
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci. , 22 August 2022
Sec. Veterinary Infectious Diseases
Volume 9 - 2022 | https://doi.org/10.3389/fvets.2022.966533
Mastitis is an economically important disease in the dairy industry, which is caused by various infectious pathogens. There is limited information known about the situation of drug resistance and virulence factors of Staphylococcus aureus (S. aureus) in mastitis bovine milk in Anhui. Therefore, a total of 125 fresh milk samples from clinically mastitis-positive bovine animals were collected. The bacteria pathogens were identified via bacterial culture, Gram staining, biochemical analysis, DNA extraction, 16s rRNA amplification, and phylogenetic analysis. Drug resistance analyses were performed through drug-resistant genes and virulence genes amplification. Results showed that a total of 24.8% (31/125) bacterial isolates were isolated and identified as S. aureus by Gram straining, biochemical reactions, and 16 s rRNA genes blasting. Multiple sequence alignment analysis found that the current isolates were highly similar (96.9–100.0%) to previous isolates. Phylogenetic analysis demonstrated that S. aureus was similar with MK809241.1 isolated from food in China and wCP030426.1 isolated from a person in the United States. The bacterial isolates were detected resistant to 11 antibiotics, such as Penicillin G, SXT, Ciprofloxacin, Norfloxacin, Polymyxin B, Levofloxacin, Chloramphenicol, Clindamycin, Clarithromycin, Erythromycin, and Spectinomycin. Drug-resistant genes of blaZ, ermC, rpoB, and ant (4')-la were successfully amplified. Virulence genes of hla, nuc, clfa, and eta were found in S. aureus bacteria. The current study isolated S. aureus from milk samples and revealed its drug-resistant situation, drug-resistant genes, and virulence genes. Hence, regular monitoring of S. aureus in milk samples from dairy cows may contribute to the prevention and treatment of public health concerns causing bacteria in this region.
Mastitis is the inflammation of the udder due to the infection of a variety of bacteria that enter the mammary glands and damage them (1). Mastitis is a serious livestock disease, which usually causes significant economic losses along with clinical and sub-clinical symptoms in highly productive dairy animals (2). The opportunistic pathogen Staphylococcus aureus (S. aureus) is a representative microflora species, which is commonly disseminated on healthy skin and mucous membranes of people and animals (3–7). Previous studies reported that over 50 species of Staphylococcus could lead to mastitis, among them S. aureus is generally recognized as one of the most frequently examined pathogens in cattle (8). Moreover, it is causing udder inflammation in cows, which leads to infections or intoxications to consumers (9, 10). In Asia, S. aureus is considered the major etiologic pathogen of mastitis in cattle and buffaloes (11). The infection of S. aureus in dairy cows not only harms the health of animals and milk production or quality (2) but also potentially transmits this pathogen to herdsmen or citizens (9).
Nowadays, various kinds of antibiotics are used for the treatment and control of mastitis in dairy animals. As a result of extensive and irrational use of antibiotics, drug-resistant bacteria are increasing day by day and becoming a major problem for human health worldwide (12–21). The public health concern is that drug-resistant or multi-drug-resistant S. aureus strains are occasionally reported to cause serious dermatitis, pneumonia, mastitis, septicemia, myelitis, and bacterial endocarditis (3, 4). Previously, the prevalence of penicillin resistance to S. aureus in mastitis cases was more than 50% in the United States of America, Ireland, and England (22). Multi-drug-resistant S. aureus strains were also reported in Bangladesh, Jordan, Ethiopia, and the Czech Republic (10, 11, 23, 24).
Anhui province is located in the Yangtze River Delta region in East China, with a northern latitude of 29°41′-34°38′ and an east longitude of 114°54′-119°37′ (Supplementary Figure S1). The warm, temperate, and subtropical climates contribute to the prosperous agricultural development in this region. In 2020, there were 948,000 head of cattle and 376,400 tons of milk produced in the Anhui province (National Bureau of Statistics, https://data.stats.gov.cn/index.htm). A previous study reported that the prevalence of S. aureus in dairy cows was 29.1% on farms in this province (25). However, limited information is available about the situation of drug resistance and virulence factors of S. aureus in mastitis bovine milk products in Anhui. Herein, this study was carried out to explore the molecular characterization of multi-drug-resistant S. aureus in mastitis bovine milk in the Anhui province.
A total of 125 milk samples were aseptically collected from 125 Holstein cows with clinical mastitis in commercial sterile tubes (Thermo Fisher Nunc™, China) by a veterinarian from a dairy farm in 2022 in Anhui, China. Each cow had local signs of inflammation, such as hot, painful, and enlarged mammary glands, which were confirmed by a veterinarian. The collected samples were shipped to a clinical laboratory of Hebei Agricultural University by dry ice (Meijie Dry Ice Technology Co., LTD, Hefei, China) for further processing.
From each milk sample, 0.1 ml raw milk was mixed with 0.9 ml Luria-Bertani (LB) broth (Qingdao Hope Bio-Technology Co., Ltd) in a 5 ml tube and incubated at 36 ± 1°C in an orbital shaker for an h for bacterial enrichment. Then, enriched bacteria were plated on Mannitol salt agar (Qingdao Hope Bio-Technology Co., Ltd) in Petri dishes and incubated at 36 ± 1°C for 24 h. The single colonies were utilized for morphology, Gram staining, and biochemical analysis (Hangzhou Microbial Reagent Co., Ltd). The biochemical reactions, including N-acetyl-glucosamine, trehalose, sucrose, urea, mannose, maltose, xylose, nitrate reduction, N-mannitol, and lactose, were performed for the identification of bacteria.
