- 1Key Laboratory of Animal Genetics, Breeding and Reproduction of Shaanxi Province, College of Animal Science and Technology, Northwest A&F University, Shaanxi, China
- 2Shaanxi Provincial Engineering and Technology Research Center of Cashmere Goats, Yulin University, Yulin, China
- 3Life Science Research Center, Yulin University, Yulin, China
- 4College of Animal Science and Technology, Hunan Agricultural University, Changsha, China
- 5Institute of Animal Husbandry and Veterinary, Fujian Academy of Agricultural Sciences, Fuzhou, China
The sorting nexin 29 (SNX29) gene, a member of the SNX family, is associated with material transport and lipid metabolism. Previous studies have shown that lipid metabolism affects reproductive function in animals. Thus, we hypothesized there is a correlation between the SNX29 gene and reproductive trait. To date, studies on the relationship between the SNX29 gene and reproductive traits are limited. Therefore, the purpose of this study was to examine the polymorphism in the SNX29 gene and its correlation with litter size. Herein, the mRNA expression levels of SNX29 were assayed in various goat tissue. Surprisingly, we found that SNX29 was highly expressed in the corpus luteum, large and small follicles. This result led us to suggest that the SNX29 gene has a critical role in reproduction. We further detected potential polymorphisms in Shaanbei white cashmere (SBWC) goats, including insertion/deletion (InDel, n = 2,057) and copy number variation (CNV, n = 1,402), which were related to fertility. The 17 bp deletion (n = 1004) and the 20 bp deletion (n = 1,053) within the SNX29 gene were discovered to be significantly associated with litter size (P < 0.05), and individuals the ID genotype of P1-Del-17 bp and the DD genotype of P2-Del-20bp had larger litter size. Additionally, the four CNV loci had significant correlations with litter size (P < 0.01) in our detected population. In CNV5, individuals with the median genotype were superior compared to those with loss or gain genotype in term of litter size, and in other three CNVs showed better reproductive trait in the gain genotype. Briefly, these findings suggest that SNX29 could be used as a candidate gene for litter size in goat breeding through marker-assisted selection (MAS).
Introduction
Litter size, one of the most important economic traits, is affected by multiple factors, among which genetic aspects are the most essential factors. However, so far there are few studies that identified the key genes affecting litter size (1). This requires us to develop modern molecular techniques, in order to find candidate genes that affect female fecundity. As a method of modern molecular breeding, marker-assisted selection (MAS) has plenty of advantages, such as efficiency, stability and reliability of outcomes, and good repeatability (2). Meanwhile, as a typical type of molecular marker, insertion/deletions (InDels) and copy number variations (CNVs) have been used in animal genetics and breeding (3), gene localization and gene cloning (4). InDel has been widely used in MAS, whereas CNV has been rarely utilized in breeding and has received attention in recent years.
The sorting nexin (SNX) family has more than 30 members, which are located in the cytoplasm and have membrane binding potential either binding to the PX domain through their lipids or through interactions between proteins and membrane-associated protein complexes (5, 6). This family is characterized by the involvement of the PX domain in cell-related functions (7), such as cell signaling and vesicle trafficking (8). Studies have shown that SNX24 has a certain regulatory effect on estrogen (9). The sorting nexin 29 (SNX29) gene is one of the members of the SNX family and can bind to phosphatidylinositol. It has a similar protein domain and function as the SNX24 gene (10). Previous studies have demonstrated that lipid metabolism affects reproductive function in animals (11), DNA methylation statuses of the SNX29 gene were under the testosterone control (12). Thus, we speculated that the SNX29 gene is an important candidate gene for reproductive trait.
Moreover, the relevant literatures describe that the SNX29 gene has been proved to influence the formation of autism (13) and the regulation of nervous system development (14). Earlier researches have also reported that the SNX29 gene is one of the translocation partners involved in the major histocompatibility complex class II transactivator CIITA (MHC2TA) in human KH-H2 cells (15) and is strongly associated with schizophrenia (16), intelligence (17), bipolar disorder (BPD) as well as major depressive disorder (MDD) (18). In addition, the SNX29 gene has been identified as a growth-related gene that was correlated with body size and body weight (19), and it plays a vital role in York pigs (20) and other species. Growth and reproduction are inextricably linked. Most studies have revealed that genes can simultaneously affect growth traits and reproductive traits in livestock, such as the PLAG1 gene (21–23). However, the effect of SNX29 on reproductive traits of goats has not been reported at present, so we assumed that the SNX29 gene is involved in reproduction.
Although SNX29 has been widely studied with respect to neurodevelopment and growth traits, relevant studies on the relationship between the SNX29 gene polymorphisms and reproductive traits are limited. Therefore, this study aimed to assay the mRNA expression level of SNX29, explore the genetic variations of the SNX29 gene in Shaanbei white cashmere (SBWC) goats by polymerase chain reaction (PCR) and quantitative PCR (qPCR) detecting system, and verify its effect on reproductive traits. These results will help us to further study the relationship in livestock reproductive traits, and provide a new method to advance animal genetic reproduction and breeding, in order to improve the DNA molecular breeding technology in SBWC goats.
Materials and methods
Animal welfare explanation
All the samples in this experiment were in accordance with the use and management of experimental animals at Northwest A&F University (NWAFU-314020038).
Sample collection
Total RNA extraction and examination
SBWC goats were randomly selected from the Yulin goat yard in Shaanxi province and were raised under the same feeding, water supply, and management conditions. Additionally, tissue samples including heart, liver, skeletal muscle, small follicles, large follicles, spleen, kidney, epididymis, testis, brain, fat, skin, ovary, and corpus luteum of adult SBWC goats were collected (n = 3) (24). The total RNA was extracted using Trizol reagent (Takara, Dalian, China) (25). The OD260/280 ratio was measured by a NanoDrop™1000 spectrophotometer (Thermo Scientific, Waltham, MA, USA) and a ratio between 1.8 and 2.0 meant qualified (26). Then, cDNA was synthesized using the PrimeScript RT Reagent Kit (Takara, Dalian, China) and temporarily stored at −20°C.
