
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci. , 08 August 2022
Sec. Veterinary Infectious Diseases
Volume 9 - 2022 | https://doi.org/10.3389/fvets.2022.938822
This article is part of the Research Topic Antimicrobials Alternatives for the Prevention and Treatment of Veterinary Infectious Diseases View all 16 articles
 Weijie Zhang1,2
Weijie Zhang1,2 Liangchuan Chen1,2
Liangchuan Chen1,2 Haiyun Feng1,2
Haiyun Feng1,2 Junlin Wang1,2
Junlin Wang1,2 Fuyuan Zeng1,2
Fuyuan Zeng1,2 Xing Xiao1,2
Xing Xiao1,2 Jichang Jian1,2
Jichang Jian1,2 Na Wang3
Na Wang3 Huanying Pang1,2*
Huanying Pang1,2*Vibrio alginolyticus, a Gram-negative bacterium, is an opportunistic pathogen of both marine animals and humans, resulting in significant losses in the aquaculture industry. Type III secretion system (T3SS) is a crucial virulence mechanism of V. alginolyticus. In this study, the T3SS regulatory gene exsA, which was cloned from V. alginolyticus wild-type strain HY9901, is 861 bp encoding a protein of 286 amino acids. The ΔexsA was constructed by homologous recombination and Overlap-PCR. Although there was no difference in growth between HY9901 and ΔexsA, the ΔexsA exhibited significantly decreased extracellular protease activity and biofilm formation. Besides, the ΔexsA showed a weakened swarming phenotype and an ~100-fold decrease in virulence to zebrafish. Antibiotic susceptibility testing showed the HY9901ΔexsA was more sensitive to kanamycin, minocycline, tetracycline, gentamicin, doxycycline and neomycin. Compared to HY9901 there were 541 up-regulated genes and 663 down-regulated genes in ΔexsA, screened by transcriptome sequencing. qRT-PCR and β-galactosidase reporter assays were used to analyze the transcription levels of hop gene revealing that exsA gene could facilitate the expression of hop gene. Finally, Danio rerio, vaccinated with ΔexsA through intramuscular injection, induced a relative percent survival (RPS) value of 66.7% after challenging with HY9901 wild type strain. qRT-PCR assays showed that vaccination with ΔexsA increased the expression of immune-related genes, including GATA-1, IL6, IgM, and TNF-α in zebrafish. In summary, these results demonstrate the importance of exsA in V. alginolyticus and provide a basis for further investigations into the virulence and infection mechanism.
Vibrio alginolyticus, which is a gram-negative halophilic bacterium, is diffusely distributed in nature occurring in both fresh and saltwater (1, 2). Obviously, Vibriosis disease outbreaks cause significant setbacks to aquaculture. V. alginolyticus was considered a serious pathogenic bacterium for many marine vertebrates, including cultured fish species such as turbot, Larimichthys crocea, grouper, as well as invertebrates, such as shrimp and shellfish (3, 4). It is also a zoonotic bacterium that can cause food poisoning in humans, including diarrheal and septicemia (5). Antibiotic treatment is the traditional measure used to treat V. alginolyticus infections, and this is considered a serious concern for the escalation of antibiotic resistant Vibrios (6). Hence, it is imperative to achieve a better understanding of the pathogenic mechanisms of V. alginolyticus to facilitate the discovery of more effective therapies.
Type III secretion system (T3SS) plays an important role in pathogenic process, and its structural components are highly conserved among bacterial species (7–9). ExsA, as is a member of the AraC/XylS family of DNA-binding proteins, is the main transcriptional activator of T3SS gene expression (10). And A previous study showed that the expression of ~40 gene products constituting the T3SS was regulated by 10 ExsA-dependent promoters (11). Qu et al. (12) showed that deletion of exsA suppressed the production of T3SS pilus (PopB/D) and effector protein (ExoT/U) in Pseudomonas aeruginosa. Moreover, T3SS1 expression in V. parahaemolyticus is genetically regulated by the ExsACDE cascade in which the master transcription factor ExsA positively regulates T3SS1 expression (13, 14). Zhou speculated that ExsA could act directly on the T3SS1 promoter sequence in view of the result that ExsA can bind the upstream region of VP1668 and VP1687 (15). To summarize, the ExsA protein exhibits significant functions during many Gram-negative bacteria pathogenesis.
When V. alginolyticus infects the host cell, it secretes effector proteins through T3SS, thus subverting host cellular functions and resulting in cell death (16). HopPmaJ, as a type III system effector (T3SE), is considered an important virulence factor in V. alginolyticus. Pang et al. (17) found HopPmaJ encoded by hop gene in V. alginolyticus is pathogenic to the orange-spotted grouper (Epinephelus coioides), which exhibits symptoms of ulceration, hemorrhage of liver and kidney, and swelling. Furthermore, a later study revealed that regulatory protein TyeA could up-regulated the expression of HopPmaj (18). While the T3SS is highly conserved, the regulators are unique proteins with very specialized functions critical to virulence. It was reported that T3SS of V. alginolyticus is similar to T3SS1 of Vibrio parahaemolyticus in genome analysis (19), but the regulator ExsA in V. alginolyticus T3SS remains poorly understood.
To better understand the function of ExsA in the V. alginolyticus T3SS, we constructed a HY9901ΔexsA mutant strain, and then studied the physiology and pathogenicity of HY9901ΔexsA. Besides, the differentially expressed genes between HY9901 and HY9901ΔexsA were investigated by transcriptome sequencing. Furthermore, we have evaluated the protective efficacy of ΔexsA, and found that the ΔexsA mutant could be used as a live attenuated vaccine to combat V. alginolyticus in zebrafish.
In this study, V. alginolyticus HY9901 was isolated from diseased red snapper (Lutjanus sanguineus) (20). E. coli S17-1 (λ Pir) and suicide plasmid pDM4 were kept in our laboratory (Table 1).
Healthy zebrafish (Danio rerio) were purchased from a commercial fish farm in Guangzhou, China, 3–4 cm long and 0.3 g in weight. The zebrafish were tested by the bacterial recovery assay, kept in freshwater in the circulatory system at 28°C for 2 weeks before the experiment.
TIANamp Bacteria DNAKit (Beijing, Tiangen Biotech Co., Ltd.); Easy PureTMQuick GelExtraction Kit and Easy PureTMPlasmid MiniPrepKit (Beijing, TransGen Biotech Co., Ltd.); pDM4 vector, ExTaqDNA polymerase, Prime START MHS DNA polymerase, BglII, SalI, and T4DNA ligase were all purchased from TaKaRa (Japan). The primers were synthesized by Guangzhou Sangon Biotech Co., Ltd.
A pair of primers exaA-F/exsA-R were designed based on the V. alginolyticus gene sequence (GenBank Number: GU074526.1) (Table 2). PCR was performed according to the previous study of Zhou et al. (18). PCR products were run on 1.5% agarose gels, visualized with ethidium bromide. Then, the recovered PCR products were cloned into the pMD18-T vector, and transformed into E. coli DH5α (Table 1). The inserted fragment was sequenced by Sangon Biological Engineering Technology & Services Co., Ltd. (Guangzhou, China). The similarity analyses were based on the method of Pang et al. (22).
The in-frame deletion of exsA in the V. alginolyticus was generated according to the previous study (23). For the construction of ΔexsA, two pairs of primers were firstly designed to obtain the exsA upstream homologous arm fragment A (Primers: exsA-A1-F and exsA-A1-R) and downstream homologous arm fragment B (Primers: exsA-A2-F and exsA-A2-R). Both fragments contained a 10 bp overlapping sequence and were used as templates for the subsequent PCR procedure, which used primers exsA-A1-F and exsA-A1-R. The PCR product was ligated into suicide vector pMD4 to generate pMD4-ΔexsA. Then, the recombinant suicide plasmid was used to transform into E. coli S17-1. The single crossover mutants were acquired by transferring the resulting plasmid directly into V. alginolyticus HY9901. Ten percentage sucrose TSA plates were used to select the deletion mutants. HY9901ΔexsA was subsequently confirmed by PCR and sequencing using primers exsA-F and exsA-R.
