
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci. , 26 August 2022
Sec. Animal Nutrition and Metabolism
Volume 9 - 2022 | https://doi.org/10.3389/fvets.2022.948545
This article is part of the Research Topic Strategies for Mitigating the Transition Period Stress in Dairy Cattle View all 7 articles
We investigated the temporal dynamics of ruminal pH and the composition and predicted functions of the rumen bacterial community in Holstein cows during the periparturient period. Eight multiparous Holstein cows (body weight; 707.4 ± 29.9 kg, parity; 3.6 ± 0.6) were used for continuous reticulo-ruminal pH measurement, among which five were also used for bacterial community analysis. Rumen fluid samples were collected at 3 weeks before and 2 and 6 weeks after parturition, and blood samples were collected 3 weeks before and 0, 2, 4, and 6 weeks after parturition. After the parturition, reduction in the 1-h mean reticulo-ruminal pH was associated with a significant (P < 0.05) increase in total volatile fatty acid concentration. However, with the exception of a significant change in an unclassified genus of Prevotellaceae (P < 0.05), we detected no significant changes in the compositions of major bacterial phyla or genera or diversity indices during the periparturient period. On the basis of predicted functional analysis, we identified a total of 53 MetaCyc pathways (45 upregulated), 200 enzyme commissions (184 upregulated), and 714 Kyoto Encyclopedia of Genes and Genomes orthologs (667 upregulated) at 6 weeks postpartum that were significantly (P < 0.05) different to those at 3 weeks prepartum. Among the 14 MetaCyc pathways (P < 0.05) in which pyruvate is an end product, PWY-3661 [log2-fold change (FC) = 5.49, false discovery rate (FDR) corrected P < 0.001] was the most highly upregulated pyruvate-producing pathway. In addition, P381-PWY [adenosylcobalamin biosynthesis II (aerobic); FC = 5.48, FDR corrected P < 0.001] was the second most upregulated adenosylcobalamin (Vitamin B12)-producing pathway. In contrast, PWY-2221 (FC = −4.54, FDR corrected P = 0.003), predominantly found in pectinolytic bacteria, was the most downregulated pathway. Our findings indicate that changes in rumen bacterial community structure are not strictly associated with transitions in parturition or diet; however, we did observe changes in reticulo-ruminal pH and the metabolic adaptation of predicted functional pathways. Consequently, predictive analysis of postpartum functional pathways may enable us to gain insights into the underlying functional adaptations of bacterial communities in Holstein cows during the periparturient period.
At around time that the lactation period commences, a high-grain postpartum diet is necessary to enhance rumen fermentation and meet the energy requirements of lactation (1). However, enhanced rumen fermentation is accompanied by a decline in ruminal pH, which is attributable to an increase in the production of volatile fatty acids (VFAs) or lactic acid. In turn, this contributes to increases in the occurrence and severity of ruminal acidosis and subacute ruminal acidosis (SARA) (2). SARA is defined as a condition characterized by ruminal pH below 5.6 for more than 3 h per day (3) or below 5.8 for more than 5 h per day (4). SARA has been established to be associated with a range of health problems in dairy cows, among which are feed intake depression, reduced fiber digestion, milk fat depression, an increased production of bacterial endotoxins, and inflammation (5, 6). In addition, under conditions of high-grain diet challenge, it has been observed that SARA (7, 8) or parturition (9–11) has the influences on the structure and diversity of the ruminal bacterial community.
The impact of the transition from a high-forage diet to one of high grain content on rumen microbial composition has previously been investigated in Holstein cows. For example, single SARA challenge has been found to promote reductions in the diversity and abundance of gram-negative bacteria (12–14), although adaptation to the repeated SARA challenge and a subsequent recovery associated with changes in community composition and diversity have also been detected in rumen bacterial communities (8). Similarly, changes dairy cow rumen bacterial and archaeal communities have also been identified during the transition period (11). Accordingly, these observations provide convincing evidence to indicate that the development of an acidic ruminal environment in response to the transition to a high-grain diet can have a significant influence on rumen bacterial community structure and vice versa. This transition is characterized by changes in rumen fermentation, including elevated total VFA production and altered constituent profiles, along with changes in the characteristics of rumen bacteria (8, 13).
Although these findings may be broadly applicable, they might not necessarily be equally relevant in Holstein cows during the transition period. For example, Mohammed et al. (15) have reported variability in the severity of ruminal acidosis among pre- and postpartum cows, which they attribute to variability among bacterial communities in the rumen digesta. Such findings thus highlight that cows are not equally susceptible to ruminal acidosis, and that bacterial community composition may shift independently of differences in the severity of postpartum ruminal acidosis, and does not differ substantially between the pre- and post-calving periods (15). Similarly, the findings of a further study indicate the parity (primiparous vs. multiparous) has a significant impact on the dynamics of the ruminal microbiome of cows during the transition period (10). Nevertheless, despite this variability in the rumen bacterial community, the fermentative activities of these organisms continue to be sufficiently efficient to provide the requisite energy for milk production after parturition (16), thereby highlighting the adaptability of the bacterial community (17) in overcoming these metabolic challenges.
With a view toward clarifying the mechanisms underlying this adaptability, we investigated changes in the composition and predicted function of the rumen bacterial community in Holstein cows during the periparturient period. Specifically, we examined the possible associations between these changes with co-occurring physiological changes, including those in ruminal pH, fermentation, and blood metabolites and hormones. We hypothesized that parturition and simultaneous dietary changes may induce changes in the structure and potential functions of the rumen bacterial community. Our findings may offer plausible clues that will enable us to gain a more comprehensive understanding of these relationships and provide a basis for further studies on postpartum SARA.
The experimental protocol devised for this study was approved by the Iwate University Laboratory Animal Care and Use Committee (A201452-2; Morioka, Japan). For the purposes of analyses, we used eight multiparous Holstein cows (body weight; 707.4 ± 29.9, parity; 3.6 ± 0.6; Supplementary Table 1), which were housed in a facility of the Hiroshima University Experimental farm (Hiroshima, Japan), in which they had free access to water throughout the study period. Prior and subsequent to parturition, these cows were fed total mixed ration containing respectively dry and lactation period diets ad libitum (Table 1). Dry and lactation period diets were supplied daily in two equal portions at 900 and 1,600 h. Feed compositions and amounts were based on the requirements of the Japanese Feeding Standard for Dairy Cattle.
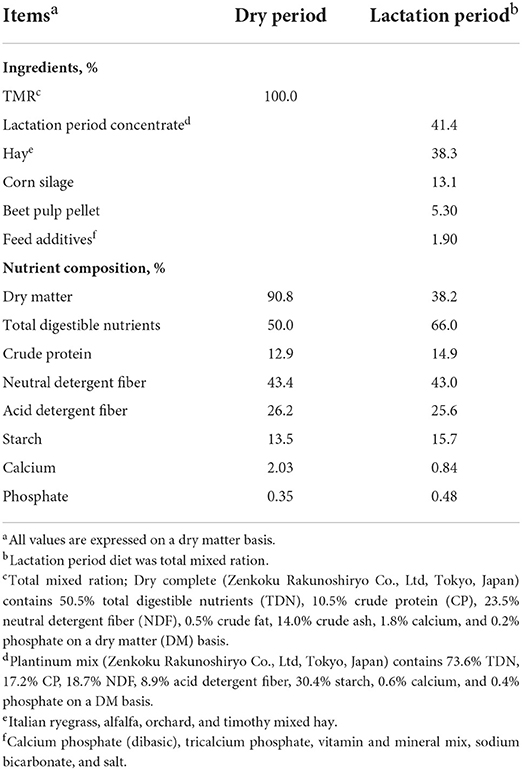
Table 1. Ingredients and nutrient composition of the dry and lactation period diets on a dry matter (DM) basis.