Genomic DNA isolation and DNA pool construction
Under the same feeding conditions, a total of 2,057 healthy SBWC goats were selected from Yulin goat farm in Shaanxi province and their ear tissues were collected. Using the phenol-chloroform method (27), genomic DNA was extracted from the goat ear tissues preserved in 70% alcohol at −80°C (28). OD values and OD260/280 ratios of DNA samples were measured by a NanoDrop™1000 Spectrophotometer (Thermo Scientific, Waltham, MA, USA). After quality detection, the DNA was diluted to 20 ng/μl and stored at −40°C for long-term preservation. After that, 30 samples were randomly selected to construct a pool of genomic DNA mixture to determine the presence of InDel sites occurred in the tested population.
Primer design
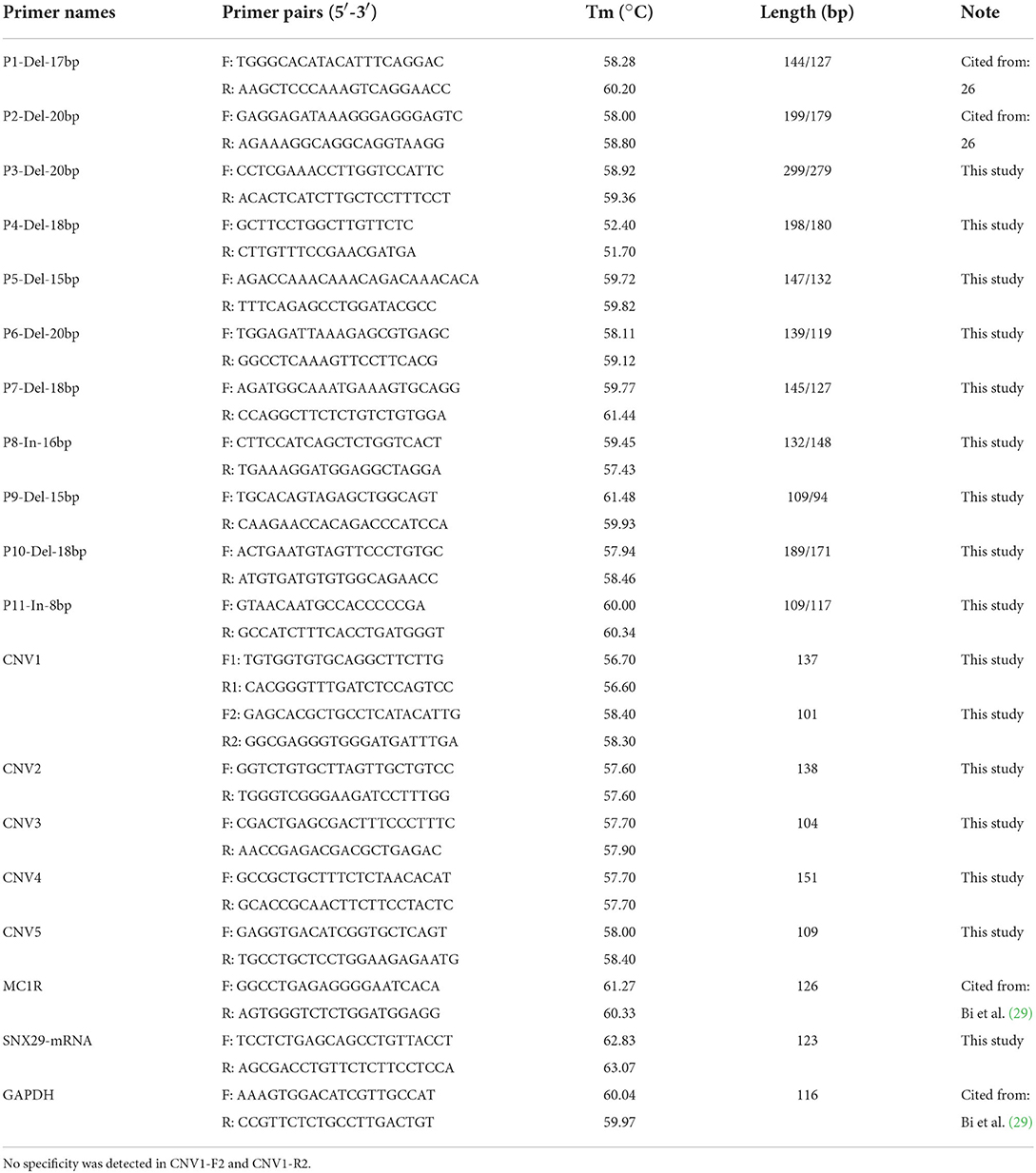
We searched the Ensembl (https://asia.ensembl.org/) and Animal Omics database (http://animal.nwsuaf.edu.cn/), and found 11 InDel and five CNV loci of the SNX29 gene in goats. Then, 16 primer pairs for amplification were designed using the Primer-BLAST tool in the NCBI database (https://www.ncbi.nlm.nih.gov/tools/primer-blast/) and the primer premier 5.0 software (Thermo Scientific, Waltham, MA, USA) (Table 1). Besides, four pairs of primers including P1-Del-17bp (Rs659002477, TAAAGGAAAGCAATGTA/-), P2-Del-20bp (Rs654310334, AGCTTCCGGTGAGCCTGTCG/-) (30), MC1R, and GAPDH (30, 31) were referred to our previous studies. MC1R and GAPDH were used as reference genes.
