- 1Department of Anatomy, School of Biomedical Sciences, University of Otago, Dunedin, New Zealand
- 2Brain Health Research Centre, University of Otago, Dunedin, New Zealand
A brain atlas is essential for understanding the anatomical relationship between neuroanatomical structures. Standard stereotaxic coordinates and reference systems have been developed for humans, non-human primates and small laboratory animals to contribute to translational neuroscience research. Despite similar neuroanatomical and neurofunctional features between the sheep and human brain, little is known of the sheep brain stereotaxy, and a detailed sheep atlas is scarce. Here, we briefly discuss the value of using sheep in neurological research and the paucity of literature concerning the coordinates system during neurosurgical approaches. Recent advancements such as computerized tomography, positron emission tomography, magnetic resonance imaging, functional magnetic resonance imaging and diffusion tensor imaging are used for targeting and localizing the coordinates and brain areas in humans. Still, their application in sheep is rare due to the lack of a 3D stereotaxic sheep atlas by which to map sheep brain structures to its human counterparts. More recently, a T1- and T2-weighted high-resolution MRI 3D stereotaxic atlas of the sheep brain has been generated, however, the journey to create a sheep brain atlas by which to map directly to the human brain is still uncharted. Therefore, developing a detailed sheep brain atlas is valuable for the future to facilitate the use of sheep as a large animal experimental non-primate model for translational neurological research.
Introduction
A brain atlas is a valuable tool containing pictures of brain sections in different anatomical orientations (three-dimensional space; coronal, sagittal and axial planes), with the coordinates of relevant brain structures to define their outlines or volumes. An atlas enables the researcher to calculate stereotaxic coordinates for a variety of stereotaxic procedures to accurately target deep brain structures for recording or lesioning (1–4). This atlas when combined with electrophysiological data helps to better understand the functional activity in brain networks across species (5). Having a highly precise and consistent atlas assures consistency in defining the boundaries of various brain structures between publications from different researchers (6). In addition, atlas and templates are necessary to improve understanding of neuroanatomical structures, leading to advancements in the field of neurology, and neurosurgery and to aid translational neuroscience research.
Among the various animal species used in neuroscience, rodents are the most common. Other used species include domestic animals such as pigs, goats, and sheep, and those that are also companion animals such as horses, dogs and cats. Stereotaxic and automatic tissue segmentation systems with varied detail have been developed for humans, non-human primates, dogs, cats and rodents, but not for sheep. A detailed brain atlas exists for humans, the Allen Human Brain Atlas; for rats, the Rat Brain in Stereotaxic Coordinates (7); for mouse, Allen Brain Atlas: Mouse Brain; for rhesus macaques, NIH Blueprint Non-Human Primate (NHP) Atlas; for long-tailed macaque monkeys (4); for domestic species such as the pig (1), dog (8–10), cat (5), and horse. Some example two-dimensional images of brain coronal and sagittal sections of various animal species is shown in Figure 1. Unfortunately, to our knowledge, no such detailed atlas is available for sheep.