The ΔexsA phenotype characterized by growth, genetic stability, extracellular protease (ECPase) activity, swarming motility and biofilm Formation. The experimental method of genetic stability was based on previous studies our research group undertook (18). In brief, ΔexsA were seeded into a Tryptic Soya agar (TSA) plate, passed randomly for 30 generations. Its genetic stability was determined by PCR. Growth curves were determined using the method of Wu et al. (24). Extracellular protease activity assay was performed referring to Windle and Kelleher (25). HY9901 and ΔexsA were inoculated onto TSA plates coated with sterile cellophane, and then cultured at 28°C for 24 h, washed with PBS, centrifuged at low temperatrue for 30 min, and filtered the supernatant using a 0.22 μm porous membrane. The protease activity of the supernatant was measured at OD442 using an azocasein trichloroacetic acid colorimetric assay solution.
Biofilm formation was measured with the method of Zhou et al. (18). Swarming motility was assayed according to the previous method described by Mathew et al. (26). LD50 of HY9901 and ΔexsA were assessed by the previous study of Chen et al. (27). In brief, a total of 330 fish were blindly separated into three groups. The zebrafish in experimental group were injected with 5 μL HY9901 and ΔexsA suspended in PBS containing 104-108 CFU/mL, while negative control group was injected with 5 μL of PBS. Fish were monitored for 14 days or until no morbidities occurred, and LD50 of HY9901 and ΔexsA was calculated. All experiments were carried out triplicate.
Antibiotic susceptibility testing was performed. Briefly, HY9901 and ΔexsA were seeded into TSA plates, and then 30 different antibiotics disks were respectly added into the plates. The plates were incubated for 16 h at 28°C and the diameters of the inhibition zones were measured by Vernier calipers. All experiments were carried out triplicate.
With reference to the method described previously (22), HY9901 and ΔexsA cells were cultured in DMEM media at 28°C. The bacterial cells were harvested after 12 h. The series of experiments, including mRNA extraction, RNA fragmentation, cDNA synthesis and RNA-Seq library construction, were conducted by Novogene Co., Ltd. The data generated from the Illumina platform were used for bioinformatics analysis using I-Sanger Cloud Platform (www.i-sanger.com). Besides, the GO terms of the different expression genes were analyzed with the Goatools tool (EMBL, European Molecular Biology Laboratory).
qRT-PCR was used to validate the different expression genes obtained from transcriptome analysis. The related-gene primers sequences were tabulated in Table 2. Besides, the 16 s rRNA gene was used as the internal reference. RNA was extracted, synthetic cDNA and real-time PCR was based on the method of previous study (28).
In order to facilitate T3SS secretion, HY9901 and HY9901ΔexsA were cultured in DMEM media for 16 h. The primers for hop are shown in Table 2. 16S rRNA is used as an internal reference. RNA was extracted, synthetic cDNA and real-time PCR was based on the method of previous study (28).
The β-galactosidase reporter assay was carried out as previously described (29). In brief, the recombinant plasmid PME6522 containing the promoter region of hop was introduced to HY9901 and HY9901ΔexsA, and a LacZ reporter assay was employed to measure the activity and regulation of promoters. The recombinant plasmid PME6522 and related-gene primers are, respectively, shown in Tables 1, 2. The experiments were carried out triplicate.
The concentration of ΔexsA in LD50 experiment is 105 cfu/mL, which is not lethal to zebrafish (Table 3). Vaccination was undertaken according to Zhou et al. (18). Briefly, zebrafish were randomly separated into two groups with 80 fish per group. The water temperature was adjusted to 28°C.Fish in the ΔexsA group were injected with 5 μL ΔexsA bacterial solution (105 cfu/mL), while control fish were injected with 5 μL PBS. The experiment was repeated three times.
The number of HY9901ΔexsA in livers and spleens of selected from three fish in HY9901ΔexsA group were investigated to further assess safety of live attenuated HY9901ΔexsA vaccine. Briefly, fish were weighed and euthanized to sample liver and spleen once daily for seven consecutive days. All the samples were weighed, homogenized in 1 mL of sterile PBS. The homogenates were diluted serially and spread on TCBS plates, and the plates were incubated at 28°C for 16 h. Colony numbers were counted visually and colonies measuring at least 50 μm were counted. The bacteria counts were calculated by dividing the weights of the tissues and from the mean of three samples. Besides, the identities of the HY9901ΔexsA isolates were confirmed by colonial morphology and PCR.
Briefly, liver and spleen samples were taken from three fish from each group, respectively, at 1 day before the challenge. The expression levels of immune-related genes were determined by qRT-PCR. Primers are shown in Table 2, and β-actin was used as internal reference. Detailed experimental steps refer to the previous study of Li et al. (28).
To evaluate the safety of the ΔexsA vaccines, livers and spleens of selected from three fish from each group were fixed in neutral buffered 10% formalin, embedded in paraffin and processed for routine histopathological examination. Detailed experimental steps refer to our previous study (30).
Fish were challenged 28 days post vaccination. zebrafish (n = 30) were given intramuscular injection of 5 μL 1 × 108 cfu mL−1 of V. alginolyticus HY9901. The relative percent survival (RPS) of post-challgenged fishes were measured per day in a 14-day time frame. The experiment was repeated three times.
Statistical analyses were conducted using SPSS 17.0 (SPSS Inc., USA). The statistical significance of differences between the wild-type strain and ΔexsA mutant, were determined using the Student's t-test. Group differences were determined by Duncan's test. Data was considered statistically significant when p < 0.05.
All animal experiments were conducted strictly based on the recommendations in the “Guide for the Care and Use of Laboratory Animals” set by the National Institutes of Health. The animal protocols were approved by the Animal Ethics Committee of Guangdong Ocean University (Zhanjiang, China).
The bacteria protocols were approved by the Biosecurity Committee of Guangdong Ocean University (Zhanjiang, China).
The exsA gene is 861 bp long, and encodes 286 amino acids (aa) with a predicted molecular weight of 32.549 kDa and a theoretical isoelectric point of 6.0 (exsA accession no. MN385414) (Figures 1A,B). To understand the potential function of ExsA in V. alginolyticus, an untagged in-frame deletion mutant strain HY9901ΔexsA was constructed by Overlap PCR and a dual selection strategy. HY9901 was determined by PCR by generating a fragment of 1,811 bp, while ΔexsA was determined by PCR by generating a fragment of 1,019 bp (Figure 2).