A single colony was cultivated in a sterile tube with 5 ml of LB broth and incubated at 36 ± 1°C in an orbital shaker for 24 h, then 1 ml medium was taken and centrifuged at 8,000 rpm for 5 min for DNA extraction. The DNA extraction of bacteria was employed through a commercial bacterial genomic DNA extraction kit (Solarbio life sciences, China) as the previous study described (1). Then, the 16s rRNA gene was amplified by using universal primers (27F, Forward, 5'-AGA GTT TGA TCM TGG CTC AG-3', reverse, 1492R, and 5'-TAC GGY TAC CTT GTT ACG ACT T-3'). The 25 μl PCR reaction mixture contains 1 μl DNA template, 1 μl of forward and reverse primers, respectively, 12.5 μl Taq PCR Master Mix (Sangon Biotech, China), and 9.5 μl distilled water. The PCR amplification contained 35 PCR cycles with 95°C for 30 s, 62°C for 35 s, and 72°C for 45 s in each cycle after an initial hot start at 95°C for 3 min and ending with 72°C for 5 min. After that, all the 16s rRNA PCR products were examined through 0.8% agarose gel electrophoresis. Then, the positive 16s rRNA PCR amplified products with the expected size were purified by using the GenElute™ Gel Extraction Kit (Catalog number NA1111, Sigma-Aldrich, The United States of America) according to the manufacturer's explanatory memorandum.
All the purified 16s rRNA PCR samples were further subjected to bidirectional gene sequencing via a 3730xl DNA Analyzer at Sangon Biotech (Shanghai, China). Multiple sequence alignments were performed between 16s rRNA of S. aureus bovine milk isolates and references genes of 16s rRNA of bacteria available in the NCBI database by piloting Lasergene (Version 7.0). These used reference strains were S. aureus strain RM_AST_SA006 (MK809241.1), S. aureus strain MRSA-5043 (MT250912.1), S. aureus strain 18BWI (KX456106.1), S. aureus strain 3-355MR (OK090920.1), S. aureus strain DSM 20231 (Type) (MN652637.1), S. aureus strain ER04320.3 (CP030426.1), S. aureus strain AR_0226 (CP029664.1), Streptococcus alactolyticus (LC632504.1), Streptococcus oricebi JCM 30719 (LC638727.1), Streptococcus oralis 4-KN-2020 (LC545467.1), Streptococcus mitis 3-KN-2020 (LC545466.1), Streptococcus loxodontisalivarius JCM 19287 (LC589217.1), Streptococcus troglodytae JCM 18038 (LC521977.1), Streptococcus dentasini JCM 17943 (LC520002.1), Streptococcus fryi JCM 16387 (LC519990.1), Streptococcus sp. Marseille-P5794 (LR597666.1), Streptococcus thermophilus YS5 (LC485981.1), and Escherichia coli JCM 16946 (LC682250.1) (out group).
The phylogenetic relationship analysis between the 16s rRNA gene of S. aureus bovine milk isolates and reference bacteria genes was performed by utilizing MEGA (Version 6.0) through neighbor-joining (NJ) methods computing the distances as described in the previous study (11). The stability of branches was assessed after bootstrapping replicates (n = 1,000).
The disk diffusion method was performed to examine the antimicrobial sensitivity profile of S. aureus isolated from bovine milk samples, according to the criteria introduced by the Clinical and Laboratory Standards Institute (CLSI, 2021). Plates with LB agar were used with 18 commercial antimicrobial agents (Hangzhou Binhe Microorganism Reagent Co., Ltd., China), namely penicillin G (10 μg, Catalog # C001), cefoxitin (30 μg, Catalog # C058), SXT (1.25 μg, Catalog # C027), ciprofloxacin (5 μg, Catalog # C045), norfloxacin (10 μg, Catalog # C033), vancomycin (30 μg, Catalog # C030), polymyxin B (300 μg, Catalog # C025), levofloxacin (5 μg, Catalog # C066), macrodantin (300 μg, Catalog # C028), tetracycline (30 μg, Catalog # 021), chloramphenicol (30 μg, Catalog # 022), oxacillin (1 μg, Catalog # C004), phosphonomycin (200 μg, Catalog # C092), clindamycin (2 μg, Catalog # C093), minocycline (30 μg, Catalog # C046), clarithromycin (15 μg, Catalog # C063), erythromycin (15 μg, Catalog # C023), and spectinomycin (30 μg, Catalog # C036), for antimicrobial sensitivity detection. Each examination was performed three times. Laboratory stored S. aureus (ATCC 25923) and E. coli (ATCC 25922) were employed as positive and negative control strains, respectively.
Genomic DNA from drug-resistant S. aureus isolates was extracted using a commercial bacterial genomic DNA extraction kit (Solarbio life sciences, China) as previous studies described (1). The resistant genes and virulence genes were amplified by piloting specific primers (Table 1) as described in a previous study (11–59). The 25 μl PCR reaction mixture contains 2 μl DNA template, 1 μl of forward and reverse primers, respectively, 12.5 μl of the Taq PCR Master Mix (Sangon Biotech, China), and 8.5 μl of distilled water. The PCR amplification contained 30 PCR cycles with 95°C for 30 s, Tm for 45 s, and 72°C for 60 s in each cycle after an initial hot start at 95°C for 5 min and ending with 72°C for 10 min. After that, all the PCR products were examined through 1.2% agarose gel electrophoresis.
A total of 24.8% (31/125) of bacteria isolates were isolated by a Mannitol salt agar plate from clinically positive samples. Gram staining showed Gram-positive purple grape-globose bacteria, and biochemical reactions revealed positive results of N-acetyl-glucosamine, trehalose, sucrose, urea, mannose, maltose, N-mannitol, and lactose, respectively, while negative results were observed for xylose and nitrate reduction. The agarose gel electrophoresis indicated ~1,500 bp bands of 16 s rRNA genes of bacteria, and then those positive PCR bands were purified, sequenced, and standard nucleotide blasting was performed with NCBI databases (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastn&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome). The confirmed 16 s rRNA gene of S. aureus was deposited into the NCBI database with accession number ON138912-ON138914.