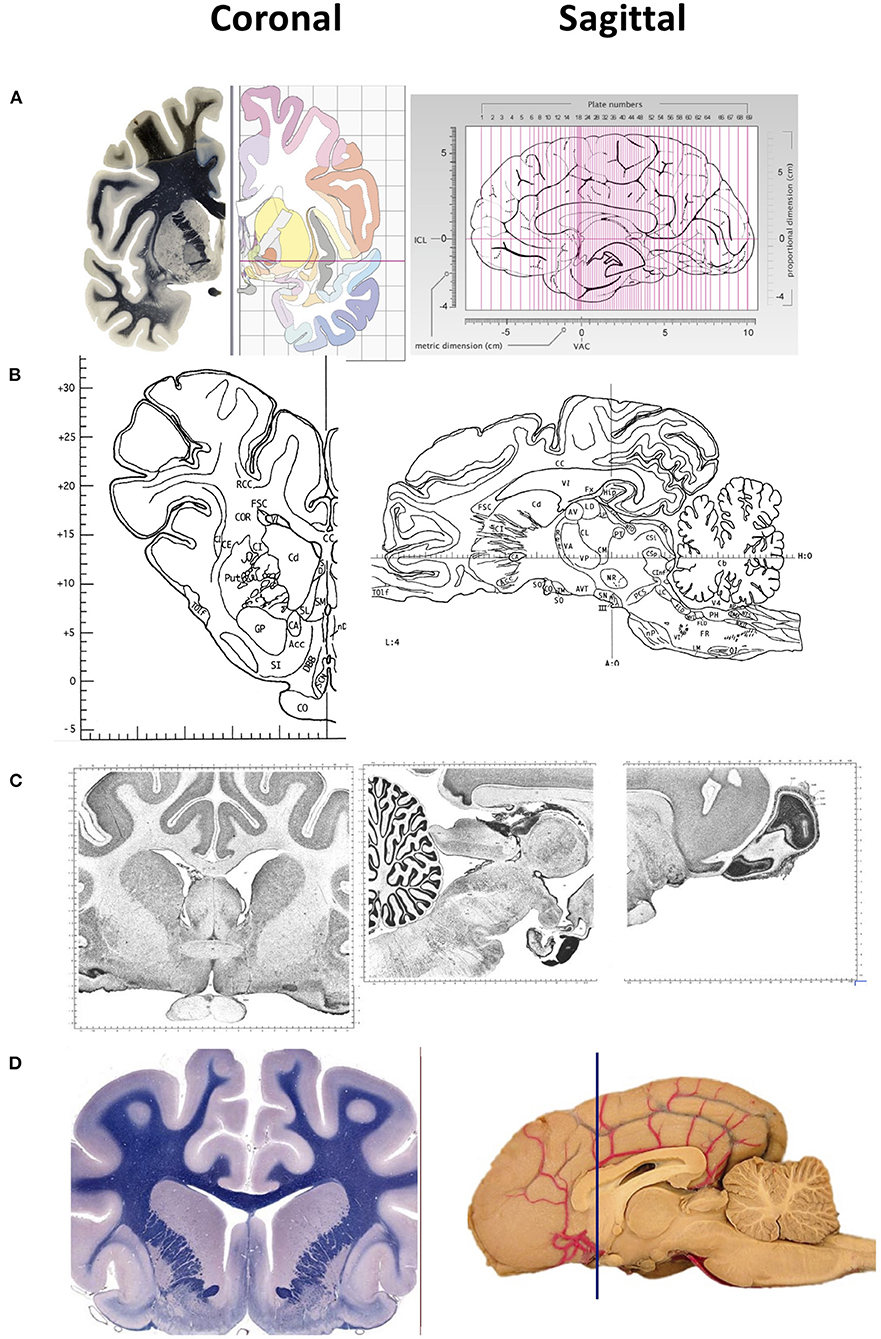
Figure 1. Representative images of two-dimensional sections of the brain, coronal (right) and sagittal (left) sectons of brains of various species. (A) Human brain. Coronal sections show the fiber tracts in the left panel and schematic diagrams in the right panel. The sagittal section shows the plane of the 69 sections depicted in the atlas. The intercommisural line (ICL) and the vertical line (VCA) pass through the center of the anterior and posterior commissure, and the center of the anterior commissure, respectively. (B) Pig brain. Coronal and sagittal sections in this example show the coordinates 14.50 mm ahead of the posterior commissure and 4.00 mm laterally from the midsagittal plane, respectively. (C) Cat brain. Coronal and sagittal sections are examples of Nissl stained sections from the adult cat (Felis Catus). (D) Dog brain. Coronal and sagittal sections are shown in the maps of the whole brain from a 5 month old dog (Canis Lupus). The blue line shows the levels at the frontal lobe Images in (A) are reproduced from the human brain website https://www.thehumanbrain.info/brain/sections.php. Retrieved May 12, 2022. Images in (B) are reproduced from the Stereotaxic atlas of the pig brain by Felix et al. (1) with permission from Elsevier. Images (C,D) are reproduced from the brain maps website http://brainmaps.org/ajax-viewer.phpdatid=32&sname=p099-100, retrieved on May 13, 2022.
