
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Vet. Sci., 12 September 2022
Sec. Veterinary Infectious Diseases
Volume 9 - 2022 | https://doi.org/10.3389/fvets.2022.935133
This article is part of the Research TopicInsights in Veterinary Infectious Diseases: 2022View all 9 articles
Mycobacterium avium subsp. paratuberculosis (MAP) causes Johne's disease or paratuberculosis (PTB), with important animal health and economic implications. There are no therapeutic strategies to control this disease, and vaccination with inactivated vaccines is limited in many countries because it can interfere with the intradermal test used for bovine tuberculosis detection. Thus, infected animals either get culled after a positive ELISA or fecal PCR result or die due to clinical disease. In this study, we review recent studies aimed to discover genetic markers which could help to identify and select cattle less susceptible and more resilient to PTB. In recent years, the genotyping and subsequent imputation to whole-genome sequence (WGS) has allowed the identification of single-nucleotide polymorphisms (SNPs), quantitative trait loci (QTL), and candidate genes in the Bos taurus genome associated with susceptibility to MAP infection. In most of these genome-wide association studies (GWAS), phenotypes were based on ante-mortem test results including serum ELISA, milk ELISA, and detection of MAP by fecal PCR and bacteriological culture. Cattle infected with MAP display lesions with distinct severity but the associations between host genetics and PTB-associated pathology had not been explored until very recently. On the contrary, the understanding of the mechanisms and genetic loci influencing pathogen resistance, and disease tolerance in asymptomatic individuals is currently very limited. The identification of long-time asymptomatic cattle that is able to resist the infection and/or tolerate the disease without having their health and milk production compromised is important for disease control and breeding purposes.
Mycobacterium avium subsp. paratuberculosis (MAP) infections affecting domestic and wild ruminants represent a global major issue on animal health, recognized by the World Organization for Animal Health (OIE) (1), which requires member countries to maintain epidemiological surveillance with notification of disease cases. Clinical signs of MAP infection in cattle include diarrhea, progressive loss of body condition in the animals, and eventually their death (2). Paratuberculosis (PTB) is responsible for significant economic losses in dairy herds worldwide due to decreased milk production, increased management costs, and premature culling or death from clinical disease (3–5). More than 50% of dairy cattle herds are positive for MAP antibodies in the USA and Europe and, therefore, the disease is endemic in these areas (6, 7). In Canada, the economic losses caused by PTB were estimated at $50 CAN per cow per year in MAP-infected herds (8). In Ireland, a profit margin reduction between €168 and €253 was estimated for a cow on a PTB-affected farm (9). The economic impact of PTB on the US dairy industry has been estimated between US $250 M per year to US $1.5 billion annually, with a net return of almost US $100 less per cow in a positive herd than in a negative herd (10). The economic impact of PTB in Europe has been estimated at US $364.31 million per year (11). There is no cure for PTB, and MAP control across the globe has proven to be difficult. MAP survives pasteurization and could enter the human food chain through meat, dairy products, and untreated water supplies (12). It has been detected in samples of patients with Crohn's disease (CD), ulcerative colitis, and idiopathic inflammatory bowel disease (IBD)-associated colorectal cancer (13–15). MAP has also been postulated as a possible trigger in several human autoimmune diseases such as rheumatoid arthritis, multiple sclerosis, and type I diabetes (16–19).
Many control programs for PTB based on vaccination or testing and culling of test-positive cows have been developed worldwide. Commercial inactivated vaccines against bovine PTB are very successful in reducing MAP presence in feces and tissues and in increasing both milk production and the productive life of cattle (20, 21). However, PTB vaccination is not allowed in most European countries due to its interference with Mycobacterium bovis detection tests (22). PTB control is currently based on testing and culling and avoiding MAP transmission to susceptible animals by enhancing on-farm biosecurity measures (23, 24). Some factors that hamper the success of such control programs include the lack of compliance with management protocols, the use of tests with low sensitivities to identify all the infected cattle, and the purchase of infected replacement animals which cause new introductions (25). MAP infection occurs primarily through the fecal-oral route, and clinical onset takes place around calving when animals are 18 months or older. According to their extension in the intestine, cellular infiltrate, and amount of MAP, PTB-associated lesions were classified into focal, multifocal, and diffuse (diffuse paucibacillary or lymphoplasmacytic, diffuse intermediate, and diffuse multibacillary or histiocytic) (26, 27). Focal lesions consist of small granulomas in the ileal and jejunal lymph nodes or the ileocecal lymphoid tissue. Multifocal lesions are middle-size granulomas that appear in the apex of some intestinal villi and are formed by groups of macrophages, surrounded by lymphocytes. The focal and multifocal lesions do not cause diffuse enteritis or modify the normal architecture of the intestine. In contrast, the diffuse lesions are associated with a diffuse infiltrate and severe enteritis affecting different intestinal locations and lymph nodes. High bacterial burden, clinical signs, and gross lesions are mainly associated with the presence of diffuse multibacillary lesions.