Eight multiparous Holstein cows were used for continuous reticulo-ruminal pH measurement, among which five were also used for bacterial community analysis (Supplementary Table 2). Throughout the study period, reticulo-ruminal pH was measured continuously at 10-min intervals using a YCOW-S radio transmission system (DKK-TOA, Yamagata, Japan), as described by Sato et al. (18). For the purposes of calibration, a pH sensor was inserted orally and calibration was performed at standard pH values of 4 and 7 prior to and after the experiment. No changes in pH were observed during the calibration period. Rumen fluid samples were collected using an oral stomach tube in the morning before feeding 3 weeks prior to the anticipated date of parturition and 2 and 6 weeks after parturition. The initial rumen fluid sample (100–200 mL) was discarded to avoid the possible contamination of rumen fluid with saliva. Rumen fluid samples were used to analyze bacterial community and concentrations of total VFAs, individual VFAs, NH3-N, lactic acid, and lipopolysaccharide (LPS). Fluid samples were immediately filtered through two layers of cheesecloth and stored at −80°C until use. Blood samples were collected in the morning before feeding 3 weeks prior to the anticipated date of parturition and 0, 2, 4, and 6 weeks after parturition. Samples were collected from the jugular vein into 10-mL evacuated serum-separating tubes and heparin-containing tubes (BD Vacutainer, Franklin Lakes, NJ, USA). These were immediately centrifuged at 1,500 × g for 15 min at 4°C to separate the serum and then preserved at −80°C until used for analysis.
For VFA analyses, 1 mL of 25% HO3P in 3 N H2SO4 was added to 5 mL of rumen fluid. Total and individual (acetic, propionic, and butyric acids) VFAs were separated and quantified by liquid chromatography (HPLC; Shimadzu, Kyoto, Japan) using a shim-pack SCR-102H packed column (Shimadzu, Kyoto, Japan). To determine the concentration of NH3-N in rumen fluid, samples were analyzed based on the steam distillation method using an NH3-N analyzer (Kjeltec Auto Sampler System 1035 Analyzer; Tecator Inc., Höganäs, Sweden). To measure rumen LPS concentration, rumen fluid samples were centrifuged at 11,000 × g for 15 min at 4°C and assayed using a kinetic Limulus amebocyte lysate assay (Pyrochrome with Glucashield; Seikagaku, Tokyo, Japan) as described previously (19). For blood biochemical analyses, the concentrations of glucose (GLU), non-esterified fatty acids (NEFAs), total ketone bodies (T-KB), cholesterol (T-CHO), triglyceride (TG), total protein (TP), albumin, blood urea nitrogen, total calcium (Ca), phosphate, aspartate transaminase (AST), and γ-glutamyl transpeptidase (GGT) were measured using an Acecut automated biochemistry analyzer (Toshiba Ltd., Tokyo, Japan). Plasma growth hormone (GH), insulin (INS), glucagon (GCG), and insulin-like growth factor-1 (IGF-1) concentrations were determined based on radioimmunoassays (20).
Total bacterial DNA was extracted from rumen fluid samples as described by Kim et al. (21). Briefly, samples were incubated with 750 μg/mL lysozyme (Sigma-Aldrich Co., St. Louis, MO, USA) at 37°C for 90 min. This was followed by the addition of 10 μL of purified achromopeptidase (Wako Pure Chemical Industries Ltd., Osaka, Japan) at 10,000 U/mL, with the resulting mixture being incubated at 37°C for 30 min. This suspension was subsequently treated with 60 μL 1% sodium dodecyl sulfate and 1 mg/mL proteinase K (Merck Japan Ltd., Tokyo, Japan) with incubation at 55°C for 5 min, and the lysate thus obtained was extracted with three rounds of phenol/chloroform/isoamyl alcohol (Wako Pure Chemical Industries Ltd.) and chloroform (Life Technologies Japan Ltd., Tokyo, Japan). The resulting DNA was precipitated using 5 M sodium chloride and 100% ethanol, followed by centrifugation at 21,900 × g for 15 min at 4°C. The DNA pellet was rinsed with 70% ethanol, dried, and dissolved in Tris-hydrochloride buffer. The Purified DNA was quantified using a Biospec-nano spectrophotometer (Shimadzu Biotech, Kyoto, Japan) and stored at −80°C until used for further analyses.
For the purposes of identifying rumen bacteria, we amplified the bacterial 16S rRNA gene using the barcoded universal primers 515F (5′-TGYCAGCMGCCGCGGTAA-3′) and 806R (5′-GGACTACNVGGGTWTCTAAT-3′) spanning the V4 hypervariable region. Polymerase chain reactions (PCR) were performed using 25-μL reaction mixture containing 12.5 μL of 2 × KAPA HiFi HotStart ReadyMix (Kapa Biosystems, Wilmington, MA, USA), 5 μL of each primer (1 μM), and 2.5 μL of template DNA (10 ng/μL). The thermal cycling conditions were as follows: an initial denaturation at 95°C for 3 min, followed by 25 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s, with a final extension of 72°C for 5 min. Amplicons were purified using AMPure XP beads (Beckman Coulter, High Wycombe, United Kingdom) according to the manufacturer's instructions. Libraries were constructed by ligating sequencing adapters and indices to the purified PCR products using the Nextera XT Sample Preparation Kit (Illumina, San Diego, CA, USA) according to the manufacturer's instructions. Paired-end sequencing (2 × 151 bp) was conducted using the Illumina MiSeq platform according to standard protocols. The sequencing data obtained have been deposited in the Sequence Read Archive of the National Center for Biotechnology Information and can be accessed via the SRA BioSample accession number SAMN19291604 (https://submit.ncbi.nlm.nih.gov/subs/sra/).
All sequencing reads were processed using the mothur program [version 1.41.1; University of Michigan; http://www.mothur.org/wiki/; Schloss et al. (22)], following the standard operating procedure for MiSeq [https://mothur.org/wiki/MiSeq_SOP; Kozich et al. (23)] with minor modifications. To obtain a non-redundant set of sequences, unique sequences were identified and aligned against the SILVA reference database [SSURef release 128; (24)], and sequence comparisons were performed using the mothur Ribosomal Database Project training set (version 16). Sequences were clustered and classified into operational taxonomic units (OTUs) using a cutoff value of 97% similarity. All samples were standardized by random subsampling (1,269 sequences/sample) using the “sub.sample” command, resulting in the elimination of a single sample (collected at 2 weeks postpartum) due to low sequence numbers. The “summary.single” command was used to analyze the OTUs, Chao1, abundance-based coverage estimator (ACE) of species richness, and Shannon, Simpson, and Heip diversity indices.
Representative sequences for each OTU were determined using the “get.oturep” command. Sequence comparisons were performed using BLASTn (https://blast.ncbi.nlm.nih.gov/Blast.cgi) against a 16S ribosomal RNA sequence database (Bacteria and Archaea; April 2021). Representative sequences and tabulated raw count data were used to predict functional features from 16S rRNA gene data using PICRUSt2 (25) with the default pipeline. Overall functional dissimilarities among the groups were examined based on the relative abundance of MetaCyc pathways (http://MetaCyc.org), enzyme commissions (EC), and Kyoto Encyclopedia of Genes and Genomes (KEGG) ortholog counts.
A mixed-model analysis of variance (accounting for repeated measures), using time as a fixed effect, followed by Dunnett's multiple comparison method was used to determine within-group differences. Principal component analysis (PCA) plots were constructed using the R package ggbiplot (R software version 3.3.2; R Foundation for Statistical Computing, Vienna, Austria), and non-metric multidimensional scaling (NMDS) plots were constructed in R (http://www.r-project.org; R Foundation for Statistical Computing, Vienna, Austria) using the “vegan” package. All numerical data were analyzed using Prism software (version 8.21). A P-value <0.05 was considered to be indicative of a significant difference and P < 0.10 was taken to indicate a significant tendency. Pearson correlation coefficient (r) values and significance level (P-value) were used to determine the relationships among rumen measurements (7-d mean ruminal pH, time with pH under 5.6, area under pH 5.6, total VFA and lactic acid concentrations, proportions of individual VFAs, and LPS concentration) and core OTUs. A heatmap was constructed using Prism software based on the Pearson correlation data. To determine whether the predicted functional pathways of the rumen bacterial community differed compared with that at 3 weeks prepartum, the raw MetaCyc, EC, and KEGG orthologs count outputs from PICRUSt2 were analyzed using DESeq2 with default parameters, after flooring fractional counts to the nearest integer (26). The inferred predicted functional pathways were considered differentially abundant (log2-fold change; FC) at a false discovery rate (FDR)-corrected significance threshold < 0.05.
