abrogates cisplatin-induced oxidative stress, p53 expression, goblet cell disintegration and apoptotic responses in the jejunum of Wistar rats. Br J Nutr. (2012) 108:1574–85. doi: 10.1017/S0007114511007239
42. García-de-la-Asunción J, Barber G, Rus D, Perez-Griera J, Belda FJ, Martí F, et al. Hyperoxia during colon surgery is associated with a reduction of xanthine oxidase activity and oxidative stress in colonic mucosa. Redox Rep. (2011) 16:121–8. doi: 10.1179/174329211X13049558293632
43. Lin HH, Chang CC, Yang SH, Chang SC, Chen WS, Liang WY, et al. Elevated serum alpha-fetoprotein in poorly differentiated adenocarcinoma with neuroendocrine differentiation of the ascending colon: a case report. World J Surg Oncol. (2016) 14:84. doi: 10.1186/s12957-016-0838-0
44. Yang SB, Chen X, Wu BY, Wang MW, Cai CH, Cho DB, et al. Immunoglobulin kappa and immunoglobulin lambda are required for expression of the anti-apoptotic molecule Bcl-xL in human colorectal cancer tissue. Scand J Gastroenterol. (2009) 44:1443–51. doi: 10.3109/00365520903369953
45. Tabuso M, Adya R, Stark R, Gopalakrishnan K, Tsang YW, James S, et al. Fibrotic phenotype of peritumour mesenteric adipose tissue in human colon cancer: a potential hallmark of metastatic properties. Int J Mol Sci. (2021) 22:2430. doi: 10.3390/ijms22052430
46. Kessel I, Leib M, Levy A, Miller-Lotan R, Waisman D, Jacobson E, et al. Does haptoglobin phenotype influence postnatal morbidity in preterm neonates? Am J Perinatol. (2016) 33:130–5. doi: 10.1055/s-0035-1560042
47. Oh MJ, Lee SH, Kim U, An HJ. In-depth investigation of altered glycosylation in human haptoglobin associated cancer by mass spectrometry. Mass Spectrom Rev. (2021) 5:1–23. doi: 10.1002/mas.21707
48. Jeong S, Oh MJ, Kim U, Lee J, Kim J-H, An HJ. Glycosylation of serum haptoglobin as a marker of gastric cancer: an overview for clinicians. Expert Rev Proteomics. (2020) 17:109–17. doi: 10.1080/14789450.2020.1740091
49. Costacou T, Levy AP. Haptoglobin genotype and its role in diabetic cardiovascular disease. J Cardiovasc Transl Res. (2012) 5:423–35. doi: 10.1007/s12265-012-9361-z
50. Wen Q, Huang L-T, Luo N, Wang Y-T, Li X-Y, Mao H-P, et al. Proteomic profiling identifies haptoglobin as a potential serum biomarker for steroid-resistant nephrotic syndrome. Am J Nephrol. (2012) 36:105–13. doi: 10.1159/000339755
51. Choi K-S, Kang J-H, Cho H-C, Yu D-H, Park J. Changes in serum protein electrophoresis profiles and acute phase proteins in calves with diarrhea. Can J Vet Res. (2021) 85:45−50.