The sheep (Ovis aries) is an attractive and convenient animal model for mapping human disorders, particularly for neurosurgical and neuropathological research. Physiological and neuroanatomical similarities between sheep and humans, such as cerebral white matter distribution (11), gyrencephalic cerebral cortexes, thick meninges, and highly distinct sulci and gyri (12–14), make sheep an acceptable large brain animal model for neurological research. Sheep cerebral cortices contain four lobes defined by external landmarks, similar to those of humans (15). Furthermore, the sub-cortical structures in particular, the dorsal striatum, are in two separate sections; caudate nucleus and putamen in sheep, similar to humans (14, 16). In addition, the relatively round skull of the sheep is comparable to the human head, unlike pigs which have a flat and thick skull (17). Therefore, the brains of sheep may have distinct anatomical advantages over small brains for translational research. A detailed review of neuroanatomy is beyond the scope of this paper, but clinically relevant areas are compared briefly in Table 1. Other benefits of using sheep such as greater acceptability to animal ethics committee compared to companion animals and primates, easily available, less expensive, reasonably outbreed, easy management, environmental enrichment not required as they live in their natural pasture, make sheep advantageous an experimental model for translational research over large animal species, in particular primates.
Despite similar neuroanatomical and neurofunctional features between sheep and human brains, and the relatively low cost of the sheep as a model for teaching human-like brain anatomy, very little is known about the ovine brain anatomical relationships, and a detailed sheep atlas is scarce. The only available atlases are either limited to specific brain areas (35), consist of a series of gross photographs of sheep brain (36), or provide labeled coronal sections (both cell and fiber stain) but do not offer standard stereotaxic coordinates and reference system (e.g., The Michigan histological sheep brain atlas; https://brains.anatomy.msu.edu/brains/sheep/index.html; retrieved on May 12, 2022). Although there is a sheep atlas with a coordinate system (covering a few brain areas), histology sections with labeled brain structures (as in the Michigan histological data atlas; but not the standard stereotaxic coordinates/reference system) and neuroimaging atlas there is no comprehensive atlas that actually combines all these features to enable defined regions to be easily localized. Therefore, the neuroscience research community would be benefitted by having a detailed sheep atlas that combines both neuroanatomical regions on both histology (as in the Michigan histological data atlas) as well as neuroimaging (MRI atlases)to enable defined regions to be easily localized.
Brief history of resources developed for the study of the sheep brain
The study of the cortico-spinal or pyramid tract fibers of the sheep brain can be dated back to the late 18th century (the literature refers to 1877 by Atruro Maracacci)(37). Researchers made strenuous efforts to map the brain and anatomical areas and to partition these based on cytoarchitectonic information available at that early stage. Most of the early studies in sheep were based on electrical or mechanical stimulation of the different brain regions and mapping with a pattern of body response to stimuli (37–39). In 1967, Richard published the first hard copy sheep atlas, “Atlas stereotaxique du cerveau de beria.” However, the atlas was written in the French language and was limited to specific brain areas, including subcortical structures such as the thalamus, hippocampus, and hypothalamus (35). The stereotaxic brain atlas in English was prepared by McKenzie and Smith in 1973 from female merino sheep and was used by others in guiding electrical stimulation of the brain, diencephalic lesions, and intracerebroventricular chemical stimulations (40). Opdam and colleagues, first reported the sheep (merino) model of focal epilepsy (penicillin-induced) in 2002 using concurrent EEG and fMRI (41). The localization of coordinates for surgical placement and electrodes was based on the anatomic atlas of the sheep brain, however, the authors did not relate the details of the stereotaxic reference systems to the distinct anatomical structures identified from their experimental findings to help map the findings to the structures found in the human brain.