During the periparturient period, we detected a decline in 1-h mean reticulo-ruminal pH values following parturition concomitant with the transition to a lactation period diet (Figure 1). Moreover, during this period, there were significant changes with respect to the 7-d mean reticulo-ruminal pH, and lengths of time during which pH values were <5.6 and <5.8 (P < 0.05). Compared with the values measured at 2 weeks prepartum, the pH values recorded at 0 to 6 weeks postpartum were significantly lower (P < 0.05), and the durations during which pH was below 5.6 (207 ± 28 min/d at 3 weeks postpartum) and below 5.8 (345 ± 76 and 307 ± 65 min/d at 1 and 2 weeks postpartum, respectively) were significantly higher (P < 0.05; Supplementary Table 3).
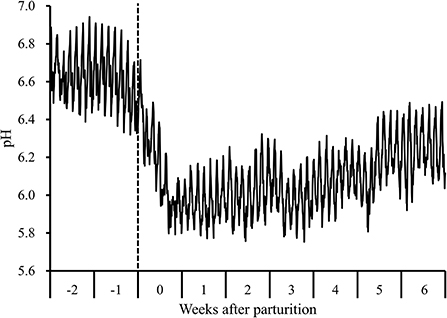
Figure 1. Diurnal changes in the 1-h mean reticulo-ruminal pH in Holstein cows (n = 8) during the periparturient period. “-2,” “-1,” “0,” “1,” “2,” “3,” “4,” “5,” and “6” denote observations at 2 and 1 weeks prepartum and 0, 1, 2, 3, 4, 5, and 6 weeks postpartum, respectively. The dotted line indicates the day of parturition.
We also detected significant changes in total VFA concentrations (P < 0.05) during the periparturient period, the values of which were found to be sig6nificantly higher (P < 0.05) at 2 and 6 weeks postpartum compared with 3 weeks prepartum (Table 2). Similarly, we detected significant changes in the proportions of acetic and propionic acids (P < 0.05) during the periparturient period, which were lower and higher, respectively, compared with those determined at 3 weeks prepartum (Table 2). Consistently, we established that ruminal LPS concentration had undergone a significant change (P < 0.05) during the periparturient period, with higher levels being recorded at 2 and 6 weeks postpartum compared with 3 weeks prepartum.
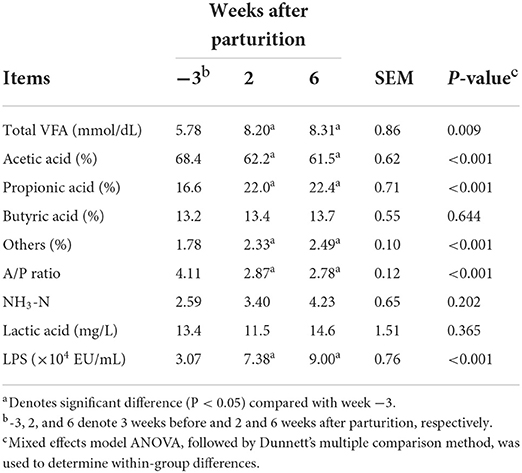
Table 2. Total volatile fatty acids (VFA), proportions of individual VFAs, acetic acid to propionic acid (A/P) ratio, and NH3-N, lactic acid, and lipopolysaccharide (LPS) concentrations in Holstein cows (n = 8) during the periparturient period.
Significant changes (P < 0.05) during the periparturient period were also detected with respect to plasma concentration of GLU, NEFA, T-KB, T-CHO, TG, TP, ALB, and Ca, and the activities of AST and GGT (Table 3). Concentrations of the hormones IGF-1, CGC, GH, and INS in the peripheral plasma were likewise significantly altered (P < 0.05) during the periparturient period (Table 3). Compared with 3 weeks prepartum, we detected significantly lower concentrations of IGF-1 (0, 2, 4, and 6 weeks postpartum) and INS (2 and 4 weeks postpartum; P < 0.05), and significantly higher concentrations of CGC (0, 2, 4, and 6 weeks postpartum) and GH (0 weeks postpartum; P < 0.05). PCA plots revealed 7-d mean ruminal pH (Figure 2A), the proportion of acetic acid (Figure 2B), and concentrations of GLU (Figure 2C) and IGF-1 (Figure 2D) to be the most influential factors at 3 weeks prepartum, whereas concentrations of GH were the most influential factor at 0 weeks postpartum (Figure 2D).
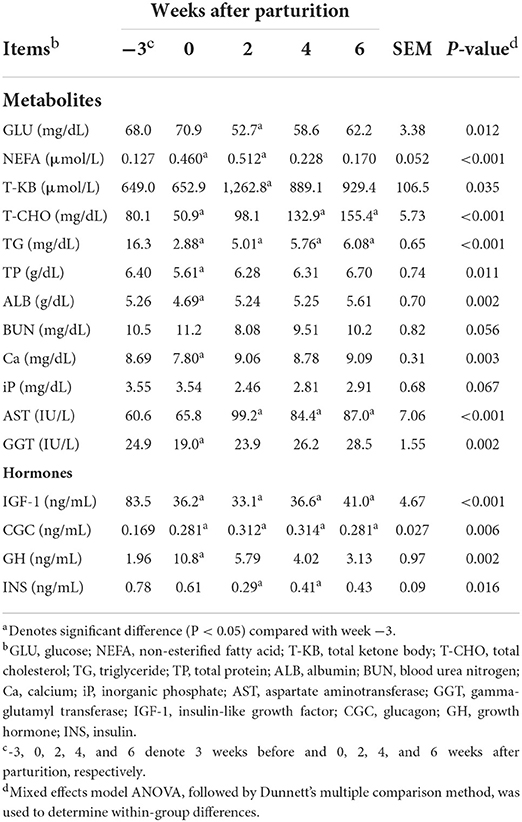
Table 3. Profiles of the peripheral blood metabolites and hormones in Holstein cows (n = 8) during the periparturient period.