Multiple sequence alignment analysis found that the current isolates ON138912, ON138913, and ON138914 were highly similar (96.9–100.0%) to MK809241.1, MT250912.1, MT023385.1, KX456106.1, OK090920.1, MN652637.1, and CP030426.1, especially ON138912 and ON138913 isolates (Figure 1). Phylogenetic analysis demonstrated that ON138912 and ON138913 clade with MK809241.1 were isolated from food in China, whereas ON138914 clade with CP030426.1 was isolated from a person from the USA (Figure 2).
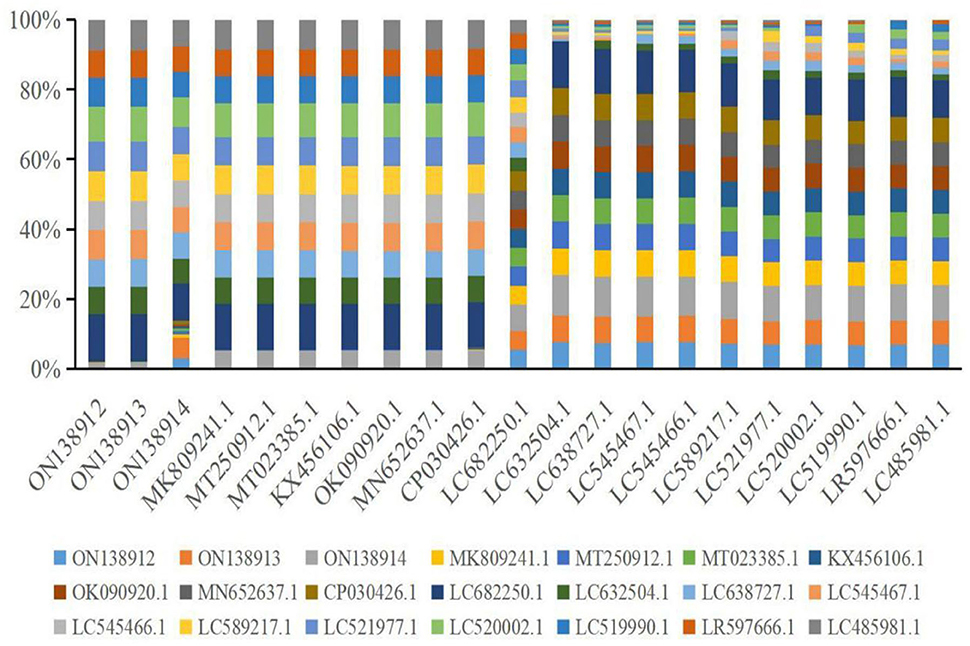
Figure 1. Multiple alignments analysis of 16s rRNA gene of Staphylococcus aureus (S. aureus) with reference strains.
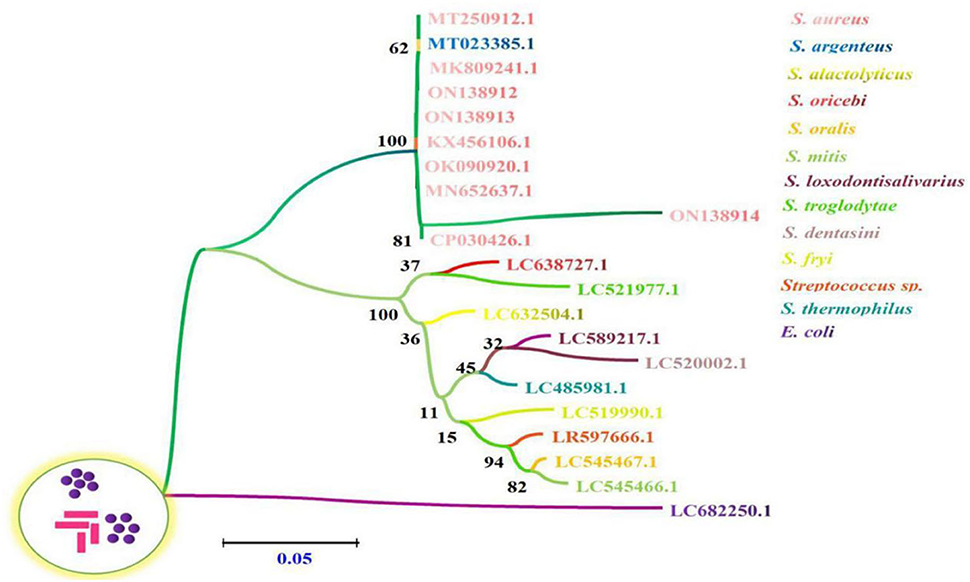
Figure 2. The phylogenetic relationships of 16s rRNA gene between S. aureus sequences derived from bovine milk samples and reference sequences by employing a Neighbor-Joining (NJ) method via Kimura two-parameter analysis. The number of nodes indicates the bootstrap values. Bootstrap values >50% from 1,000 replicates are shown on the nodes.
The present bacterial isolates were detected resistant to 11 antibiotics, namely penicillin G, SXT, ciprofloxacin, norfloxacin, polymyxin B, levofloxacin, chloramphenicol, clindamycin, clarithromycin, erythromycin, and spectinomycin (Figure 3), with drug resistance rate ranging from 3.22 (1/31) to 100% (31/31). In these drug resistance S. aureus, double antibiotic resistance to eight antibiotics was examined with the prevalence of 29.03 (9/31) to 93.55% (29/31) (Figure 4). Then, according to the antibiotic-resistant results, the amplification of the 14 commonly known drug-resistant genes (blaZ, ermC, rpoB, and ant(4')-la) was successfully amplified (Figure 5). The virulence genes of hla, nuc, clfa, and eta were found in the current S. aureus bacteria (Figure 5).