The response to MAP infection is complex and heritable which leads to differences between individuals. Some animals are susceptible to the infection and develop clinical signs while others are resilient and long-term asymptomatic animals. Host defense strategies against infectious diseases are comprised of pathogen avoidance, disease tolerance, and resistance to the infection (Figure 1). Pathogen avoidance refers to the behaviors that animals use to avoid infection caused by pathogens. Resistance is defined as “the ability of the host to prevent invasion (i.e., absence of a target receptor) or to clear the pathogen at the early stage of infection by mounting a protective innate immune response” (28). Disease tolerance is defined as “the mechanisms that decrease host susceptibility to tissue damage, or other fitness costs caused by pathogens or by the immune response” (29). Unlike resistance, disease tolerance does not necessarily imply changes in the pathogen load. Therefore, three major outcomes following exposure to MAP infection can be established: (i) Susceptible host: individuals who progress to clinical disease, (ii) Resistant host: Individuals who can prevent bacterial entry or eliminate the bacteria by inducing innate immune responses at the early stage of the infection, (iii) Tolerant host: If innate immunity is unable to eliminate MAP, the host might initiate disease tolerance mechanisms to prevent and repair tissue damage. The contribution of host genetics is one of the fundamental issues in understanding disease tolerance and susceptibility and resistance to MAP infection.
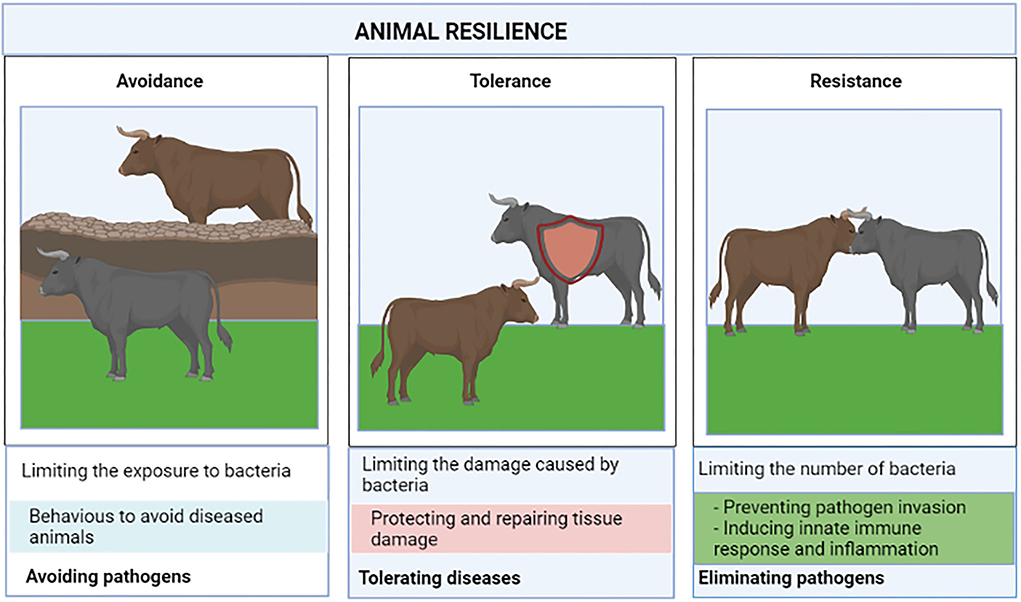
Figure 1. Resilient animals defend themselves against pathogens using the complimentary strategies of avoidance, disease tolerance, and pathogen resistance.
The application of animal genetics in breeding programs is currently an important motor for efficient livestock production, not only to increase performance and productivity but also to ensure the resilience and health of livestock while improving their longevity. A widely accepted strategy to reduce or eliminate animal diseases is through the selective breeding of animals with reduced susceptibility and/or enhanced resilience against specific pathogens. Over the last decade, specific candidate genes provided evidence for the existence of single nucleotide polymorphisms (SNPs) associated with differences in susceptibility to bovine PTB (30). The identification of SNPs, quantitative trait loci (QTLs), and candidate genes associated with PTB susceptibility is extremely important not just for breeding strategies but also to understand the mechanisms involved in the pathogenesis of the disease. Previous studies established that the heritability (h2) estimates of susceptibility to MAP in Friesian cattle ranges from 0.03 to 0.27 (31, 32). The genotyping of SNPs with chips of low (LD), medium (MD), and high density (HD) and subsequent imputation to whole-genome sequences (WGS) have allowed the identification of QTLs and candidate genes in the Bos taurus genome associated with susceptibility to MAP infection. Table 1 shows recent studies designated to identify SNPs associated with susceptibility to MAP infection, and Supplementary Table 1 summarizes the candidate genes identified in these studies. Although the cellular immune response is considered the protective effector arm of the immune system for intracellular infections including MAP infection, there have not been GWAS studies published with hereditability estimates based on skin tests or IFN gamma. In most of these GWAS, phenotypes were based on ante-mortem test results such as serum ELISA, milk ELISA, and MAP detection by culture or PCR from fecal samples. We recently identified SNPs, QTLs, and candidate genes associated with ante-mortem (serum ELISA) and post-mortem (tissue PCR and culture) diagnostic definitions in a common set of Spanish Holstein cattle (N = 983) using WGS data (46). We observed that the combination of diagnostic tests increased the h2 estimates, with the highest h2 obtained for the combination of ELISA-tissue PCR-tissue culture (h2 = 0.139).