The mRNA expression levels of the SNX29 gene
The relative mRNA expression levels of the SNX29 gene was assayed by performing qPCR. The reaction system contained 5 μl of 2 × SYBR Premix Ex Taq (Takara, Dalian, China), 0.5 μl of DNA, 0.5 μl of each upstream and downstream primers, up to 10 μl with ddH2O (32). The data was analyzed by 2−ΔΔCt method (33).
InDel genotyping of the SNX29 gene
After testing the designed primers with DNA mixing pools, PCR amplification was performed in 13 μl of PCR reaction mixture, containing 6.5 μl of 2 × Taq Master mix (BioLinker, Shanghai, China), 0.5 μl of F and R primers, 0.5 μl of genomic DNA, and finally made up to 13 μl with ddH2O. The touch-down PCR procedure was utilized for PCR amplification (34, 35). Then PCR amplification products (P1 = 144 bp, P2 = 199 bp) were detected by 2% agarose gel electrophoresis.
CNV genotyping of the SNX29 gene
The primers were tested to ensure the that designed primers could amplify the target fragment (Supplementary Figure S1). Then, a total of 1,402 samples were used for testing. The total qPCR reaction system consisted of 10 μl comprised 5 μl of 2 × SYBR Premix Ex Taq (Takara, Dalian, China), 0.5 μl of DNA, 0.5 μl of each primer, and 3.5 μl of ddH2O. Previously explored experimental procedures were as follows, 95°C for 45 s, 40 cycles of 15 s at 95°C and 45 s at 60°C. Finally, the result was processed by method 2*2−ΔCt·2*2−ΔCt>2 was Gain, it < 2 was Loss, it = 2 was Median genotype (29).
Statistical analyses
The genotype and allele frequencies, heterozygosity (He), homozygosity (Ho), polymorphism information content (PIC), Hardy-Weinberg equilibrium (HWE) and linkage disequilibrium (LD) were calculated using the SHEsis platform (http://analysis.bio-x.cn). Furthermore, analysis of variance (ANOVA) and χ2 test were used to explore the association between the variants and litter size in SPSS 26.0 (IBM, USA), when P < 0.05, all results were correlated with litter size, and all statistical tests were conducted by two-sided tests. The line model was reference previous study (36).
Results
The mRNA expression levels of the goat SNX29 gene
The results of the gene expression profile results indicated that the mRNA expression level of SNX29 was higher in small follicles, large follicles and corpus luteum than in other tissues. Among 16 tissues, the relative expression level of the corpus luteum was the highest. In addition, more abundant expression was found in large and small follicles (Figure 1). Thus, this result suggests that the SNX29 gene play an important role in goat follicle development.
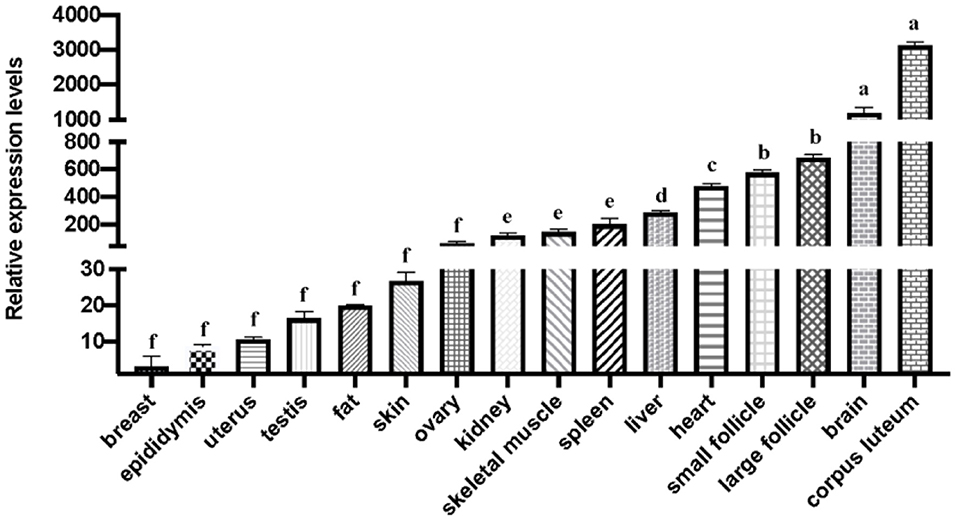
Figure 1. The mRNA expression of the goat SNX29 gene. The values with different letters (a, b, d, e, f) indicates differ significantly at P < 0.05.
InDel detection: Genotype frequency, linkage disequilibrium and haplotype analyses of the goat SNX29 gene
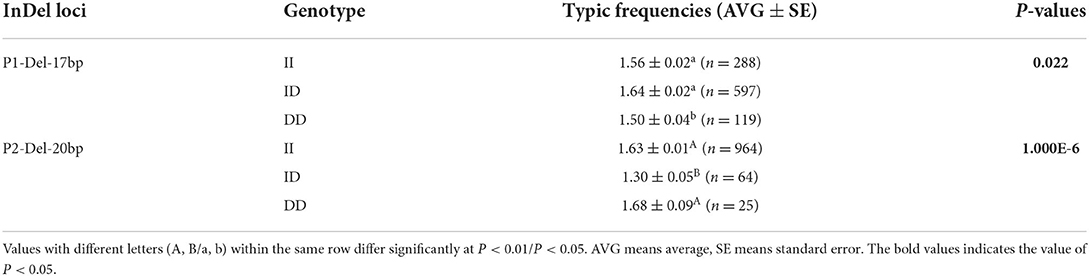
To further investigate the effect of SNX29 on reproduction, we screened the genetic variations of SNX29 and found that two InDel sites were polymorphic (P1-Del-17bp, P2-Del-20bp) among 11 pairs of primers. In the 17 bp deletion (n = 1,004), the frequency of ID genotype was much higher than that of II and DD. In the 20 bp deletion (n = 1,053), the number individuals with II genotype were much higher than the others. Besides, the P1-Del-17bp and P2-Del-20bp identified in the SNX29 gene were not consistent the HWE (P < 0.05). According to PIC values, the detected SNX29 P1-Del-17bp showed medium genetic diversity, while P2-Del-20bp exhibited low polymorphism in SBWC goats.
