
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci. , 05 August 2022
Sec. Livestock Genomics
Volume 9 - 2022 | https://doi.org/10.3389/fvets.2022.948873
This article is part of the Research Topic Decoding the Genetics of Milk Fat Traits View all 9 articles
 Jiqing Wang*
Jiqing Wang* Zhiyun Hao
Zhiyun Hao Liyan Hu
Liyan Hu Lirong Qiao
Lirong Qiao Yuzhu Luo
Yuzhu Luo Jiang Hu
Jiang Hu Xiu Liu
Xiu Liu Shaobin Li
Shaobin Li Fangfang Zhao
Fangfang Zhao Jiyuan Shen
Jiyuan Shen Mingna Li
Mingna Li Zhidong Zhao
Zhidong ZhaoIn our previous study, microRNA (miR)-199a-3p was found to be the most upregulated miRNA in mammary gland tissue during the non-lactation period compared with the peak-lactation period. However, there have been no reports describing the function of miR-199a-3p in ovine mammary epithelial cells (OMECs) and the biological mechanisms by which the miRNA affects cell proliferation and milk fat synthesis in sheep. In this study, the effect of miR-199a-3p on viability, proliferation, and milk fat synthesis of OMECs was investigated, and the target relationship of the miRNA with very low-density lipoprotein receptor (VLDLR) was also verified. Transfection with a miR-199a-3p mimic increased the viability of OMECs and the number of Edu-labeled positive OMECs. In contrast, a miR-199-3p inhibitor had the opposite effect with the miR-199a-3p mimic. The expression levels of three marker genes were also regulated by both the miR-199a-3p mimic and miR-199-3p inhibitor in OMECs. Together, these results suggest that miR-199a-3p promotes the viability and proliferation of OMECs. A dual luciferase assay confirmed that miR-199a-3p can target VLDLR by binding to the 3′-untranslated regions (3'UTR) of the gene. Further studies found a negative correlation in the expression of miR-199a-3p with VLDLR. The miR-199a-3p mimic decreased the content of triglycerides, as well as the expression levels of six milk fat synthesis marker genes in OMECs, namely, lipoprotein lipase gene (LPL), acetyl-CoA carboxylase alpha gene (ACACA), fatty acid binding protein 3 gene (FABP3), CD36, stearoyl-CoA desaturase gene (SCD), and fatty acid synthase gene (FASN). The inhibition of miR-199a-3p increased the level of triglycerides and the expression of LPL, ACACA, FABP3, SCD, and FASN in OMECs. These findings suggest that miR-199a-3p inhibited milk fat synthesis of OMECs. This is the first study to reveal the molecular mechanisms by which miR-199a-3p regulates the proliferation and milk fat synthesis of OMECs in sheep.
The mammary gland is a special exocrine gland in mammals and is of economic importance. Mammary epithelial cells (MECs) around alveoli absorb nutrients from the blood and then convert them into milk. It is well-known that the viability, proliferation number, and secretory ability of MECs are responsible for milk yield and milk components, which in turn affect the survival rate, development, and pre-weaning growth rate of lambs, especially for non-dairy, multiple-born-lambs sheep breeds, such as Small-tailed Han sheep and Hu sheep farmed in China. The two breeds have average lambing rates of more than 250% per ewe. In this context, an in-depth knowledge of the molecular mechanisms that regulate MECs activities offers an opportunity to improve milk yield and quality, eventually resulting in an increase in the commercial return to sheep farmers.
It was found that the activity of MECs and mammary gland development are regulated by many functional genes and non-coding RNAs, including microRNAs (miRNAs). The length of miRNAs ranges from 18 to 25 nucleotides. They can complementally bind to the 3′-untranslated regions (3′UTR) of the target mRNAs to either induce degradation or inhibit translation, with an accompanying decrease of the target genes in expression (1). Increasing studies showed crucial roles of miRNAs in proliferation, apoptosis, and survival of MECs, as well as milk fat and milk protein synthesis in MECs (2). For example, miR-143 increased the content of triglycerides by targeting SMAD family 3 (SMAD3) (3). The miR-204-5p and miR-211 were found to coordinately inhibit the content of α-S1 casein (CSN1S1) (4). The effect of other miRNAs on MECs has also been reported, including miR-193a-5p (5), miR-128 (6), miR-106b (7), miR-221 (8), miR-16a (9), and miR-26a/b (10). However, these studies have been focused on dairy cows and dairy goats.
In sheep, the expression profiles of miRNAs in ovine mammary gland collected from different development periods or different breeds have been widely reported (11, 12), and some differentially expressed miRNAs were then screened. Although the functions of the differentially expressed miRNAs were investigated by enrichment analysis of their target genes, they have not been verified by experimental assays. At present, to the best of our knowledge, there are very few studies on the role of single miRNA in ovine mammary epithelial cells (OMECs) or mammary gland development in sheep. Hao et al. (13) found an inhibited effect of miR-432 on milk fat synthesis by targeting lipoprotein lipase (LPL) and stearoyl-CoA desaturase (SCD) genes. Nonetheless, it should be legitimately suggested that miRNAs are worthy of further investigation for a better understanding of molecular mechanisms underlying mammary gland development in sheep.
In our previous study, miR-199a-3p was found to be the most upregulated miRNA in the mammary gland during the non-lactation period when compared with the mammary gland at peak-lactation period, with the expression levels being 3.81-fold higher during the non-lactation (12). However, there have been no reports on the molecular mechanism of miR-199a-3p regulating mammary gland development and milk fat synthesis in sheep. Accordingly in this study, we investigated the effect of miR-199a-3p on viability, proliferation, and milk fat synthesis of OMECs, as well as the expression levels of some marker genes. We also confirmed the target binding of miR-199a-3p with very low-density lipoprotein receptor (VLDLR). These results will uncover the regulatory mechanism of miR-199a-3p underlying mammary gland development and milk fat synthesis in sheep.
Sample collection and cell treatment involved in this study were approved by the Animal Experiment Ethics Committee of Gansu Agricultural University, Lanzhou, China (Approval number GSAU-ETH-AST-2021-027). A healthy 3-year-old Small-tailed Han ewe was slaughtered at peak lactation (21 days postpartum), and mammary gland parenchyma samples were then collected.