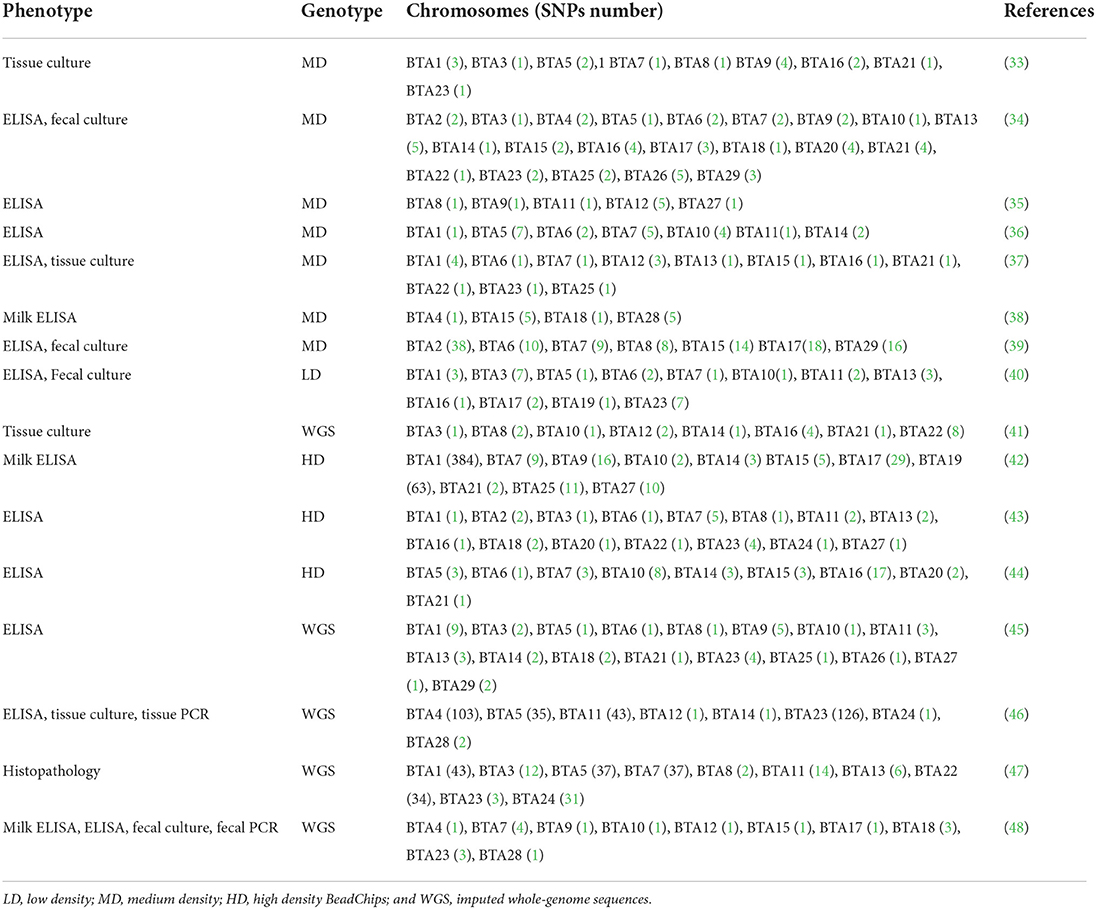
Table 1. Genome-wide association studies designed to identify SNPs associated with susceptibility to MAP infection.