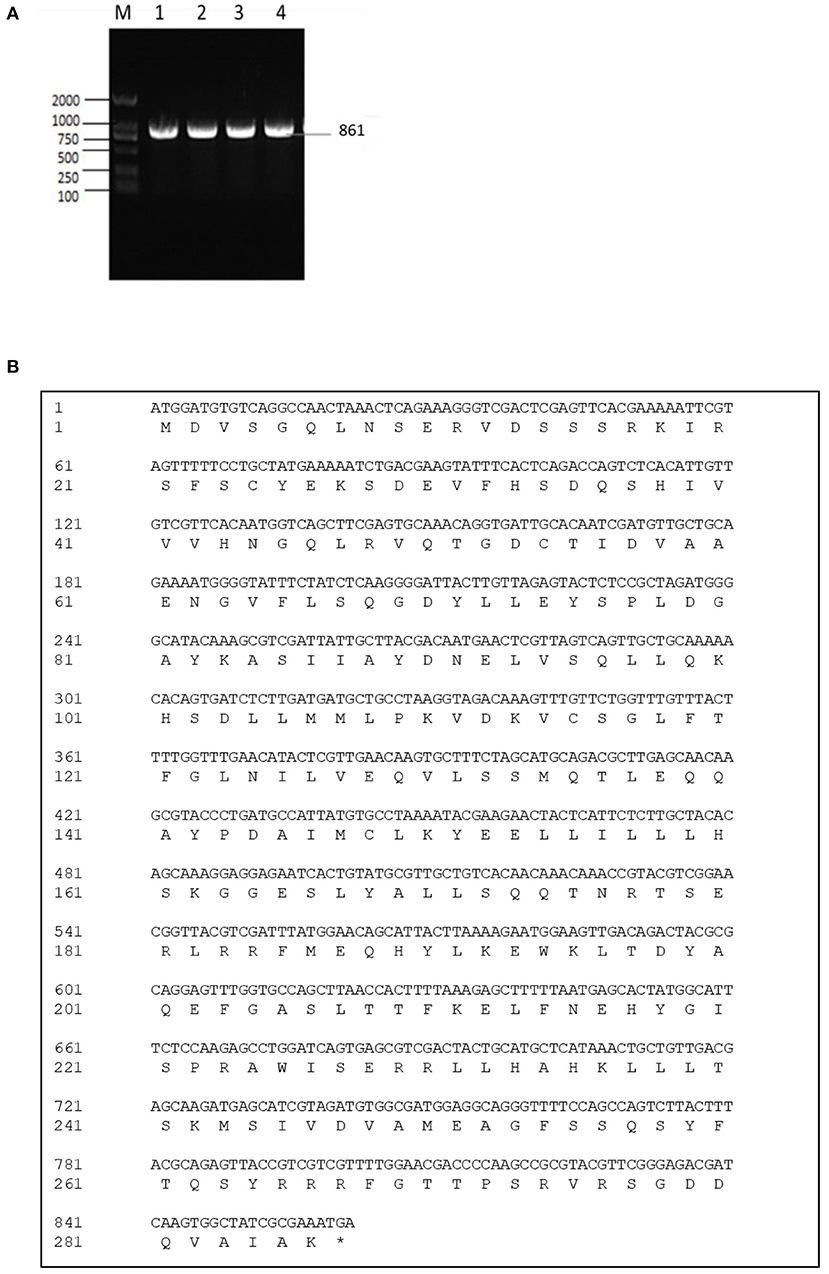
Figure 1. (A) Cloning of exsA gene. M: DL2000 marker. Lane 1-4: The 861 bp fragment amplified from genomic DNAs of HY9901 using primer pairs of ExsA-F/ExsA-R. (B) exsA gene nucleotide and its encoded amino acid sequence.
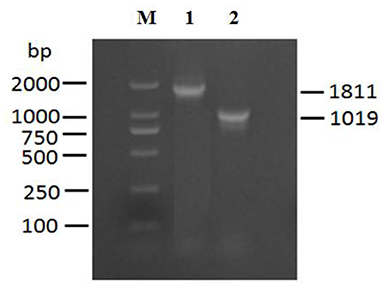
Figure 2. Construction and confirmation of the knockout mutant strain HY9901ΔexsA. M: DL2000 marker, Lane 1. A fragment of 1,811 bp is obtained for HY9901 using primer pairs of exsA-A1-F/exsA-A2-R. Lane 2. A fragment of 11,019 bp is obtained for HY9901ΔesxA using primer pairs of exsA-A1-F/exsA-A2-R.
After 30 generations of continuous blind transmission of HY9901ΔexsA, the PCR genome of HY9901ΔexsA and wild-type strain HY9901 using primers ExsA-F/ExsA-R, a fragment of 861 bp was obtained for HY9901, while Δ exsA is negative. This result indicates that ΔesxA has deleted the gene esxA, which can stabilize the inheritance (Figure 3A).
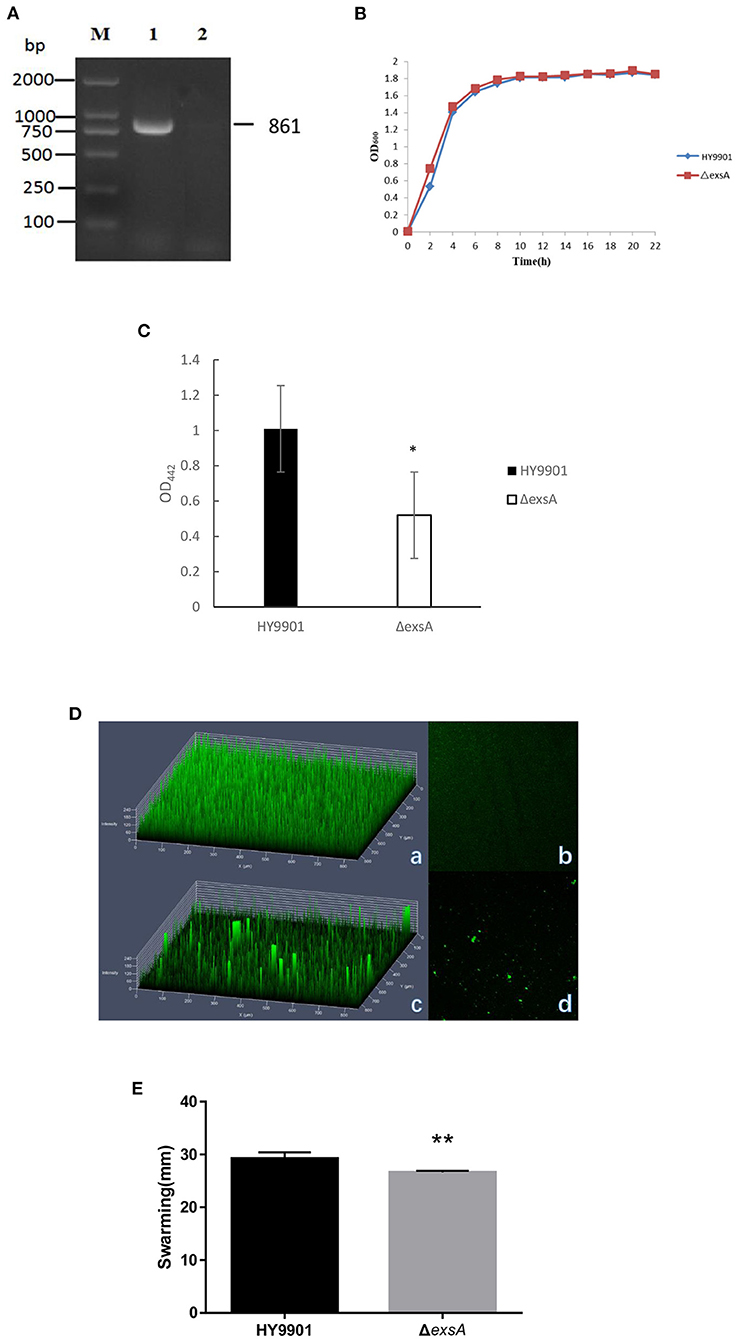
Figure 3. Characteristics of different strains. (A) Genetic stability detection of HY9901ΔexsA deletion mutantt. M: DL2000 marker; Lane 1. The 861 bp fragment amplified from genomic DNAs of HY9901 using primer pairs of ExsA-F/ExsA-R; Lane 2. The negative result amplified from genomic DNAs of ΔexsA using primer pairs of ExsA-F/ExsA-R. (B) Growth rates of HY9901ΔexsA and wild-type strain HY9901 (wt). Aliquots of cell culture were obtained at various time points and measured for cell density at OD600. (C) Activity of extracellaluar proteases. The extracellular protease activity was measured at OD442. (D) Measurement of biofilm by LSCM. (a) HY9901 2.5d diagram. (b)HY9901ΔexsA 2.5d diagram. (c) HY9901 2d diagram. (d) HY9901ΔexsA 2d diagram; HY9901 Biofilm thickness: 60 ± 10 μm, HY9901ΔexsA Biofilm thickness:90 ± 20 μm. (E) Swarming diameters were measured after 24 h incubation on TSA containing 0.3% agar plates. **p < 0.01.
There were no significant differences in growth rates between wild-type strain HY9901 and HY9901ΔexsA (p > 0.05). The exponential growth phase of the two bacterial strains was from 0 to 4 h, with growth stationary at 16 h, OD600 ≈ 1.8 (Figure 3B).
ECP is a crucial virulence factor, and have a variety of protease activities. After deletion of exsA gene, the extracellular protease activity of ΔexsA was decreased compared to the wild-type strain HY9901 (p < 0.01) (Figure 3C).
Biofilms are organized groups of microorganisms, usually formed on abiotic or natural surfaces, where organisms are embedded into a matrix composed of extracellular polymers. In comparison to the development observed in HY9901 wild type strain by confocal microscopy, ΔexsA showed a decline in biofilm formation capability (p < 0.01) (Figure 3D; Table 4).