The OMECs were isolated according to the method of Hao et al. (13). Briefly, cells were cultured in a basal DMEM/F12 medium (Invitrogen, CA, USA) consisting of 10% fetal bovine serum, 5 μg/ml hydrocortisone, 5 μl/ml insulin-transferrin-sodium selenium, 10 ng/ml epidermal growth factor 1, and 1 μg/ml hydrocortisone. To induce lactation of OMECs, they were then maintained in a lactogenic medium with the basal DMEM/F12 medium described above supplemented with 2 μg/ml prolactin (USBiological, MA, USA) for 48 h.
A miR-199a-3p mimic, a miR-199a-3p inhibitor, and their negative controls (miR-199a-3p mimic NC and miR-199a-3p inhibitor NC) were synthesized by RiboBio Co. Ltd. (Guangzhou, China). When the confluence of OMECs reached ~80%, either the miR-199a-3p mimic (50 nM) or the miR-199a-3p inhibitor (100 nM) or their negative controls were transfected into the cells using INVI DNA & RNA Transfection Reagent™ (Invigentech, CA, USA). To investigate the effect of miR-199a-3p on the viability of OMECs, a 30 μl aliquot of CCK8 (Vazyme, Nanjing, China) was added to each well 46 h after transfection and then continued to incubate for 2 h. A Thermo Scientific Microplate Reader (Thermo Scientific, MA, USA) was used to measure the absorbance of OMECs at 450 nm. Four wells were selected from each group of cells treated to count their cell viability.
To detect the proliferation of OMECs regulated by miR-199a-3p, an Edu assay was performed 44 h after transfection with the miR-199a-3p mimic and miR-199a-3p inhibitor. Briefly, 100 ml of 50 mM Cell-Light™ Edu reagent (Beyotime, Shanghai, China) was added to each well and then cultured for 4 h. A microscope IX73 (Olympus, Tokyo, Japan) was used to observe the Edu staining result, and the percentage of Edu-labeled positive OMECs was calculated.
To investigate the transfection efficiency of miR-199a-3p, as well as its roles in proliferation and milk fat synthesis of OMECs, reverse transcription-quantitative PCR (RT-qPCR) analysis was performed. Briefly, when OMECs grew to 70–80%, the cells were transfected with the miR-199a-3p mimic, miR-199a-3p inhibitor, miR-199a-3p mimic NC, and miR-199a-3p inhibitor NC. Total RNA was isolated from transfected OMECs using TRIzol reagent (Invitrogen, CA, USA), and then reverse transcribed to produce cDNA using SuperScriptTM II reverse transcriptase kit (Invitrogen, CA, USA). The RT-qPCR was carried out in triplicate using an SYBR qPCR Master Mix (Vazyme, Nanjing, China). GAPDH was chosen as an internal reference for standardization as suggested by Jiao et al. (8). A 2−ΔΔCt method was used to calculate the relative expression levels of miR-199a-3p and 10 genes, namely, VLDLR, BCL2 apoptosis regulator (BCL-2), BCL2-associated X (Bax), Caspase3, LPL, acetyl-CoA carboxylase alpha (ACACA), fatty acid binding protein 3 (FABP3), CD36, SCD, and fatty acid synthase (FASN). Their PCR primers are listed in Table 1.
The VLDLR was predicted to be a target gene of miR-199a-3p using the Targetscan 3.1 and miRanda 3.3a. To verify the target interaction of miR-199a-3p with VLDLR, a dual luciferase reporter assay was performed. Briefly, a 573-bp fragment in the 3′UTR of VLDLR containing the target site of miR-199a-3p predicted was amplified from OMECs cDNA using PCR primers (Table 1). The amplified product was inserted into the SacI and Xhol restriction sites of a pmirGLO vector to produce a wild-type pmirGLO dual luciferase reporter vector. Among the 573-bp 3′UTR of VLDLR, the target binding sequences for miR-199a-3p were complementarily mutated from “ACTACTG” to “TGATGAC,” thereby producing a mutant pmirGLO dual luciferase reporter vector. The two vectors were validated by electrophoresis in 1.5% agarose gels and Sanger sequencing.
A total of 0.20 μg plasmid was extracted from the wild-type or mutant pmirGLO dual luciferase reporter vector, and 20 pmol of miR-199a-3p mimic or miR-199a-3p mimic NC were co-transfected into the HEK293 T cells using INVI DNA & RNA Transfection Reagent™ (Invigentech, CA, USA). After transfection for 48 h, the Renilla and firefly luciferase activities of VLDLR were detected using the Dual-Luciferase Reporter Kit (Promega, WI, USA) by a Varioskan LUX Microplate Reader (Thermo Lifetech, MA, USA).
For the OMECs transfected with the miR-199a-3p mimic, miR-199a-3p inhibitor, miR-199a-3p mimic NC, and miR-19a-3p inhibitor NC, the supernatant was collected by centrifugation from 300 μl of lysate solution added 48 h after transfection. The content of triglycerides in OMECs was measured in quadruplicate using a triglyceride content assay kit (Solarbio, Beijing, China) by a microplate reader (Thermo Lifetech, MA, USA).
The difference between the two groups was compared using two-tailed Student's t-test in SPSS 22.0 (IBM, NY, USA). Data are indicated as mean ± standard error of the mean (SEM). All P-values were considered statistically significant when P < 0.05.
To explore the effect of miR-199a-3p on viability, proliferation, and milk fat synthesis of OMECs, the miR-199a-3p mimic, miR-199a-3p inhibitor, and their NC were transfected into OMECs. The RT-qPCR analysis revealed that the expression of miR-199a-3p was 80-fold higher in the miR-199a-3p mimic group compared with miR-199a-3p mimic NC. In contrast, compared with miR-199a-3p inhibitor NC, miR-199a-3p had a significant decrease in the expression of OMECs transfected with miR-199a-3p inhibitor (Figure 1A). The results suggest that miR-199a-3p mimic and miR-199a-3p inhibitor were successfully transfected into OMECs.