In the linkage disequilibrium (LD) analysis, the D' and r2 values for the linkage between P1-Del-17bp and P2-Del-20bp in the SNX29 gene were 0.27 and 0.01, respectively, which indicated that these two polymorphisms were not strongly linked. The haplotype analysis results for the SNX29 gene generated four haplotypes, and I1I2 had the highest frequency (Figure 2). Between the litter size and the two different combination genotypes of two InDels showed that they differed significantly (P < 0.05) in least-squares mean and standard error (Figure 3).
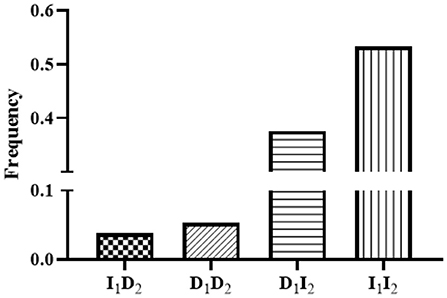
Figure 2. Haplotype frequencies of the two InDels within the goat SNX29 gene. The 1 means P1-Del-17bp, 2 means P2-Del-20bp.
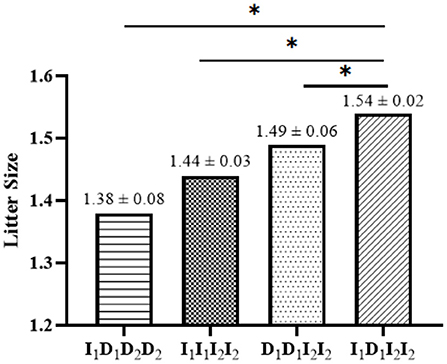
Figure 3. Least squares mean and standard error for litter size of different combination genotypes of the two InDels within the SNX29 gene in SBWC goats. The 1 means P1-Del-17bp, 2 means P2-Del-20bp; The * means P < 0.05.
CNV detection: Frequency of the goat SNX29 gene genotypes
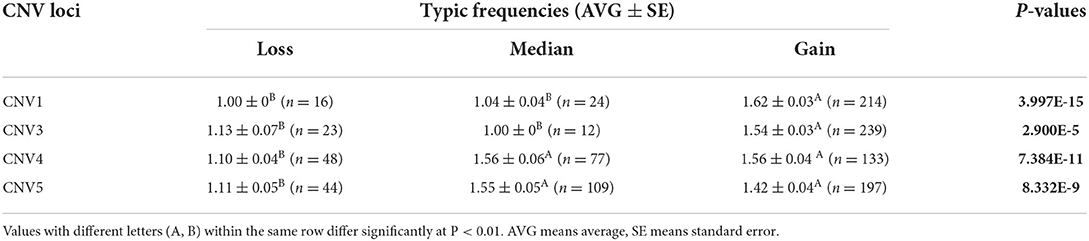
There were five CNV loci in the goat genome, and four of them (CNV1, CNV3, CNV4, CNV5) showed three variant types, while CNV2 had one variant type in SBWC goats. In four loci, the individual number of Gain genotype (CNV1 = 214, CNV3 = 239, CNV4 = 133, CNV5 = 197) was higher than Loss and Median, and the Gain frequencies in four sites (CNV1 = 0.834, CNV3 = 0.872, CNV4 = 0.516, CNV5 = 0.563) were significantly higher (Table 2).
Association analyses of InDel and CNV of the goat SNX29 gene
It has been found that two InDel loci were related to litter size by association analyses. The 17 bp deletion (P = 0.022) and the 20 bp deletion (P = 1.000E-6) within the SNX29 gene were discovered to be significantly associated with litter size, in which individuals the DD genotype of P2-Del-20bp (P < 0.01) and the ID genotype of P1-Del-17bp (P < 0.05) had larger litter size (Table 3; Figure 4). Moreover, the χ2 test showed that the genotype distribution of P1-Del-17bp (χ2 =10.870, P = 0.004) and P2-Del-20bp (χ2 = 27.924, P = 8.640E-7) were distinct between mothers of single-lamb and multi-lamb in 2,057 SBWC goats (Table 4; Figure 5).
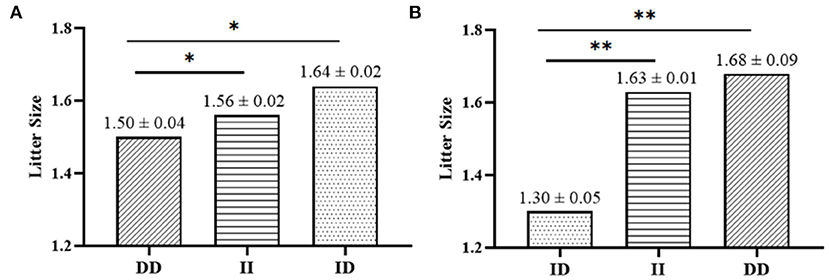
Figure 4. Association analyses between litter size and the two InDels in SBWC goats; (A) P1-Del-17bp, (B) P2-Del-20bp. * means P < 0.05; ** means P < 0.01.