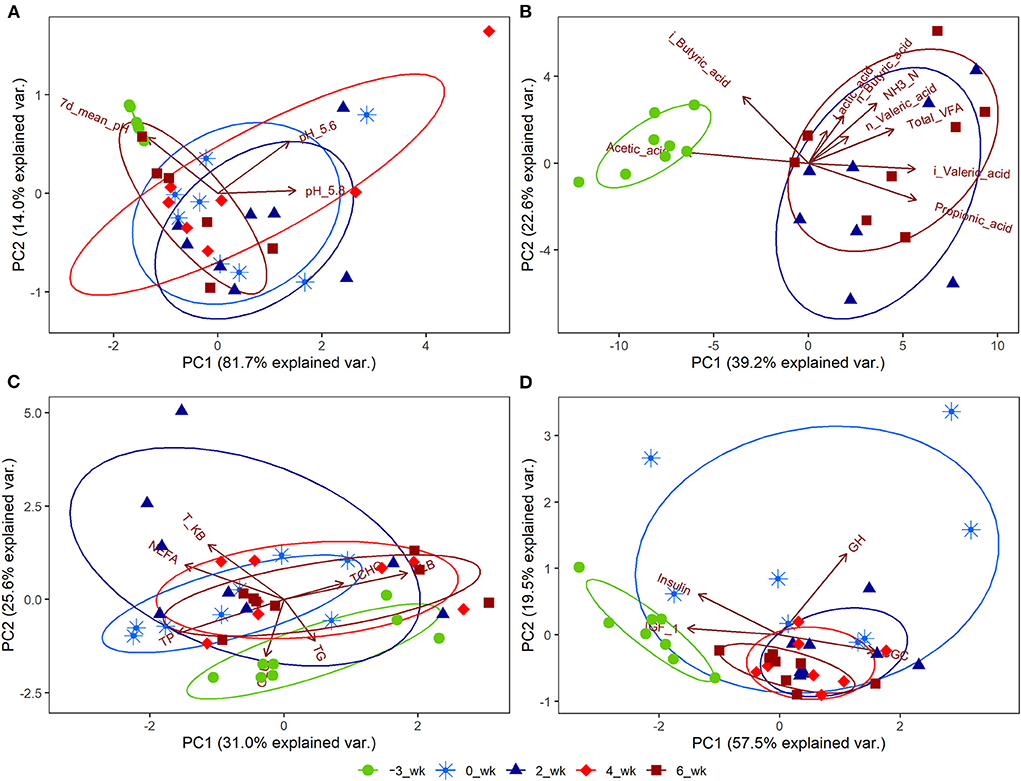
Figure 2. Principal component analysis (PCA) plots for Holstein cows (n = 8) during the periparturient period. The PCA plots were generated for (A) ruminal pH, (B) rumen fermentation measurements, (C) peripheral blood metabolites, and (D) hormones. The circle shows the correlation among the variables in each period. PC1 and PC2 represent principal components 1 and 2, respectively. 7d_mean_pH, 7-d mean reticulo-ruminal pH; pH_5.6, duration at which reticulo-ruminal pH was <5.6; pH_5.8, duration at which reticulo-ruminal pH was <5.8; GLU, glucose; NEFA, non-esterified fatty acid; T-KB, total ketone bodies; T-CHO, total cholesterol; TG, triglyceride; TP, total protein; ALB, albumin; BUN, blood urea nitrogen; Ca, calcium; iP, phosphate; AST, aspartate aminotransferase; GGT, gamma-glutamyl transferase; IGF_1, insulin-like growth factor-1; GCG, glucagon; GH, growth hormone.
Although during the periparturient period we detected no significant changes (P > 0.1) in the relative abundances of different bacterial phyla (Table 4), at the genus level, there was a significant change in the relative abundance of an unclassified Prevotellaceae (P < 0.05), and tendencies toward significant change in the relative abundances of unclassified Bacteroidetes (P = 0.078) and Ruminococcus (P = 0.055; Table 4). Moreover, compared with 3 weeks prepartum, we observed significantly higher relative abundance of unclassified Prevotellaceae at 6 weeks postpartum (P < 0.05). At the OTU level, the total richness of all groups was determined to be 981, and a total of 37 core bacterial OTUs were identified in all samples (Supplementary Table 4). These 37 core bacterial OTUs and the positive or negative correlations among rumen measurements are shown in Supplementary Figure 1.
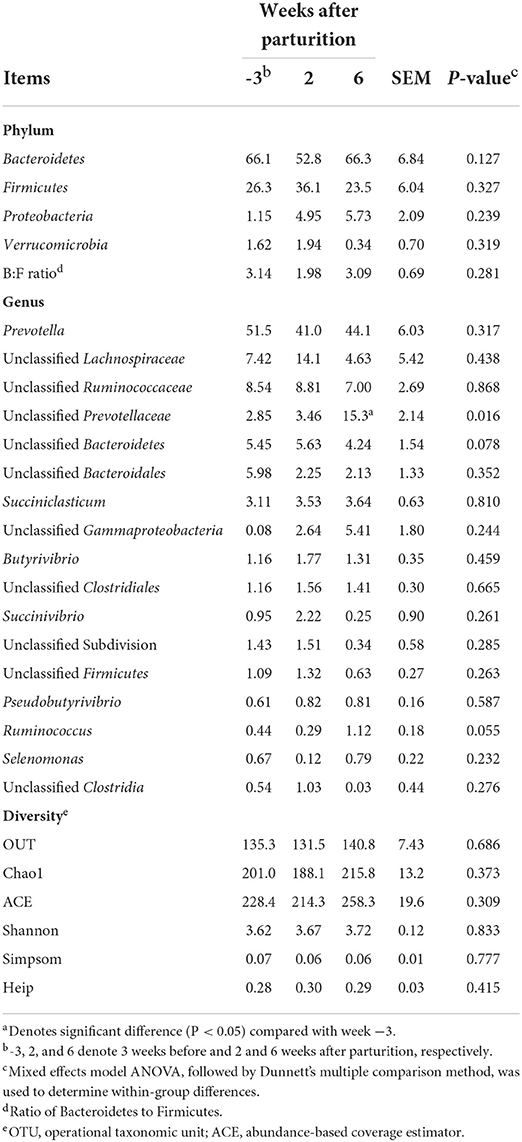
Table 4. Relative abundances of major bacterial phyla and genera (>0.05% of total sequences) and bacterial diversity indices in Holstein cows (n = 5) during the periparturient period.
Using the PICRUSt2 analysis pipeline, we identified a total of 457 MetaCyc, 2,808 ECs, and 9,955 KEGG orthologs counts. Among the analysis outputs, we established that a total of 53 MetaCyc pathways (45 upregulated; Table 5), 200 ECs (184 upregulated; Supplementary Table 5), and 714 KEGG orthologs (667 upregulated; Supplementary Table 6) at 6 weeks postpartum were significantly different (P < 0.05) from those at 3 weeks prepartum. However, compared with 3 weeks prepartum, we detected no significant changes among the PICRUSt2 outputs (P > 0.1) at 2 weeks postpartum. NMDS based on OTUs (Figure 3A) revealed that MetaCyc (Figure 3B), EC (Figure 3C), and KEGG ortholog (Figure 3D) counts at 3 weeks prepartum and 6 weeks postpartum were partitioned into two separated clusters, whereas plots constructed for 2 weeks postpartum showed a pattern of dispersed distribution. Stress values of 0.09, 0.11, 0.10, and 0.09 were obtained for out-, MetaCyc-, EC-, and KEGG ortholog-based ordinations, respectively.
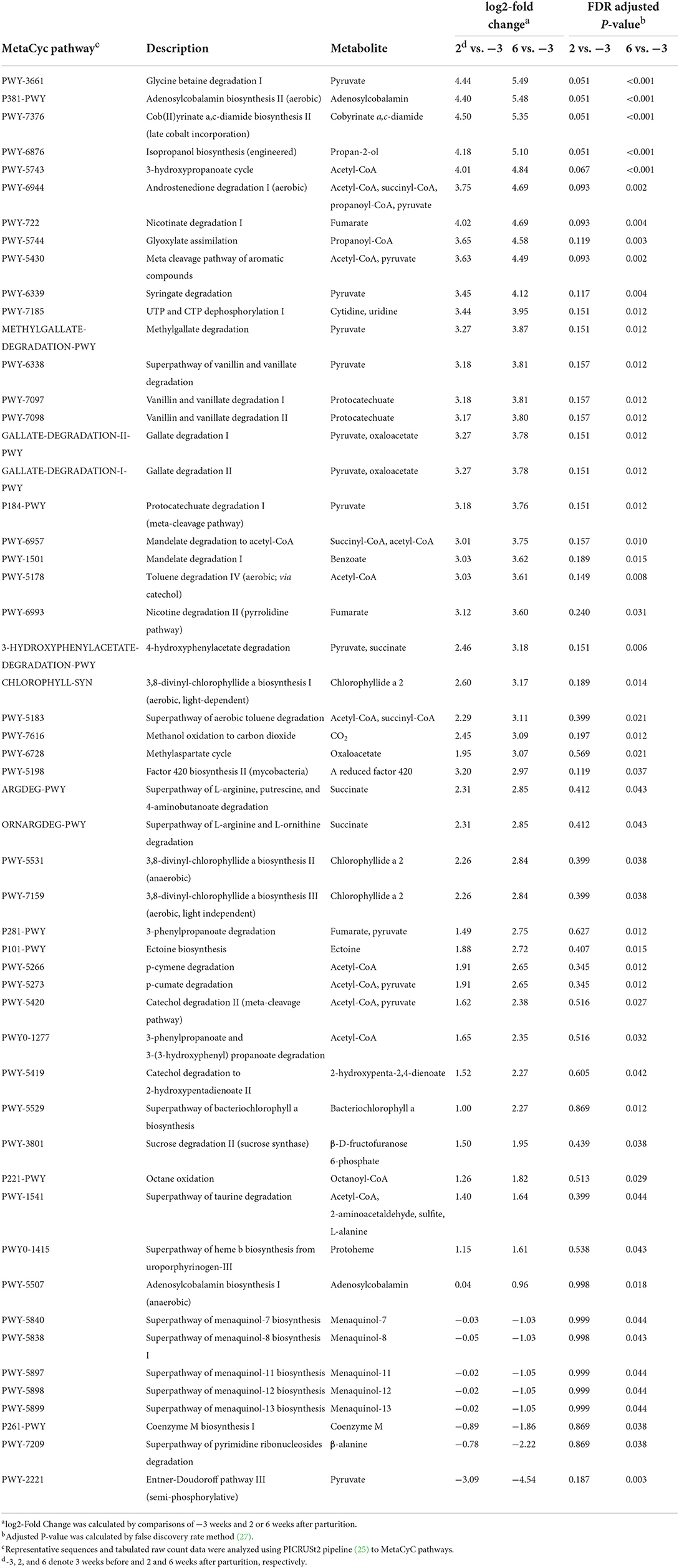
Table 5. Predicted functional pathway analysis using the PICRUSt2 pipeline to identify MetaCyc pathways in Holstein cows (n = 5) during the periparturient period.