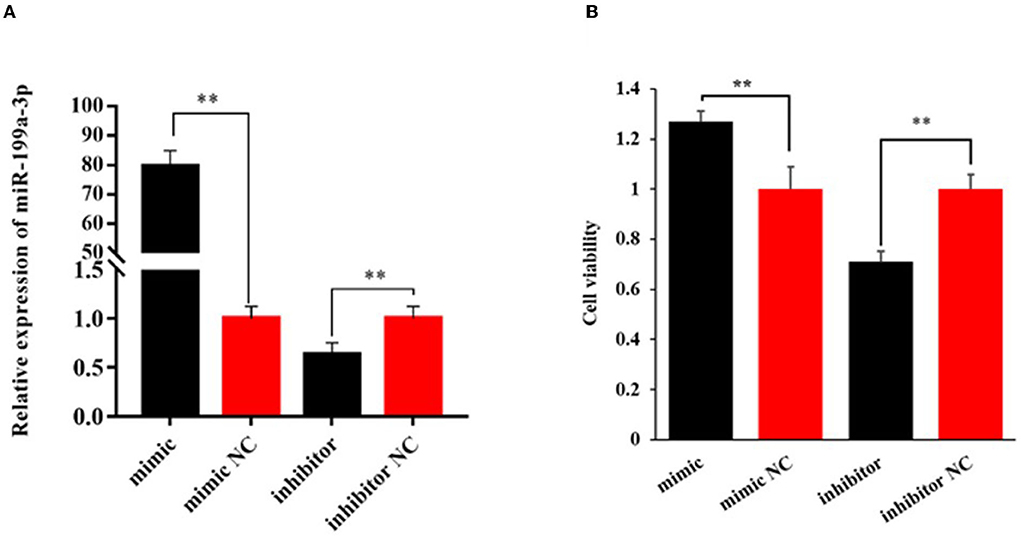
Figure 1. Relative expression level of miR-199a-3p (A) and its effect on the viability of ovine mammary epithelial cells (OMECs) (B) when miR-199a-3p mimic and miR-199a-3p inhibitor were transfected into OMECs. **P < 0.01.
The CCK8 assay found that cell viability significantly increased in OMECs transfected with miR-199a-3p mimic when compared with miR-199a-3p mimic NC, while miR-199a-3p inhibitor resulted in a significant decrease in cell viability compared with its NC group (P < 0.01, Figure 1B).
The Edu analysis revealed that the percentage of Edu-labeled positive OMECs was increased by 20.43% in the miR-199a-3p mimic group compared with its NC group (P < 0.05), while the percentage was decreased by 30.29% in the miR-199a-3p inhibitor group (P < 0.01, Figures 2A,B).
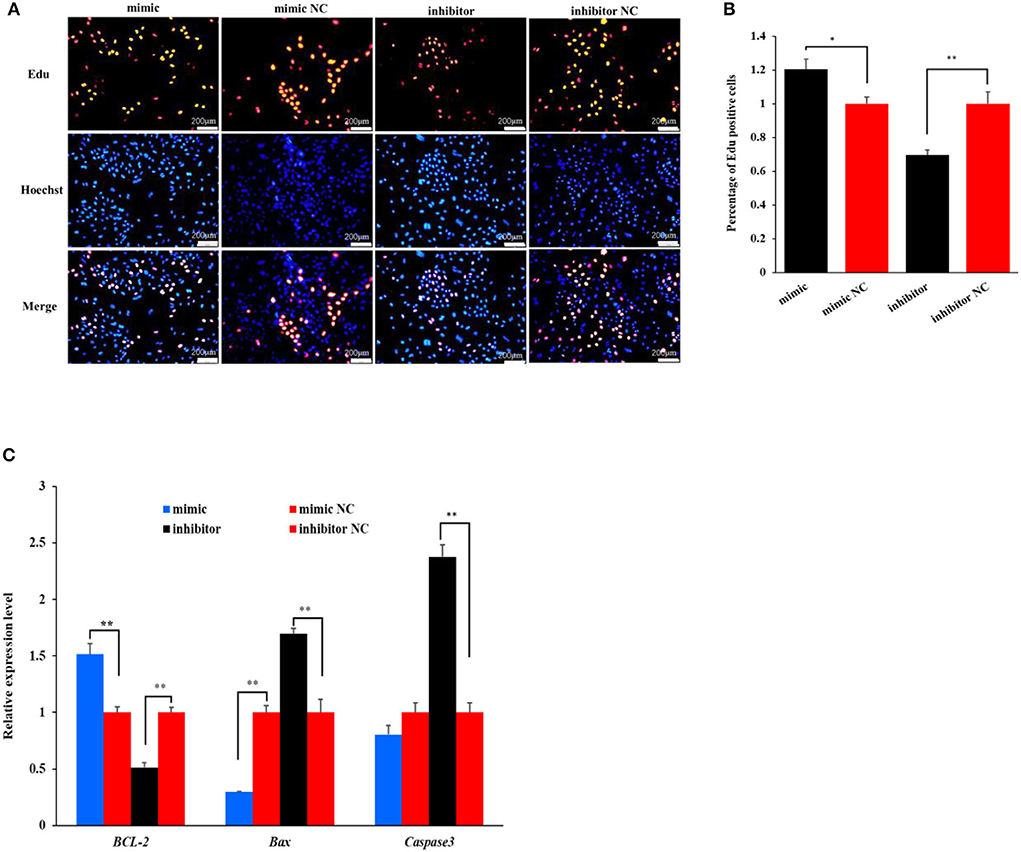
Figure 2. The effect of miR-199a-3p on the proliferation of ovine mammary epithelial cells (OMECs) when miR-199a-3p mimic and miR-199a-3p inhibitor were transfected into OMECs. (A) The proliferation of OMECs detected using an Edu assay. (B) The percentage of Edu-labeled positive OMECs counted by the ImageJ software. (C) The expression levels of BCL-2, Bax, and Caspase 3. **P < 0.01 and *P < 0.05.