Recent advancement
Recent advances in mapping technology have made it possible to study neuroanatomical features and localization of brain areas in a variety of animals, including sheep (3, 42–44). The established techniques for targeting brain areas and localizing the coordinates such as CT scan, PET, MRI, fMRI, and DTI are widely used in humans. The neuroimaging techniques are being applied to sheep, albeit not extensively because they fit into conventional scanners and MRI units due to their comparable body size, skulls and brain volumes to humans (14, 28, 43, 45).
In 2014, frameless MRI guided stereotactic access to the ovine brainstem was developed and validated using the modified BrainsightTM stereotactic system (46). Nevertheless, the approach was limited to the midbrain and pons on post-mortem imaging on sheep heads. Nitzsche et al. (44) published an MRI-based ovine brain template with tissue probability maps offering a detailed stereotaxic reference frame to localize brain areas and anatomical features. However, this ovine brain atlas was limited to cerebral morphology and tissue volume, mainly gray matter, white matter, cerebrospinal fluid (CSF), cerebral peduncle, and pons. Other critical anatomical structures such as the cerebellum, medulla oblongata, olfactory bulb and subcortical brain areas were not included.
Russell et al. (47) used CT scanning and 3D reconstruction techniques to study intracranial volume loss as well as regional neurodegeneration over time (between 5 and 13 months and 11 and 15 months for CLN5 and CLN6 sheep Baten disease models, respectively) in occipital lobes and propagation throughout the cerebral cortex. The CT images included olfactory bulbs in the cerebellum. Nevertheless, details of inner and external sheep brain structures was again absent from these works.
3D high-resolution MRI sheep brain templates have been constructed for multi-institution neuroimaging studies using living animals. More recently, Ella and colleagues generated the first complete T1 and T2-weighted high-resolution MRI 3D stereotaxic atlas of an in vivo sheep brain (2, 48). This elegant 3D atlas defined an MRI stereotaxic coordinate system, probability maps and templates of CSF, gray and white matter. In addition, the 3D atlas also demonstrated 25 cortical and 28 subcortical brain structures. However, the journey of mapping the coordinates of sheep brain structures identified in atlases such as this to that of the human brain still needs to advance, thereby providing a missing tool for translational research.
Clarke and Whitteridge, (49) made their own atlases for the cortex to study the cortical visual areas (Visual I and II) of the sheep for the investigation of controlled eye movement. In their work, they considered the coordinates used were relative to the interaural plane in the usual way but the details of the coordinates for targeting the visual cortex were missing. However, they have referred to the Richard atlas for the mid-brain. The visual cortex was found to be thin in sheep (1.488 mm) compared to those of macaques (1.657 mm), and chimpanzees (2.109 mm) (50).
Bombardi et al. (51) studied the organization of the lateral nucleus of sheep amygdaloid complex which is little known in ruminants. Due to the lack of a proper sheep brain atlas, the delineation of the nuclear boundaries of the amygdala was based on the image sections found in the University of Wisconsin and Michigan State Comparative Mammalian brain collections.
More recently, DTI, mathematical and image-analysis techniques, of the intracranial pyramidal and extrapyramidal tracts, were applied to study the laminar organization and projections of the sheep motor cortex. The results showed the comparable thickness of the cortex and other morphometric values to other mammalian species including the chimpanzee. Somatotopic mapping however was taken from early studies by Simpson and King due to the lack of a currently detailed sheep atlas (52). Mapping orbitofrontal cortex (OFC) and its connection to brain areas of the sheep with the chimpanzee and humans was investigated by Tommaso et al. using DTI. The authors found a higher number of cortico-cortical fibers connecting the visual areas with OFC similar to that of the human brain (53).