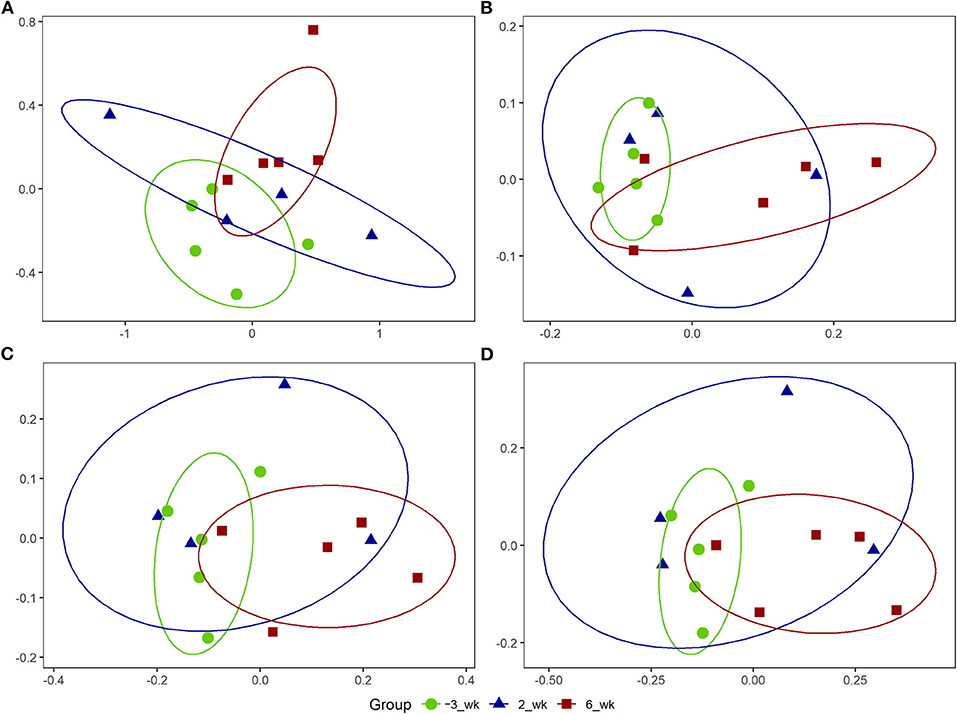
Figure 3. Non-metric multidimensional scaling (NMDS) plots for Holstein cows (n = 5) during the periparturient period. The NMDS plots were generated for (A) bacterial operational taxonomic units (OTUs), (B) MetaCyc pathways, (C) enzyme commissions (EC), and (D) Kyoto Encyclopedia of Genes and Genomes (KEGG) ortholog counts. The circle shows the correlation among the variables in each period. The stress values for OTU, MetaCyc, EC, and KEGG orthologs-based ordinations were 0.09, 0.11, 0.10, and 0.09, respectively.
Among the MetaCyc pathways (P < 0.05), 14 and 11 pathways associated with the production of pyruvate and acetyl-CoA as end products, respectively, were identified as being involved in pyruvate metabolism and various metabolic process (acetyl-CoA), respectively. PWY-3661 (glycine betaine degradation I; FC = 5.49, FDR corrected P < 0.001) was the most upregulated pyruvate metabolism-involved pathway, and P381-PWY [adenosylcobalamin biosynthesis II (aerobic); FC = 5.48, FDR corrected P < 0.001] was the second most upregulated adenosylcobalamin-involved pathway, and PWY-7376 [cob(II)yrinate a,c-diamide biosynthesis II (late cobalt incorporation); FC = 5.35, FDR corrected P < 0.001] was the third most upregulated cobalamin-involved pathway. Among the multiple MetaCyc pathways identified for different end products, we identified three metabolic pathways for each of succinate, succinyl-CoA, propanoyl-CoA, oxaloacetate, fumarate, and chlorophyllide a 2, and three pathways for each of protocatechuate and adenosylcobalamin that were significantly upregulated (P < 0.05). In contrast, PWY-2221 [Entner-Doudoroff pathway III (semi-phosphorylative); FC = −4.54, FDR corrected P = 0.003] involved in pyruvate metabolism was identified as the most downregulated pyruvate metabolism-involved pathway. In addition, the superpathways of menaquinol-7 (FC = −1.03, FDR corrected P = 0.044), −8 (FC = −1.03, FDR corrected P = 0.043), −11 (FC = −1.05, FDR corrected P = 0.044), −12 (FC = −1.05, FDR corrected P = 0.044), and −13 (FC = −1.05, FDR corrected P = 0.044) biosynthesis, P261-PWY (coenzyme M biosynthesis I; FC = −1.86, FDR corrected P = 0.038), and PWY-7209 (superpathway of pyrimidine ribonucleosides degradation, FC = −2.22, FDR corrected P = 0.038) were significantly downregulated.
During the transition period, the diets of dairy cows are frequently modified to meet the energy requirement for milk production (1). However, abrupt transition from a low-energy dry diet to a high-energy lactating diet may render cows susceptible to rumen acidosis or SARA (16). In the present study, we detected notable reductions in 7-d and 1-h mean reticulo-ruminal pH values after parturition, concomitant with the transition to a lactation period diet, leading to a prolongation of the periods during which pH values were <5.6 and 5.8 postpartum. These observations are sufficient to diagnose SARA at 1–3 weeks postpartum, given that this condition is characterized by an extended period during which ruminal pH is below 5.6 (3) or below 5.8 (4). Furthermore, the higher total VFA and LPS concentrations, proportions of propionic and butyric acids, and a lower proportion of acetic acid during the postpartum period is consistent with the responses previously observed using high-grain diet challenge models (7, 28, 29).
A postpartum depression in reticulo-ruminal pH is considered to have a less severe impact on the blood metabolites, as no distinct differences have been detected in cows with and without SARA with respect to energy metabolism indicators (GLU, NEFA, T-KB, T-CHO, and TG) (16). In the present study, similar patterns were identified among peripheral blood indicators, such as significantly higher (NEFA, T-KB, and T-CHO) and lower (GLU and TG) values after parturition. The low levels of GLU detected after parturition tend to indicate that rumen fermentation is unable to satisfy the energy requirements of lactation, whereas elevated levels of NEFA and T-KB imply a negative energy balance due to fat mobilization for gluconeogenesis in the liver (30).