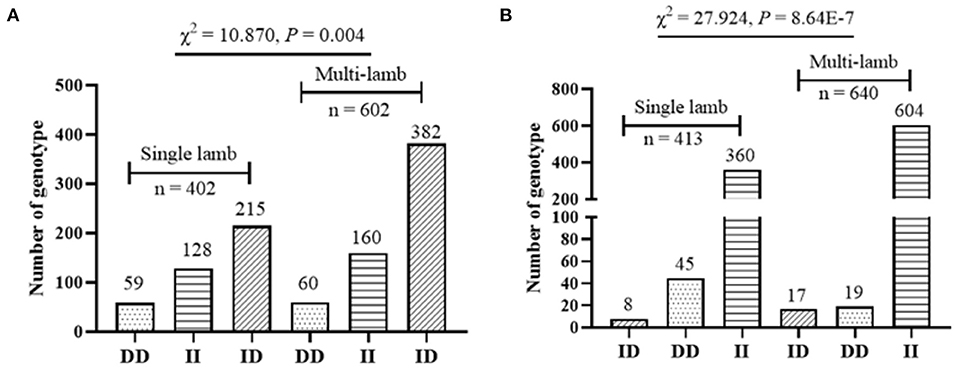
Figure 5. Genotype distribution between mothers of single-lamb and multi-lamb in SBWC goats; (A) P1-Del-17bp; (B) P2-Del-20bp.
Additionally, four CNV loci also had significant correlations with litter size (CNV1 = 3.997E-15, CNV3 = 2.900E-5, CNV4 = 7.387E-11, CNV 5 = 8.332E-9) in our detected population (n = 1,402). In CNV5 individuals with median genotype were superior to those with loss or Gain genotypes in term of litter size, and in the other three CNVs, the gain genotype performed better reproductive traits (Table 5; Figure 6). Besides, the χ2 analysis results demonstrated the two groups of goats with different litter types were significantly different (P < 0.01) (Table 6).
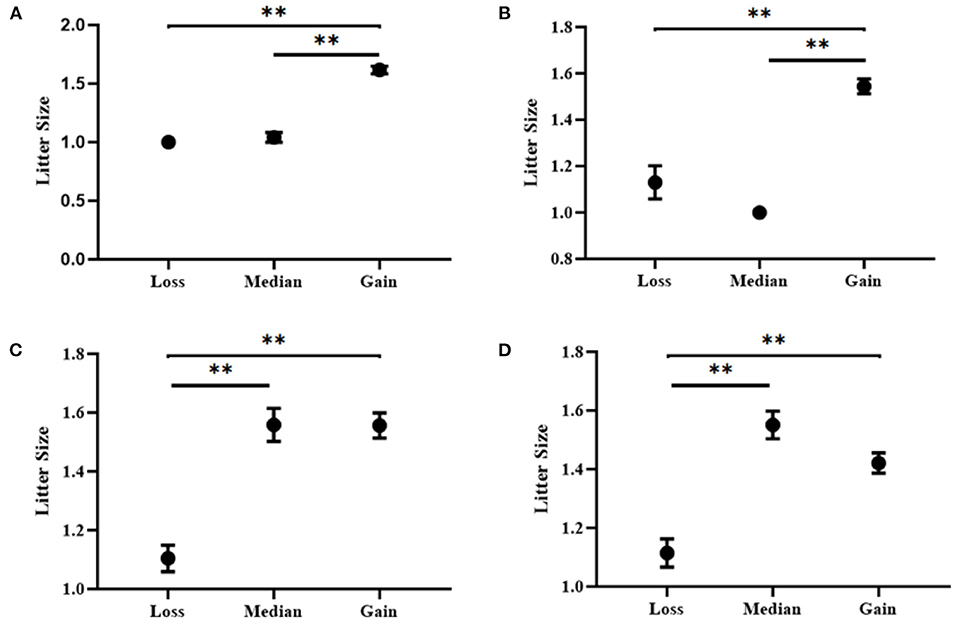
Figure 6. Association analyses between litter size and copy number variation types in SBWC goats. (A) CNV1; (B) CNV3; (C) CNV4; (D) CNV5. ** means P < 0.01.
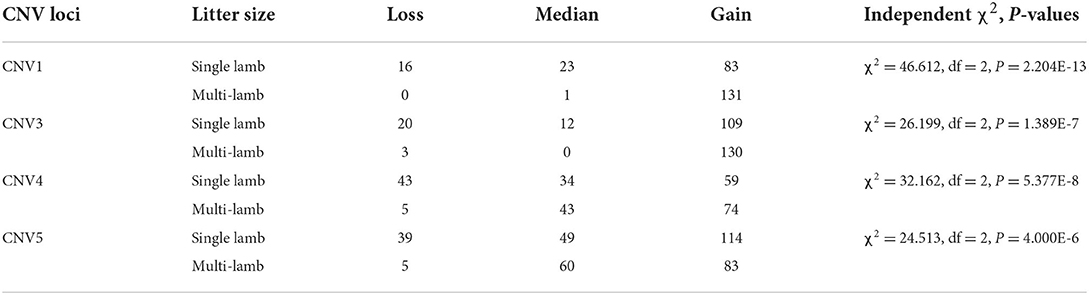
Table 6. The distribution of different CNVs between mothers of single-lamb and multi-lamb population.
Discussion
To explore the function of the SNX29 gene, the mRNA expression of this gene was first detected in different goat tissues, and we found that it was highly expressed in the corpus luteum and other reproductive organs. Previous studies have shown that SNX24 regulates the level of estrogen (9). These results suggested that the SNX29 gene may have a fertility-related function and plays a crucial role in reproductive performance.
As the main types of genetic variations, InDels and CNVs have contributed greatly to MAS. Although previous studies have reported that CNV27 in the SNX29 gene significantly affects the growth traits in goats (37), and that the two InDels within this gene are significantly correlated with chest width, chest depth, chest circumference and hip width in goats (30), the relationship between InDel and CNV and litter size of goats has not been studied. Thus, in this study, 11 InDel loci and five CNV loci were retrieved from the database. The results showed that only two InDel loci and four CNV loci were polymorphic in SBWC goats, whereas other loci exhibited only a single mutation type. Despite the fact that all variants are located in intronic regions, the mutations are located at non-coding regions on chromosomes may affect the binding of transcription factors (38).