Conclusion
At least 80–90% of novel therapeutic agents tested on many rodents are found to be ineffective when translating therapies into clinical research. One of the factors could be the inappropriate selection of a suitable experimental animal model for translational research. In addition, regulatory authorities expect therapeutic agents and medical device testing on at least one small rodent and large non-rodent gyrencephalic species prior to clinical trials. As neuroscience becomes increasingly advanced, highly accurate and consistent brain atlases are needed for high-precision neuroscience experimentation. The neuroscience research community would be benefitted by having a detailed ovine brain atlas with coordinates that combine both neuroanatomical regions on both histology (as in the Michigan histological data atlas) as well as neuroimaging (MRI atlases) to enable defined regions to be easily localized. Although there are some still existing limits such as the scarcity of neurochemical and electrophysiological data, as well as the difficulty and high cost of performing transgenesis experiments, we believe that creating a detailed ovine brain atlas consisting of well-defined brain regions (easily identifiable) and somatotopic organization (similar to the available human and rodents atlas) is necessary for clinically relevant translational neuroscience and neurological research. Such an atlas would allow for detailed regional analysis and easier surgical manipulation, facilitate the generalizability and comparability of experimental results across studies and laboratories and reduce the requirement for higher resolution imaging. This would be a significant piece of research. Such a tool would be extremely valuable for research in the next 5 to 10 years using sheep as a large animal non-primate model for translational neurological research.
Author contributions
Conceptualization and writing—original draft: AB. Visualization and writing—review and editing: AB and JR. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Neurological Foundation New Zealand grant 2130PRG.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Felix B, Leger ME, Albe-Fessard D, Marcilloux JC, Rampin O, Laplace JP. Stereotaxic atlas of the pig brain. Brain Res Bull. (1999) 49:1–137. doi: 10.1016/S0361-9230(99)00012-X
2. Ella A, Barriere DA, Adriaensen H, Palmer DN, Melzer TR, Mitchell NL, et al. The development of brain magnetic resonance approaches in large animal models for preclinical research. Anim Front. (2019) 9:44–51. doi: 10.1093/af/vfz024
3. Liyanage KA, Steward C, Moffat BA, Opie NL, Rind GS, John SE, et al. Development and implementation of a corriedale ovine brain atlas for use in atlas-based segmentation. PLoS One. (2016) 11:e0155974. doi: 10.1371/journal.pone.0155974
4. Lanciego JL, Vazquez A. The basal ganglia and thalamus of the long-tailed macaque in stereotaxic coordinates. A template atlas based on coronal, sagittal and horizontal brain sections. Brain Struct Funct. (2012) 217:613–66. doi: 10.1007/s00429-011-0370-5
5. Stolzberg D, Wong C, Butler BE, Lomber SG. Catlas: An magnetic resonance imaging-based three-dimensional cortical atlas and tissue probability maps for the domestic cat (Felis catus). J Comp Neurol. (2017) 525:3190–206. doi: 10.1002/cne.24271
6. Azimi N, Yadollahikhales G, Argenti JP, Cunningham MG. Discrepancies in stereotaxic coordinate publications and improving precision using an animal-specific atlas. J Neurosci Methods. (2017) 284:15–20. doi: 10.1016/j.jneumeth.2017.03.019