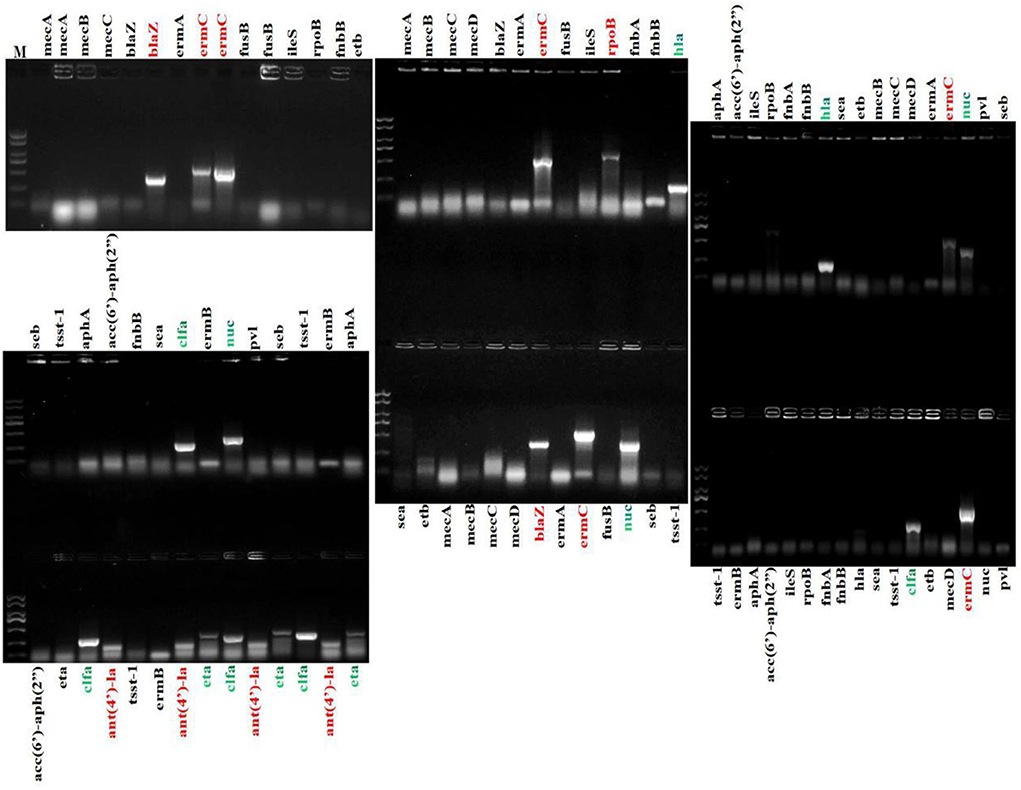
Figure 5. Amplification of drug resistance genes and virulence genes of S. aureus. Marker ladder: 2,000, 1,500, 1,000, 750, 500, 250, and 100 bp.
Mastitis is one of the major problems that severely affect the dairy sector globally (1). It has a prominent deteriorated impact on milk production, milk quality, and ruminant health and welfare with considerable economic damage (11). S. aureus is recognized as the third most important worldwide food-borne pathogen which produces bacterial enterotoxins (32). The polluted milk products were frequently mixed up with the S. aureus contamination (33). Milk and its related products are important macro-nutrients for consumers, which makes them vulnerable to pathogens contamination (34–40). With the emergence of anti-microbial S. aureus increasing day by day, it is important to regularly perform the monitoring and examination of the prevalence, and the molecular characterization of S. aureus in mastitis suspected milk samples.
In the current study, S. aureus was isolated from milk samples, which was identified by Gram staining and biochemical reactions. This was then confirmed by the amplification of 16 s rRNA genes of bacteria, sequencing, and blasting results. Multiple sequence alignment analysis found that the current isolates (ON138912, ON138913, and ON138914) were highly similar (96.9–100.0%) to previous isolates (MK809241.1, MT250912.1, MT023385.1, KX456106.1, OK090920.1, MN652637.1, and CP030426.1). Phylogenetic analysis demonstrated that ON138912 and ON138913 clade with MK809241.1 were isolated from food in China, whereas ON138914 clade with CP030426.1 was isolated from a person from the United States of America.
The prevalence of S. aureus isolated from milk samples was 24.8% (31/125), which is in line with the prevalence of S. aureus in cow bulk tank milk samples in the region of Balikesir in Turkey (28%) (4) and milk samples obtained from acute mastitis cases in Jordan (23). The prevalence was lower than in bovine mastitis milk samples in the US (46.6%), Indonesia (57%), and Bangladesh (72.7%) (11, 32, 41), but higher than in milk samples obtained from culled cows in Jordan (13.7%) (23), milk samples in cows in Ethiopia (12.5%) (10), and composite milk samples in dairy cows in South Africa (42). This variability between different study results could be attributed to the differences in geographical, study population, farm hygienic and management practices (such as breeds, farm size, the practice of hand milking, and absence of dry cow therapy), milking methods, and instruments employed (11, 42).