We have recently demonstrated that the post-mortem examination of gut tissues and regional lymph nodes improves the accuracy of the classification of naturally infected animals and provides higher h2 estimates (47). A total of 192 and 92 SNPs defining 13 and 9 distinct QTLs were associated (P ≤ 5 × 10−7) with the multifocal (h2 = 0.075) and the diffuse (h2 = 0.189) lesions, respectively. No overlap was seen in the SNPs associated with each type of lesion which suggested that distinct genetic variants might control the multifocal and diffuse lesions and that these lesions represent divergent disease outcomes. Pathway analysis with the candidate genes overlapping the identified QTLs revealed a significant enrichment of the keratinization pathway and cholesterol metabolism in the animals with multifocal and diffuse lesions, respectively. The keratin family (KRT), the major subgroup among the intermediate filament family of cytoskeletal proteins, is responsible for maintaining the integrity of the gastrointestinal epithelium, providing resilience against many agents, and regulating various cellular functions such as cellular proliferation, and inflammatory and immune responses. In recent years, it has been demonstrated that the presence of epithelioid granulomas with multifocal distribution in leprosy controls M. leprae replication and its dissemination (49). Similarly, we hypothesized that the PTB-associated multifocal granulomas might prevent MAP dissemination and limit tissue damage, representing a signature of disease tolerance. The KRTs might be playing an important role in controlling tissue resilience and bacterial dissemination in animals with this specific type of lesion which might even recover from infection. In contrast, the diffuse lesions represent a disseminated form of the disease characterized by a diffuse inflammatory infiltrate composed mainly of foamy macrophages loaded with cholesterol and large numbers of MAP (Figure 2).
Although GWAS have allowed the identification of SNPs associated with the susceptibility to MAP infection, the genes through which these variants exert their effects are unknown, and only a few functional mutations for PTB have been identified (50, 51). Expression quantitative trait loci (eQTLs) are genetic variants located in gene regulatory regions that alter gene expression. eQTLs can be considered as functional links between genomic variants, gene expression, and ultimately phenotype. Recently, the integration of gene expression data (RNA-Seq) and genotypes (54,609 SNPs per animal) from a cohort of cows naturally exposed to MAP has allowed the identification of 192 and 48 cis-eQTLs associated with the expression of 145 and 43 genes in peripheral blood (PB) and ileocecal valve (ICV), respectively (52). Three of these cis-eQTLs regulating the expression of the MECOM, eEF1A2, and U1 spliceosomal RNA expression were significantly associated with PTB susceptibility (Figure 2). This study identified the first cis-eQTLs involved in gene transcription regulation and PTB susceptibility by integrating genotypes and RNA-seq data.
Although we identified a total of 380 SNPs associated (FDR ≤ 0.05; P < 5 × 10−7) with PTB susceptibility using ante-mortem and post-mortem definitions in a common set of Spanish Holstein cattle (N = 983) (46, 47), no conclusions could be drawn regarding host resistance. While test-positive cows are very likely infected, animals could test negative due to the lack of sensitivity of the diagnostic methods or lack of exposure to MAP. In other words, a negative test does not always reflect resistance, which precludes the identification of loci associated with resistance. Few studies have attempted to identify genetic variants associated with resistance to MAP infection in cattle. In 2010, Pant et al. (53) genotyped 232 Canadian dairy Holstein cattle with MAP infection status assessed by ELISA and milk ELISA. Using this approach, eleven SNPs on BTA1, 5, 6, 7, 10, 11, and 14 were associated with resistance to MAP infection. However, the population size was small (232 animals) and the classification of the animals based only on ELISA test results could lead to misclassification due to the lack of sensitivity of the ELISA. More recently, Sanchez et al. (54) carried out a multi-breeding GWAS study of 1,644 Holstein Friesian and 649 Normande cows with imputed WGS. In this study, control cows without clinical signs were tested by serum ELISA and fecal PCR at least four times. Clinical cases were confirmed with both ELISA and PCR tests. Thus, animals were classified as controls, cases without clinical signs, and cases with clinical signs. With this approach, a total of 2,827 resistance-associated SNPs distributed in 20 quantitative trait loci (QTLs) were identified. However, the threshold used to determine the significance of the identified variants was very low (P < 10−6), and the multiple positive correction was limited only to 50,000 independent tests.
Regarding disease tolerance, genetic variants that confer greater fitness displaying positive or advantageous selection can potentially be observed in the host genome (55). Recently, we have searched for genetic loci associated with tolerance to PTB by using WGS data from infected Spanish Holstein cows with MAP detected by tissue PCR and bacteriological culture but without lesions in gut tissues and regional lymph nodes. A PTB tolerant animal was PCR and culture-positive (infected) but no lesions could be observed in the histopathological analysis of gut tissues (no disease) (56). Although an earlier paper described a GWAS for PTB tolerance (57) with a subsequent refinement of genetic regions associated with MAP tissue infection and tolerance to PTB (58), our GWAS is the first to complete an analysis of the genetic markers associated with tolerance to PTB using WGS data and epidemiological data based on three diagnostic tests; histopathology, tissue PCR, and bacteriological culture. Results from our study do not corroborate those of Zanella et al. (57, 58), who identified a tolerance-associated region of 6.5 Kb on BTA15 which we did not identify. In the studies by Zanella et al., tolerance measured the relationship between MAP infection intensity (level of MAP in gut tissues) and fitness (level of MAP fecal shedding). The amount of MAP in feces was used in this study as a proxy for fitness but to define tolerance, direct disease outcomes measurements are critical. In our study, PTB-tolerant cows are infected animals with positive PCR and bacteriological culture results but without lesions in gut tissues. Our results suggest that there is genetic variation associated with PTB tolerance (h2 = 0.55) and that this variation is indicative of an immunogenic profile in the PTB tolerant animals designed to control bacterial growth, modulate inflammation, and limit tissue damage (Figure 2) (56). Some of the identified QTLs overlapped with QTLs previously associated with PTB, bovine tuberculosis, mastitis, somatic cell score, bovine diarrhea virus persistent infection, tick resistance, and length of productive life. However, the need to slaughter animals to measure MAP load in tissues as an indicator of tolerance limits its employment. Thus, novel tools to measure PTB tolerance must be developed.