7. Paxinos G, Watson C. The Rat Brain in Sterotaxic Coordinates. 6th ed. London: Academic Press (2007).
8. Datta R, Lee J, Duda J, Avants BB, Vite CH, Tseng B, et al. A digital atlas of the dog brain. PLoS ONE. (2012) 7:e52140. doi: 10.1371/journal.pone.0052140
10. Johnson PJ, Luh WM, Rivard BC, Graham KL, White A, FitzMaurice M, et al. Stereotactic cortical atlas of the domestic canine brain. Sci Rep. (2020) 10:4781. doi: 10.1038/s41598-020-61665-0
11. Back SA, Riddle A, Dean J, Hohimer AR. The instrumented fetal sheep as a model of cerebral white matter injury in the premature infant. Neurotherapeutics. (2012) 9:359–70. doi: 10.1007/s13311-012-0108-y
12. Piumatti M, Palazzo O, La Rosa C, Crociara P, Parolisi R, Luzzati F, et al. Non-newly generated, “immature" neurons in the sheep brain are not restricted to cerebral cortex. J Neurosci. (2018) 38:826–42. doi: 10.1523/JNEUROSCI.1781-17.2017
13. Rose JE. The cytoarchitectural study of the sheep cortex. J Comp Neurol. (1942) 76:102. doi: 10.1002/cne.900760102
14. Morton AJ, Howland DS. Large genetic animal models of Huntington's disease. J Huntingtons Dis. (2013) 2:3–19. doi: 10.3233/JHD-130050
15. Sun T, Hevner RF. Growth and folding of the mammalian cerebral cortex: from molecules to malformations. Nat Rev Neurosci. (2014) 15:217–32. doi: 10.1038/nrn3707
16. McKenzie JS, Chai SY, Giles ME, McKinley MJ. Dopaminergic System in the Basal Ganglia of Sheep. Boston, MA: Springer (2002). doi: 10.1007/978-1-4615-0715-4_12
17. Lee W, Lee SD, Park MY, Foley L, Purcell-Estabrook E, Kim H, et al. Functional and diffusion tensor magnetic resonance imaging of the sheep brain. BMC Vet Res. (2015) 11:262. doi: 10.1186/s12917-015-0581-8
18. Snyder JM, Hagan CE, Bolon B, Keene CD. Nervous System. In: Treuting PM, Dintzis SM, Montine KM, editors. Comparative Anatomy and Histology:A Mouse, Rat, and Human Atlas. 2nd ed: Elseveir (2018). p. 403–44. doi: 10.1016/B978-0-12-802900-8.00020-8
19. Pelekanos M, Leinenga G, Odabaee M, Odabaee M, Saifzadeh S, Steck R, et al. Establishing sheep as an experimental species to validate ultrasound-mediated blood-brain barrier opening for potential therapeutic interventions. Theranostics. (2018) 8:2583–602. doi: 10.7150/thno.22852
20. Glatzle M, Hoops M, Kauffold J, Seeger J, Fietz SA. Development of deep and upper neuronal layers in the domestic cat, sheep and pig neocortex. Anat Histol Embryol. (2017) 46:397–404. doi: 10.1111/ahe.12282
21. John SE, Lovell TJH, Opie NL, Wilson S, Scordas TC, Wong YT, et al. The ovine motor cortex: A review of functional mapping and cytoarchitecture. Neurosci Biobehav Rev. (2017) 80:306–15. doi: 10.1016/j.neubiorev.2017.06.002
22. Murray SJ, Black BL, Reid SJ, Rudiger SR, Simon Bawden C, Snell RG, et al. Chemical neuroanatomy of the substantia nigra in the ovine brain. J Chem Neuroanat. (2019) 97:43–56. doi: 10.1016/j.jchemneu.2019.01.007