The result of swarming assay was showed in the Table 3. As for swarming activity, the swarming circle of the wild strain HY9901was 29.2 ± 1.2 mm, and ΔexsA was 26.6 ± 0.3 mm. The result indicated that the swarming ability of HY9901ΔexsA was significantly weakened (Figure 3E; Table 4).
In this study, zebrafish were used as models to evaluate the virulence of the HY9901 and ΔexsA. The results showed that the 50% lethal dose of ΔexsA was 100 times higher than that of wild strain (Tables 3, 4). All of the dead fish exhibited the clinical symptoms of Vibriosis such as ulcers on the skin, hemorrhagic and swelling in the liver and kidney. The results showed that the virulence of HY9901ΔexsA was significantly decreased when compared with the HY9901 (p < 0.01).
The susceptibility of the wild strain HY9901 and ΔexsA to 30 antibiotics was determined by disc diffusion method. The result was as follows. The HY9901 was highly resistant to chloramphenicol, while the ΔexsA had a zone of inhibition against chloramphenicol. Besides, the ΔexsA was more sensitive to kanamycin, minocycline, tetracycline, gentamicin, doxycycline and neomycin (Table 5).
The transcriptome sequencing analysis identified a total of 541 upregulated and 663 downregulated differentially expressed genes (DEGs) in ΔexsA compared to HY9901 (Figure 4A; Supplementary Table S1). These DEGs were enriched by GO function. Go analysis showed that DEGs of HY9901 and strain ΔexsA were related to locomotion, movement of cell or subcellular component and bacterial-type flagellum-dependent cell motility (Figure 4B). We also identified the virulence genes in HY9901 and ΔexsA in the transcriptome sequencing data. Compared with HY9901, genes in biofilm formation (crp, fis, flrA, ompT), bacterial secretion (gspC, lip), two-component system (epsP, pfeR, phoA,) and lipopolysaccharide biosynthesis (lpxK,) were at lower expression level in ΔexsA.
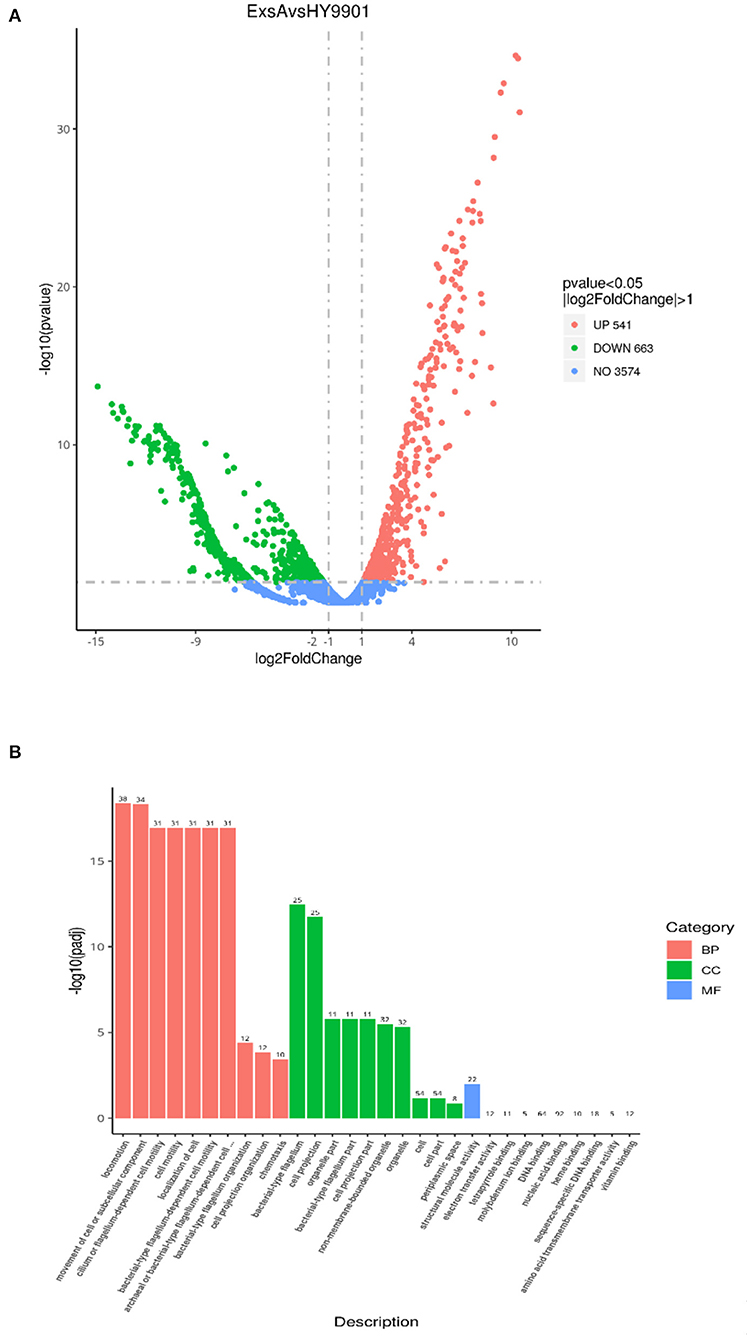
Figure 4. (A) Differential analysis volcano map. (B) The most enriched GO terms of differentially expressed genes.
To verify our transcriptome results, a total of 8 candidate genes (including flaE, flaG, cheA, fliN, fliS, kdsA, minD and cysN) were analyzed by qRT-PCR. qRT-PCR assay displayed that the changes in mRNA expression of ΔexsA were consistent with those of RNA-Seq (Figure 5). These results further validate the reliability of the transcriptome data.
The results of qRT-PCR showed that compared with HY9901, ΔexsA decreased the expression of hop at 18, 24, 36, 48 and 72 h (p < 0.01) (Figure 6A). Besides, expression of the LacZ reporter gene was measured with a β-galactosidase enzymatic assay. As shown in Figure 6B, HY9901 wild type strain exhibited high β-galactosidase activity, and ΔexsA produced low β-galactosidase activity (p < 0.05). These results suggested that exsA gene could facilitate the expression of hop gene.
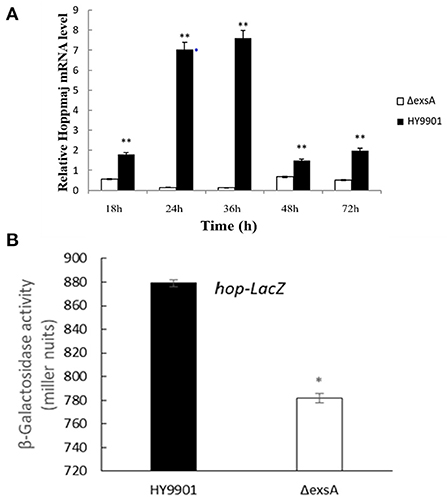
Figure 6. hop gene expression analysis. (A) Expression of hop genes by DMEM between HY9901 wild type and HY9901ΔexsA. (B) Expression of the LacZ reporter gene between HY9901 wild type and HY9901ΔexsA.
As shown in Figure 7, ΔexsA could survive momently in fish liver and spleen then was progressively excreted from the host body. The highest bacterial number was detected in spleen on day 2, followed by the liver.
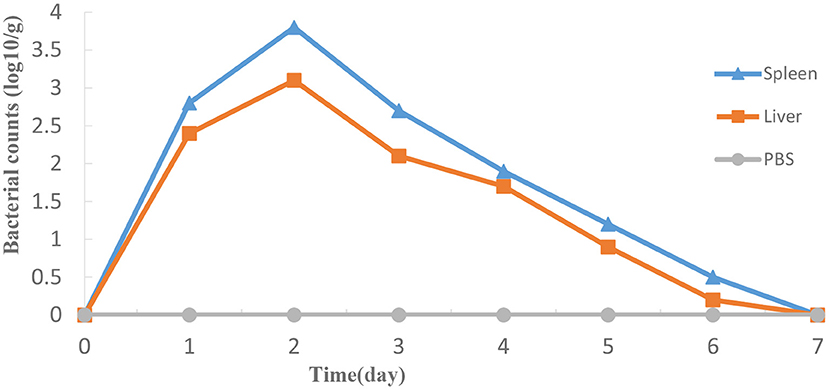
Figure 7. Propagation of HY9901ΔexsA in grouper liver (A) and spleen (B) following i.m. injection with 5 μL 1 × 105 cfu mL−1 ΔexsA. Control fish were i.m. injection with 5 μL sterile PBS. The number of viable bacteria was shown as the mean ± standard of three samples.