Since it is well recognized that not all asymptomatic animals will progress into clinical cases during their productive life, the identification of genetic markers associated with PTB tolerance might help farmers or animal health managers to select which infected cows should not be culled which in turn should increase the benefit/cost of their control program. In addition, the introduction of novel genetic variants associated with PTB susceptibility and tolerance into marker-assisted breeding programs would help producers to select less susceptible cattle, and more tolerant to PTB and likely to other bovine diseases as well, ultimately preventing economic losses and reducing antimicrobial use. Preventing endemic and chronic diseases such as PTB by selecting resilient cattle is important for sustainable and efficient dairy farming and the maintenance of the rural economy. Although this is a long-term control strategy, the benefits of breeding resilient animals could be permanent and transferred to subsequent generations.
MAH conceived and coordinated the project and wrote the manuscript. All authors contributed to the article and approved the submitted version.
Financial support for this study was provided by a grant (RTI2018-094192-R-C21) funded by MCIN/AEI/10.13039/501100011033 and by FEDER, Una manera de hacer Europa. MC and GBB have been awarded fellowships from MCIN/AEI/10.13039/501100011033 and FSE Invierte en tu futuro; grants FPI2016-00041 and PRE2019-090562, respectively.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2022.935133/full#supplementary-material
2. Clarke CJ. The pathology and pathogenesis of paratuberculosis in ruminants and other species. J Comp Path. (1997) 116:217–61.
3. Ott SL, Wells SJ, Wagner BA. Herd-level economic losses associated with Johne's disease on US dairy operations. Prev Vet Med. (1999) 40:179–92.
4. Hasonova L, Pavlik I. Economic impact of paratuberculosis in dairy cattle herds: a review. Vet Med. (2006) 51:193–211. doi: 10.17221/5539-VETMED
5. Smith RL, Strawderman RL, Schukken YH, Wells SJ, Pradhan AK, Espejo LA, et al. Effect of Johne's disease status on reproduction and culling in dairy cattle. J Dairy Sci. (2010) 93:3513–24. doi: 10.3168/jds.2009-2742
6. Nielsen SS, Toft N. A review of prevalences of paratuberculosis in farmed animals in Europe. Prev Vet Med. (2009) 88:1–14. doi: 10.1016/j.prevetmed.2008.07.003
7. Lombard JE. Epidemiology and economics of paratuberculosis. Vet Clin N Am Food Anim Pract. (2011) 27:525–35. doi: 10.1016/j.cvfa.2011.07.012
8. Stott AW, Jones GM, Humphry RW, Gunn GJ. The financial incentive to control paratuberculosis (Johne's disease) on UK dairy farms. Vet Rec. (2005) 156:825–31. doi: 10.1136/vr.156.26.825
9. Barrett DJ, Good M, Hayes M, More SJ. The economic impact of Johne's disease in an Irish dairy herd: a case study. Ir Vet J. (2006) 59:282–8.