Furthermore, the result of the LD analysis showed no linkage between the two InDel loci, which indicates that the two mutation loci are physically far apart on the chromosomes (39). Haplotype frequencies and different genotype combinations of the two InDels were significant in goats. Besides, the PIC of P2-Del-20bp exhibited low polymorphism and the PIC of P1-Del-17bp showed medium polymorphism, indicating that their solid potential value for selection in goat breeds (40). Moreover, these variants did not conform to HWE. This could be due to the fact that artificial intervention results in individuals not being able to naturally mate with each other. We also found that the two InDel loci had multiple genotypes. Interestingly, the individual number of three genotypes was not equal. The individuals with ID genotype were more than those with II and DD genotype in P1-Del-17bp. The number of II genotypes was more than the ID and DD genotype in P2-Del-20bp. Many factors can change the frequency of the three genotypes, such as isolation, genetic drift and directional mating (41). These outcomes suggest that the ID genotype of 17 bp deletion and the II genotype of 20 bp InDel have a positive effect on litter size (42). Meanwhile, in CNV5, individuals with median genotype were superior than those with loss or gain genotypes in term of litter size, and in other three CNVs, the gain genotype showed better reproductive traits, which could be due to genetic drift migration, mutation, gene recombination, and selection (43). Thus, we concluded that the individuals with the Gain genotype perform better in reproductive performance, and more individuals with Gain genotype are preserved in breeding. The InDel loci and CNV loci may also have a remarkable influence on the SNX29 mRNA expression in the corpus luteum, small follicles and large follicles. Therefore, further researches should be conducted to explore the relationship between variants and reproduction.
The association between the SNX29 gene variants and litter size was analyzed. Notably, association analysis showed that two InDel loci and four CNV loci were observably associated with the litter size (P < 0.01; P < 0.05), which supports our conjecture, while no previous studies have demonstrated that the SNX29 gene is involved in goat reproduction. Referring to the relevant literature, some genes have affected both neural and reproductive development in animals. For instance, it has been reported that DSCAML1 affects both neural development and reproductive traits in animals (44, 45). At the same time, growth traits and reproductive traits are inextricably linked. Some genes play a role in both reproductive traits and growth traits in livestock. For instance, the mutation of the PLAG1 gene has affected growth traits and reproductive traits in animals (21–23). Although the mechanisms of these genes that simultaneously affect growth, nerves, reproduction are not well understood, we speculated that the SNX29 gene also has the same function as the aforementioned-genes, which are involved in the development of animal growth, nerve, reproduction and other traits.
Collectively, goat SNX29 mRNA expression in the corpus luteum, small follicles and large follicles significantly differed from other tissues. Two InDels (P1-Del-17bp and P2-Del-20bp) and four CNVs (CNV1, CNV3, CNV4 and CNV5) were significantly association with litter size. This finding reveals the reproductive function of the SNX29 gene in livestock, and the InDel and CNV variations of the SNX29 gene in this study will make the use of MAS more widespread in goat breeding, providing a theoretical guidance for goat breed selection.
Conclusion
The SNX29 gene was highly expressed in the corpus luteum and other reproductive tissues, and the SNX29 gene loci (InDels and CNVs) were associated with litter size in SBWC goats. Thus, this gene may be an essential functional candidate gene for reproductive traits.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
Ethics statement
The animal study was reviewed and approved by Northwest A&F University.
Author contributions
Sample collection: QW, YB, ZW, and HZ. Experimental operation: QW, YB, and ZW. Data collation and analysis: QW, YB, HZ, and XW. Article writing: QW. Paper revision and editing: QW, YB, CP, and XW. Project management: CP, XW, and ML. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China [No. 32002166].
Acknowledgments
We sincerely thank Shaanxi Yulin goat farm for providing us with samples; We also would like to thank the Dr. Lei Qu, Dr. Hailong Yan, Dr. Jinwang Liu from Yulin University for their help in sample collection. We also thank for Dr. ML for her help in the College of Animal Science and Technology of Northwest A&F University, Hunan Agricultural University, as well as the staff of the Life Science Research Core Services (LSRCS) for their support.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2022.981315/full#supplementary-material
References
1. Liu N, Cui W, Chen M, Zhang X, Song X, Pan CA, et al. 21-bp indel within the LLGL1 gene is significantly associated with litter size in goat. Anim Biotechnol. (2021) 32:213–8. doi: 10.1080/10495398.2019.1677682
2. Cui Y, Chen R, Lv X, Pan C. Detection of coding sequence, mRNA expression and three insertions/deletions (indels) of KDM6A gene in male pig. Theriogenology. (2019) 133:10–21. doi: 10.1016/j.theriogenology.2019.04.023
3. Zhang S, Jiang E, Wang K, Zhang Y, Yan H, Qu L, et al. Two insertion/deletion variants within SPAG17 gene are associated with goat body measurement traits. Animals. (2019) 9:379. doi: 10.3390/ani9060379
4. Tan CT Yu H, Yang Y, Xu X, Chen M, Rudd JC, et al. Development and validation of KASP markers for the greenbug resistance gene Gb7 and the Hessian fly resistance gene H32 in wheat. Theor Appl Genet. (2017) 130:1867–84. doi: 10.1007/s00122-017-2930-4
5. Hao X, Wang Y, Ren F, Zhu S, Ren Y, Jia B, et al. SNX25 regulates TGF-β signaling by enhancing the receptor degradation. Cell Signal. (2011) 23:935–46. doi: 10.1016/j.cellsig.2011.01.022
6. Lin YJ, Chang JS, Liu X, Lin TH, Huang SM, Liao CC, et al. Sorting nexin 24 genetic variation associates with coronary artery aneurysm severity in Kawasaki disease patients. Cell Biosci. (2013) 3:44. doi: 10.1186/2045-3701-3-44
7. Worby CA, Dixon JE. Sorting out the cellular functions of sorting nexins. Nat. Rev. Mol. Cell Biol. (2002) 3:919–31. doi: 10.1038/nrm974
8. Chandra M, Collins BM. The phox homology (PX) domain. Adv Exp Med Biol. (2019) 1111:1–17. doi: 10.1007/5584_2018_185
9. Wright PK, May FE, Darby S, Saif R, Lennard TW, Westley BR, et al. Estrogen regulates vesicle trafficking gene expression in EFF-3, EFM-19 and MCF-7 breast cancer cells. Int J Clin Exp Pathol. (2009) 2:463–75.