To further validate the effect of miR-199a-3p on cell proliferation, the expression levels of three marker genes were detected using RT-qPCR. As shown in Figure 2C, miR-199a-3p mimic increased the expression of BCL-2, while the inhibition of miR-199a-3p had the opposite effect on the gene in expression (P < 0.05). Meanwhile, miR-199a-3p mimic inhibited the expression of Bax (P < 0.05), and 199a-3p inhibitor led to an increase in the expression level of Caspase3 (P < 0.05, Figure 2C). Taken together, these results suggest that miR-199a-3p promotes the proliferation of OMECs in sheep.
The agarose gel electrophoresis result revealed that amplicons of the expected size (~573 bp) were successfully ligated into dual luciferase reporter vectors (Figure 3A). The result from Sanger sequencing further confirmed the presence of the binding sequence in the 3′UTR of VLDLR with miR-199a-3p in the wild-type pmirGLO vector, as well as mutated sequences in the mutant vector (Figure 3B). This suggests that the two vectors were constructed as expected.
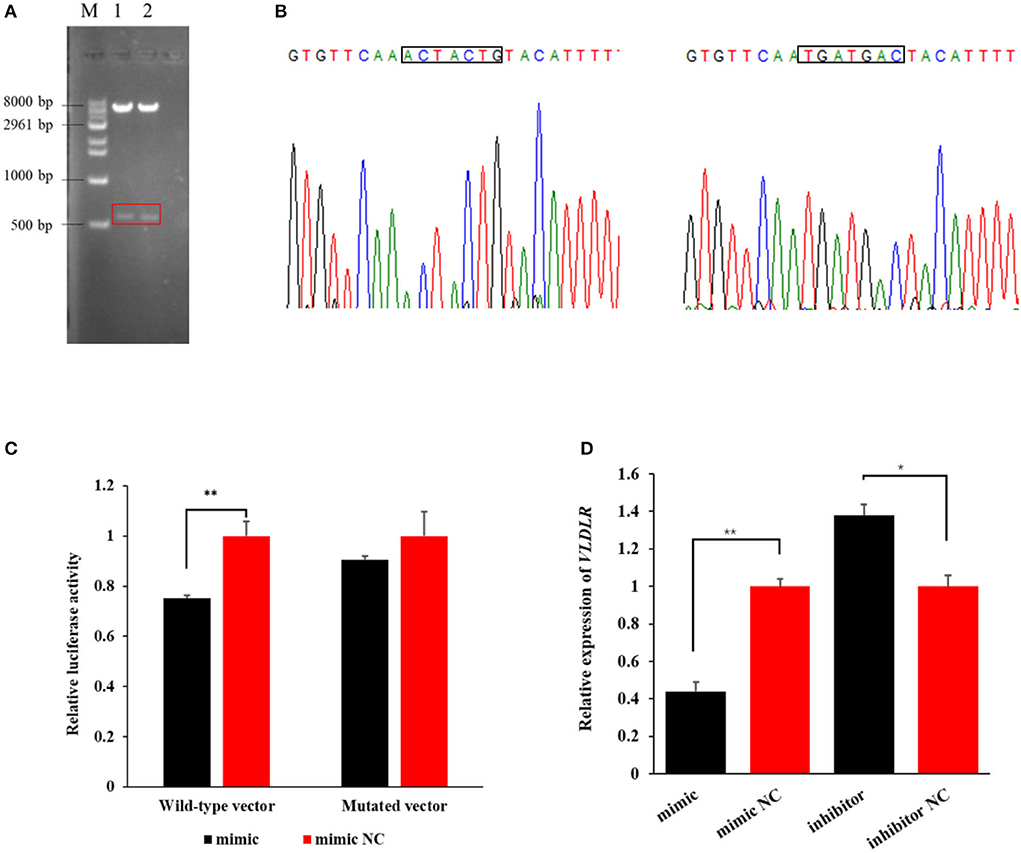
Figure 3. The target relationship verification of miR-199a-3p with VLDLR using a dual-luciferase reporter assay. (A) Dual luciferase reporter vectors after restriction endonucleases digestion were visualized using agarose gel electrophoresis. M: marker; 1: wild-type vector; 2: mutant vector. (B) The Sanger sequencing result of sequences of wild-type and mutant vectors. (C) The luciferase activity of VLDLR was detected when the miR-199a-3p mimic or miR-199a-3p mimic NC, and wild-type or mutant dual luciferase reporter vectors were co-transfected into HEK293T cells. (D) Effect of miR-199a-3p mimic and miR-199a-3p inhibitor on the expression level of VLDLR in ovine mammary epithelial cells (OMECs). **P < 0.01 and *P < 0.05.
In the group of co-transfections with wild-type pmirGLO vector and miR-199a-3p mimic, the luciferase activity of VLDLR significantly decreased to 75% of the negative control group (P < 0.01). However, in the HEK293 T cells transfected with the mutant pmirGLO vector, there was no significant difference in the luciferase activity of VLDLR between miR-199a-3p mimic and miR-199a-3p mimic NC (P > 0.05, Figure 3C). This suggests that miR-199a-3p targets VLDLR by binding to its 3′UTR.
The RT-qPCR analysis revealed that miR-199a-3p mimic decreased the expression of VLDLR (P < 0.01), while the inhibition of miR-199a-3p led to the upregulation of VLDLR expression in OMECs (P < 0.05, Figure 3D).
To investigate the effect of miR-199a-3p on milk fat synthesis, both the content of triglycerides and the expression levels of six milk fat synthesis marker genes, namely, LPL, ACACA, FABP3, CD36, SCD, and FASN, were detected in OMECs transfected with miR-199a-3p mimic and miR-199a-3p inhibitor.
The miR-199a-3p mimic decreased the level of triglycerides by 48.02% (P < 0.01). In contrast, the inhibition of the miRNA increased the content of triglycerides by 45.27% in OMECs (P < 0.05, Figure 4).