23. Fowke TM, Galinsky R, Davidson JO, Wassink G, Karunasinghe RN, Prasad JD, et al. Loss of interneurons and disruption of perineuronal nets in the cerebral cortex following hypoxia-ischaemia in near-term fetal sheep. Sci Rep. (2018) 8:17686. doi: 10.1038/s41598-018-36083-y
24. Brus M, Meurisse M, Gheusi G, Keller M, Lledo PM, Levy F. Dynamics of olfactory and hippocampal neurogenesis in adult sheep. J Comp Neurol. (2013) 521:169–88. doi: 10.1002/cne.23169
25. Keller M, Levy F. The main but not the accessory olfactory system is involved in the processing of socially relevant chemosignals in ungulates. Front Neuroanat. (2012) 6:39. doi: 10.3389/fnana.2012.00039
26. Clarke PGH, Donaldson IML, Whitteridge D. Binocular visual mechanism in cortical areas I and II of the sheep. J Physiol. (1976) 256:509–26. doi: 10.1113/jphysiol.1976.sp011336
27. Pieri V, Trovatelli M, Cadioli M, Zani DD, Brizzola S, Ravasio G, et al. In vivo diffusion tensor magnetic resonance tractography of the sheep Brain: an atlas of the ovine white matter fiber bundles. Front Vet Sci. (2019) 6:345. doi: 10.3389/fvets.2019.00345
28. Vink R. Large animal models of traumatic brain injury. J Neurosci Res. (2018) 96:527–35. doi: 10.1002/jnr.24079
29. Hoffmann A, Stoffel MH, Nitzsche B, Lobsien D, Seeger J, Schneider H, et al. The ovine cerebral venous system: comparative anatomy, visualization, and implications for translational research. PLoS ONE. (2014) 9:e92990. doi: 10.1371/journal.pone.0092990
30. Low VF, Faull RL, Bennet L, Gunn AJ, Curtis MA. Neurogenesis and progenitor cell distribution in the subgranular zone and subventricular zone of the adult sheep brain. Neuroscience. (2013) 244:173–87. doi: 10.1016/j.neuroscience.2013.04.006
31. Halliday GM, Törk I. Comparative anatomy of the ventromedial mesencephalic tegmentum in the rat, cat, monkey and human. J Comp Neurol. (1986) 252:423–45. doi: 10.1002/cne.902520402
32. Wilke H, Kettler A, Wenger KH, Claes L. Anatomy of the sheep spine and its comparison to the human Spine. Anat Rec. (1997) 247:542–55. doi: 10.1002/(SICI)1097-0185(199704)247:4<542::AID-AR13>3.0.CO;2-P
33. Azevedo RB, Leonardo AS, Nogueira K, Okiyama FTM, Santos AC, Lima EMM. Origin and distribution of the sciatic nerve in Santa Inês ovines. Aust J Basic & Appl Sci. (2014) 8:257–61. Available online at: https://www.researchgate.net/publication/263534757
34. Prabhavathi M, Basha SH, Paramasivan S, Venkatesan S, Ramesh G. Histomorphology of the pineal gland in sheep. Indian Vet J. (2010) 87:698–700. Available online at: https://www.researchgate.net/publication/263544047
35. Richard P. Atlas Stereotaxique Du Cerveau De Beris. Paris, France: Institut National de la Recherche Agronomique (INRA) (1967).