Herein, the four virulence genes of hla, nuc, clfa, and eta were examined in the current S. aureus bacteria, which differed from the six genes named sea, seb, seg, sei, selp, and tsst1 in a previous study in S. aureus isolated from tank milk samples in Turkey (43), and twenty-two genes (aur, splA, hlgC, hlb, lukE, seg, sei, sem, sen, seo, seu, seb, sec, sed, seh, lukD, sek, splB, hlgA, sel, seq, and hlgB) from milk samples in the US (32). Exfoliative toxin (eta) genes were rarely detected in S. aureus isolated from mastitis positive dairy animals. The present study and previous studies found eta gene in S. aureus from cows in Belgium and Poland (44, 45), which illustrated that the bovine S. aureus evolutionary precursors clones had an important relationship with human S. aureus clones (46). Hemolysins of staphylococcal were considered pivotal factors related to bacterial invasion and evading host immunity (47). Of these, alpha hemolysin (hla) toxin is commonly accepted as the most emphasized and characterized virulence factor in S. aureus (48). ClfA is one of the important S. aureus genes for adhesion to extracellular matrix proteins to colonize and establish infections (48). S. aureus nuclease (nuc) is highly related to the trapping of bacteria biofilm (49). Those four virulence genes examined in S. aureus isolated from milk samples may combine to make the bacterium a versatile pathogen (50).
For the prevention and control of mastitis in cattle, a wide range of antimicrobials are commonly used (51), however inappropriate and excessive use of antibiotics leads to resistant bacteria. In the present results, a high antibiotic resistance rate was found, which is in line with previous studies of drug resistant S. aureus in South Africa and Turkey (4, 42). The resistance of S. aureus to penicillin G, SXT, ciprofloxacin, norfloxacin, polymyxin B, clindamycin, clarithromycin, and erythromycin was significant, which indicates that it is unsuitable to employ those antibiotics in this region. In current findings, blaZ, ermC, rpoB, and ant(4')-Ia are successfully amplified, which is not in agreement with previous studies in which S. aureus harboring norA, aph (3′)-Ia, mecA, ant (6)-la, aph (3′) III, mph(C), msr(A), norA, and blaZ (32). β-lactamases (blaZ) is one of the important enzymes which resist penicillin (48, 52); the current results are in line with a previous study about blaZ in S. aureus (50). The drugs (clarithromycin, erythromycin, spectinomycin, clindamycin, and streptogramin B) resistant gene ermC, which is detected in S. aureus from milk samples in the present study are commonly found in staphylococci from mastitis positive animals in Brazil (53, 54). A gene point mutation associated with resistance against rifampicin of rpoB is likely found in S. aureus (55). Ant(4′)Ia is one of the important enzymes causing aminoglycoside resistance in S. aureus (56). Those four drug resistance genes found in S. aureus may further confirm multiple drug resistance bacteria in the study area.
Infected cattle at dairy farms may transmit pathogens to other animals and farm workers who have close contact with cows (57). The bacteria from raw milk may also cause food-poisoning infection through the ingestion of S. aureus contaminated milk (34). Therefore, the multi-resistance of S. aureus isolates found in cow milk samples is an important concern for animals and public health. The prevention and control strategy of mastitis caused by bacteria S. aureus could be achieved by pathogens isolation and characterization, infected animals' segregation, dry cow therapy, and timely treatment of infected cases (11). S. aureus is a potential reservoir for toxins that bring harmful effects on human health (26, 27, 45, 58). Therefore, there is a need to establish countermeasures to promote careful utilization of antibiotics to reduce drug resistance to S. aureus development in dairy cattle.
The current study isolated S. aureus from milk samples and revealed its drug-resistant situation, drug-resistant genes, and virulence genes. So, regular monitoring of S. aureus in milk samples from dairy cows may contribute to the prevention and treatment of public health concerns causing bacteria in this region.
The mastitis bovine milk samples were collected from a dairy farm, which is the limitation of this study. Therefore, it is recommended to conduct this study throughout the province for a better indication of multi-drug-resistant Staphylococcus aureus.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, ON138912.1, ON138913.1, ON138914.1.
All the procedures of the study were performed under the approval of Laboratory Animals Research Center of Hebei and Anhui province in P. R. China, and the Ethics Committee of Hebei Agricultural University.
JL: methodology and writing original draft. CB and ZH: supervision and visualization. XW, KM, and JQ: reagents, materials, and analysis of tools. JL, XW, KM, and FA: writing review and editing. CB, XW, and ZH: conceptualization, funding, and resources. All authors contributed to the article and approved the submitted version.
The current research was supported by the establishment and application of a new mode for prevention and control of mixed infection of main animal diseases, Shandong Province agricultural major application technology innovation project (SD2019XM007); the evaluation test of animal disease detection reagents of Hangzhou Bori Technology Co., Ltd. (LYDX-BIOER-202011); the development and application of animal disease monitoring and early warning system (LYDX-SPRING-202107).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2022.966533/full#supplementary-material
1. Dabele DT, Borena BM, Admasu P, Gebremedhin EZ. Marami LM. Prevalence and Risk Factors of Mastitis and Isolation, Identification and Antibiogram of Staphylococcus Species from Mastitis Positive Zebu Cows in Toke Kutaye, Cheliya, and Dendi Districts, West Shewa Zone, Oromia, Ethiopia. Infect Drug Resist. (2021) 14:987–98. doi: 10.2147/IDR.S295257