Zebrafish were vaccinated with PBS and ΔexsA by intramuscular injection for 28 days, challenged with the wild type HY9901. As shown in Figure 8, the mortality rate in the control group injected with sterile PBS was 90%, the mortality rate in the injection immunization group was 30%, and the relative percentage survival was 66.7%.
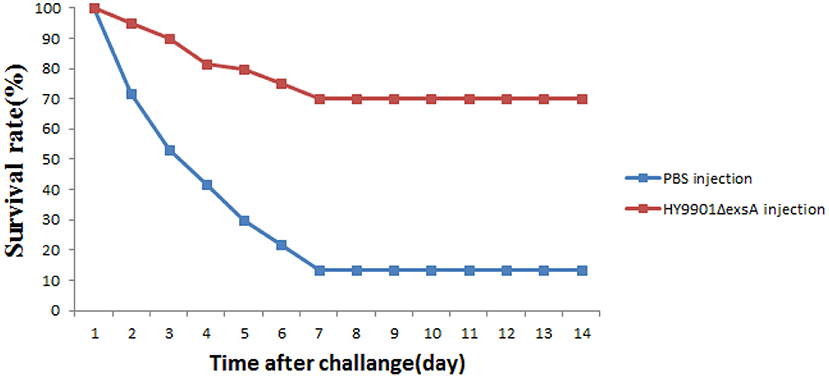
Figure 8. Survival in groups vaccinated with HY9901ΔexsA and PBS following challenge with V. alginolyticus HY9901.
qRT-PCR was used to analyze the expression of proinflammation and anti-inflammation genes. The results indicated that compared to PBS group, vaccination with ΔexsA had significantly increased expression of IL6, IL8, IL-6R, IL-1β, TNF-α, tlr5, rag-1, gata-1, IgM, genes in liver and gata-1, IL6, IL-6R, IgM, TNF-α, rag-1, TNF-α genes in spleen (p < 0.01) (Figure 9).
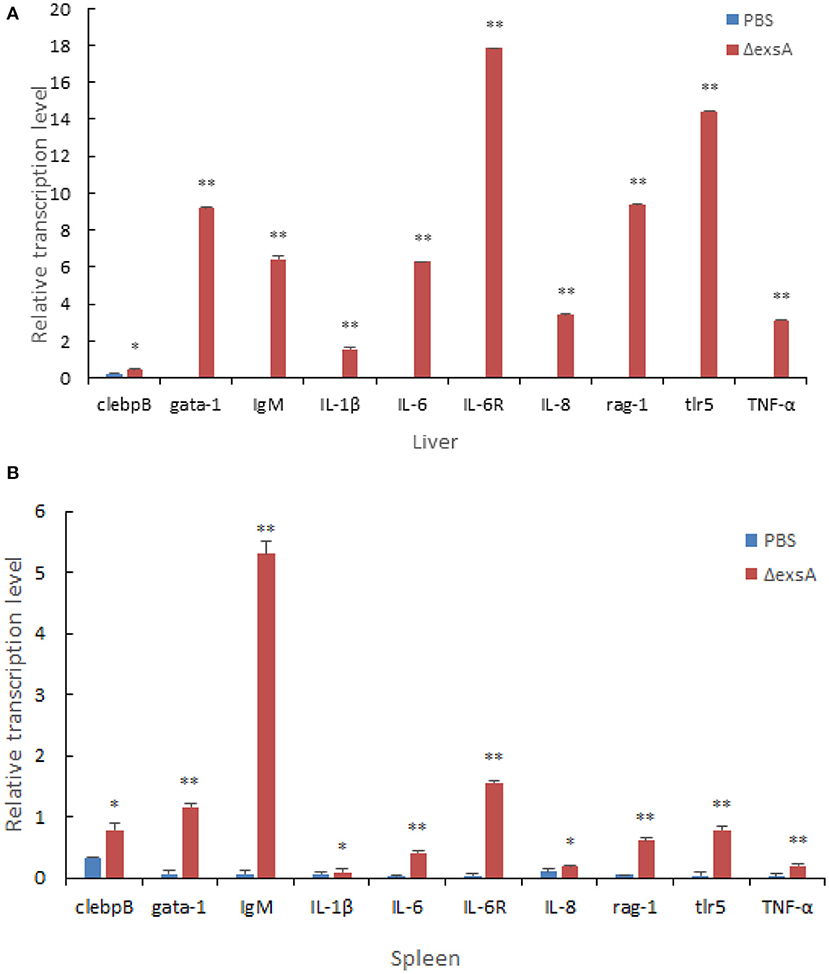
Figure 9. Comparative analysis of the expression of immune-related genes in liver (A) and spleen (B) of zebrafish given the live attenuated vaccine and unvaccinated zebrafish. The liver and spleen of zebrafish were sampled at 1 day before challenge, and the mRNA level of each immune-related gene was normalized to that of β-actin. Bars represent the mean relative expression of three biological replicates and error bars represent standard deviation.
As shown in Figure 10, pathological features, such as liver congestion and the boundaries between the lymphocytes in the spleen, were observed visually in the tissue sections of the zebrafish injected wild-type strain HY9901. There was a little number of bleeding spots and congestion in the zebrafish immunized by ΔexsA for 28 days, but not in PBS group.
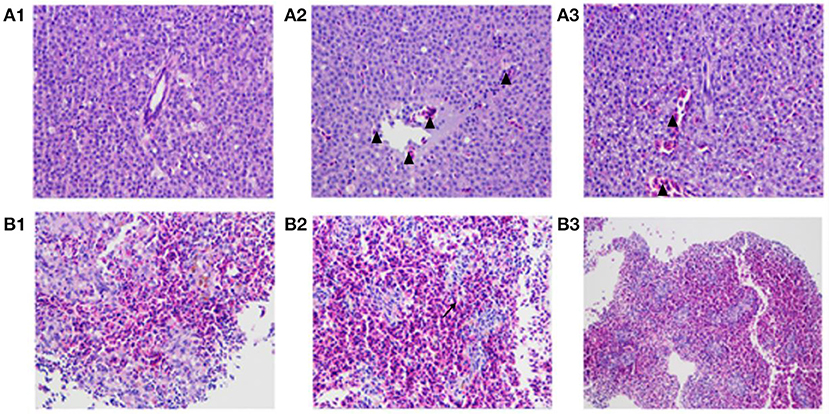
Figure 10. The pathological changes of vaccinated zebrafish. Zebrafish liver tissue section, 400× magnification (A1: injected with PBS A2: injected with HY9901 A3: injected with ΔexsA). Zebrafish spleen tissue section, 200 × magnification (B1: injected with PBS B2: injected with HY9901 B3: injected with ΔexsA). The triangle (▴) represents hyperemia, and the arrow (↗) represents the blurred boundary of lymphocytes in the figure.
In recent years, vibriosis outbreaks in China have caused serious economic losses, and overuse of antibiotics has driven the emergence and spread of resistance. In this context, the development of a vibrio-based vaccine might be a valuable alternative. Live attenuated vaccines closely mimic natural infection and, via specific antibody and cell-mediated immune responses, usually provide excellent protection against infection (31). Recently, attenuated virus strains can be constructed by knockout of the virulence genes. In this study, we knocked out the T3SS gene exsA of V. alginolyticus, probed into its biology and pathogenicity, and evaluated its effect as a live attenuated vaccine. And the genetic stability result indicated that the genetic information of the ΔexsA was stably inherited to offspring after 30 generations.