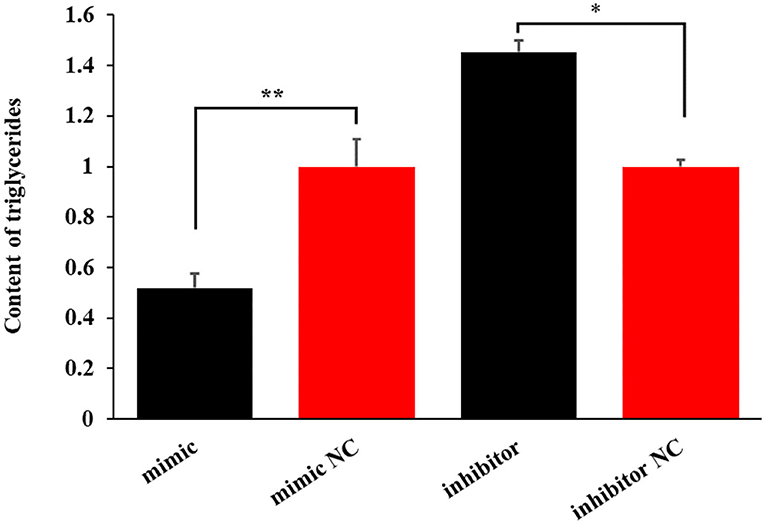
Figure 4. The effect of miR-199a-3p on the content of triglycerides in ovine mammary epithelial cells (OMECs). **P < 0.01 and *P < 0.05.
Similarly, miR-199a-3p mimic decreased the expression levels of the six genes, while miR-199a-3p inhibitor increased the expression of LPL, ACACA, FABP3, SCD, and FASN in OMECs (Figure 5). Taken together, these results suggest that miR-199a-3p inhibits milk fat synthesis of OMECs in sheep.
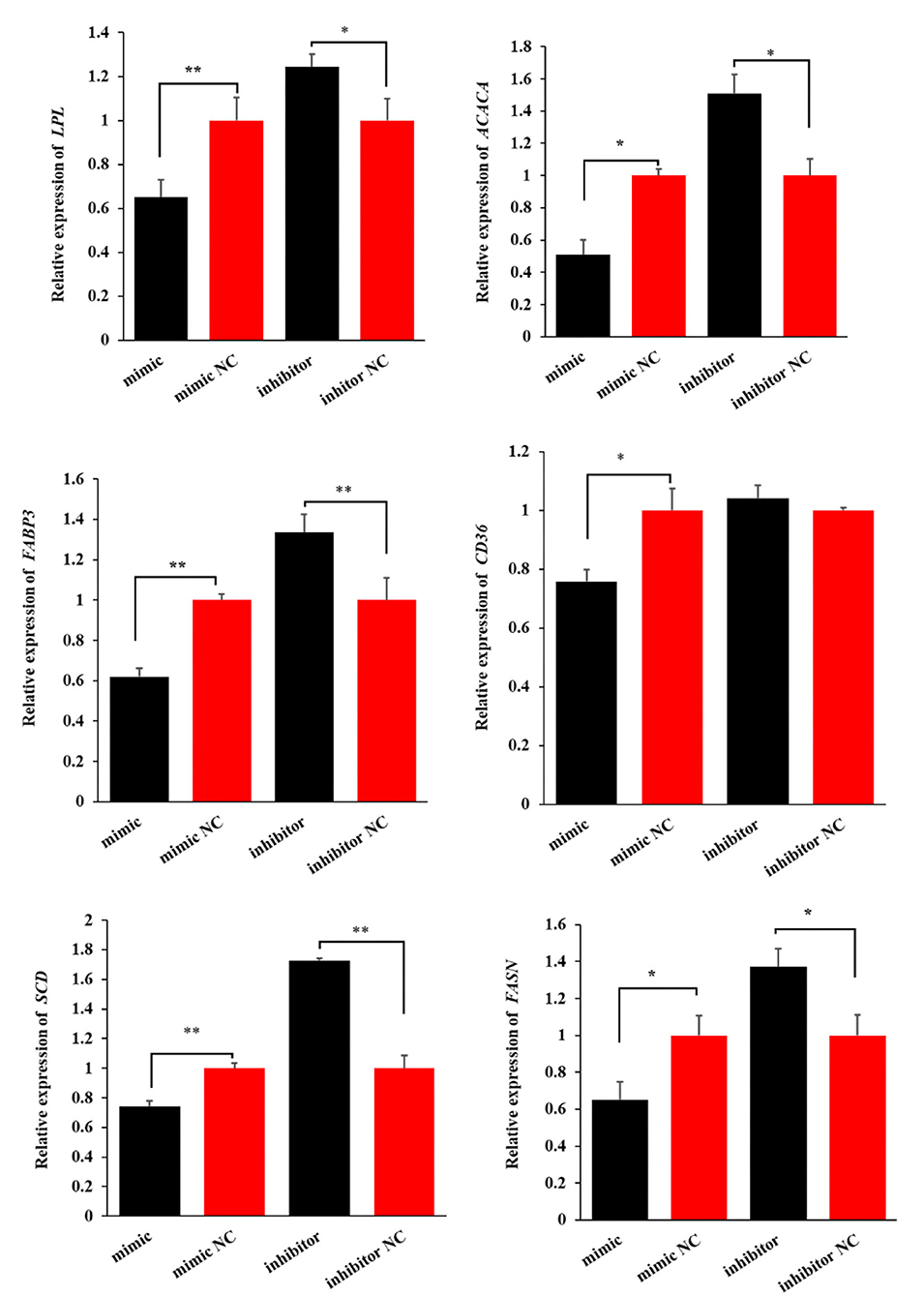
Figure 5. The expression levels of six milk fat synthesis marker genes, namely, LPL, ACACA, FABP3, CD36, SCD, and FASN, when the miR-199a-3p mimic and miR-199a-3p inhibitor were transfected into ovine mammary epithelial cells (OMECs). **P < 0.01 and *P < 0.05.