10. Harris NB, Barletta RG. Mycobacterium avium subsp. paratuberculosis in veterinary medicine. Clin Microbiol Rev. (2001) 14:489–512. doi: 10.1128/CMR.14.3.489-512.2001
11. Rasmussen P, Barkema HW, Mason S, Beaulieu E, Hall DC. Economic losses due to Johne's disease (paratuberculosis) in dairy cattle. J Dairy Sci. (2021) 104:3123–43. doi: 10.3168/jds.2020-19381
12. Alonso-Hearn M, Molina E, Geijo M, Vázquez P, Sevilla I, Garrido JM, et al. Isolation of Mycobacterium avium subsp. paratuberculosis from muscle tissue of naturally infected cattle. Foodborne Pathog Dis. (2009) 6:513–8. doi: 10.1089/fpd.2008.0226
13. Feller M, Huwiler K, Stephan R, Altpeter E, Shang A, Furrer H, et al. Mycobacterium avium subspecies paratuberculosis and Crohn's disease: a systematic review and meta-analysis. Lancet Infect Dis. (2007) 7:607–13. doi: 10.1016/S1473-3099(07)70211-6
14. Jeyanathan M, Boutros-Tadros O, Radhi J, Semret M, Bitton A, Behr MA. Visualization of Mycobacterium avium in Crohn's tissue by oil-immersion microscopy. Microbes Infect. (2007) 9:1567–73. doi: 10.1016/j.micinf.2007.09.001
15. Juste RA, Elguezabal N, Garrido JM, Pavon A, Geijo MV, Sevilla I, et al. On the prevalence of M. avium subspecies paratuberculosis DNA in the blood of healthy individuals and patients with inflammatory bowel disease. PLoS ONE. (2008) 3:3–8. doi: 10.1371/journal.pone.0002537
16. Mameli G, Cocco E, Frau J, Marrosu MG, Sechi LA. Epstein barr virus and Mycobacterium avium subsp. paratuberculosis peptides are recognized in sera and cerebrospinal fluid of MS patients. Sci Rep. (2016) 6:1–10. doi: 10.1038/srep22401
17. Niegowska M, Rapini N, Piccinini S, Mameli G, Caggiu E, Bitti MLM, et al. Type 1 Diabetes at-risk children highly recognize Mycobacterium avium subspecies paratuberculosis epitopes homologous to human Znt8 and Proinsulin. Sci Rep. (2016) 6:2–11. doi: 10.1038/srep22266
18. Bo M, Erre GL, Niegowska M, Piras M, Taras L, Longu MG, et al. Interferon regulatory factor 5 is a potential target of autoimmune response triggered by Epstein-Barr virus and Mycobacterium avium subsp. paratuberculosis in rheumatoid arthritis: investigating a mechanism of molecular mimicry. Clin Exp Rheumatol. (2018) 36:0376–81.
19. Yokoyama K, Cossu D, Hoshino Y, Tomizawa Y, Momotani E, Hattori M. Anti-mycobacterial antibodies in paired cerebrospinal fluid and serum samples from Japanese patients with multiple sclerosis or neuromyelitis optica spectrum disorder. J Clin Med. (2018) 7:522. doi: 10.3390/jcm7120522
20. Juste RA, Alonso-Hearn M, Molina E, Geijo M, Vázquez P, Sevilla I, et al. Significant reduction in bacterial shedding and improvement in milk production in dairy farms after the use of a new inactivated paratuberculosis vaccine in a field trial. BMC Res Notes. (2009) 2:1–6. doi: 10.1186/1756-0500-2-233
21. Alonso-Hearn M, Molina E, Geijo M, Vázquez P, Sevilla IA, Garrido JM, et al. Immunization of adult dairy cattle with a new heat-killed vaccine is associated with longer productive life prior to cows being sent to slaughter with suspected paratuberculosis. J Dairy Sci. (2012) 95:618–29. doi: 10.3168/jds.2009-2860
22. Garrido JM, Vázquez P, Molina E, Plazaola JM, Sevilla IA, Geijo MV, et al. Paratuberculosis vaccination causes only limited cross-reactivity in the skin test for diagnosis of bovine tuberculosis. PLoS ONE. (2013) 8:2–8. doi: 10.1371/journal.pone.0080985
23. Groenendaal H, Nielen M, Jalvingh AW, Horst SH, Galligan DT, Hesselink JW, et al. simulation of Johne's disease control. Prev Vet Med. (2002) 54:225–45. doi: 10.1016/S0167-5877(02)00027-2
24. Garry F. Control of paratuberculosis in dairy herds. Vet Clin N Am Food Anim Pract. (2011) 27:599–607. doi: 10.1016/j.cvfa.2011.07.006
25. Barkema HW, Orsel K, Nielsen SS, Koets AP, Rutten VPMG, Bannantine JP, et al. Knowledge gaps that hamper prevention and control of Mycobacterium avium subspecies paratuberculosis infection. Transbound Emerg Dis. (2018) 65:125–48. doi: 10.1111/tbed.12723
26. González J, Geijo MV, García-Pariente C, Verna A, Corpa JM, Reyes LE, et al. Histopathological classification of lesions associated with natural paratuberculosis infection in cattle. J Comp Pathol. (2005) 133:184–96. doi: 10.1016/j.jcpa.2005.04.007
27. Balseiro A, Perez V, Juste RA. Chronic regional intestinal inflammatory disease: a trans-species slow infection? Comput Immunol Microbiol Infect Dis. (2019) 62:88–100. doi: 10.1016/j.cimid.2018.12.001