Biofilms are an important protective mechanism in which microbes form a strong shield against several antimicrobials (32). Vibrio polysaccharide (VPS) is critical for the formation of biofilm architecture in Vibrio cholera (33, 34). Flagella-mediated motility is required for the initial stages of biofilm formation (35), and deletion of V. parahaemolyticus polar flagellum genes leads to defective biofilm formation (36). Although there was no significant difference in growth between HY9901 and ΔexsA, the biofilm formation ability test showed that the mutant strain ΔexsA was significantly decreased compared to the wild-type strain HY9901. It is tempting to speculate that exsA may represses the expression of flagellum-related genes (flaA and fliG) or genes related to biofilm matrix polysaccharides, resulted in reduced ability to form mature biofilms. Extracellular products (ECP) are mainly thought to be characteristics of the virulent strain of V. alginolyticus (20). In this study, the extracellular protease activity of ΔexsA was decreased compared to the wild-type strain HY9901, which suggest exsA gene may be a positive contributor to activity of ECP in V. alginolyticus. ExsA may promote related gene expression of ECP such as vhh and ptlh, but this needs further investigation. Flagellar motility has long been recognized as a major colonization and virulence factor in many bacterial pathogens, including Pseudomonas aeruginosa (37) and Vibrio cholera (38). The flagella contribute to swarming motility and enable bacteria to invade into host cells to maintain their ecological niches in vivo (39). In this study, ΔexsA had suppressed swarming motility, which indicated that exsA was a positive contributor to swarming motility in V. alginolyticus. We could speculate that exsA might play an important role in regulating the expression level of flagella via an unclear mechanism. However, the regulatory mechanism is still unknown and also needs further investigation. Compared to HY9901, the LD50 of ΔexsA was decreased by 20 orders. From these results, we can find that exsA is related to the pathogenesis of V. alginolyticus.
The origin and molecular basis of bacterial resistance is the presence of antibiotic resistance genes (ARGs). An organism can acquire antibiotic resistance through mutation, horizontal gene transfer, or inheriting resistance genes from other organisms (40, 41). Besides, it is also possible to acquire antimicrobial resistance by mutation, for example changing the antimicrobial target site (42). V. alginolyticus has been reported to be resistant to ampicillin, vancomycin, and cephalothin (43). The results of Raissy et al. (44) showed that strB, tetS, and ermB genes, which encode streptomycin, tetracycline and erythromycin, respectively, could be detected in several strains of V. alginolyticus from seafood in Persian Gulf. Further, the author found that some strain not containing tesT gene was resistant to tetracycline and induced that there were other genes causing tetracycline resistance (44). In this study, HY9901 was highly resistant to chloramphenicol, while ΔexsA was sensitive. Besides, the ΔexsA was more sensitive to kanamycin, minocycline, tetracycline, gentamicin, doxycycline and neomycin. It could be guessed that the exsA gene might be related to drug resistance genes and regulate the expression of drug resistance-related genes. Nevertheless, it needs to be further studied.
The transcriptome is a collection of all RNA molecules in a cell, which reflects the expression status of the entire genome. It is essential for deciphering the functional complexity of the genome and to obtain a better understanding of cellular activities in organisms, including growth, development, disease, and immune defense (45–47). In the present study, a total of 541 upregulated and 663 downregulated differentially expressed genes in ΔexsA compared to HY9901. The outcome of GO analysis revealed that these genes were mainly enriched in a number of GO terms, including locomotion process, movement of cell or subcellular component process, non-membrane-bounded organelle process, organelle process, cilium or flagellum-dependent cell motility process and cell motility process. These results further suggest that exsA gene can regulate the expression of flagella related genes to affect the virulence of V. alginolyticus. Nonetheless, its specific regulatory mechanism needs further investigation.
The T3SS is a membrane-embedded nanomachine and it can deliver T3SEs from pathogen to host. The T3SS secretion in vitro and in vivo is complex, and its pathway is modulated by a single regulatory protein or several interacting regulatory proteins. For example, In Pseudomonas savastanoi HrpR and HrpS form a hetero-hexamer, which activates the expression of HrpL, inducing all T3SS genes by binding to a “hrp box” in promoters (48). ExsA is an activator of transcription in T3SS. A previous study indicates that in P. aeruginosa impA gene encoding an extracellular metalloprotease is under the regulation of ExsA, and ExsA can directly regulate the transcription of impA by binding to its upstream region, demonstrated in electrophoretic mobility shift assay (EMSA) (49). The results of Liu et al. (21) indicated that in V. alginolyticus ExsA was a positive regulator in effector proteins Va1686 and Va1687. HopPmaJ is one of the T3SEs from V. alginolyticus, which has been reported to attack the host efficiently (50). In this study, the deletion of exsA is responsible for the down-regulation of hop gene for various time periods, which was demonstrated in qRT-PCR assay. Furthermore, we also found that exsA gene could facilitate the expression of hop gene as well as acting as positive regulators. From these results, we can draw a conclusion that exsA can promote the expression of hop gene by binding the promoter regions, which may facilitate HopPmaJ protein secretion.
As a model of good vertebrate animals, zebrafish have been widely used for the study of immunity from a unique perspective. The low cost, rapid development and high fecundity of zebrafish makes it ideal as a vaccine-screening tool (24). It is reported that the V. parahaemolyticus can infect zebrafish larvae and activate the innate immune response (51). A previous study indicated that Th17 cells were activated following vaccination of zebrafish (52). Therefore, it is achievable to use zebrafish to investigate the effectiveness of a Vibrio vaccine. In the present study, the RPS of zebrafish vaccinated with ΔexsA reached 77.6%, and it was significantly higher than that of the control group.
TLR5 can activate the innate immune system by recognition of bacterial flagellin (53). IgM of fish plays an important role in resisting diseases, similar function as IgM in mammals (54). IgM plays a vital role in immunity, driving direct antimicrobial functions including complement activation, opsonophagocytosis, and agglutination. IL-1β and IL-6 are important cytokines involved in regulating inflammatory response (55, 56). TNF-α and IL-8 are important inflammatory cytokines that mediate the body inflammatory responses and have important roles in the physiological functions and pathological processes (57). In the present study, the elevated expression of immune-related genes (IgM, IL-1β, IL-6, and TNF-α), confirmed that ΔexsA can effectively induce the protective immune response of zebrafish associated with proinflammatory and immunoglobulin activity. Tissue sections results indicated a favorable safety profile of ΔexsA vaccines. In summary, these results showed that the ΔexsA could provide protection against V. alginolyticus and has the potential as an attenuated live vaccine. In addition to antibiotics, vaccines are one of the most important means to prevent and control of V. alginolyticus, which has great significance for aquaculture industries.
Taken together, we have successfully constructed an in-frame deletion strain of ΔexsA and investigated its physiology and pathogenicity. We found that ΔexsA could decrease ECPase activity and swarming motility, and the exsA gene regulates hop genes expression. Moreover, ΔexsA exhibited a high level of protection against V. alginolyticus challenge, and could induce protective immune responses in zebrafish. These results may provide experimental evidence for the importance of exsA in V. alginolyticus and provide a reference for further study into this virulence and infection mechanism.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
The animal study was reviewed and approved by the Animal Ethics Committee of Guangdong Ocean University (Zhanjiang, China).