36. Vanderwolf CH, Cooley RK. The Sheep Brain: A Photographic Series. London, Ontario: AJ Kirby Co. (2002).
37. Simpson S, King JL. Localization of the motor area in sheep. Q. J. Exp. Physiol. (1911) 4:53–64. doi: 10.1113/expphysiol.1911.sp000083
38. Grovum WL, Gonzalez JS. Electrical stimulation of the medial frontal lobe elicits a pattern of body movements in sheep. Brain Res. (1999) 18:252–3. doi: 10.1016/S0006-8993(99)02083-1
39. King JL. Localisation of the motor area in the sheep's brain by the histological method. J Comp Neurol. (1911) 21:311–21. doi: 10.1002/cne.900210305
41. Opdam HI, Federico P, Jackson GD, Buchanan J, Abbott DF, Fabinyi GCA, et al. A sheep model for the study of focal epilepsy with concurrent intracranial EEG and functional MRI. Epilepsia. (2002) 43:779–87. doi: 10.1046/j.1528-1157.2002.04202.x
42. Ella A, Keller M. Construction of an MRI 3D high resolution sheep brain template. Magn Reson Imaging. (2015) 33:1329–37. doi: 10.1016/j.mri.2015.09.001
43. Hurtig MB, Buschmann MD, Fortier LA, Hoemann CD, Hunziker EB, Jurvelin JS, et al. Preclinical studies for cartilage repair: recommendations from the international cartilage repair society. Cartilage. (2011) 2:137–52. doi: 10.1177/1947603511401905
44. Nitzsche B, Frey S, Collins LD, Seeger J, Lobsien D, Dreyer A, et al. A stereotaxic, population-averaged T1w ovine brain atlas including cerebral morphology and tissue volumes. Front Neuroanat. (2015) 9:69. doi: 10.3389/fnana.2015.00069
45. Sorby-Adams AJ, Vink R, Turner RJ. Large animal models of stroke and traumatic brain injury as translational tools. Am J Physiol Regul Integr Comp Physiol. (2018) 315:R165–R90. doi: 10.1152/ajpregu.00163.2017
46. Staudacher A A. O, Stoffel MH, Gorgas D. Validation of a magnetic resonance imaging guided stereotactic access to the ovine brainstem. BMC Vet Res. (2014) 10:216. doi: 10.1186/s12917-014-0216-5
47. Russell KN, Mitchell NL, Anderson NG, Bunt CR, Wellby MP, Melzer TR, et al. Computed tomography provides enhanced techniques for longitudinal monitoring of progressive intracranial volume loss associated with regional neurodegeneration in ovine neuronal ceroid lipofuscinoses. Brain Behav. (2018) 8:e01096. doi: 10.1002/brb3.1096
48. Ella A, Delgadillo JA, Chemineau P, Keller M. Computation of a high-resolution MRI 3D stereotaxic atlas of the sheep brain. J Comp Neurol. (2017) 525:676–92. doi: 10.1002/cne.24079
49. Clarke PG, Whitteridge D. The cortical visual areas of the sheep. J Physiol. (1976) 256:497–508. doi: 10.1113/jphysiol.1976.sp011335
50. Graic JM, Peruffo A, Corain L, Finos L, Grisan E, Cozzi B. The primary visual cortex of Cetartiodactyls: organization, cytoarchitectonics and comparison with perissodactyls and primates. Brain Struct Funct. (2022) 227:1195–225. doi: 10.1007/s00429-021-02392-8
51. Bombardi C, Grandis A, Chiocchetti R, Lucchi ML. Distribution of calbindin-D28k, neuronal nitric oxide synthase, and nicotinamide adenine dinucleotide phosphate diaphorase (NADPH-d) in the lateral nucleus of the sheep amygdaloid complex. Anat Embryol (Berl). (2006) 211:707–20. doi: 10.1007/s00429-006-0133-x
52. Peruffo A, Corain L, Bombardi C, Centelleghe C, Grisan E, Graic JM, et al. The motor cortex of the sheep: laminar organization, projections and diffusion tensor imaging of the intracranial pyramidal and extrapyramidal tracts. Brain Struct Funct. (2019) 224:1933–46. doi: 10.1007/s00429-019-01885-x
Keywords: sheep, brain, atlas, template, translational neuroscience, stereotaxic coordinates
Citation: Banstola A and Reynolds JNJ (2022) Mapping sheep to human brain: The need for a sheep brain atlas. Front. Vet. Sci. 9:961413. doi: 10.3389/fvets.2022.961413
Received: 04 June 2022; Accepted: 12 July 2022;
Published: 29 July 2022.
Edited by:
Adalberto Merighi, University of Turin, ItalyReviewed by:
Annabel Sorby-Adams, University of Adelaide, AustraliaCristiano Bombardi, University of Bologna, Italy
Copyright © 2022 Banstola and Reynolds. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ashik Banstola, ashik.banstola@otago.ac.nz
 Ashik Banstola
Ashik Banstola John N. J. Reynolds
John N. J. Reynolds