28. Medzhitov R, Schneider DS, Soares MP. Disease tolerance as a defense strategy. Science. (2012) 335:936–41. doi: 10.1126/science.1214935
29. Divangahi M, Khan N, Kaufmann E. Beyond killing Mycobacterium tuberculosis: disease tolerance. Front Immunol. (2018) 9:2976. doi: 10.3389/fimmu.2018.02976
30. Purdie AC, Plain KM, Begg DJ, de Silva K, Whittington RJ. Candidate gene and genome-wide association studies of Mycobacterium avium subsp. paratuberculosis infection in cattle and sheep: a review. Comput Immunol Microbiol Infect Dis. (2011) 34:197–208. doi: 10.1016/j.cimid.2010.12.003
31. Koets AP, Adugna G, Janss LLJ, van Weering HJ, Kalis CHJ, Wentink GH, et al. Genetic variation of susceptibility to Mycobacterium avium subsp. paratuberculosis infection in dairy cattle. J Dairy Sci. (2000) 83:2702–8. doi: 10.3168/jds.S0022-0302(00)75164-2
32. Gonda MG, Kirkpatrick BW, Shook GE, Collins MT. Identification of a QTL on BTA20 affecting susceptibility to Mycobacterium avium ssp. paratuberculosis infection in US Holsteins. Anim Genet. (2007) 38:389–96. doi: 10.1111/j.1365-2052.2007.01627.x
33. Settles M, Zanella R, McKay SD, Schnabel RD, Taylor JF, Whitlock R, et al. A whole genome association analysis identifies loci associated with Mycobacterium avium subsp. paratuberculosis infection status in US holstein cattle. Anim Genet. (2009) 40:655–62. doi: 10.1111/j.1365-2052.2009.01896.x
34. Kirkpatrick BW, Shi X, Shook GE, Collins MT. Whole-genome association analysis of susceptibility to paratuberculosis in Holstein cattle. Anim Genet. (2011) 42:149–60. doi: 10.1111/j.1365-2052.2010.02097.x
35. Minozzi G, Buggiotti L, Stella A, Strozzi F, Luini M, Williams JL. Genetic loci involved in antibody response to Mycobacterium avium ssp. paratuberculosis in cattle. PLoS ONE. (2010) 5:e11117. doi: 10.1371/journal.pone.0011117
36. Pant SD, Verschoor CP, Skelding AM, Schenkel FS, You Q, Biggar GA, et al. Bovine IFNGR2, IL12RB1, IL12RB2, and IL23R polymorphisms and MAP infection status. Mamm Genome. (2011) 22:583–8. doi: 10.1007/s00335-011-9332-8
37. Minozzi G, Williams JL, Stella A, Strozzi F, Luini M, Settles ML, et al. Meta-analysis of two genome-wide association studies of bovine paratuberculosis. PLoS ONE. (2012) 7:1–8. doi: 10.1371/journal.pone.0032578
38. van Hulzen KJE, Schopen GCB, van Arendonk JAM, Nielen M, Koets AP, Schrooten C, et al. Genome-wide association study to identify chromosomal regions associated with antibody response to Mycobacterium avium subspecies paratuberculosis in milk of Dutch Holstein-Friesians. J Dairy Sci. (2012) 95:2740–8. doi: 10.3168/jds.2011-5005
39. Alpay F, Zare Y, Kamalludin MH, Huang X, Xi X, Shook GE, et al. Genome-wide association study of susceptibility to infection by Mycobacterium avium subspecies paratuberculosis in Holstein cattle. PLoS ONE. (2014) 9:1–14. doi: 10.1371/journal.pone.0111704
40. Zare Y, Shook GE, Collins MT, Kirkpatrick BW. Genome-wide association analysis and genomic prediction of Mycobacterium avium subspecies paratuberculosis infection in US Jersey cattle. PLoS ONE. (2014) 9:e88380. doi: 10.1371/journal.pone.0088380
41. Kiser JN, White SN, Johnson KA, Hoff JL, Taylor JF, Neibergs HL. Identification of loci associated with susceptibility to Mycobacterium avium subspecies paratuberculosis (Map) tissue infection in cattle. J Anim Sci. (2017) 95:1080–91. doi: 10.2527/jas.2016.1152
42. Brito LF, Mallikarjunappa S, Sargolzaei M, Koeck A, Chesnais J, Schenkel FS, et al. The genetic architecture of milk ELISA scores as an indicator of Johne's disease (paratuberculosis) in dairy cattle. J Dairy Sci. (2018) 101:10062–75. doi: 10.3168/jds.2017-14250
43. Gao Y, Jiang J, Yang S, Cao J, Han B, Wang W, et al. Genome-wide association study of Mycobacterium avium subspecies paratuberculosis infection in Chinese Holstein. BMC Genom. (2018) 19:1–10. doi: 10.1186/s12864-018-5385-3
44. Mallikarjunappa S, Sargolzaei M, Brito LF, Meade KG, Karrow NA, Pant SD. Short communication: uncovering quantitative trait loci associated with resistance to Mycobacterium avium ssp. paratuberculosis infection in Holstein cattle using a high-density single nucleotide polymorphism panel. J Dairy Sci. (2018) 101:7280–6. doi: 10.3168/jds.2018-14388