WZ and HP designed the experiment. FZ, HF, LC, and XX generated experimental data. WZ and JW wrote manuscripts. HP, JJ, and NW conceived the work and critically reviewed the manuscript. WZ, LC, HF, JW, FZ, XX, JJ, NW, and HP have made extensive contributions to the work in this manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the National Natural Science Foundation of China (No. 32073015), Natural Science Foundation of Guangdong Province (No. 2021A1515011078), Undergraduate Innovation Team of Guangdong Ocean University (No. CCTD201802), Innovation and Entrepreneurship Training Program for College Students (No. CXXL2022005), Science and Technology Innovation Cultivation Program for College Student (No. pdjh2021b0239), and Guangdong Postgraduate Education Innovation Project.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2022.938822/full#supplementary-material
1. Millikan DS, Ruby EG. Vibrio fischeri flagellin A is essential for normal motility and for symbiotic competence during initial squid light organ colonization. J Bacteriol. (2004) 186:4315–25. doi: 10.1128/JB.186.13.4315-4325.2004
2. Kitaoka M, Nishigaki T, Ihara K, Nishioka N, Kojima S, Homma M. A novel dnaJ family gene, sflA, encodes an inhibitor of flagellation in marine Vibrio species. J Bacteriol. (2013) 195:816–22. doi: 10.1128/JB.01850-12
3. Jun JW, Kim HJ, Yun SK, Chai JY, Park SC. Eating oysters without risk of vibriosis: application of a bacteriophage against Vibrio parahaemolyticus in oysters. Int J Food Microbiol. (2014) 188:31–45. doi: 10.1016/j.ijfoodmicro.2014.07.007
4. Letchumanan V, Yin WF, Lee LH, Chan KG. Prevalence and antimicrobial susceptibility of Vibrio parahaemolyticus isolated from retail shrimps in Malaysia. Front Microbiol. (2015) 6:33. doi: 10.3389/fmicb.2015.00033
5. Ahmed HA, El Bayomi RM, Hussein MA, Khedr MHE, Abo Remela EM, El-Ashram AMM. Molecular characterization, antibiotic resistance pattern and biofilm formation of Vibrio parahaemolyticus and V. cholerae isolated from crustaceans and humans. Int J Food Microbiol. (2018) 274:31–7. doi: 10.1016/j.ijfoodmicro.2018.03.013
6. Matamp N, Bhat SG. Phage endolysins as potential antimicrobials against multidrug resistant vibrio alginolyticus and vibrio parahaemolyticus: current status of research and challenges ahead. Microorganisms. (2019) 7:84. doi: 10.3390/microorganisms7030084
7. Cornelis GR. The type III secretion injectisome. Nat Rev Microbiol. (2006) 4:811–25. doi: 10.1038/nrmicro1526
8. Galán JE, Wolf-Watz H. Protein delivery into eukaryotic cells by type III secretion machines. Nature. (2006) 444:567–73. doi: 10.1038/nature05272
9. Büttner D. Protein export according to schedule: architecture, assembly, and regulation of type III secretion systems from plant- and animal-pathogenic bacteria. Microbiol Mol Biol Rev. (2012) 76:262–310. doi: 10.1128/MMBR.05017-11
10. Brutinel ED, King JM, Marsden AE, Yahr TL. The distal ExsA-binding site in Pseudomonas aeruginosa type III secretion system promoters is the primary determinant for promoter-specific properties. J Bacteriol. (2012) 194:2564–72. doi: 10.1128/JB.00106-12
11. Frank DW. The exoenzyme S regulon of Pseudomonas aeruginosa. Mol Microbiol. (1997) 26:621–9. doi: 10.1046/j.1365-2958.1997.6251991.x
12. Qu J, Prasad NK, Yu MA, Chen S, Lyden A, Herrera N, et al. Modulating pathogenesis with mobile-CRISPRi. J Bacteriol. (2019) 201:e00304–19. doi: 10.1128/JB.00304-19
13. Gu D, Zhang Y, Wang Q, Zhou X. S-nitrosylation-mediated activation of a histidine kinase represses the type 3 secretion system and promotes virulence of an enteric pathogen. Nat Commun. (2020) 11:5777. doi: 10.1038/s41467-020-19506-1
14. Zhou X, Konkel ME, Call DR. Regulation of type III secretion system 1 gene expression in Vibrio parahaemolyticus is dependent on interactions between ExsA, ExsC, and Exs D. Virulence. (2010) 1:260–72. doi: 10.4161/viru.1.4.12318
15. Zhou X, Shah DH, Konkel ME, Call DR. Type III secretion system 1 genes in Vibrio parahaemolyticus are positively regulated by ExsA and negatively regulated by ExsD. Mol Microbiol. (2008) 69:747–764. doi: 10.1111/j.1365-2958.2008.06326.x
16. Darwinata AE, Gotoh K, Mima T, Yamamoto Y, Yokota K, Matsushita O. Vibrio alginolyticus VepA induces lysosomal membrane permeability and cathepsin-independent cell death. Acta Med Okayama. (2018) 72:231–9. doi: 10.18926/AMO/56068
17. Pang H, Qiu M, Zhao J, Hoare R, Monaghan SJ, Song D, et al. Construction of a Vibrio alginolyticus hopPmaJ (hop) mutant and evaluation of its potential as a live attenuated vaccine in orange-spotted grouper (Epinephelus coioides). Fish Shellfish Immunol. (2018) 76:93–100. doi: 10.1016/j.fsi.2018.02.012
18. Zhou S, Tu X, Pang H, Hoare R, Monaghan SJ, Luo J, et al. A T3SS regulator mutant of Vibrio alginolyticus affects antibiotic susceptibilities and provides significant protection to Danio rerio as a live attenuated vaccine. Front Cell Infect Microbiol. (2020) 10:183. doi: 10.3389/fcimb.2020.00183
19. Wu C, Zhao Z, Liu Y, Zhu X, Liu M, Luo P, et al. Type III secretion 1 effector gene diversity among vibrio isolates from coastal areas in China. Front Cell Infect Microbiol. (2020) 10:301. doi: 10.3389/fcimb.2020.00301
20. Cai SH, Wu ZH, Jian JC, Lu YS. Cloning and expression of the gene encoding an extracellular alkaline serine protease from Vibrio alginolyticus strain HY9901, the causative agent of vibriosis in Lutjanus erythopterus (Bloch). J Fish Dis. (2007) 30:493–500. doi: 10.1111/j.1365-2761.2007.00835.x
21. Liu J, Lu SY, Orfe LH, Ren CH, Hu CQ, Call DR, et al. ExsE is a negative regulator for T3SS gene expression in Vibrio alginolyticus. Front Cell Infect Microbiol. (2016) 6:177. doi: 10.3389/fcimb.2016.00177
22. Pang H, Chang Y, Zheng H, Tan H, Zhou S, Zeng F, et al. A live attenuated strain of HY9901ΔvscB provides protection against Vibrio alginolyticus in pearl gentian grouper (♀Epinephelus fuscoguttatus x ♂Epinephelus lanceolatu). Aquaculture. (2022) 546:737353. doi: 10.1016/j.aquaculture.2021.737353
23. Luo P, He X, Liu Q, Hu C. Developing universal genetic tools for rapid and efficient deletion mutation in vibrio species based on suicide T-vectors carrying a novel counterselectable marker, vmi480. PLoS ONE. (2015) 10:e0144465. doi: 10.1371/journal.pone.0144465
24. Wu P, Zhou S, Su Z, Liu C, Zeng F, Pang H, et al. Functional characterization of T3SS C-ring component VscQ and evaluation of its mutant as a live attenuated vaccine in zebrafish (Danio rerio) model. Fish Shellfish Immunol. (2020) 104:123–32. doi: 10.1016/j.fsi.2020.05.063
25. Windle HJ, Kelleher D. Identification and characterization of a metalloprotease activity from Helicobacter pylori. Infect Immun. (1997) 65:3132–237. doi: 10.1128/iai.65.8.3132-3137.1997
26. Mathew JA, Tan YP, Srinivasa Rao PS, Lim TM, Leung KY. Edwardsiella tarda mutants defective in siderophore production, motility, serum resistance and catalase activity. Microbiology. (2001) 147:449–57. doi: 10.1099/00221287-147-2-449
27. Chen Y, Cai S, Jian J. Protection against Vibrio alginolyticus in pearl gentian grouper (♀Epinephelus fuscoguttatus × ♂Epinephelus lanceolatu) immunized with an acfA-deletion live attenuated vaccine. Fish Shellfish Immunol. (2019) 86:875–81. doi: 10.1016/j.fsi.2018.12.030