Although the occurrence of postpartum SARA is likely to attenuate the hepatic transcriptomic responsiveness to postpartum metabolic load and hormones (16), we observed no distinguishable changes from the general pattern of peripheral blood plasma hormones. Plasma GH levels, associated with the stimulation of carbohydrate and lipid metabolism and gluconeogenesis for milk production (31), were found to be elevated soon after parturition, thereby suppressing hepatic IGF-1 production via a negative feedback loop (32). In addition, we found that levels of INS and GCG, considered to be the primary regulators of gluconeogenesis in ruminants (33), were significantly increased and reduced after parturition, respectively. In periparturient cows, these hormones are assumed play important roles in the metabolism of a range of nutrients, and the observed changes are considered to reflect a natural adaptation designed to improve postpartum negative energy balance for milk production, leading to enhanced liver transcriptomic responsiveness to the periparturient hormonal changes (16).
With respect to ruminal bacterial community composition, unclassified Prevotellaceae has been identified as the only taxon that responds or adapts to dietary and reticulo-ruminal pH alterations, and in sheep, its proportional occurrence in the ruminal microbiota has been found to be higher in high-grain diet-fed animals than in those fed a low-grain diet (34). However, in the present study, we detected no significant changes in bacterial richness and diversity indices during the periparturient period. Among the core bacterial OTUs, we observed alterations in relatively few bacterial species during the periparturient period. Those that did show changes include OTU2 (Prevotella ruminicola), OTU37 (Prevotella brevis), OTU39 (Ruminococcus albus), OTU52 (Prevotella melaninogenica), and OTU57 (Eubacterium ruminantium), and we can thus speculate that these bacteria might play important roles in the response or adaptation to dietary and reticulo-ruminal pH changes during the periparturient period. Generally, however, it appears that the structural composition of the rumen bacterial community in Holstein cows undergoes only minor changes during the periparturient period, which is consistent with the conclusions drawn in previous studies on bacterial variability (10, 15).
In the present study, we used 16S rRNA gene sequences of representative OTUs to predict functional features using PICRUSt2. We accordingly identified a total of 53 MetaCyc pathways, 200 ECs, and 714 KEGG orthologs that were significantly upregulated at 6 weeks postpartum compared with 3 weeks prepartum, whereas in contrast, no significant changes in PICRUSt2 outputs were detected at 2 weeks postpartum, thereby tending to indicate delayed responses among members of the bacterial community. Furthermore, our NMDS analysis of OTUs revealed similar patterns among MetaCyc, EC, and KEGG orthologs, with clustering into two separate groups at 3 weeks prepartum and 6 weeks postpartum and a dispersed distribution at 2 weeks postpartum.
MetaCyc is a universal database describing scientifically verified metabolic pathways and enzymes from all domains of life (35), and recent studies have frequently used this database to identify functional changes in ruminal bacteria metabolism (36, 37). We therefore assessed the predicted functional pathways of the rumen bacterial community by focusing on MetaCyc pathways, given that changes in PICRUSt2 outputs showed similar patterns during the periparturient period. We accordingly established that 26.4 and 20.8% of the identified MetaCyc pathways produce pyruvate and acetyl-CoA as an end product, respectively. Pyruvate is a central intermediate metabolite that can enter numerous pathways supplying energy to bovine ruminal cells (2), and rumen microbial species generate energy via the oxidation of pyruvate to VFAs (38, 39). Within the rumen, hexose (glucose, fructose, and galactose) glycolysis and the pentose (xylose) phosphate pathway are the metabolic pathways used to convert plant carbohydrates to pyruvate (38). Acetyl-CoA, the product of 11 of the identified MetaCyc pathways, is an intermediate metabolite in a range of ATP-generating metabolic pathways (39).
Glycine betaine, a component of the most highly expressed functional pathway (PWY-3661) identified in the present study, is a naturally occurring compound produced as a metabolic by-product of choline oxidation (40). Supplementation of the diets of primiparous lactating dairy goats with glycine betaine has been shown to induce the production of higher levels of short-chain fatty acids in milk and also higher milk production and levels of milk fat during late lactation (41). The second most highly expressed functional pathway (P381-PWY) produces adenosylcobalamin (a biologically active form of Vitamin B12) as an end product, which is synthesized exclusively by mainly anaerobic microorganisms (42, 43). Gut-synthesized adenosylcobalamin is absorbed through the gastrointestinal tract, and transported via the blood to body tissues and fluids including the liver, muscles, and milk (44). Intramuscular injections of vitamin B12 have been shown to increase the yields energy-corrected milk, and levels of milk solids, fat, and lactose in early lactation primiparous dairy cows (45). The third most highly expressed functional pathway identified in the present study (PWY-7376) produces cobyrinate a,c-diamide. This pathway, referred to as the “late cobalt insertion” pathway (46), is associated also associated with vitamin B12. Cobalt is a key ionic component the vitamin B12, in the structure of which, it is bound to the center of tetrapyrrole-derived macrocycle (46), and consequently, a source of cobalt is essential for the formation of vitamin B12 by rumen microorganisms (47). The PWY-7376 pathway would thus appear to represent an important step involved in the incorporation of cobalt ions in the structure of vitamin B12, and is accordingly connected with the sub-pathway P381-PWY in the rumen bacterial community. Collectively, the findings of our predictive analyses would thus tend to indicate that the high expression of these three functional pathways, along with that of the multiple pathways identified for certain other products, might represent a delayed adaptive response of the bacterial community to an abrupt transition in the diet of Holstein dairy cows after parturition.
In addition to the upregulated responses of the aforementioned pathways, we also identified a number of bacterial functional pathways characterized by downregulated responses to dietary transition and the associated low reticulo-ruminal pH. Among these, the most notable is the Entner–Doudoroff pathway, which in gram-negative bacteria, certain gram-positive bacteria, and archaea plays a prominent role in the catabolism of glucose to pyruvate (48). In this regard, previous studies have demonstrated high activity of 2-keto-3-deoxy-6-phosphogluconate aldolase (the final enzyme of the Entner–Douforoff pathway) in rumen pectinolytic bacteria, including Butyrivibrio fibrisolvens 787 and Prevotella ruminicola AR29, and is present among bacteria grown on a pectin substrate, although not in those grown on a glucose substrate (49). Although we failed in the present study to detect any significant changes among bacteria in the genera Prevotella and Butyrivibrio, we did, nevertheless, establish that the transition from a high-forage to low-forage (high-grain) diet might induce the suppression of pectinolytic activity (Entner–Douforoff pathway) among the predicted responses of the rumen bacterial community. With respect to the PWY-7209 pathway, degradation of purines and pyrimidines, releasing small amount of NH3, is a part of the urea recycling system in lactating dairy cows (50). Although the P261-PWY was postulated based on methane-producing bacteria, downregulation of this pathway is consistent with reduced methanogenesis associated with the consumption of a high-grain diet compared with the high-forage diet feeding in beef steers (51). Furthermore, long-chain menaquinone species (5 to 13), referred to as Vitamin K2 are synthesized by bacteria (52), and we found that the bacterial function of menaquinol-7, 8, 11, 12, and 13 biosynthesis was reduced in response to transition to a high-grain diet after parturition.
On the basis of our observations, it would appear that parturition and postpartum dietary changes do not induce any marked alterations in rumen bacterial community of Holstein cows during the periparturient period. However, dietary transition may promote changes in the functional pathways of certain bacteria to satisfy energy requirements and/or facilitate adaptation to a change in diet and the associated lower reticulo-rumen pH in these cows.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
The animal study was reviewed and approved by Iwate University Laboratory Animal Care and Use Committee.
TS and SS: conceptualization. AK, TS, and SS: investigation. Y-HK: data curation and writing—original draft preparation. Y-HK, AK, and SS: writing—review and editing. All authors have read and agreed to the published version of the manuscript.
This research was financially supported by a grant from the Ito Foundation, Japan.