This study investigates the effect of miR-199a-3p on the proliferation and milk fat synthesis of OMECs, as well as its regulatory mechanism underlying the cells. At present, research into the effect of miR-199a-3p on cell proliferation has primarily been focused on cancer cells and stem cells in humans, but the findings obtained were controversial. Most studies found that miR-199a-3p inhibited the proliferation of hepatocellular carcinoma, colorectal cancer, and prostate cancer cells (14). An inhibition role of miR-199a-3p in the proliferation of human embryonic stem cells has also been reported (15). However, there are exceptions. Shatseva et al. (16) found that miR-199a-3p expression increased the viability and proliferation of both breast cancer cells and endothelial cells. The promoted effect of miR-199a-3p on cell proliferation has also been reported in gastric cancer cells (17) and adipocytes of mice (18). The last findings supported our results about the promotion of miR-199a-3p in cell viability and proliferation of OMECs. It has been reported that the proliferation number and viability of mammary epithelial cells are positively related to their milk yield in mammals (11). In this context, it was speculated that miR-199a-3p may improve milk yield in sheep. The contention is supported by the observation that miR-199a-3p was found to be upregulated in Kazakh horse with higher milk yield (19).
It was notable that miR-199a-3p regulated the expression of BCL-2, Bax, and Caspase 3 in OMECs. These three genes are related to cell survival or apoptosis. BCL-2 is an anti-apoptotic protein and is able to inhibit apoptosis of MECs induced by a variety of stimuli. In a gain-of-function transgenic mouse model, BCL-2 gain of function significantly decreased the number of apoptotic MECs but increased the survival rate of MECs during mammary gland involution periods (20). Meanwhile, over-expressed BCL-2 was found to increase the number of MECs in transgenic mice (21). In this respect, the result that miR-199a-3p enhanced the expression of BCL-2 further supported the promoted effect of miR-199a-3p on the proliferation of OMECs in the study. Bax and Caspase 3 are pro-apoptotic effector members. Over-expressed Bax has been demonstrated to induce apoptosis of MECs (22). Caspase 3, an endoprotease, is a crucial regulator of programmed cell death. The knockout of Caspase 3 led to increases in proliferation in vivo and hyperproliferation after mitogenic stimulation in vitro (23). These suggest a negative correlation between the expression of Caspase 3 with cell proliferation. Upon our joint analysis of the Targetscan 3.1 and miRanda 3.3a, no target relationships between miR-199a-3p and the three marker genes were predicted. Additionally, although miR-199a-3p can target VLDLR, the protein VLDLR mainly plays a biological role in fat synthesis. It has not been reported that VLDLR or the pathway via VLDLR regulated cell proliferation or viability. It is, therefore, impossible that miR-199a-3p regulates the proliferation and viability of OMECs by targeting VLDLR. Taken together, it was, therefore, inferred that miR-199a-3p promoted the proliferation and viability of OMECs by regulating the expression of BCL-2, Bax, and Caspase 3. However, the molecular mechanism underlying the process would need to be investigated.
Triglycerides are the main component of milk fat globules located in mammary epithelial cells, eventually forming milk fat. In this study, miR-199a-3p decreased the content of triglycerides in OMECs. The result was in accordance with the observation that over-expressed miR-199a-3p had the capability to weaken lipid accumulation and adipogenic gene expression in preadipocytes (24) and 3T3-L1 adipocytes (18) in mice. The miRNA was found to be one of the upregulated miRNAs identified in the bovine mammary gland, for Holstein dairy cows fed diets supplemented with either linseed or safflower oil that significantly decreased milk fat (25). The miR-199a-3p has been reported to exhibit a higher expression level during the non-lactating period than at lactation in ewes (12) and dairy cows (26). Given that more fat droplets originated from triglycerides at lactation when compared with the non-lactation period, these were also consistent with our observation. However, it was reported that over-expressed miR-199a-3p accelerated adipogenesis of bone narrow-derived mesenchymal stem cells (27). The inconsistent results, therefore, suggest that the effect of miR-199a-3p on lipogenesis should be further investigated in different kinds of cells.
This is the first study to report the target binding of miR-199a-3p with VLDLR. VLDLR was found to be expressed in the bovine mammary gland (28), which was in agreement with our results. The protein VLDLR is a member of the low-density lipoprotein receptor family. VLDLR can bind apolipoprotein E (apoE) triglyceride-rich lipoproteins and then accelerate subsequent triglyceride hydrolysis by LPL, resulting in the entry of lipid into MECs (29). Meanwhile, VLDLR also strengthens the activity of LPL that releases long-chain fatty acids from chylomicron and very low-density lipoprotein. Long-chain fatty acids are used as raw materials for synthesizing triglycerides in MECs (13). In addition, knockout mice of VLDLR exhibited a decrease in adipocyte triglyceride storage (30). Taken together, these studies suggest that VLDLR may be responsible for triglyceride synthesis. In this study, it was observed that over-expression of miR-199a-3p decreased VLDLR expression in OMECs. These results confirmed again the inhibited effect of miR-199a-3p on triglyceride content of OMECs by targeting VLDLR.
Besides VLDLR, miR-199a-3p reduced the expression levels of FASN, ACACA, CD36, LPL, FABP3, and SCD of OMECs in this study. These genes were all associated with milk fat synthesis. Milk fat synthesis in OMECs consisted of exogenous fatty acid absorption, de novo synthesis of fatty acid, and synthesis and secretion of triglycerides. The rate-limiting enzymes, FASN and ACACA, are involved in de novo lipogenesis in OMECs. CD36 is a fatty acid transferase, and it contributes to the absorption of long-chain fatty acids in OMECs when entering the extracellular environment (31). Non-esterified fatty acids carried by chylomicrons, very low-density lipoprotein, and albumin are hydrolyzed into long-chain fatty acids by LPL (32), followed by intracellular transport for esterification by FABP3 (33). SCD is a key factor in the synthesis of monounsaturated fatty acids, which is further used for producing triglycerides in OMECs. It was, therefore, concluded that downregulation of FASN, ACACA, CD36, LPL, FABP3, and SCD in the expression by miR-199a-3p was responsible for the inhibition of milk fat synthesis in OMECs.
In addition to mammary gland tissues, miR-199a-3p was found to be expressed in hair follicles (16), cardiac myocytes (34), ovary (35), and adipose tissues (24). The miRNA has been verified to play biological functions in the regulation of hair follicles development, tumorigenesis, and adipocyte differentiation by targeting caveolin 2 (CAV2), zinc fingers and homeoboxes 1 (ZHX1), SCD, and mechanistic target of rapamycin kinase (mTOR) (16–18, 24). These studies affirm that a single miRNA appears to target a range of different mRNAs and suggest that miR-199a-3p may play pleiotropic roles in a variety of cellular activities and biological processes by targeting different functional genes.