28. Li W, Yao Z, Sun L, Hu W, Cao J, Lin W, et al. Proteomics analysis reveals a potential antibiotic cocktail therapy strategy for Aeromonas hydrophila infection in biofilm. J Proteome Res. (2016) 15:1810–20. doi: 10.1021/acs.jproteome.5b01127
29. Thibault G, Ismail N, Ng DT. The unfolded protein response supports cellular robustness as a broad-spectrum compensatory pathway. Proc Natl Acad Sci USA. (2011) 108:20597–602. doi: 10.1073/pnas.1117184109
30. Joseph J, Ametepe ES, Haribabu N, Agbayani G, Krishnan L, Blais A, et al. Inhibition of ROS and upregulation of inflammatory cytokines by FoxO3a promotes survival against Salmonella typhimurium. Nat Commun. (2016) 7:12748. doi: 10.1038/ncomms12748
31. Jang YH, Byun YH, Lee KH, Park ES, Lee YH, Lee YJ, et al. Host defense mechanism-based rational design of live vaccine. PLoS ONE. (2013) 8:e75043. doi: 10.1371/journal.pone.0075043
32. Wang Y, Wang Y, Liu B, Wang S, Li J, Gong S, et al. pdh modulate virulence through reducing stress tolerance and biofilm formation of Streptococcus suis serotype 2. Virulence. (2019) 10:588–99. doi: 10.1080/21505594.2019.1631661
33. Absalon C, Van Dellen K, Watnick PI. A communal bacterial adhesin anchors biofilm and bystander cells to surfaces. PLoS Pathog. (2011) 7:e1002210. doi: 10.1371/journal.ppat.1002210
34. Berk V, Fong JC, Dempsey GT, Develioglu ON, Zhuang X, Liphardt J, et al. Molecular architecture and assembly principles of Vibrio cholerae biofilms. Science. (2012) 337:236–9. doi: 10.1126/science.1222981
35. Yildiz FH, Visick KL. Vibrio biofilms: so much the same yet so different. Trends Microbiol. (2009) 17:109–18. doi: 10.1016/j.tim.2008.12.004
36. Enos-Berlage JL, Guvener ZT, Keenan CE, McCarter LL. Genetic determinants of biofilm development of opaque and translucent Vibrio parahaemolyticus. Mol Microbiol. (2005) 55:1160–82. doi: 10.1111/j.1365-2958.2004.04453.x
37. Demirdjian S, Hopkins D, Sanchez H, Libre M, Gerber SA, Berwin B. Phosphatidylinositol-(3,4,5)-trisphosphate induces phagocytosis of nonmotile Pseudomonas aeruginosa. Infect Immun. (2018) 86:e00215–18. doi: 10.1128/IAI.00215-18
38. Correa NE, Lauriano CM, McGee R, Klose KE. Phosphorylation of the flagellar regulatory protein FlrC is necessary for Vibrio cholerae motility and enhanced colonization. Mol Microbiol. (2000) 35:743–55. doi: 10.1046/j.1365-2958.2000.01745.x
39. Echazarreta MA, Klose KE. Vibrio flagellar synthesis. Front Cell Infect Microbiol. (2019) 9:131. doi: 10.3389/fcimb.2019.00131
40. Davies J, Davies D. Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev. (2010) 74:417–33. doi: 10.1128/MMBR.00016-10
41. Cox G, Wright GD. Intrinsic antibiotic resistance: mechanisms, origins, challenges and solutions. Int J Med Microbiol. (2013) 303:287–92. doi: 10.1016/j.ijmm.2013.02.009
42. Agersø Y, Bjerre K, Brockmann E, Johansen E, Nielsen B, Siezen R, et al. Putative antibiotic resistance genes present in extant Bacillus licheniformis and Bacillus paralicheniformis strains are probably intrinsic and part of the ancient resistome. PLoS ONE. (2019) 14:e0210363. doi: 10.1371/journal.pone.0210363
43. Kang CH, Shin Y, Jang S, Jung Y, So JS. Antimicrobial susceptibility of Vibrio alginolyticus isolated from oyster in Korea. Environ Sci Pollut Res Int. (2016) 23:21106–12. doi: 10.1007/s11356-016-7426-2
44. Raissy M, Moumeni M, Ansari M, Rahimi E. Antibiotic resistance pattern of some Vibrio strains isolated from seafood. Iranian J Fish Sci. (2012) 11:618–26. doi: 10.4103/0972-124X.99267
45. Xiang LX, He D, Dong WR, Zhang YW, Shao JZ. Deep sequencing-based transcriptome profiling analysis of bacteria-challenged Lateolabrax japonicus reveals insight into the immune-relevant genes in marine fish. BMC Genomics. (2010) 11:472. doi: 10.1186/1471-2164-11-472
46. Zhang B, Luo G, Zhao L, Huang L, Qin Y, Su Y, et al. Integration of RNAi and RNA-seq uncovers the immune responses of Epinephelus coioides to L321_RS19110 gene of Pseudomonas plecoglossicida. Fish Shellfish Immunol. (2018) 81:121–129. doi: 10.1016/j.fsi.2018.06.051
47. Tang Y, Xin G, Zhao LM, Huang LX, Qin YX, Su YQ, et al. Novel insights into host-pathogen interactions of large yellow croakers (Larimichthys crocea) and pathogenic bacterium Pseudomonas plecoglossicida using time-resolved dual RNA-seq of infected spleens. Zool Res. (2020) 41:314–27. doi: 10.24272/j.issn.2095-8137.2020.035
48. Wang J, Shao X, Zhang Y, Zhu Y, Yang P, Yuan J, et al. HrpS is a global regulator on type III secretion system (T3SS) and non-T3SS genes in Pseudomonas savastanoi pv. phaseolicola. Mol Plant Microbe Interact. (2018) 31:1232–43. doi: 10.1094/MPMI-02-18-0035-R
49. Tian Z, Cheng S, Xia B, Jin Y, Bai F, Cheng Z, et al. Pseudomonas aeruginosa ExsA regulates a metalloprotease, ImpA, that inhibits phagocytosis of macrophages. Infect Immun. (2019) 87:e00695–19. doi: 10.1128/IAI.00695-19
50. Zhuang Q, Dai F, Zhao X, Shao Y, Guo M, Lv Z, et al. Cloning and characterization of the virulence factor Hop from Vibrio splendidus. Microb Pathog. (2020) 139:103900. doi: 10.1016/j.micpath.2019.103900
51. Ji C, Guo X, Dong X, Ren J, Zu Y, Li W, et al. Notch1a can widely mediate innate immune responses in zebrafish larvae infected with Vibrio parahaemolyticus. Fish Shellfish Immunol. (2019) 92:680–689. doi: 10.1016/j.fsi.2019.06.058
52. Zhang H, Fei C, Wu H, Yang M, Liu Q, Wang Q, et al. Transcriptome profiling reveals Th17-like immune responses induced in zebrafish bath-vaccinated with a live attenuated Vibrio anguillarum. PLoS ONE. (2013) 8:e73871. doi: 10.1371/journal.pone.0073871
53. Voogdt CGP, Wagenaar JA, van Putten MJP. Duplicated TLR5 of zebrafish functions as a heterodimeric receptor. Proc Natl Acad Sci USA. (2018) 115:E3221–9. doi: 10.1073/pnas.1719245115
54. Solem ST, Stenvik J. Antibody repertoire development in teleosts–a review with emphasis on salmonids and Gadus morhua L. Dev Comp Immunol. (2006) 30:57–76. doi: 10.1016/j.dci.2005.06.007
55. Lin H, Gao D, Hu MM, Zhang M, Wu XX, Feng L, et al. MARCH3 attenuates IL-1β-triggered inflammation by mediating K48-linked polyubiquitination and degradation of IL-1RI. Proc Natl Acad Sci USA. (2018) 115:12483–12488. doi: 10.1073/pnas.1806217115
56. Alaridah N, Lutay N, Tenland E, Rönnholm A, Hallgren O, Puthia M, et al. Mycobacteria manipulate G-protein-coupled receptors to increase mucosal Rac1 expression in the lungs. J Innate Immun. (2017) 9:318–29. doi: 10.1159/000453454
Keywords: type III secretory system, characteristics, live attenuated vaccine, Vibrio alginolyticus, exsA gene
Citation: Zhang W, Chen L, Feng H, Wang J, Zeng F, Xiao X, Jian J, Wang N and Pang H (2022) Functional characterization of Vibrio alginolyticus T3SS regulator ExsA and evaluation of its mutant as a live attenuated vaccine candidate in zebrafish (Danio rerio) model. Front. Vet. Sci. 9:938822. doi: 10.3389/fvets.2022.938822
Received: 08 May 2022; Accepted: 14 July 2022;
Published: 08 August 2022.
Edited by:
Kun Li, Nanjing Agricultural University, ChinaCopyright © 2022 Zhang, Chen, Feng, Wang, Zeng, Xiao, Jian, Wang and Pang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huanying Pang, cGh5aW5nMTIxOEAxNjMuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.