The authors thank Kawashima, Chiba Prefectural Livestock Research Center, for VFA analyses and Kushibiki, Institute of Livestock and Grassland Science, for hormone measurement.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2022.948545/full#supplementary-material
Supplementary Figure 1. Correlation analyses between relative operational taxonomic unit (OTU) abundance and rumen measurements. Cells are colored based on Pearson correlation analyses, where blue and red represent negative and positive correlations, respectively. * denotes a significant correlation (P < 0.05) between rumen measurements and OTUs. 7-d mean pH, 7-d mean reticulo-ruminal pH; pH <5.6, period during which reticulo-ruminal pH was <5.6; pH <5.8, period during which reticulo-ruminal pH was <5.8; EU, endotoxin units.
Supplementary Table 1. Changes in body weight, body condition score, dry matter intake, milk yield, and milk fat composition in Holstein cows (n = 8) during the periparturient period.
Supplementary Table 2. The profile of sample collection in experimental cows (n = 8) during the periparturient period.
Supplementary Table 3. The 7-d mean reticulo-ruminal pH and duration of times during which pH was <5.6 and 5.8 in Holstein cows (n = 8) during the periparturient period.
Supplementary Table 4. Relative abundances and taxonomic classification of operational taxonomic units (OTUs; shared by all samples) in Holstein cows (n = 5) during the periparturient period.
Supplementary Table 5. Predicted functional pathway analysis using the PICRUSt2 pipeline to identify Enzyme Commissions (EC) in Holstein cows (n = 5) during the periparturient period.
Supplementary Table 6. Predicted functional pathway analysis using PICRUSt2 pipeline to identify Kyoto Encyclopedia of Genes and Genomes (KEGG) orthologs in Holstein cows (n = 5) during the periparturient period.
1. Ingvartsen KL. Feeding-and management-related diseases in the transition cow: physiological adaptations around calving and strategies to reduce feeding-related diseases. Anim Feed Sci Technol. (2006) 126:175–213. doi: 10.1016/j.anifeedsci.2005.08.003
2. Penner GB, Taniguchi M, Guan LL, Beauchemin KA, Oba M. Effect of dietary forage to concentrate ratio on volatile fatty acid absorption and the expression of genes related to volatile fatty acid absorption and metabolism in ruminal tissue. J Dairy Sci. (2009) 92:2767–81. doi: 10.3168/jds.2008-1716
3. Gozho GN, Plaizier JC, Krause DO, Kennedy AD, Wittenberg KM. Subacute ruminal acidosis induces ruminal lipopolysaccharide endotoxin release and triggers an inflammatory response. J Dairy Sci. (2005) 88:1399–403. doi: 10.3168/jds.S0022-0302(05)72807-1
4. Penner GB, Beauchemin KA, Mutsvangwa T. Severity of ruminal acidosis in primiparous Holstein cows during the periparturient period. J Dairy Sci. (2007) 90:365–75. doi: 10.3168/jds.S0022-0302(07)72638-3
5. Plaizier JC, Krause DO, Gozho GN, McBride BW. Subacute ruminal acidosis in dairy cows: the physiological causes, incidence and consequences. Vet J. (2008) 176:21–31. doi: 10.1016/j.tvjl.2007.12.016
6. Plaizier JC, Li S, Danscher AM, Derakshani H, Andersen PH, Khafipour E. Changes in microbiota in rumen digesta and feces due to a grain-based subacute ruminal acidosis (SARA) challenge. Microb Ecol. (2017) 74:485–95. doi: 10.1007/s00248-017-0940-z
7. Petri RM, Schwaiger T, Penner GB, Beauchemin KA, Forster RJ, McKinnon JJ, et al. Changes in the rumen epimural bacterial diversity of beef cattle as affected by diet and induced ruminal acidosis. Appl Environ Microbiol. (2013) 79:3744–55. doi: 10.1128/AEM.03983-12
8. Nagata R, Kim YH, Ohkubo A, Kushibiki S, Ichijo T, Sato S. Effects of repeated subacute ruminal acidosis challenges on the adaptation of the rumen bacterial community in Holstein bulls. J Dairy Sci. (2018) 101:4424–36. doi: 10.3168/jds.2017-13859
9. Wang X, Li X, Zhao C, Hu P, Chen H, Liu Z, et al. Correlation between composition of the bacterial community and concentration of volatile fatty acids in the rumen during the transition period and ketosis in dairy cows. Appl Environ Microbiol. (2012) 78:2386–92. doi: 10.1128/AEM.07545-11
10. Pitta DW, Kumar S, Vecchiarelli B, Shirley DJ, Bittinger K, Baker LD, et al. Temporal dynamics in the ruminal microbiome of dairy cows during the transition period. J Anim Sci. (2014) 92:4014–22. doi: 10.2527/jas.2014-7621
11. Zhu Z, Kristensen L, Difford GF, Poulsen M, Noel SJ, Al-Soud WA, et al. Changes in rumen bacterial and archaeal communities over the transition period in primiparous Holstein dairy cows. J Dairy Sci. (2018) 101:9847–62. doi: 10.3168/jds.2017-14366
12. Khafipour E, Li S, Plaizier JC, Krause DO. Rumen microbiome composition determined using two nutritional models of subacute ruminal acidosis. Appl Environ Microbiol. (2009) 75:7115. doi: 10.1128/AEM.00739-09
13. Hook SE, Steele MA, Northwood KS, Dijkstra J, France J, Wright ADG, et al. Impact of subacute ruminal acidosis (SARA) adaptation and recovery on the density and diversity of bacteria in the rumen of dairy cows. FEMS Microbiol Ecol. (2011) 78:275–84. doi: 10.1111/j.1574-6941.2011.01154.x
14. Mao SY, Zhang RY, Wang DS, Zhu WY. Impact of subacute ruminal acidosis (SARA) adaptation on rumen microbiota in dairy cattle using pyrosequencing. Anaerobe. (2013) 24:12–9. doi: 10.1016/j.anaerobe.2013.08.003
15. Mohammed R, Stevenson DM, Weimer PJ, Penner GB, Beauchemin KA. Individual animal variability in ruminal bacterial communities and ruminal acidosis in primiparous Holstein cows during the periparturient period. J. Dairy Sci. (2012) 95:6716–30. doi: 10.3168/jds.2012-5772
16. Tsuchiya Y, Chiba E, Sugino T, Kawashima K, Kushibiki S, Kizaki K, et al. Liver transcriptome response to periparturient hormonal and metabolic changes depends on the postpartum occurrence of subacute ruminal acidosis in Holstein cows. Physiol Genomics. (2021) 53:285–94. doi: 10.1152/physiolgenomics.00048.2021
17. Ogata T, Makino H, Ishizuka N, Iwamoto E, Masaki T, Ikuta K, et al. Long-term high-grain diet altered the ruminal pH, fermentation, and composition and functions of the rumen bacterial community, leading to enhanced lactic acid production in Japanese Black beef cattle during fattening. PLoS ONE. (2019) 14:e0225448. doi: 10.1371/journal.pone.0225448
18. Sato S, Mizuguchi H, Ito K, Ikuta K, Kimura A, Okada K. Technical note: development and testing of a radio transmission pH measurement system for continuous monitoring of ruminal pH in cows. Prev Vet Med. (2012) 103:274–9. doi: 10.1016/j.prevetmed.2011.09.004
19. Hirabayashi H, Kawashima K, Okimura T, Tateno A, Suzuki A, Asakuma S, et al. Effect of nutrient levels during the far-off period on postpartum productivity in dairy cows. Anim Sci J. (2017) 88:1162–70. doi: 10.1111/asj.12743
20. Kushibiki S, Hodate K, Shingu H, Obara Y, Touno E, Shinoda M, et al. Metabolic and lactational responses during recombinant bovine tumor necrosis factor-alpha treatment in lactating cows. J Dairy Sci. (2003) 86:819–27. doi: 10.3168/jds.S0022-0302(03)73664-9
21. Kim YH, Nagata R, Ohtani N, Ichijo T, Ikuta K, Sato S. Effects of dietary forage and calf starter diet on ruminal pH and bacteria in Holstein calves during weaning transition. Front Microbiol. (2016) 7:1575. doi: 10.3389/fmicb.2016.01575
22. Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform- independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. (2009) 75:7537–41. doi: 10.1128/AEM.01541-09
23. Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol. (2013) 79:5112–20. doi: 10.1128/AEM.01043-13
24. Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. (2007) 35:7188–96. doi: 10.1093/nar/gkm864
25. Douglas GM, Maffei VJ, Zaneveld JR, Yurgel SN, Brown JR, Taylor CM, et al. PICRUSt2 for prediction of metagenome functions. Nat Biotechnol. (2020) 38:685–8. doi: 10.1038/s41587-020-0548-6
26. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. (2014) 15:1–21. doi: 10.1186/s13059-014-0550-8
27. Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc B. (1995) 57:289–300.