2. Gebremedhin EZ, Ararso AB, Borana BM, Kelbesa KA, Tadese ND, Marami LM, et al. Isolation and identification of Staphylococcus aureus from milk and milk products, associated factors for contamination, and their antibiogram in Holeta, Central Ethiopia. Vet Med Int. (2022) 2022:6544705. doi: 10.1155/2022/6544705
3. Cuny C, Köck R, Witte W. Livestock associated MRSA (LA-MRSA) and its relevance for humans in Germany. Int J Med Microbiol. (2013) 303:331–7. doi: 10.1016/j.ijmm.2013.02.010
4. Nisanur E, Mukadderat G, Çibik R. The Prevalence and antibiotic resistance of methicillin-resistant Staphylococcus aureus (MRSA) in milk and dairy products in Balikesir, Turkey. J Hell Vet Med Soc. (2017) 4:613–20. doi: 10.12681/jhvms.16062
5. Abdeen EE, Mousa WS, Abdel-Tawab AA, El-Faramawy R Abo-Shama UH. Phenotypic, genotypic and antibiogram among Staphylococcus aureus isolated from bovine subclinical mastitis. Pak Vet J. (2021) 41: 289-293. doi: 10.29261/pakvetj/2021.008
6. Du XX, SA Sherein, P Liu, MA Haque and A Khan. Bovine Mastitis: Behavioral Changes, Treatment and Control. Vet J. (2022) 2:15-23. Available online at: file:///C:/Users/DELL/Downloads/1656491917_2%20CVJ-21-0103%202(1)pp% 2015-23.pdf
7. Ijaz M, Manzoor A, Mohy-ud-Din MT, Hassan F, Mohy-ud-Din Z, Ans M, Saleem MI, Khan HH and Khanum F. An economical non-antibiotic alternative to antibiotic therapy for subclinical mastitis in cows. Pak Vet J. (2021) 41: 475-480. doi: 10.29261/pakvetj/2021.059
8. Cosandey A, Boss R, Luini M, Artursson K, Bardiau M, Breitenwieser F, et al. Staphylococcus aureus genotype B and other genotypes isolated from cow milk in European countries. J Dairy Sci. (2016) 99:529–40. doi: 10.3168/jds.2015-9587
9. Abebe R, Hatiya H, Abera M, Megersa B, Asmare K. Bovine mastitis: prevalence, risk factors and isolation of Staphylococcus aureus in dairy herds at Hawassa milk shed, South Ethiopia. BMC Vet Res. (2016) 12:270. doi: 10.1186/s12917-016-0905-3
10. Kalayu AA, Woldetsadik DA, Woldeamanuel Y, Wang S, Gebreyes WA, Teferi T. Burden and antimicrobial resistance of S. aureus in dairy farms in Mekelle, Northern Ethiopia. BMC Vet Res. (2020) 16:1–8. doi: 10.1186/s12917-020-2235-8
11. Hoque MN, Das ZC, Rahman A, Haider MG, Islam MA. Molecular characterization of Staphylococcus aureus strains in bovine mastitis milk in Bangladesh. Int J Vet Sci Med. (2018) 6:53–60. doi: 10.1016/j.ijvsm.2018.03.008
12. Al-Sarraj FMB, A. Review on the impacts of Azadirachta indica on multi-drug resistant extended spectrum beta lactamasepositive of Escherichia coli and Klebsiella pneumonia. Int J Adv Life Sci. (2021) 8:228–32. Available online at: https://www.als-journal.com/833-21/
13. Iqbal A, Irshad S, Saeed S, Tareen A. Probiotic potential of encapsulated Lactobacillus species in yogurt formation indigenously isolated from dairy source. Int J Adv Life Sci. 8:267–74. Available online at: https://www.als-journal.com/8311-21/
14. Jabeen R, Yasmin M, Dar HR, Siddiqui RT, Ullah I. Characterization of mutations linked with second line anti-TB drug resistance in Pakistan. Int J Adv Life Sci. (2021) 8:137–42. Available online at: http://www.als-journal.com/828-21/
15. Javed MU, Ijaz M, Fatima Z, Anjum AA, Aqib AI, Ali MM, et al. Frequency and antimicrobial susceptibility of methicillin and vancomycin-resistant Staphylococcus aureus from bovine milk. Pak Vet J. (2021) 41:463–8. doi: 10.29261/pakvetj/2021.060
16. Kumari N, Patoli BB, Patoli AA, Jabeen S. Biocontrol of MRSA and E. coli using bacteriophages from cow manure. Int J Adv Life Sci. (2020) 7:264–9. Available online at: http://www.als-journal.com/7413-20/
17. Nazir I, Rabbani M, Sheikh AA, Firyal S, Raza S. and Riaz MI. Antibacterial activity of medicinal flowers against multi drug resistant E Coli. Pak Vet J. (2021) 41:166–8. doi: 10.29261/pakvetj/2021.019
18. Riaz S, Hussain A, Sohail M, Rehman S, Javed N, Abbas Z. Isolation and characterization of Vancomycin resistant Staphylococcus aureus (VRSA) from Intensive Care Units (ICU) of different hospitals in Lahore, Pakistan. Int J Adv Life Sci. (2021) 8:339–44. Available online at: http://www.als-journal.com/846-21/
19. Sebbar G, Fellahi S, Filali-Maltouf A and Belkadi B. Detection of colistin resistance in Mannheimia haemolytica and Pasteurella multocida isolates from ruminants in Morocco. Pak Vet J. (2020) 41: 127-131. doi: 10.29261/pakvetj/2020.077
20. Sharif M, Tunio SA, Bano S. Synergistic effects of Zinc oxide nanoparticles and conventional antibiotics against methicillin resistant Staphylococcus aureus. (2021). Int J Adv Life Sci. (2021) 8:167–71. Available online at: http://www.als-journal.com/8213-21/
21. Swar SO. RZ Abbas, R Asrar, S Yousuf, A Mehmood, B Shehzad, HR Farhan, MT Aleem, LA Marcelino and M Mohsin. Milk adulteration and emerging health issues in humans and animals (a review) Continental. Vet J. (2021) 1:1–8. Available online at: http://www.als-journal.com/8213-21/
22. Vintov J, Aarestrup FM, Zinn CE, Olsen JE. Association between phage types and antimicrobial resistance among bovine Staphylococcus aureus from 10 countries. Vet Microbiol. (2003) 95:133–47. doi: 10.1016/S0378-1135(03)00156-1
23. Ismail ZB. Molecular characteristics, antibiogram and prevalence of multi-drug resistant Staphylococcus aureus (MDRSA) isolated from milk obtained from culled dairy cows and from cows with acute clinical mastitis. Asian Pac J Trop Biomed. (2017) 7:694–7. doi: 10.1016/j.apjtb.2017.07.005
24. Klimešová M, Manga I, Nejeschlebová L, Horáček J, PoníŽil A, Vondrušková E. Occurrence of Staphylococcus aureus in cattle, sheep, goat, and pig rearing in the Czech Republic. Acta Vet Brno. (2017) 86:3–10. doi: 10.2754/avb201786010003
25. Liu JJ, Wang X, Bi CL, Mehmood K, Ashfaq H, Qin JH, et al. Epidemiological investigation of Staphylococcus aureus infection in dairy cattle in Anhui, China. Pak Vet J. (2022).