10. Yong X, Hu W, Zhou X, Wang J, Burstein E, Jia D, et al. Expression and purification of the SNX1/SNX6 complex. Protein Expr Purif. (2018) 151:93–8. doi: 10.1016/j.pep.2018.06.010
11. de Andrade Melo-Sterza F, Poehland R. Lipid metabolism in bovine oocytes and early embryos under in vivo, in vitro, stress conditions. Int J Mol Sci. (2021) 22:3421. doi: 10.3390/ijms22073421
12. Ito S, Hirabayashi K, Moriishi K, Matsui Y, Moriya K, Koike K, et al. Novel sex-dependent differentially methylated regions are demethylated in adult male mouse livers. Biochem Biophys Res Commun. (2015) 462:332–8. doi: 10.1016/j.bbrc.2015.04.137
13. Iossifov I, O'Roak BJ, Sanders SJ, Ronemus M, Krumm N, Levy D, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. (2014) 515:216–21. doi: 10.1038/nature13908
14. Xia L, Ou J, Li K, Guo H, Hu Z, Bai T, et al. Genome-wide association analysis of autism identified multiple loci that have been reported as strong signals for neuropsychiatric disorders. Autism Research. (2020) 13:382–96. doi: 10.1002/aur.2229
15. Steidl C, Shah SP, Woolcock BW, Rui L, Kawahara M, Farinha P, et al. MHC class II transactivator CIITA is a recurrent gene fusion partner in lymphoid cancers. Nature. (2011) 471:377–81. doi: 10.1038/nature09754
16. Børglum AD, Demontis D, Grove J, Pallesen J, Hollegaard MV, Pedersen CB, et al. Genome-wide study of association and interaction with maternal cytomegalovirus infection suggests new schizophrenia loci. Mol Psychiatry. (2014) 19:325–33. doi: 10.1038/mp.2013.2
17. Hill WD, Marioni RE, Maghzian O, Ritchie SJ, Hagenaars SP, McIntosh AM, et al. combined analysis of genetically correlated traits identifies 187 loci and a role for neurogenesis and myelination in intelligence. Mol Psychiatry. (2019) 24:169–81. doi: 10.1038/s41380-017-0001-5
18. Chen JH, Zhao Y, Khan RAW Li ZQ, Zhou J, Shen JW, et al. SNX29, a new susceptibility gene shared with major mental disorders in Han Chinese population. World J Biol Psychiatry. (2021) 22:526–34. doi: 10.1080/15622975.2020.1845793
19. Wang Z, Ma H, Xu L, Zhu B, Liu Y, Bordbar F, et al. Genome-wide scan identifies selection signatures in Chinese wagyu cattle using a high-density SNP array. Animals. (2019) 9:296. doi: 10.3390/ani9060296
20. Dong Q, Liu H, Li X, Wei W, Zhao S, Cao JA, et al. genome-wide association study of five meat quality traits in Yorkshire pigs. Front Agric Sci Eng. (2014) 1:137–43. doi: 10.15302/J-FASE-2014014
21. Li Z, Wu M, Zhao H, Fan L, Zhang Y, Yuan T, et al. The PLAG1 mRNA expression analysis among genetic variants and relevance to growth traits in Chinese cattle. Anim Biotechnol. (2020) 31:504–11. doi: 10.1080/10495398.2019.1632207
22. Wei Z, Wang K, Wu H, Wang Z, Pan C, Chen H, et al. Detection of 15-bp deletion mutation within PLAG1 gene and its effects on growth traits in goats. Animals. (2021) 11:2064. doi: 10.3390/ani11072064
23. Engle BN, Hayes BJ. Genetic variation in PLAG1 is associated with early fertility in Australian Brahman cattle. J Anim Sci. (2022) 2022:100. skac084. doi: 10.1093/jas/skac084
24. Zhang X, Zhang S, Tang Q, Jiang E, Wang K, Lan X, et al. Goat sperm associated antigen 17 protein gene (SPAG17): Small and large fragment genetic variation detection, association analysis, and mRNA expression in gonads. Genomics. (2020) 112:5115–21. doi: 10.1016/j.ygeno.2020.09.029
25. Wu J, Li A, Cai H, Zhang C, Lei C, Lan X, et al. Intron retention as an alternative splice variant of the cattle ANGPTL6 gene. Gene. (2019) 709:17–24. doi: 10.1016/j.gene.2019.05.031
26. Zhang X, Yu S, Yang Q, Wang K, Zhang S, Pan C, et al. Goat boule: isoforms identification, mRNA expression in testis and functional study and promoter methylation profiles. Theriogenology. (2018) 116:53–63. doi: 10.1016/j.theriogenology.2018.05.002
27. Li J, Zhu X, Ma L, Xu H, Cao X, Luo R, et al. Detection of a new 20-bp insertion/deletion (indel) within sheep PRND gene using mathematical expectation (ME) method. Prion. (2017) 11:143–50. doi: 10.1080/19336896.2017.1300740
28. Bi Y, He L, Feng B, Lan X, Song X, Qu L, et al. 5-bp mutation within MSTN/GDF8 gene was significantly associated with growth traits in Inner Mongolia white cashmere goats. Anim Biotechnol. (2021) 32:610–5. doi: 10.1080/10495398.2020.1736088
29. Bi Y, Feng W, Kang Y, Wang K, Yang Y, Qu L, et al. Detection of mRNA expression and copy number variations within the goat FecB gene associated with litter size. Front Vet Sci. (2021) 8:758705. doi: 10.3389/fvets.2021.758705