52. Kurdiova T, Balaz M, Kovanicova Z, Zemkova E, Kuzma M, Belan V, et al. Serum afamin a novel marker of increased hepatic lipid content. Front Endocrinol. (2021) 12: 670425. doi: 10.3389/fendo.2021.670425
53. Juhász I, Ujfalusi S, Seres I, Lorincz H, Varga VE, Paragh G, et al. Afamin levels and their correlation with oxidative and lipid parameters in non-diabetic, obese patients. Biomolecules. (2022) 12:116. doi: 10.3390/biom12010116
54. Kollerits B, Lamina C, Huth C, Marques-Vidal P, Kiechl S, Seppälä I, et al. Plasma concentrations of afamin are associated with prevalent and incident type 2 diabetes: a pooled analysis in more than 20,000 individuals. Diabetes Care. (2017) 40:1386–93. doi: 10.2337/dc17-0201
55. Yao W, Yang C, Wen Y, Zhang W, Zhang X, Ma Q, et al. Treatment effects and mechanisms of Yujin Powder on rat model of large intestine dampness-heat syndrome. J Ethnopharmacol. (2017) 202:265–80. doi: 10.1016/j.jep.2017.03.030
56. Humphries JM, Penno MA, Weiland F, Klingler-Hoffmann M, Zuber A, Boussioutas A, et al. Identification and validation of novel candidate protein biomarkers for the detection of human gastric cancer. Biochim Biophys Acta. (2014) 1844:1051–8. doi: 10.1016/j.bbapap.2014.01.018
57. Köninger A, Mathan A, Mach P, Frank M, Schmidt B, Schleussner E, et al. Is afamin a novel biomarker for gestational diabetes mellitus? A pilot study. Reprod Biol Endocrinol. (2018) 16:30. doi: 10.1186/s12958-018-0338-x
58. Qin S, Zhou Y, Gray L, Kusebauch U, McEvoy L, Antoine DJ, et al. Identification of organ-enriched protein biomarkers of acute liver injury by targeted quantitative proteomics of blood in acetaminophen- and carbon-tetrachloride-treated mouse models and acetaminophen overdose patients. J Proteome Res. (2016) 15:3724–40. doi: 10.1021/acs.jproteome.6b00547
59. Lewis LD, Phillips RW, Elliott CD. Changes in plasma glucose and lactate concentrations and enzyme activities in the neonatal calf with diarrhea. Am J Vet Res. (1975) 36(4 Pt.1):413–6.
60. Yuan P, Zhang H, Cai C, Zhu S, Zhou Y, Yang X, et al. Chondroitin sulfate proteoglycan 4 functions as the cellular receptor for Clostridium difficile toxin B. Cell Res. (2015) 25:157–68. doi: 10.1038/cr.2014.169
61. Tayyeb JZ, Popeijus HE, Mensink RP, Plat J. Butyric acid added apically to intestinal Caco-2 cells elevates hepatic apoa-I transcription and rescues lower apoa-I expression in inflamed HepG2 cells co-cultured in the basolateral compartment. Biomolecules. (2021) 11:71. doi: 10.3390/biom11010071
62. Zhang J, Zhao D, Yi D, Wu M, Chen H, Wu T, et al. Microarray analysis reveals the inhibition of intestinal expression of nutrient transporters in piglets infected with porcine epidemic diarrhea virus. Sci Rep. (2019) 9:19798. doi: 10.1038/s41598-019-56391-1
Keywords: biomarkers, calf, dampness heat diarrhea, parallel reaction monitoring, proteomic
Citation: Yan Z, Zhang K, Wang G, Wang L, Zhang J, Qiu Z, Guo Z, Zhang K and Li J (2022) Differential proteomic of plasma provides a new perspective on scientific diagnosis and drug screening for dampness heat diarrhea in calves. Front. Vet. Sci. 9:986329. doi: 10.3389/fvets.2022.986329
Received: 05 July 2022; Accepted: 23 August 2022;
Published: 20 September 2022.
Edited by:
Shengfa F. Liao, Mississippi State University, United StatesReviewed by:
Ruchika Bhawal, Cornell University, United StatesAna Mar Bhawal, University of Murcia, Spain
Copyright © 2022 Yan, Zhang, Wang, Wang, Zhang, Qiu, Guo, Zhang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianxi Li, bHpqaWFueGlsQDE2My5jb20=; Kai Zhang, Znlqb2huQHNpbmEuY29t
†These authors have contributed equally to this work and share first authorship
 Zunxiang Yan
Zunxiang Yan Kang Zhang
Kang Zhang Guibo Wang
Guibo Wang Lei Wang
Lei Wang Jingyan Zhang
Jingyan Zhang Zhengying Qiu
Zhengying Qiu Zhiting Guo
Zhiting Guo Kai Zhang*
Kai Zhang* Jianxi Li
Jianxi Li