26. Tegegne DT, Mamo G, Waktole H, Messele YE. Molecular characterization of virulence factors in Staphylococcus aureus isolated from bovine subclinical mastitis in central Ethiopia. Ann Microbiol. (2021) 71. doi: 10.1186/s13213-021-01639-3
27. Liu BG, Dong Y, Sun HR, Lu SF, Wang BY, Hu GZ, Xu EP. Isolation, identification and drug resistance analysis of Staphylococcus aureus. J Northeast Agric Sci. (2021) 1–6. Available online at: https://kns.cnki.net/kcms/detail/22.1376.S.20211029.1619.004.html
28. Hu JN, Zhong PP, Yang PC, Zhao RL, Qin SY, Sun YF, et al. Study on drug resistance characteristics and typing of methicillin-resistant Staphylococcus aureus milk isolates. Heilongjiang Anim Husb Vet. (2022) 2022:82–91. (Chinese).
29. Ming Y, Qiu Y, Zhou JL, Tan RF, Wei GL, Xu RJ, et al. Isolation, identification and drug resistance analysis of Staphylococcus aureus from different animals. Heilongjiang Anim Husb Vet. (2020) 2020:101–6. (Chinese).
30. Fu XR, Kang BP, Xu XL, Ma YY, Zhou L. Analysis of drug resistance and drug resistance genes of staphylococcus aureus isolated from patients in burn department. Lab Med Clin Pract. (2020) 17:1655–8. (Chinese).
31. Liu BG, Dong Y, Sun HR, Lu SF, Wang BY, Hu GZ, et al. Isolation, identification and drug resistance analysis of Staphylococcus aureus. Northeast Agric Sci. (2021) 2021:1–6.
32. Patel K, Godden SM, Royster EE, Crooker BA, Johnson TJ, Smith EA, et al. Prevalence, antibiotic resistance, virulence and genetic diversity of Staphylococcus aureus isolated from bulk tank milk samples of U.S. dairy herds. BMC Genomics. (2021) 22:1–138. doi: 10.1186/s12864-021-07603-4
33. De Buyser M, Dufour B, Maire M, Lafarge V. Implication of Milk and Milk Products in Food-Borne Diseases in France and in Different Industrialized Countries. Amsterdam: Elsevier B.V (2001). p. 1-17.
34. Sadat A, Shata RR, Farag AMM, Ramadan H, Alkhedaide A, Soliman MM, et al. Prevalence and characterization of PVL-Positive Staphylococcus aureus isolated from raw cow's milk. Toxins. (2022) 14:97. doi: 10.3390/toxins14020097
35. Balbin MM, Lazaro JV, Candelaria CR, Cuasay JG, Abes NS. and Mingala CN. Evaluation of physico-chemical properties and nutrient components of dairy water buffalo (Bubalus bubalis) milk collected during early lactation. Int J Vet Sci. (2020) 9:24–9. Available online at: http://www.ijvets.com/.../24-29.pdf
36. Ibrahim AS, Saad MF. and Hafiz NM. Toxic elements in dried milk and evaluation of their dietary intake in infant formula. Int J Vet Sci. (2020) 9:563–7. doi: 10.37422/IJVS/20.070
37. Sarwar I, Ashar A, Mahfooz A, Aqib AI, Saleem MI, Butt AA, et al. Evaluation of antibacterial potential of raw turmeric, nano-turmeric, and NSAIDs against multiple drug resistant Staphylococcus aureus and E. coli isolated from animal wounds. Pak Vet J. (2021) 41:209–14. doi: 10.29261/pakvetj/2021.014
38. Goksoy EO, Kirkan S, Bardakcioglu HE, Sekkin S, Beyaz D, Parin U, et al. Evaluation of the effects of milking hygiene and sanitation education on total bacterial and somatic cell number of bulk tank milk in dairy cattle breeding. Int J Vet Sci. (2021) 10:37–42. doi: 10.47278/journal.ijvs/2020.009
39. Roshdy SE, Omar LM, Sayed RH, Hassan H, Hanafy MH and Soliman R. Reduction of milk contamination with aflatoxin-M1 through vaccination of dairy cattle with aflatoxin-B1 vaccine. Int J Vet Sci. (2020) 9:528–33. doi: 10.37422/IJVS/20.069
40. Youssif NH. NM Hafiz, MA Halawa, HM Aziz, MF Saad. Impact of subclinical mastitis on milk quality in different seasons. Int J Vet Sci. (2020) 9:313–6. doi: 10.37422/IJVS/20.020
41. Qolbaini EN, Khoeri MM, Salsabila K, Paramaiswari WT, Tafroji W, Artika IM, et al. Identification and antimicrobial susceptibility of methicillin-resistant Staphylococcus aureus-associated subclinical mastitis isolated from dairy cows in Bogor, Indonesia. Vet World. (2021) 14:1180–4. doi: 10.14202/vetworld.2021.1180-1184
42. Mphahlele MP, Oguttu JW, Petzer I, Qekwana DN. Prevalence and antimicrobial drug resistance of Staphylococcus aureus isolated from cow milk samples. Veterinary World. (2020) 13:2736–42. doi: 10.14202/vetworld.2020.2736-2742
43. Fursova KK, Shchannikova MP, Loskutova IV, Shepelyakovskaya AO, Laman AG, Boutanaev AM, et al. Exotoxin diversity of Staphylococcus aureus isolated from milk of cows with subclinical mastitis in Central Russia. J Dairy Sci. (2018) 101:4325–31. doi: 10.3168/jds.2017-14074