45. McGovern SP, Purfield DC, Ring SC, Carthy TR, Graham DA, Berry DP. Candidate genes associated with the heritable humoral response to Mycobacterium avium ssp. paratuberculosis in dairy cows have factors in common with gastrointestinal diseases in humans. J Dairy Sci. (2019) 102:4249–63. doi: 10.3168/jds.2018-15906
46. Canive M, González-Recio O, Fernández A, Vázquez P, Badia-Bringué G, Lavin JL, et al. Identification of loci associated with susceptibility to Mycobacterium avium subsp. paratuberculosis infection in Holstein cattle using combinations of diagnostic tests and imputed whole-genome sequence data. PLoS ONE. (2021) 16:1–17. doi: 10.1371/journal.pone.0256091
47. Canive M, Badia-Bringué G, Vázquez P, González-Recio O, Fernández A, Garrido JM, et al. Identification of loci associated with pathological outcomes in Holstein cattle infected with Mycobacterium avium subsp paratuberculosis using whole-genome sequence data. Sci Rep. (2021) 11:1–13. doi: 10.1038/s41598-021-99672-4
48. Kirkpatrick BW, Cooke ME, Frie M, Sporer KRB, Lett B, Wells SJ, et al. Genome-wide association analysis for susceptibility to infection by Mycobacterium avium ssp. paratuberculosis in US Holsteins. J Dairy Sci. (2022) 105:4301–13. doi: 10.3168/jds.2021-21276
49. Leal-Calvo T, Avanzi C, Mendes MA, Benjak A, Busso P, Pinheiro RO, et al. A new paradigm for leprosy diagnosis based on host gene expression. PLoS Pathog. (2021) 17:1–27. doi: 10.1371/journal.ppat.1009972
50. Kiser JN, Wang Z, Zanella R, Scraggs E, Neupane M, Cantrell B, et al. Functional variants surrounding endothelin 2 are associated with Mycobacterium avium Subspecies paratuberculosis infection. Front Vet Sci. (2021) 8:1–10. doi: 10.3389/fvets.2021.625323
51. Aryngaziyev B, Beltramo C, Dondo A, Karymsakov T, Varello K, Goria M, et al. Polymorphisms associated to bovine paratuberculosis: investigation of their role in dna-protein interactions and transcriptional regulation. Vet Ital. (2020) 56:109–14. doi: 10.12834/VetIt.2325.13205.1
52. Canive M, Fernandez-Jimenez N, Casais R, Vázquez P, Lavin JL, Bilbao JR, et al. Identification of loci associated with susceptibility to bovine paratuberculosis and with the dysregulation of the MECOM, eEF1A2, and U1 spliceosomal RNA expression. Sci Rep. (2021) 11:1–14. doi: 10.1038/s41598-020-79619-x
53. Pant SD, Schenkel FS, Verschoor CP, You Q, Kelton DF, Moore SS, et al. A principal component regression based genome wide analysis approach reveals the presence of a novel QTL on BTA7 for MAP resistance in holstein cattle. Genomics. (2010) 95:176–82. doi: 10.1016/j.ygeno.2010.01.001
54. Sanchez MP, Guatteo R, Davergne A, Saout J, Grohs C, Deloche MC, et al. Identification of the ABCC4, IER3, and CBFA2T2 candidate genes for resistance to paratuberculosis from sequence-based GWAS in Holstein and Normande dairy cattle. Genet Sel Evol. (2020) 52:1–17. doi: 10.1186/s12711-020-00535-9
55. Barreiro LB, Quintana-Murci L. From evolutionary genetics to human immunology: how selection shapes host defence genes. Nat Rev Genet. (2010) 11:17–30. doi: 10.1038/nrg2698
56. Canive M, Badia-Bringué G, Vázquez P, Garrido JM, Juste RA, Fernández A, et al. A genome-wide association study for tolerance to paratuberculosis identifies candidate genes involved in DNA packaging, DNA damage repair, innate immunity, and pathogen persistence. Front Immunol. (2022) 13:1–14. doi: 10.3389/fimmu.2022.820965
57. Zanella R, Settles ML, McKay SD, Schnabel R, Taylor J, Whitlock RH, et al. Identification of loci associated with tolerance to Johne's disease in Holstein cattle. Anim Genet. (2011) 42:28–38. doi: 10.1111/j.1365-2052.2010.02076.x
Keywords: paratuberculosis, genome-wide association study, disease tolerance, susceptibility, animal breeding, resistance
Citation: Alonso-Hearn M, Badia-Bringué G and Canive M (2022) Genome-wide association studies for the identification of cattle susceptible and resilient to paratuberculosis. Front. Vet. Sci. 9:935133. doi: 10.3389/fvets.2022.935133
Received: 03 May 2022; Accepted: 22 August 2022;
Published: 12 September 2022.
Edited by:
Mujeeb Ur Rehman, Livestock and Dairy Development Department, PakistanReviewed by:
Brian Kirkpatrick, University of Wisconsin-Madison, United StatesCopyright © 2022 Alonso-Hearn, Badia-Bringué and Canive. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marta Alonso-Hearn, bWFsb25zb0BuZWlrZXIuZXVz
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.