30. Bi Y, Chen Y, Xin D, Liu T, He L, Kang Y, et al. Effect of indel variants within the sorting nexin 29 (SNX29) gene on growth traits of goats. Anim Biotechnol. (2020) 19:1–6. doi: 10.1080/10495398.2020.1846547
31. Cui Y, Yan H, Wang K, Xu H, Zhang X, Zhu H, et al. Insertion/Deletion within the KDM6A gene is significantly associated with litter size in goat. Front Genet. (2018) 9:91. doi: 10.3389/fgene.2018.00091
32. Jia W, Wu X, Li X, Xia T, Lei C, Chen H, et al. Novel genetic variants associated with mRNA expression of signal transducer and activator of transcription 3 (STAT3) gene significantly affected goat growth traits. Small Ruminant Research. (2015) 129:25–36. doi: 10.1016/j.smallrumres.2015.05.014
33. Zhang S, Kang Z, Sun X, Cao X, Pan C, Dang R, et al. Novel lncRNA lncFAM200B: molecular characteristics and effects of genetic variants on promoter activity and cattle body measurement traits. Front Genet. (2019) 10:968. doi: 10.3389/fgene.2019.00968
34. Yang Q, Zhang S, Li J, Wang X, Peng K, Lan X, et al. Development of a touch-down multiplex PCR method for simultaneously rapidly detecting three novel insertion/deletions (InDels) within one gene: an example for goat GHR gene. Anim Biotechnol. (2019) 30:366–71. doi: 10.1080/10495398.2018.1517770
35. Li J, Zhang S, Erdenee S, Sun X, Dang R, Huang Y, et al. Nucleotide variants in prion-related protein (testis-specific) gene (PRNT) and effects on Chinese and Mongolian sheep phenotypes. Prion. (2018) 12:185–96. doi: 10.1080/19336896.2018.1467193
36. Akhatayeva Z, Cao C, Huang Y, Zhou Q, Zhang Q, Guo Z, et al. Newly reported 90-bp deletion within the ovine BMPRIB gene: Does it widely distribute, link to the famous FecB (pQ249R) mutation, and affect litter size? Theriogenology. (2022) 189:222–9. doi: 10.1016/j.theriogenology.2022.06.020
37. Liu M, Woodward-Greene J, Kang X, Pan MG, Rosen B, Van Tassell CP, et al. Genome-wide CNV analysis revealed variants associated with growth traits in African indigenous goats. Genomics. (2020) 112:1477–80. doi: 10.1016/j.ygeno.2019.08.018
38. Michael AK, Thomä NH. Reading the chromatinized genome. Cell. (2021) 184:3599–611. doi: 10.1016/j.cell.2021.05.029
39. Huang Y, Su P, Akhatayeva Z, Pan C, Zhang Q, Lan X, et al. Novel InDel variations of the Cry2 gene are associated with litter size in Australian white sheep. Theriogenology. (2022) 179:155–61. doi: 10.1016/j.theriogenology.2021.11.023
40. Yang Y, Hu H, Mao C, Jiang F, Lu X, Han X, et al. Detection of the 23-bp nucleotide sequence mutation in retinoid acid receptor related orphan receptor alpha (RORA) gene and its effect on sheep litter size. Anim Biotechnol. (2020) 33:70–8. doi: 10.1080/10495398.2020.1770273
41. Zhang Y, Wang K, Liu J, Zhu H, Qu L, Chen H, et al. An 11-bp indel polymorphism within the CSN1S1 gene is associated with milk performance and body measurement traits in Chinese goats. Animals. (2019) 9:1114. doi: 10.3390/ani9121114
42. Zhou T, Wei H, Li D, Yang W, Cui Y, Gao J, et al. novel missense mutation within the domain of lysine demethylase 4D (KDM4D) gene is strongly associated with testis morphology traits in pigs. Anim Biotechnol. (2020) 31:52–8. doi: 10.1080/10495398.2018.1531880
43. Wang K, Hui Y, Zhang S, Wang M, Yan H, Zhu H, et al. Deletion mutation within the ATBF1 gene is strongly associated with goat litter size. Anim Biotechnol. (2020) 31:174–80. doi: 10.1080/10495398.2018.1561459
44. Wang K, Kang Z, Jiang E, Yan H, Zhu H, Liu J, et al. Genetic effects of DSCAML1 identified in genome-wide association study revealing strong associations with litter size and semen quality in goat (capra hircus). Theriogenology. (2020) 146:20–5. doi: 10.1016/j.theriogenology.2020.01.079
Keywords: sorting nexin 29 (SNX29), mRNA expression, insertion/deletion (INDEL), copy number variation (CNV), goats, litter size
Citation: Wang Q, Bi Y, Wang Z, Zhu H, Liu M, Wu X and Pan C (2022) Goat SNX29: mRNA expression, InDel and CNV detection, and their associations with litter size. Front. Vet. Sci. 9:981315. doi: 10.3389/fvets.2022.981315
Received: 29 June 2022; Accepted: 20 July 2022;
Published: 10 August 2022.
Edited by:
Sebastián Demyda-Peyrás, Universidad Nacional de La Plata, ArgentinaReviewed by:
Guangxin E., Southwest University, ChinaZhuanjian Li, Henan Agricultural University, China
Copyright © 2022 Wang, Bi, Wang, Zhu, Liu, Wu and Pan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chuanying Pan, Y2h1YW55aW5ncGFuQDEyNi5jb20=; Xianfeng Wu, d3V4aWFuZmVuZzMwODBAMTYzLmNvbQ==
 Qian Wang1
Qian Wang1 Yi Bi
Yi Bi Haijing Zhu
Haijing Zhu Xianfeng Wu
Xianfeng Wu Chuanying Pan
Chuanying Pan