44. Kot B, Szweda P, Frankowska-Maciejewska A, Piechota M, Wolska K. Virulence gene profiles in Staphylococcus aureus isolated from cows with subclinical mastitis in eastern Poland. J Dairy Res. (2016) 83:228–35. doi: 10.1017/S002202991600008X
45. Ote I, Taminiau B, Duprez J, Dizier I, Mainil JG. Genotypic characterization by polymerase chain reaction of Staphylococcus aureus isolates associated with bovine mastitis. Vet Microbiol. (2011) 153:285–92. doi: 10.1016/j.vetmic.2011.05.042
46. Herron-Olson L, Fitzgerald JR, Musser JM, Kapur V, Ahmed N. Molecular correlates of host specialization in Staphylococcus aureus. PLoS ONE. (2007) 2:e1120. doi: 10.1371/journal.pone.0001120
47. Pereyra EAL, Picech F, Renna MS, Baravalle C, Andreotti CS, Russi R, et al. Detection of Staphylococcus aureus adhesion and biofilm-producing genes and their expression during internalization in bovine mammary epithelial cells. Vet Microbiol. (2016) 183:69–77. doi: 10.1016/j.vetmic.2015.12.002
48. Wang D, Zhang L, Zhou X, He Y, Yong C, Shen M, et al. Antimicrobial susceptibility, virulence genes, and randomly amplified polymorphic DNA analysis of Staphylococcus aureus recovered from bovine mastitis in Ningxia, China. J Dairy Sci. (2016) 99:9560–9. doi: 10.3168/jds.2016-11625
49. Bhattacharya M, Berends E, Zheng X, Hill PJ, Chan R, Torres VJ, et al. Leukocidins and the nuclease nuc prevent neutrophil-mediated killing of Staphylococcus aureus biofilms. Infect Immun. (2020) 88. doi: 10.1128/IAI.00372-20
50. Zaatout N, Ayachi A, Kecha M, Kadlec K. Identification of Staphylococci causing mastitis in dairy cattle from Algeria and characterization of Staphylococcus aureus. J Appl Microbiol. (2019) 127:1305–14. doi: 10.1111/jam.14402
51. Stevens M, Piepers S, Supré K, De Vliegher S. Antimicrobial consumption on dairy herds and its association with antimicrobial inhibition zone diameters of non-aureus staphylococci and Staphylococcus aureus isolated from subclinical mastitis. J Dairy Sci. (2018) 101:3311–22. doi: 10.3168/jds.2017-13365
52. Olsen JE, Christensen H, Aarestrup FM. Diversity and evolution of blaZ from Staphylococcus aureus and coagulase-negative staphylococci. J Antimicrob Chemoth. (2006) 57:450–60. doi: 10.1093/jac/dki492
53. Feßler A, Kadlec K, Wang Y, Zhang W, Wu C, Shen J, et al. Small antimicrobial resistance plasmids in livestock-associated methicillin-resistant Staphylococcus aureus CC398. Front Microbiol. (2018) 9. doi: 10.3389/fmicb.2018.02063
54. Pérez VKC, Custódio DAC, Silva EMM, de Oliveira J, Guimarães AS, Brito MAVP, et al. Virulence factors and antimicrobial resistance in Staphylococcus aureus isolated from bovine mastitis in Brazil. Braz J Microbiol. (2020) 51:2111–22. doi: 10.1007/s42770-020-00363-5
55. Cui L, Isii T, Fukuda M, Ochiai T, Neoh H, Camargo ILBD, et al. An RpoB mutation confers dual heteroresistance to daptomycin and vancomycin in Staphylococcus aureus. Antimicrob Agents Ch. (2010) 54:5222–33. doi: 10.1128/AAC.00437-10
56. López Díaz MC, Ríos E, Rodríguez-Avial I, Simaluiza RJ, Picazo JJ, Culebras E. In-vitro activity of several antimicrobial agents against methicillin-resistant Staphylococcus aureus (MRSA) isolates expressing aminoglycoside-modifying enzymes: potency of plazomicin alone and in combination with other agents. Int J Antimicrob Ag. (2017) 50:191–6. doi: 10.1016/j.ijantimicag.2017.01.039
57. Smith TC. Livestock-Associated Staphylococcus aureus: the United States Experience. PLoS Pathog. (2015) 11:e1004564. doi: 10.1371/journal.ppat.1004564
Keywords: Staphylococcus aureus, mastitis, milk, PCR, bovine
Citation: Liu J, Wang X, Bi C, Mehmood K, Ali F, Qin J and Han Z (2022) Molecular characterization of multi-drug-resistant Staphylococcus aureus in mastitis bovine milk from a dairy farm in Anhui, China. Front. Vet. Sci. 9:966533. doi: 10.3389/fvets.2022.966533
Received: 13 June 2022; Accepted: 14 July 2022;
Published: 22 August 2022.
Edited by:
Ilias Giannenas, Aristotle University of Thessaloniki, GreeceReviewed by:
Chrysa Voidarou, University of Ioannina, GreeceCopyright © 2022 Liu, Wang, Bi, Mehmood, Ali, Qin and Han. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chongliang Bi, YmljaG9uZ2xpYW5nQGx5dS5lZHUuY24=; Zhaoqing Han, aGFuemhhb3FpbmdAbHl1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.