Our results suggest that miR-199a-3p promotes the viability and proliferation of OMECs, while it inhibits milk fat synthesis of OMECs by targeting VLDLR. This study provides a better understanding of the functions of miR-199a-3p in ovine mammary gland development and milk fat synthesis.
The original contributions presented in the study are publicly available. This data can be found here: NCBI [accession: ON572243 and ON572244].
The animal study was reviewed and approved by Animal Experiment Ethics Committee of Gansu Agricultural University, Lanzhou, China (Approval Number GSAU-ETH-AST-2021-027). Written informed consent was obtained from the owners for the participation of their animals in this study.
JW contributed to the data analysis and wrote the manuscript. ZH, LH, LQ, YL, and JH performed the investigation and collected the samples. XL, SL, FZ, JS, ML, and ZZ performed the formal analysis, methodology, and software analysis. JW did the project administration and revised the manuscript. All authors contributed to the article and approved the submitted version.
This study was financially supported by the National Natural Science Foundation of China (32060746 and 31860635), the Yong Supervisor Support Fund of Gansu Agricultural University (GAU-QDFC-2020-01), the Fuxi Young Talents Fund of Gansu Agricultural University (Gaufx-02Y02), and the Science and Technology Project of Lanzhou City (2021-1-162).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. (2004) 116:281–97. doi: 10.1016/s0092-8674(04)00045-5
2. Wang J, Zhou H, Hickford JGH, Hao Z, Gong H, Hu J, et al. Identification and characterization of circular RNAs in mammary gland tissue from sheep at peak lactation and during the nonlactating period. J Dairy Sci. (2021) 104:2396–409. doi: 10.3168/jds.2020-18911
3. Zhang L, Wu ZQ, Wang YJ, Wang M, Yang WC. MiR-143 regulates milk fat synthesis by targeting Smad3 in bovine mammary epithelial cells. Animals. (2020) 10:1453. doi: 10.3390/ani10091453
4. Song N, Luo J, Huang L, Tian H, Chen Y, He Q. miR-204-5p and miR-211 synergistically downregulate the αS1-casein content and contribute to the lower allergy of goat milk. J Agric Food Chem. (2021) 69:5353–62. doi: 10.1021/acs.jafc.1c01147
5. Fan Y, Arbab AAI, Zhang H, Yang Y, Lu X, Han Z, et al. MicroRNA-193a-5p regulates the synthesis of polyunsaturated fatty acids by targeting fatty acid desaturase 1 (FADS1) in bovine mammary epithelial cells. Biomolecules. (2021) 11:157. doi: 10.3390/biom11020157
6. Chen Z, Lu Q, Liang Y, Cui X, Wang X, Mao Y, et al. Circ11103 interacts with miR-128/PPARGC1A to regulate milk fat metabolism in dairy cows. J Agric Food Chem. (2021) 69:4490–500. doi: 10.1021/acs.jafc.0c07018
7. Chen Z, Chu S, Wang X, Fan Y, Zhan T, Arbab AAI, et al. MicroRNA-106b regulates milk fat metabolism via ATP binding cassette subfamily a member 1 (ABCA1) in bovine mammary epithelial cells. J Agric Food Chem. (2019) 67:3981–90. doi: 10.1021/acs.jafc.9b00622
8. Jiao BL, Zhang XL, Wang SH, Wang LX, Luo ZX, Zhao HB, et al. MicroRNA-221 regulates proliferation of bovine mammary gland epithelial cells by targeting the STAT5a and IRS1 genes. J Dairy Sci. (2019) 102:426–35. doi: 10.3168/jds.2018-15108
9. Chen Z, Chu S, Wang X, Sun Y, Xu T, Mao Y, et al. MiR-16a regulates milk fat metabolism by targeting large tumor suppressor kinase 1 (LATS1) in bovine mammary epithelial cells. J Agric Food Chem. (2019) 67:11167–78. doi: 10.1021/acs.jafc.9b04883
10. Wang H, Luo J, Zhang T, Tian H, Ma Y, Xu H, et al. MicroRNA-26a/b and their host genes synergistically regulate triacylglycerol synthesis by targeting the INSIG1 gene. RNA Biol. (2016) 13:500–10. doi: 10.1080/15476286.2016.1164365
11. Hao ZY, Wang JQ, Luo YL, Liu X, Li SB, Zhao ML, et al. Deep small RNA-Seq reveals microRNAs expression profiles in lactating mammary gland of 2 sheep breeds with different milk performance. Domest Anim Endocrinol. (2021) 74:106561. doi: 10.1016/j.domaniend.2020.106561
12. Wang J, Hao Z, Hu J, Liu X, Li S, Wang J, et al. Small RNA deep sequencing reveals the expressions of microRNAs in ovine mammary gland development at peak-lactation and during the non-lactating period. Genomics. (2021) 113:637–46. doi: 10.1016/j.ygeno.2020.09.060
13. Hao Z, Luo Y, Wang J, Hickford JGH, Zhou H, Hu J, et al. MicroRNA-432 inhibits milk fat synthesis by targeting SCD and LPL in ovine mammary epithelial cells. Food Funct. (2021) 12:9432–42. doi: 10.1039/d1fo01260f
14. Li SQ, Wang ZH, Mi XG, Liu L, Tan Y. MiR-199a/b-3p suppresses migration and invasion of breast cancer cells by downregulating PAK4/MEK/ERK signaling pathway. IUBMB Life. (2015) 67:768–77. doi: 10.1002/iub.1433
15. Chen HP, Wen J, Tan SR, Kang LM, Zhu GC. MiR-199a-3p inhibition facilitates cardiomyocyte differentiation of embryonic stem cell through promotion of MEF2C. J Cell Physiol. (2019) 234:23315–25. doi: 10.1002/jcp.28899
16. Shatseva T, Lee DY, Deng Z, Yang BB. MicroRNA miR-199a-3p regulates cell proliferation and survival by targeting caveolin-2. J Cell Sci. (2011) 124:2826–36. doi: 10.1242/jcs.077529
17. Wang Z, Ma X, Cai Q, Wang X, Yu B, Cai Q, et al. MiR-199a-3p promotes gastric cancer progression by targeting ZHX1. FEBS Lett. (2014) 588:4504–12. doi: 10.1016/j.febslet.2014.09.047
18. Tan Z, Du J, Shen L, Liu C, Ma J, Bai L, et al. miR-199a-3p affects adipocytes differentiation and fatty acid composition through targeting SCD. Biochem Biophys Res Commun. (2017) 492:82–8. doi: 10.1016/j.bbrc.2017.08.030
19. Liu L, Fang C, Sun Y, Liu W. Evaluation of key miRNAs during early pregnancy in Kazakh horse using RNA sequencing. Peer J. (2021) 9:e10796. doi: 10.7717/peerj.10796
20. Schorr K, Li M, Bar-Peled U, Lewis A, Heredia A, Lewis B, et al. Gain of Bcl-2 is more potent than bax loss in regulating mammary epithelial cell survival in vivo. Cancer Res. (1999) 59:2541–5.