28. Watanabe Y, Kim YH, Kushibiki S, Ikuta K, Ichijo T, Sato S. Effects of active dried Saccharomyces cerevisiae on ruminal fermentation and bacterial community during the short-term ruminal acidosis challenge model in Holstein calves. J Dairy Sci. (2019) 102:6518–31. doi: 10.3168/jds.2018-15871
29. Mizuguchi H, Ikeda T, Watanabe Y, Kushibiki S, Ikuta K, Kim YH, et al. Anti-lipopolysaccharide antibody administration mitigates ruminal lipopolysaccharide release and depression of ruminal pH during subacute ruminal acidosis challenge in Holstein bull cattle. J Vet Med Sci. (2021) 83:905–10. doi: 10.1292/jvms.21-0037
30. Kato J, Odate T, Kim YH, Ichijo T, Sato S. Effects of feeding management on disease incidence and blood metabolites in dairy herds in Iwate Prefecture, Japan. J Vet Med Sci. (2019) 81:958–67. doi: 10.1292/jvms.18-0742
31. Etherton TD, Bauman DE. Biology of somatotropin in growth and lactation of domestic animals. Physiol Rev. (1998) 78:745–61. doi: 10.1152/physrev.1998.78.3.745
32. Lucy MC, Jiang H, Kobayashi Y. Changes in the somatotrophic axis associated with the initiation of lactation. J Dairy Sci. (2001) 84:E113–9. doi: 10.3168/jds.S0022-0302(01)70205-6
33. Greenfield RB, Cecava MJ, Donkin SS. Changes in mRNA expression for gluconeogenic enzymes in liver of dairy cattle during the transition to lactation. J. Dairy Sci. (2000) 83:1228–36. doi: 10.3168/jds.S0022-0302(00)74989-7
34. Trabi EB, Seddik HE, Xie F, Wang X, Liu J, Mao S. Effect of pelleted high-grain total mixed ration on rumen morphology, epithelium-associated microbiota and gene expression of proinflammatory cytokines and tight junction proteins in Hu sheep. Anim Feed Sci Technol. (2020) 263:114453. doi: 10.1016/j.anifeedsci.2020.114453
35. Caspi R, Altman T, Billington R, Dreher K, Foerster H, Fulcher CA, et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome Databases. Nucleic Acids Res. (2014) 42:459–71. doi: 10.1093/nar/gkt1103
36. Bu D, Zhang X, Ma L, Park T, Wang L, Wang M, et al. Repeated inoculation of young calves with rumen microbiota does not significantly modulate the rumen prokaryotic microbiota consistently but decreases diarrhea. Front Microbiol. (2020) 2020:1403. doi: 10.3389/fmicb.2020.01403
37. Taxis TM, Wolff S, Gregg SJ, Minton NO, Zhang C, Dai J, et al. The players may change but the game remains: network analyses of ruminal microbiomes suggest taxonomic differences mask functional similarity. Nucleic Acids Res. (2015) 43:9600–12. doi: 10.1093/nar/gkv973
39. Van Houtert MFJ. The production and metabolism of volatile fatty acids by ruminants fed roughages: a review. Anim Feed Sci Technol. (1993) 43:189–225. doi: 10.1016/0377-8401(93)90078-X
40. Barak AJ, Tuma DJ. Betaine, metabolic by-product or vital methylating agent? Life Sci. (1983) 32:771–4. doi: 10.1016/0024-3205(83)90311-9
41. Fernández C, Sánchez-Seiquer P, Sánchez A, Contreras A, De La Fuente JM. Influence of betaine on milk yield and composition in primiparous lactating dairy goats. Small Rumin Res. (2004) 52:37–43. doi: 10.1016/S0921-4488(03)00202-5
42. Watanabe F. Vitamin B12 sources and bioavailability. Exp Biol Med. (2007) 232:1266–74. doi: 10.3181/0703-MR-67
43. Gille D, Schmid A. Vitamin B12 in meat and dairy products. Nutr Rev. (2015) 73:106–15. doi: 10.1093/nutrit/nuu011
44. Ortigues-Marty I, Micol D, Prache S, Dozias D, Girard CL. Nutritional value of meat: the influence of nutrition and physical activity on vitamin B12 concentrations in ruminant tissues. Reprod Nutr Dev. (2005) 45:453–67. doi: 10.1051/rnd:2005038
45. Girard CL, Matte JJ. Effects of intramuscular injections of vitamin B12 on lactation performance of dairy cows fed dietary supplements of folic acid and rumen-protected methionine. J Dairy Sci. (2005) 88:671–6. doi: 10.3168/jds.S0022-0302(05)72731-4
46. Warren MJ, Raux E, Schubert HL, Escalante-Semerena JC. The biosynthesis of adenosylcobalamin (vitamin B12). Nat Prod Rep. (2002) 19:390–412. doi: 10.1039/b108967f
47. González-Montaña JR, Escalera-Valente F, Alonso AJ, Lomillos JM, Robles R, Alonso ME. Relationship between vitamin B12 and cobalt metabolism in domestic ruminant: an update. Animals. (2020) 10:1855. doi: 10.3390/ani10101855
48. Conway T. The Entner-Doudoroff pathway: history, physiology and molecular biology. FEMS Microbiol Rev. (1992) 9:1–27. doi: 10.1111/j.1574-6968.1992.tb05822.x
49. Marounek M, Dušková D. Metabolism of pectin in rumen bacteria Butyrivibrio fibrisolvens and Prevotella ruminicola. Lett Appl Microbiol. (1999) 29:429–33. doi: 10.1046/j.1472-765X.1999.00671.x
50. Stentoft C, Røjen BA, Jensen SK, Kristensen NB, Vestergaard M, Larsen M. Absorption and intermediary metabolism of purines and pyrimidines in lactating dairy cows. Br J Nutr. (2015) 113:560–73. doi: 10.1017/S0007114514004000
51. Kim SH, Lee C, Pechtl HA, Hettick JM, Campler MR, Pairis-Garcia MD, et al. Effects of 3-nitrooxypropanol on enteric methane production, rumen fermentation, and feeding behavior in beef cattle fed a high-forage or high-grain diet. J Anim Sci. (2019) 97:2687–99. doi: 10.1093/jas/skz140
Keywords: rumen bacterial community, predicted bacterial function, periparturient period, ruminal pH, Holstein cows
Citation: Kim Y-H, Kimura A, Sugino T and Sato S (2022) Parturition and postpartum dietary change altered ruminal pH and the predicted functions of rumen bacterial communities but did not alter the bacterial composition in Holstein cows. Front. Vet. Sci. 9:948545. doi: 10.3389/fvets.2022.948545
Received: 20 May 2022; Accepted: 02 August 2022;
Published: 26 August 2022.
Edited by:
Mohanned Naif Alhussien, Technical University of Munich, GermanyReviewed by:
Adham Al-Sagheer, Zagazig University, EgyptCopyright © 2022 Kim, Kimura, Sugino and Sato. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shigeru Sato, c3NoaWdlcnVAaXdhdGUtdS5hYy5qcA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.