21. Jäger R, Herzer U, Schenkel J, Weiher H. Overexpression of Bcl-2 inhibits alveolar cell apoptosis during involution and accelerates c-myc-induced tumorigenesis of the mammary gland in transgenic mice. Oncogene. (1997) 15:1787–95. doi: 10.1038/sj.onc.1201353
22. Strange R, Metcalfe T, Thackray L, Dang M. Apoptosis in normal and neoplastic mammary gland development. Microsc Res Tech. (2001) 52:171–81. doi: 10.1002/1097-0029(20010115)52:2<171::AID-JEMT1003>3.0.CO;2-T
23. Woo M, Hakem R, Furlonger C, Hakem A, Duncan GS, Sasaki T, et al. Caspase-3 regulates cell cycle in B cells: a consequence of substrate specificity. Nat Immunol. (2003) 4:1016–22. doi: 10.1038/ni976
24. Gao Y, Cao Y, Cui X, Wang X, Zhou Y, Huang F, et al. miR-199a-3p regulates brown adipocyte differentiation through mTOR signaling pathway. Mol Cell Endocrinol. (2018) 476:155–64. doi: 10.1016/j.mce.2018.05.005
25. Li R, Beaudoin F, Ammah AA, Bissonnette N, Benchaar C, Zhao X, et al. Deep sequencing shows microRNA involvement in bovine mammary gland adaptation to diets supplemented with linseed oil or safflower oil. BMC Genomics. (2015) 16:884. doi: 10.1186/s12864-015-1965-7
26. Li Z, Lan X, Guo W, Sun J, Huang Y, Wang J, et al. Comparative transcriptome profiling of dairy goat microRNAs from dry period and peak lactation mammary gland tissues. PLoS ONE. (2012) 7:e52388. doi: 10.1371/journal.pone.0052388
27. Shuai Y, Yang R, Mu R, Yu Y, Rong L, Jin L. MiR-199a-3p mediates the adipogenic differentiation of bone marrow-derived mesenchymal stem cells by regulating KDM6A/WNT signaling. Life Sci. (2019) 220:84–91. doi: 10.1016/j.lfs.2019.01.051
28. Heegaard CW, Simonsen AC, Oka K, Kjøller L, Christensen A, Madsen B, et al. Very low density lipoprotein receptor binds and mediates endocytosis of urokinase-type plasminogen activator-type-1 plasminogen activator inhibitor complex. J Biol Chem. (1995) 270:20855–61. doi: 10.1074/jbc.270.35.20855
29. Nguyen A, Tao H, Metrione M, Hajri T. Very low density lipoprotein receptor (VLDLR) expression is a determinant factor in adipose tissue inflammation and adipocyte-macrophage interaction. J Biol Chem. (2014) 289:1688–703. doi: 10.1074/jbc.M113.515320
30. Boycott KM, Flavelle S, Bureau A, Glass HC, Fujiwara TM, Wirrell E, et al. Homozygous deletion of the very low density lipoprotein receptor gene causes autosomal recessive cerebellar hypoplasia with cerebral gyral simplification. Am J Hum Genet. (2005) 77:477–83. doi: 10.1086/444400
31. Doege H, Stahl A. Protein-mediated fatty acid uptake: novel insights from in vivo models. Physiology. (2006) 21:259–68. doi: 10.1152/physiol.00014.2006
32. Wang J, Zhou H, Hickford JGH, Hao Z, Shen J, Luo Y, et al. Comparison of the transcriptome of the ovine mammary gland in lactating and non-lactating Small-tailed han sheep. Front Genet. (2020) 11:472. doi: 10.3389/fgene.2020.00472
33. Yao D, Yang C, Ma J, Chen L, Luo J, Ma Y, et al. cAMP response element binding protein 1 (CREB1) promotes monounsaturated fatty acid synthesis and triacylglycerol accumulation in goat mammary epithelial cells. Animals. (2020) 10:1871. doi: 10.3390/ani10101871
34. Song XW, Li Q, Lin L, Wang XC, Li DF, Wang GK, et al. MicroRNAs are dynamically regulated in hypertrophic hearts, and miR-199a is essential for the maintenance of cell size in cardiomyocytes. J Cell Physiol. (2010) 225:437–43. doi: 10.1002/jcp.22217
Keywords: microRNA-199a-3p, VLDLR, ovine mammary epithelial cells, proliferation, triglyceride
Citation: Wang J, Hao Z, Hu L, Qiao L, Luo Y, Hu J, Liu X, Li S, Zhao F, Shen J, Li M and Zhao Z (2022) MicroRNA-199a-3p regulates proliferation and milk fat synthesis of ovine mammary epithelial cells by targeting VLDLR. Front. Vet. Sci. 9:948873. doi: 10.3389/fvets.2022.948873
Received: 20 May 2022; Accepted: 11 July 2022;
Published: 05 August 2022.
Edited by:
Zhi Chen, Yangzhou University, ChinaCopyright © 2022 Wang, Hao, Hu, Qiao, Luo, Hu, Liu, Li, Zhao, Shen, Li and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiqing Wang, d2FuZ2pxQGdzYXUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.